The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif (original) (raw)
Abstract
Fragile X syndrome is caused by the absence of protein FMRP, the function of which is still poorly understood. Previous studies have suggested that FMRP may be involved in various aspects of mRNA metabolism, including transport, stability and/or translatability. FMRP was shown to interact with a subset of brain mRNAs as well as with its own mRNA; however, no specific RNA-binding site could be identified precisely. Here, we report the identification and characterization of a specific and high affinity binding site for FMRP in the RGG-coding region of its own mRNA. This site contains a purine quartet motif that is essential for FMRP binding and can be substituted by a heterologous quartet-forming motif. The specific binding of FMRP to its target site was confirmed further in a reticulocyte lysate through its ability to repress translation of a reporter gene harboring the RNA target site in the 5′-untranslated region. Our data address interesting questions concerning the role of FMRP in the post-transcriptional control of its own gene and possibly other target genes.
Keywords: FMRP/fragile X syndrome/purine quartet/RNA-binding site
Introduction
Fragile X syndrome is the most frequent cause of inheritable mental retardation and is also among the most common single gene disorders (Hagerman, 1996). The gene affected by the syndrome, FMR1, is transcriptionally inactivated by the expansion and the methylation of a trinucleotide (CGG) repeat, located in the 5′-untranslated region (5′-UTR) of the gene (Imbert et al., 1998). Thus, fragile X syndrome results from the inhibition of expression of the FMR1 gene, and the subsequent absence of its product, the protein FMRP. FMRP is found predominantly in neurons, particularly those of the hippocampus and in Purkinje cells of the cerebellum (Devys et al., 1993). In the cell, FMRP is predominantly cytoplasmic, but the nuclear localization of isoforms lacking exon 14-encoded sequence (Sittler et al., 1996), together with the identification of a functional nuclear localization signal and nuclear export signal (Eberhart et al., 1996), suggested that it shuttles between both cellular compartments (Tamanini et al., 1999). In the cytoplasm, FMRP forms part of mRNP particles that interact with translating ribosomes (Siomi et al., 1996; Corbin et al., 1997; Feng et al., 1997). Immunoprecipitation experiments have shown that FMRP is associated with the homologous proteins FXR1P and FXR2P, and the RNA-binding protein nucleolin (Ceman et al., 1999). FMRP is also able to interact with three additional proteins, NUFIP1, CYFIP1 and CYFIP2, which could modulate its function (Bardoni et al., 1999; Schenck et al., 2001).
Another main characteristic of FMRP is its ability to interact with RNA. This property was suggested by the identification of two KH domains and an RGG box (Gibson et al., 1993), by homopolymer binding (Siomi et al., 1994) and by the apparent binding selectivity in vitro of FMRP for a fraction of mRNAs expressed in the brain, including its own message (Ashley et al., 1993a; Sung et al., 2000). The association of FMRP with its own mRNA in vivo was also suggested by the identification of FMR1 mRNA in FMRP-associated mRNPs (Ceman et al., 1999). The precise function of FMRP in the cell is still poorly understood. In particular, it is as yet unclear whether FMRP acts on a specific subset of mRNA in the brain (Ashley et al., 1993a), or has a broader action on mRNA metabolism and protein synthesis. The various properties of FMRP, however, suggest that it could be involved in one or more steps of nuclear export, cytoplasmic transport and translational control of target mRNAs (Imbert et al., 1998; Jin and Warren, 2000). Recently, FMRP has been proposed to be a translation inhibitor (Laggerbauer et al., 2001; Li et al., 2001). In particular, FMRP could play a role in the regulation of protein synthesis at the postsynaptic site of dendrites since its synthesis, in response to neurotransmitter activation (Weiler et al., 1997), may be important for normal maturation of the synaptic connections (Comery et al., 1997).
Since the RNA binding properties of FMRP appear to be critical for its function, we wished to characterize its RNA binding specificity and define its RNA target site(s). Here we provide evidence that FMRP binds specifically and with high affinity to FMR1 mRNA in vitro, and we identified the RNA motif responsible for this binding. A 100 nucleotide fragment was identified within the 3′-terminal part of the FMR1 coding sequence that retains the ability to bind FMRP specifically. Within this fragment, a peculiar structure involving purine quartets was demonstrated to be responsible for the interaction with FMRP. Furthermore, this RNA motif can promote recognition of a heterologous mRNA by FMRP, as indicated by the translation repression of a chimeric reporter transcript bearing the FMRP-binding site (FBS) in its 5′-UTR in a rabbit reticulocyte lysate. Altogether our data support evidence that this RNA target could play an important role in the function of FMRP.
Results
FMR1 mRNA contains a specific binding site for FMRP in its coding sequence
To determine whether FMRP exhibits some binding specificity for its own mRNA, we compared the efficiency of its binding to FMR1 mRNA and to various heterologous RNAs.
We first measured the retention of various 32P-labeled RNAs on GST–FMRP immobilized on glutathione– Sepharose beads. However, the retention of Iso-1 RNA corresponding to the coding sequence of the longest spliced isoform (Ashley et al., 1993b), without (Iso-1) or with its 3′-UTR (Iso-1 + 3′-UTR), did not differ significantly from the retention of an unrelated RNA such as the 3′-UTR of bicoid mRNA from Drosophila (60, 56 and 70% retention, respectively). Shorter unrelated RNAs such as a fragment of Escherichia coli 16S rRNA (f16S rRNA) or the human tRNA3Lys were also retained rather efficiently (30 and 20%, respectively), indicating that FMRP is capable of binding RNA non-specifically.
Since unspecific binding might introduce bias in direct measurement of binding affinities, we used competition assays that are more sensitive and allow alleviation of the contribution of the unspecific binding properties of FMRP. Therefore, we measured the competition strength of different RNAs, derived either from FMR1 mRNA or from heterologous RNAs, when challenged with Iso-1 RNA. In a first set of experiments, a constant and very low concentration of uniformly labeled Iso-1 RNA was incubated with a fixed amount of GST–FMRP immobilized on glutathione–Sepharose beads, in the presence of increasing concentrations of unlabeled competitor RNAs. Under these conditions, significant differences were observed between the different RNAs, thus revealing different binding strengths. Indeed, Iso-1 mRNA was competed more efficiently by itself than by any of the heterologous RNAs (bicoid 3′-UTR, f16S rRNA and tRNA) of various lengths and origins (Figure 1A and C). This result indicates that GST–FMRP possesses an apparent affinity for its own mRNA that is at least one order of magnitude higher than that for other unrelated RNAs. Furthermore, the competition efficiency was unaffected by the presence or absence of the FMR1 3′-UTR. Consistently, the 3′-UTR alone displayed a competition efficiency reduced by at least 10-fold. Thus, the site responsible for the high affinity binding appeared to be located within the coding region of FMR1 mRNA.
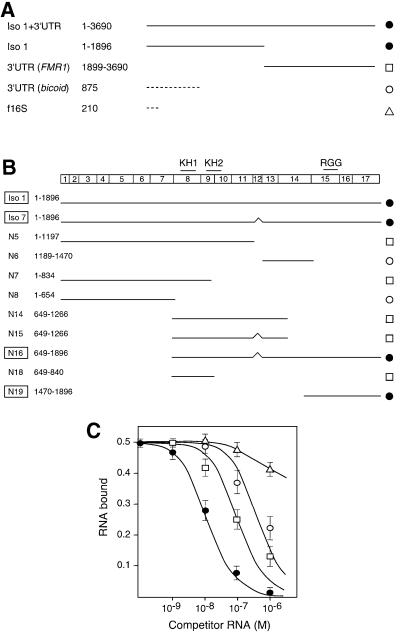
Fig. 1. Competition experiments to determine the relative affinity of GST–FMRP for various RNA sequences. (A) RNA sequences from FMR1 mRNA or from various origins; their lengths are indicated. (B) Subdomains of FMR1 mRNA with an indication of the corresponding exons and domains in FMRP (upper line). Nucleotide positions are given with respect to the coding sequence start (+1). (C) Fraction of bound Iso-1 RNA (32P labeled), retained on immobilized GST–FMRP, plotted against competitor RNA concentration. Each point reflects at least four independent experiments for each RNA species. For clarity, RNA constructs displaying similar competition strengths and falling within the same standard deviation have been grouped and are represented by four types of symbols as indicated in (A) and (B).
To localize more precisely the sequence(s) responsible for the specific binding, various RNA fragments derived from Iso-1 RNA were tested in competition assays with Iso-1 RNA. First, isoform 7 RNA (Iso-7), which codes for the most frequently spliced isoform of FMRP and corresponds to FMR1 RNA deleted of exon 12 (Ashley et al., 1993b), displayed a similar competition strength to that of Iso-1 RNA. Secondly, among the different truncated subfragments tested, only two overlapping fragments (N16 and N19) were able to compete efficiently with Iso-1 RNA (Figure 1B and C). The shortest fragment, N19 (nucleotides 1470–1896), encompasses the 427 last nucleotides of the coding region of FMR1 mRNA. All other fragments showed at least 10-fold decreased competition strengths, comparable with that of heterologous RNAs.
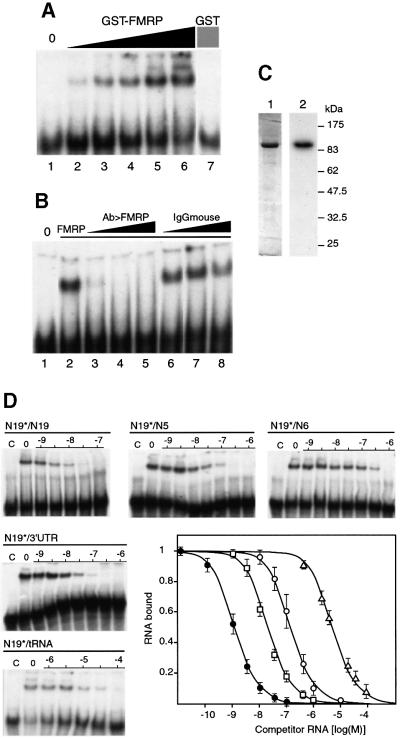
In a second set of experiments, interaction between N19 RNA and GST–FMRP was tested by gel retardation experiments to avoid any bias related to the use of an immobilized protein. GST–FMRP was able to retard the migration of N19 RNA on polyacrylamide gel electrophoresis, in a concentration-dependent manner (Figure 2A). In the same conditions, the GST protein alone was not able to produce any shift. Furthermore, pre-incubation of an anti-FMRP monoclonal antibody 1C3 (Devys et al., 1993) with GST–FMRP (Figure 2B), or its incubation with the pre-formed complex (data not shown), abolished gel shift at a concentration as low as 10 µg/ml. At the same concentration and up to 150 µg/ml, mouse IgGs had no effect. Finally, a northwestern experiment was used to verify that the purified GST–FMRP interacts directly with N19 RNA, and not via an unidentified product that might contaminate the preparation (Figure 2C). Altogether, these data strongly support the direct and specific interaction between FMRP and N19 RNA.
Fig. 2. Binding specificity of GST–FMRP for N19 RNA. (A) Labeled N19 RNA was incubated in the absence (lane 1) or presence of increasing amounts of recombinant GST–FMRP (lanes 2–6: 0.01, 0.05, 0.1, 0.2 and 0.5 pmol, respectively). A control is shown with GST alone (lane 7, 20 pmol). (B) Inhibition of the formation of an N19 RNA–GST–FMRP complex by monoclonal antibody 1C3. Lane 1 is without FMRP, lanes 2–8 are in the presence of 0.1 pmol FMRP, and antibodies were added at a concentration of 10, 50 and 150 µg/ml (lanes 3, 4 and 5 for anti-FMRP, and lanes 6, 7 and 8 for anti-mouse IgG). (C) Northwestern with GST–FMRP and N19 RNA. Lane 1, Coomassie staining of the GST–FMRP on a 10% SDS–polyacrylamide gel; lane 2, corresponding autoradiograph of the membrane hybridized with labeled N19 RNA. (D) Competition experiment to determine the relative binding strength of the various subfragments of FMR1 mRNA (Figure 1B) by gel retardation experiments. 32P-labeled N19* RNA was incubated with GST–FMRP (0.1 pmol) in the presence of increasing concentrations of unlabeled competitor. Lanes C, control without protein; lanes 0, control without competitor RNA; numbers are the log of competitor concentrations. The graph depicts the fraction of bound labeled N19 RNA plotted against unlabeled competitor RNA concentrations. Each point is the mean with standard deviation of at least three independent experiments. As in Figure 1C, the different RNA constructs are sorted into distinct classes represented by different symbols: N19 (filled circles), N5, N7, N18 and 3′-UTR (open squares), N6 and N8 (open circles), tRNA (open triangles).
Binding of labeled N19 RNA to FMRP was then challenged by increasing amounts of various unlabeled subfragments of FMR1 mRNA using the same gel shift assay (Figure 2D). Again, N19 RNA showed the highest competition efficiency. The apparent _K_d of N19 RNA was estimated to be 1 (±0.5) nM according to the concentration of unlabeled RNA displacing 50% of the bound labeled RNA. It should be noted that in the competition experiments on immobilized GST–FMRP, the amount of FMRP (∼10–7 M) was much higher than the _K_d value, thus explaining the displacement of the competition curves, and preventing simple _K_d determination by analysis of the competition curves. Using gel shift assay, we also showed that the other fragments derived from FMR1 mRNA, including the 5′- and 3′-UTRs, all display reduced competition efficiencies, similar to that of unrelated RNAs (e.g. bicoid 3′-UTR). Altogether, these experiments suggested that FMRP possesses a strong and specific binding site located in the 3′-terminal part of the coding region of its own mRNA.
Determination of a minimal RNA-binding site
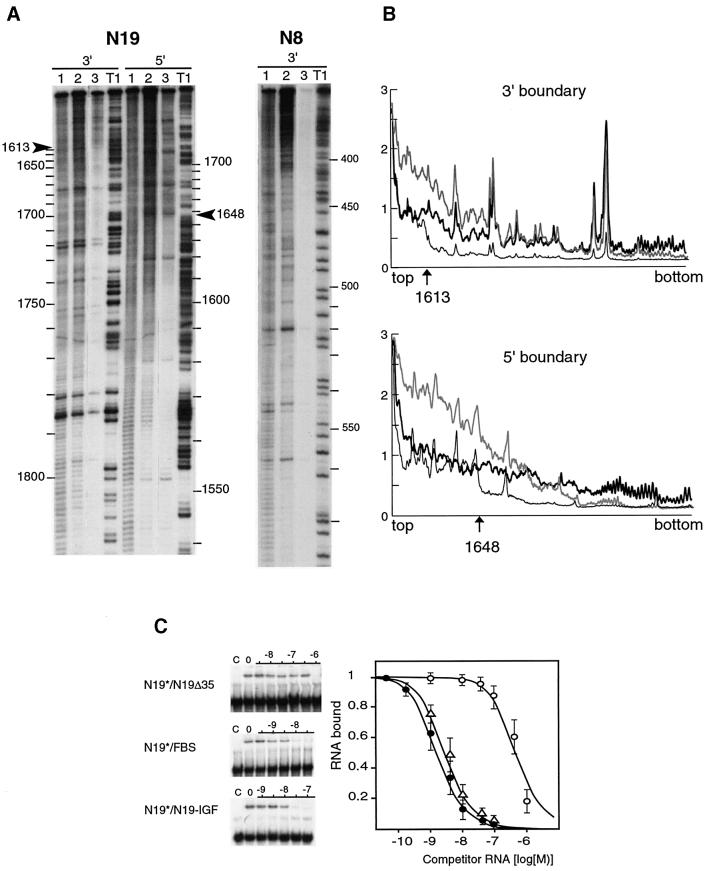
The boundaries of the FBS within the N19 RNA were determined by ladder selection experiments. In these experiments, the N19 RNA subfragments retaining FMRP binding were selected on immobilized GST– FMRP among a pool of RNAs generated by mild alkaline hydrolysis of 3′- or 5′-end labeled N19 RNA. After washing out unbound RNAs, bound RNAs were phenol– chloroform extracted and analyzed by electrophoresis on a denaturing polyacrylamide gel (Figure 3A). While the selection was essentially size-dependent in the absence of competitor RNA, a precise cut in the size of molecules retained was observed in the presence of 10–8 M unlabeled N19 RNA as competitor (Figure 3A and B). The cut positions are 1613 and 1648 for 3′- and 5′-end labeled fragments, respectively. They defined the boundaries of a 35 nucleotide region (1613–1648) that potentially was sufficient for FMRP binding. When non-specific N8 RNA (nucleotides 1–654 of FMR1 mRNA) was treated in the same conditions, no RNA was retained (Figure 3A). Similarly, no N19 RNA subfragment was retained when GST alone was used instead of GST–FMRP (data not shown).
Fig. 3. Determination of the boundaries of the FBS. (A) Determination of the 5′ and 3′ boundaries on N19 RNA (left) with a control performed on N8 RNA (right). 5′ and 3′ = RNA 32P-labeled at its 5′ or 3′ end, respectively. Lanes 1 are RNAs statistically digested with alkali. Lanes 2 are RNAs statistically digested with alkali that have been retained on immobilized GST–FMRP. Lanes 3, same as lanes 2 but in the presence of 10–8 M competitor N19 RNA. Lane T1, RNA submitted to statistical RNase T1 digestion. 5′ and 3′ boundaries are indicated by arrows. (B) Densitometer trace of lanes 1 (thick line), 2 (gray line) and 3 (thin line); the ordinate is in arbitrary units, the abscissa gives nucleotide positions. (C) Effect of deletions or domain exchange in the FBS. 32P-labeled N19* RNA was incubated with GST–FMRP (0.1 pmol) in the presence of increasing concentrations of unlabeled RNA: N19-Δ35 (open circles), FBS (filled circles), N19–IGF (open triangles). The fraction of bound labeled N19* RNA, as visualized by gel shift assays (left), was plotted against competitor RNA concentrations (right). Each point is the mean with standard deviation of at least three independent experiments.
To test its role in FMRP binding, the 35 nucleotide region was deleted in N19 RNA (N19-Δ35) and its effect was tested by gel retardation assay. As a result, N19-Δ35 RNA showed a dramatic diminution of its competition efficiency (Figure 3C). However, the 35 nucleotide RNA fragment had negligible binding efficiency when tested in similar conditions. Altogether, these data suggested that the 35 nucleotide region was necessary but not sufficient by itself to sustain efficient FMRP binding. A possible explanation is that the isolated 35 nucleotide fragment is not folded correctly. On the contrary, the presence of additional (unspecific?) interactions might be needed to provide stability. By increasing the size of the minimal RNA region (56 and 10 nucleotides on the 5′ and 3′ end, respectively), the binding affinity of FMRP was restored to that of the N19 RNA (Figure 3C). This 101 nucleotide RNA (1557–1658) has been called the FBS.
The FBS contains a purine quartet structure
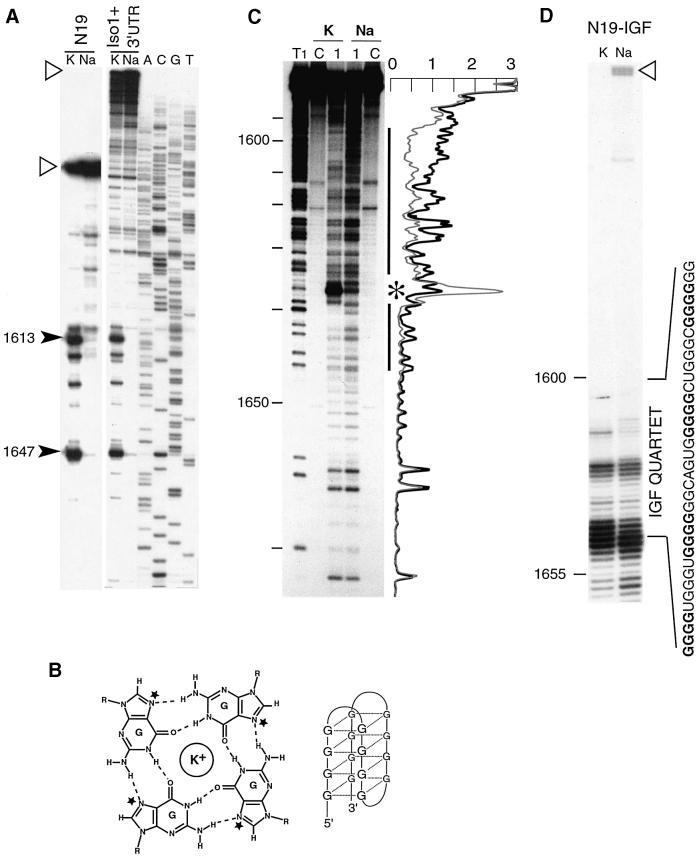
The sequence of the FBS is remarkable due to the presence of a long tract of almost uninterrupted purines (nucleotides 1603–1647) containing the 35 nucleotide region identified above. The existence of a stable structure within the FBS in N19 RNA was indicated by unusually strong pauses of reverse transcriptase progression immediately before and within the purine-rich region (Figure 4A). Purine-rich regions are potential candidates to form quadruplex structures (G or A quartets), the formation of which is cation dependent. Indeed, the planar layers of G quartets are stabilized preferentially by K+ in comparison with Na+ or Li+ (Sundquist, 1989). Since avian myeloblastosis virus (AMV) reverse transcriptase has no monovalent cation requirement (Fujinaga et al., 1970), we analyzed the influence of the nature of the cation on the pauses of reverse transcription. Thus, when NaCl (100 mM) was substituted for KCl in the reverse transcription buffer, the strong pauses disappeared (Figure 4A). These results strongly suggest the occurrence of a purine quartet structure in the FBS. Furthermore, the same pauses of reverse transcriptase were obtained using either the N19 RNA or the Iso-1 FMR1 mRNA with its 3′-UTR (Figure 4A), suggesting that the same structure also exists in the natural mRNA.
Fig. 4. Indication of the presence of a quadruplex structure in the FBS. (A) Cation-dependent termination of reverse transcription at the purine-rich domain in the N19 subfragment (N19) and in the full-length Iso-1 RNA with its 3′-UTR (Iso1 + 3′ UTR). Strong pauses of reverse transcriptase are indicated by arrowheads with their position within the FMR1 mRNA sequence. Full-length extension products are indicated by open triangles. (B) Hydrogen bonding scheme for the G quartet alongside its schematic model. The bases are hydrogen bonded via Hoogsteen base pairs in a square-planar symmetric array. N7 positions are indicated by stars. The quadruplex is stabilized by coordination of a monovalent ion lying between or within the planes of the guanine tetrads (Laughlan et al., 1994). (C) Chemical probing of N7 positions of guanines of 3′-end labeled FBS RNA with DMS. RNA was incubated in buffer containing 150 mM KCl (K) or NaCl (Na) prior to treatment. Lane T1, statistical RNase T1 digestion; lanes C, controls without DMS; lanes 1, RNAs treated with DMS. The black line indicates an area of protected guanines in the presence of KCl; the asterisk indicates a hyper-reactivity. The densitometer traces of lanes 1 are shown alongside the autoradiograph (thick line is Na, thin line is K); the ordinate is in arbitrary units; the abscissa coincides with nucleotide positions on the autoradiograph. (D) Termination of reverse transcription at the beginning of the IGF II motif inserted in the chimeric N19–IGF RNA. The sequence of the inserted motif is indicated, with tracts of Gs forming the quadruplex in bold. The full-length product is indicated as in (A).
To test the hypothesis of the presence of purine quartets in the FBS, we probed the accessibility of position N7 of the guanines with dimethyl sulfate (DMS) in the presence of KCl and NaCl. In a standard A-helix or in an unpaired region, the N7 position of guanine is usually accessible to DMS modification. However, in G quartet structures, this position is hydrogen bonded with the amino-2 position of a co-planar G, and is therefore not accessible (Figure 4B). The results clearly revealed a strong reduction in the reactivity of guanines at N7 inside a nucleotide window corresponding to positions 1591–1646 when probing was done in the presence of KCl, as compared with NaCl (Figure 4C). The only exception is the hyper-reactivity of G1637 that occurred in the presence of KCl. The window of protected nucleotides encompasses exactly the purine-rich region and overlaps with the region of reverse transcription pauses. Therefore, these results strongly support the presence of a structure involving G quartets in the FBS.
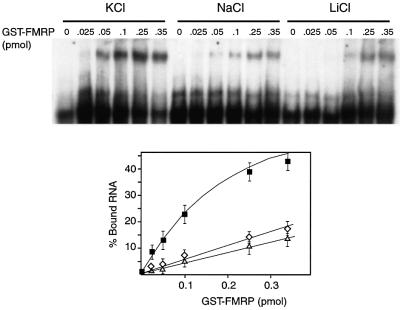
To determine whether these structural features are involved in FMRP binding, we tested the influence of cations (K+, Na+ and Li+) on GST–FMRP binding to N19 RNA using gel shift assay (Figure 5). The binding of FMRP was clearly dependent on the presence of K+ and was strongly reduced in the presence of Na+ and Li+, compared with K+. Thus, the fact that GST–FMRP binding shows the same cation dependence as that observed for purine quartet formation supports the idea that the quartet structure is essential for protein binding.
Fig. 5. Cation-dependent binding of FMRP on its RNA-binding site. Labeled N19 RNA was incubated in the absence (lane 0) or presence of increasing amounts of GST–FMRP in binding buffer containing 150 mM KCl (filled squares), NaCl (open diamonds) or LiCl (open triangles). The percentage of bound RNA (as determined by phosphoimager quantification) is plotted against the amount of GST–FMRP (error bars are reflect at least three independent experiments).
To stress further the importance of the purine quartet structure for FMRP binding, we addressed the question of whether the FBS of FMR1 mRNA could be exchanged with a different sequence capable of promoting a purine quartet structure in vitro. Thus, we constructed a chimera (N19–IGF RNA) in which we substituted the 35 nucleotides of the FBS of N19 RNA by a 36 nucleotide fragment from the 3′-UTR of human insulin-like growth factor II (IGF II) mRNA, shown to adopt a guanosine quartet structure (Christiansen et al., 1994). Interestingly, the IGF II motif induced a complete arrest of reverse transcriptase in the presence of K+ (Figure 4D), thus revealing an extremely stable quartet. In the presence of Na+, the stability of the quartet is slightly decreased as shown by the presence of full-length elongation product. The cation-dependent protection of the N7 Gs of the IGF II motif was also tested. A reduction of reactivity of the Gs was indeed observed when substituting K+ for Na+, but the reactivity decrease was not as pronounced as in wild-type N19 RNA, due to an already weak level of reactivity in the presence of Na+ (result not shown). Thus, the quartet formed by the IGF II motif appears to be even more stable (and less sensitive to the nature of cation) than that formed by the FBS motif.
Whereas the deletion of 35 nucleotides from the core of FBS abolished the binding of FMRP (see above), their replacement with the IGF II structure in chimeric N19–IGF RNA restored binding to an almost wild-type level (Figure 3B). This result suggests that the quartet structure per se, rather than the sequence of the RNA itself, was important for FMRP binding.
FMRP–FBS interaction occurring in chimeric mRNAs is capable of inhibiting translation in vitro
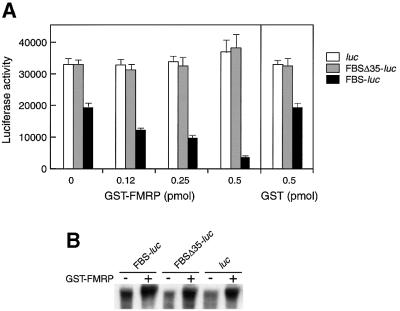
To verify that the FMRP–RNA interaction takes place under more physiological conditions (a cell lysate), we used an indirect assay that allowed the testing of FMRP binding through its ability to inhibit translation when the FBS is inserted in the 5′-UTR of a reporter gene. This test is based on the ability of a protein to inhibit translation when bound to a well-structured motif in the 5′-UTR, as shown for iron regulatory protein binding to the iron regulatory element in the case of translational regulation of ferritin mRNA (Gray and Hentze, 1994). Thus, we inserted a fragment derived from the FBS (nucleotides 1603–1658) 50 nucleotides downstream from the cap structure of the luciferase mRNA transcript and tested whether GST–FMRP could affect translation of the hybrid mRNA in a rabbit reticulocyte lysate. Control reporter mRNAs, either lacking the FBS (luc) or containing incomplete FBS (FBSΔ35-luc) in their 5′-UTR, were tested in parallel. Translation efficiency was determined by measuring the luciferase activity. In the absence of added GST–FMRP, translation of mRNA containing the complete FBS (FBS-luc) was decreased by 1.5-fold in comparison with both control mRNAs (luc and FBSΔ35-luc) (Figure 6A). This effect was most probably due to the influence of the purine quartet structure on translation as the presence of a stable structure in the 5′-UTR has been demonstrated to impede translation initiation (Kozak, 1986, 1989). The translation of mRNAs that contained an incomplete FBS (FBSΔ35-luc) or an unrelated sequence (luc) in their 5′-UTR was insensitive to GST–FMRP (up to 50 nM). These results contrasted with data reporting that FMRP could have a general inhibitory effect on translation independently of the nature of the mRNA (Laggerbauer et al., 2001). This lack of apparent specificity may be due to experimental conditions (presence of an excess of FMRP) or the influence of post-translational modifications (as yet unknown), absent in bacterially synthesized proteins, that would alter its specificity. In addition, more recent data (Li et al., 2001) reported that translation inhibition by FMRP depends on the nature of the mRNA template, suggesting discriminatory interactions with the mRNA.
Fig. 6. GST–FMRP inhibits translation of a chimeric FBS-luc transcript in vitro. Translation reaction mixture (rabbit reticulocyte lysate) was incubated with luc, FBSΔ35-luc or FBS-luc mRNAs and increasing amounts of GST–FMRP. (A) Graph depicting the dose–response relationship between the amount of GST–FMRP and the luciferase activity (error bars reflect six independent experiments). Control with GST alone is shown. (B) Stability of the mRNA after translation in the presence or absence of GST–FMRP (0.5 pmol). Labeled mRNA substrates were extracted from the reticulocyte lysate mixture after translation and analyzed by denaturing gel electrophoresis.
When we analyzed the effect of FMRP on the translation of an mRNA containing the FBS (FBS-luc), the luciferase synthesis from this mRNA decreased in a dose-dependent manner and reached 75% inhibition with the highest protein concentration tested (Figure 6A). As a control, addition of GST protein had no effect in the same concentration range (Figure 6A). We also verified that the reduced levels of luciferase were due to FMRP binding and not to a degradation of mRNAs during incubation with FMRP. Indeed, exposed mRNAs were not found reduced in comparison with those which were unexposed (Figure 6B). Rather, the presence of FMRP appeared to protect them against RNA degradation, as reported by Li et al. (2001). Thus, our data demonstrate that FMRP binding to the FBS is strong enough to repress, specifically and significantly, the translation initiation in vitro of a reporter gene when located in its 5′-UTR. Furthermore, binding does occur in a cell lysate, and appears to proceed independently of the RNA context.
Discussion
A specific RNA motif for FMRP binding
Previous experiments have shown that FMRP can interact with its own mRNA in vitro (Ashley et al., 1993a; Brown et al., 1998; Sung et al., 2000) and had been found within its mRNP particle in vivo (Ceman et al., 1999). Here we have demonstrated that although FMRP has a high propensity to interact with RNA in a non-specific manner, it has a specific and high affinity binding site on its own mRNA. The FBS, which contains 100 nucleotides, was identified in the 3′-terminal part of the FMR1 mRNA coding region. This RNA fragment interacts with FMRP with high affinity (1 ± 0.5 nM), similarly to the longest spliced isoform of FMR1 mRNA. The FBS confers to the FMR1 mRNA an apparent affinity at least one order of magnitude higher than other tested RNAs of comparable length. The FBS corresponds to nucleotides 1557–1658 and codes for the RGG box region of FMRP. This location of the FBS within the coding region of FMR1 mRNA may appear surprising, since the FBS previously was proposed to be situated rather in the 3′-UTR of FMR1 mRNA (Brown et al., 1998). The apparent discrepancy between our results and those of Brown et al. (1998) can probably be accounted by the non-specific RNA binding properties of FMRP when interactions are performed in the absence of competitor RNA. The observed affinity difference (at least 10-fold) between specific and unspecific binding falls in a range that is quite biologically relevant. As an example, a well documented study correlating affinity constants measured in vitro and translational regulation in vivo demonstrated that a 10-fold reduction in affinity between the repressor and its mRNA target induced a complete loss of control (Romby et al., 1996).
The FBS consists of a very stable structure involving a purine-rich sequence capable of forming a purine quartet both in a 100-nucleotide fragment and in the longest spliced isoform of FMR1 mRNA (3690 nucleotides). This structural feature was revealed by monovalent cation-dependent pauses of reverse transcriptase as well as by probing with DMS the N7 positions of guanines of the FBS in conditions known to stabilize (with K+) or destabilize (with Na+) a quadruplex structure (Sundquist and Heaphy, 1993). The fact that most N7 G positions of the purine-rich region of FBS are protected in a buffer containing K+ versus Na+ strongly argues for their involvement in Hoogsteen G–G pairings. The exact structure and length of the purine quartet structure remain to be defined more precisely; however, the formation of intermolecular quartets was excluded by the absence of the dimeric form of the RNA containing the FBS on native gels (data not shown).
FMRP is a protein that was shown to be capable of self-association (Zhang et al., 1995). Northwestern experiments, by demonstrating the direct interaction between the target RNA and FMRP, suggested that the purine quartet recognition was an intrinsic property of monomeric FMRP. It will be interesting to study whether the FMRP homologs FXR1P and FXR2P (that can heteromerize with FMRP) share the binding specificity for the quartet structure, and whether the binding of FMRP to the FBS is modulated by the presence of other FMRP interactors.
Interestingly, a chimeric RNA in which the natural FBS is replaced by a heterologous motif from the 3′-UTR of IGF II mRNA, also able to fold as a quadruplex structure (Christiansen et al., 1994), still binds FMRP with high affinity. This IGF II motif was shown to form a highly stable quartet, with a reduced sensitivity to the nature of the cation, when inserted in N19 RNA. This is most likely due to the fact that the IFG II quartet is built with regular stretches of Gs, while the FBS contains both guanines and adenines (Figure 7). Thus, because guanines of the FBS are interspersed with adenines, the FBS structure probably involves tetrads of adenines. In fact, RNAs containing tracts of guanines and adenines are able to form tetrads in vitro (Katahira et al., 1995). As the tetrad of adenines is isomorphous with that of guanine (Lee, 1990), the overall structure can be conserved between the two types of purine quartets. Therefore, the structure per se rather than the sequence is important for FMRP binding. It is thus tempting to propose that FMRP will bind to any quadruplex structure.
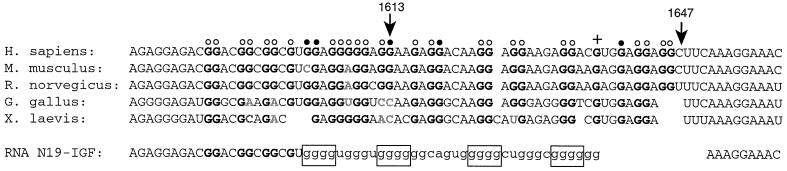
Fig. 7. Sequence alignment of the FBS purine-rich region from vertebrate FMR1 genes. The two arrows indicate the strong K+-dependent pauses of reverse transcriptase shown in Figure 4A. Protected N7 positions in the presence of K+ are indicated by dots (strong protection is represented by filled circles; moderate protection by open circles), enhanced reactivity towards DMS is indicated by a + sign. Conserved Gs in homologous FMR1 genes are depicted in bold. The sequence of RNA N19–IGF containing the insertion of the purine quartet region of IGF II mRNA (in lower case) is given. The four G tracts involved in G quartet formation are boxed.
Purine quartets in other systems
The presence of inter- and intramolecular quadruplex structures in DNA is well documented (Sundquist and Klug, 1989; Williamson et al., 1989; Sen and Gilbert, 1990; Kang et al., 1992; Smith and Feigon, 1992) and although the direct proof of their existence in vivo is still lacking, their implication in several cellular processes, including transcription, replication and recombination, has been put forward in several instances (Zahler et al., 1991; Simonsson et al., 1998). They also present a potential role as potent therapeutic agents or targets (Jing et al., 2000; Rangan et al., 2001). For RNA, tetramerization of poly(rG) and GGGG-containing oligonucleotides has also been demonstrated (Kim et al., 1991; Cheong and Moore, 1992). The intramolecular quadruplex evidenced in vitro in the 3′-UTR of IGF II mRNA (Christiansen et al., 1994) is located just downstream of an endonucleolytic cleavage site, and although its role in the cleavage is uncertain, it could play a role in the unusual stability of the 3′ product of the cleavage reaction (Scheper et al., 1995). In fact, the effect of a purine-rich sequence on in vivo stabilization of mRNA by preventing exonuclease progression via the potential formation of a quadruplex structure is a well known phenomenon (Muhlrad et al., 1995). Another example of such structures is provided by the translational repression of gene 2 mRNA of the filamentous bacteriophage fd by gene 5 protein. The mRNA sequence required for repression was shown to form a guanine tetraplex in vitro and to form a nucleation site for binding gene 5 protein to adjacent single-stranded regions (Oliver et al., 2000).
The 5′-UTR of FMR1 mRNA also contains a purine-rich region since an average of 30 CGG repeats is usually present in the human population (Imbert et al., 1998). In fact, the formation of a quadruplex structure in the CGG repeat region with the positive strand of DNA has been postulated to form and to be involved in the process of repeat amplification (Fry and Loeb, 1994). In the RNA, this region could also potentially adopt a quadruplex structure. However, we did not observe any reverse transcription pauses that could have revealed such a structure in a 5′-UTR RNA fragment containing 30 CGG repeats (data not shown). Furthermore, FMRP did not exhibit significant binding on this region of the RNA. Nevertheless, it cannot be excluded that increasing the number of repeats would promote formation of a quadruplex structure in the 5′-UTR of FMR1.
Implications for FMRP functions
Support for the functional significance of the FMRP–FBS interaction is provided by the high conservation of the sequence among all known FMRP genes of vertebrates (Figure 7). In the protein, this site corresponds to the RGG box domain. This site, however, is partially conserved in FXR1P and FXR2P and is absent in the Drosophila dFMR/FXR homolog (Wan et al., 2000). Our finding that FMRP is able to repress the translation of a heterologous gene in a reticulocyte lysate, when the FBS RNA was inserted in the 5′-UTR of the mRNA, provides support for the existence of a very stable interaction under biological conditions. However, the existence of a high affinity binding site for FMRP close to the end of the coding region of its own mRNA raises unanswered questions. It is tempting to assume that FMRP could play a role in the negative regulation of its own mRNA. Indeed, proteins were shown to control their own translation by binding to the 3′-UTR (Fu and Benchimol, 1997) or even to the C-terminal part of their own mRNA (Chu et al., 1993; Ercikan-Abali et al., 1997; Lin et al., 2000). Interactive cross-talk between the 5′ end and distant region of mRNAs has been proposed to occur in such a type of control (Wells et al., 1998). The mechanism by which binding of FMRP to the C-terminal part of the FMR1 mRNA might interfere with translation remains to be demonstrated, however.
Another possibility is that FMRP controls FMR1 mRNA decay. XRN1 (also named SEP1/DST2/KEM1/RAR5) is a 5′ to 3′ exonuclease that binds preferentially to G4 quadruplex RNA and DNA structures (Bashkirov et al., 1997) and is able to induce cuts in a single-stranded region 5′ to the G4 structure (Liu et al., 1995). It has been proposed that the role of the G4 quadruplex RNA structure could be to attract XRN1 nuclease and degrade the mRNA. Thus, another possible model for FMRP binding could be to interfere with or modulate the degration of its own mRNA. Other roles have been evoked for FMRP, such as the control of the subcellular localization of mRNAs containing a specific target site (Imbert et al., 1998). Thus, FMRP is likely to be a pluripotent protein such as, for instance, hnRNP A1, an RNA-binding protein involved in many steps of mRNA expression control, including pre-mRNA splicing, transport (Dreyfuss et al., 1993), modulation of mRNA turnover and translation (Hamilton et al., 1997). Another function identified for general RNA-binding proteins, including hnRNP A1, La autoantigen, pyrimidine tract-binding protein (hnRNP I/PTB) and the major core protein of cytoplasmic mRNP (p50), is to promote ribosome binding to mRNAs by a cap-mediated mechanism, thereby preventing spurious initiation at aberrant translation start sites (Svitkin et al., 1996).
The identification of a specific binding motif for FMRP in its own mRNA will now enable us to test these various hypotheses within the more physiological conditions of protein synthesis in transfected cells, or even in transgenic mice. This should also help to identify other potential mRNA targets of FMRP.
Materials and methods
Plasmid constructs and recombinant FMRP expression
Plasmids and primers used in this study to prepare the different RNAs are described in the Supplementary data, available at The EMBO Journal Online. The FMRP full-length protein (Iso-1) was prepared as a fusion protein with GST in a baculovirus system according to Bardoni et al. (1999). GST–FMRP was eluted from glutathione–Sepharose 4B (Pharmacia) in elution buffer (50 mM Tris–HCl pH 7.5, 100 mM KCl, 20 mM reduced glutathione).
RNA preparation
The different fragments derived from FMR1 RNA were synthesized by in vitro transcription with T7 RNA polymerase from pTL1 (Sittler et al., 1996) derivative plasmids linearized with _Pst_I, except for RNA N6 and N11 for which linearization was with _Xho_I and _Kpn_I, respectively. The RNAs were purified on a TSK G2000 column (Bio-Rad). Elution was performed with buffer consisting of 0.2 M sodium acetate and 1% methanol. RNAs were then ethanol precipitated and resuspended in the appropriate buffer. For binding assays, RNAs were labeled co-transcriptionally by incorporation of [α-32P]ATP. The 3′-end labeling of RNA was performed using T4 RNA ligase and [32P]pCp. For 5′-end labeling, RNAs with free 5′-OH groups were prepared by in vitro transcription using T7 RNA polymerase in the presence of 4 mM ApG and 1 mM NTPs according to Pitulle et al. (1992). 5′-end labeling was performed with T4 polynucleotide kinase and [γ-32P]ATP. Labeled RNAs were purified on a 6% polyacrylamide gel in 8 M urea, except for RNAs longer than 1000 bases, which were purified on a 1% low-melting agarose gel (Nusieve). Capped luciferase mRNA transcripts for in vitro translation were transcribed from pFlashSV40 constructs linearized with _Spe_I using the kit mMessage mMachine T7 (Ambion).
RNA binding assays
Assay on immobilized FMRP. Labeled RNAs (80 000 c.p.m., 5 fmol) were renatured for 10 min at 40°C in 40 µl of binding buffer [50 mM Tris–HCl pH 7.4 at 4°C, 1 mM MgCl2, 1 mM EDTA, 150 mM KCl, 1 mM dithiothreitol (DTT)] with 8 U of RNasin (Promega), 10 µg of E.coli total tRNA and 0.01% bovine serum albumin (BSA). The RNA was then added to 10 µl of GST–FMRP bound to glutathione–Sepharose beads (50% in binding buffer). RNA–protein complexes were formed for 15 min at 0°C with constant mild agitation. After incubation, beads were pelleted (30 s at 1000 r.p.m.) and supernatants transferred to new tubes. The beads were then washed twice with 50 µl of ice-cold binding buffer. The amount of radioactivity on beads, in washes and in supernatants was measured by Cerenkov counting.
Gel retardation assay. Labeled RNAs (80 000 c.p.m., 5 fmol), renatured for 10 min at 40°C in binding buffer were incubated for 10 min at 0°C in 10 µl of binding buffer with 0–0.5 pmol of GST–FMRP with 2 µg of E.coli total tRNA, 1% BSA and 8 U of RNasin. After addition of 2 µl of glycerol, the mixture was loaded on a 4% polyacrylamide gel (bisacrylamide:acrylamide 1:37.5, 2.5% glycerol, 0.5× TBE) and migration was performed for 1.5 h at 300 V at 4°C. The gel was then subjected to phosphoimaging and autoradiography. For competition experiments, unlabeled RNA (the amounts indicated in the figure legends) renatured for 15 min at 40°C in binding buffer was added simultaneously with labeled RNA.
Northwestern analysis
GST–FMRP (0.5 µg) was separated on a 10% SDS–polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Hybond-P Amersham) at 1 mA/cm2 for 90 min using a semi-dry blotting appartus (Bio-Rad). The membrane was then incubated in 20 ml of blocking buffer (10 mM Tris–HCl pH 7.8, 150 mM NaCl, 5 mg/ml yeast total tRNA) for 1 h, washed twice with 50 ml of 10 mM Tris–HCl pH 7.8 for 5 min and incubated with the N19 RNA (5 × 105 c.p.m./ml) in 10 ml of NWB buffer (10 mM Tris–HCl pH 7.8, 1 mM EDTA, 50 mM NaCl, 0.02% Ficoll, 0.02% polyvinyl pyrrolidone, 0.02% BSA). Excess RNA probe was removed by four successive 30 min washes with NWB buffer. The RNA probe bound to the protein was then detected by autoradiography.
RNA-binding site boundary determination
RNAs, either 5′- or 3′-end labeled (80 000 c.p.m.) were subjected to a limited hydrolysis by incubation in 50 mM NaCO3 pH 9 with 1 µg of total E.coli tRNA at 90°C for 4 min in 10 µl. Alkali ladders were precipitated by ethanol and then incubated in the binding buffer with GST–FMRP immobilized on glutathione–Sepharose beads, as described above. After one wash with 50 µl of binding buffer, bound RNAs were eluted from the beads by 50 µl of 7 M urea, 1 mM EDTA, 20 mM sodium acetate pH 5.8. After sedimentation of the beads (30 s at 1000 r.p.m.), the supernatant was submitted to a phenol/chloroform (v/v) extraction and the RNAs were ethanol precipitated. Recovered RNAs were then analyzed on an 8% polyacrylamide–urea gel followed by autoradiography.
Primer extension
RNA (2 pmol) and 105 c.p.m. of [γ-32P]ATP-labeled primer were heated to 95°C for 1 min and annealed at 25°C for 5 min in 6 µl of 50 mM HEPES-Na pH 7.0, 100 mM NaCl or KCl as indicated, and 50 mM EDTA. Extensions were carried out for 30 min at 37°C by adding 9 µl of a mixture containing 50 mM Tris–HCl pH 8.3 at 37°C, 6 mM MgCl2, 40 mM NaCl or KCl, 0.3 mM of each dNTP and 5 U of AMV reverse transcriptase (Appligene). Reactions were stopped by ethanol precipitation and reaction products were analyzed on 8 or 15% polyacrylamide– 8 M urea gels followed by autoradiography.
Probing with DMS
3′-end labeled RNA (150 000 c.p.m.) was renatured at 40°C for 15 min in 50 mM sodium cacodylate, 1 mM MgCl2, 1 mM EDTA, 1 mM DTT and 150 mM KCl or NaCl, as indicated in the text. Modifications were performed for 5 min at 20° C in a 25 µl final volume with 1 µl of DMS (Fluka) diluted 1:5 (v/v) in ethanol in the presence of 1 µg of E.coli total tRNA. Reactions were stopped by ethanol precipitation, and modified RNA was then treated and analyzed according to Brunel and Romby (2000).
In vitro translation
GST–FMRP or GST (0–50 ng) were incubated for 10 min at 0°C in binding buffer with 2 ng of the respective mRNA in the presence of 43 ng of the 3′-UTR of bicoid RNA as competitor RNA. Translation reactions were then performed using a rabbit reticulocyte lysate system (Promega). Reaction mixtures (final volume 9 µl) containing rabbit reticulocyte lysate (6.25 µl), 20 µM amino acids and 7 U of RNasin were incubated with the RNA–protein mixture at 30°C for 60 min. Protein elution buffer was added in all reaction assays to equalize with protein input volumes. Luciferase assays (Promega) were performed according to the manufacturer’s instructions with a Lumat (Berthold) luminometer.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Pascale Romby for her valuable suggestions and critical reading of the manuscript, Annette Schenck for discussions and help with FMRP preparation, and Isabelle Kolb-Cheynel for help with baculovirus culture. This work was supported by funding from the Centre National de la Recherche Scientifique and by Fraxa Foundation Inc. to J.L.M. and B.B.
References
- Ashley C.T. Jr, Wilkinson,K.D., Reines,D. and Warren,S.T. (1993a) FMR1 protein: conserved RNP family domains and selective RNA binding. Science, 262, 563–566. [DOI] [PubMed] [Google Scholar]
- Ashley C.T., Sutcliffe,J.S., Kunst,C.B., Leiner,H.A., Eichler,E.E., Nelson,D.L. and Warren,S.T. (1993b) Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nature Genet., 4, 244–251. [DOI] [PubMed] [Google Scholar]
- Bardoni B., Schenck,A. and Mandel,J.L. (1999) A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Hum. Mol. Genet., 8, 2557–2566. [DOI] [PubMed] [Google Scholar]
- Bashkirov V.I., Scherthan,H., Solinger,J.A., Buerstedde,J.M. and Heyer,W.D. (1997) A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol., 136, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V., Small,K., Lakkis,L., Feng,Y., Gunter,C., Wilkinson,K.D. and Warren,S.T. (1998) Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J. Biol. Chem., 273, 15521–15527. [DOI] [PubMed] [Google Scholar]
- Brunel C. and Romby,P. (2000) Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol., 318, 3–21. [DOI] [PubMed] [Google Scholar]
- Ceman S., Brown,V. and Warren,S.T. (1999) Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X related proteins as components of the complex. Mol. Cell. Biol., 19, 7925–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C. and Moore,P.B. (1992) Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Biochemistry, 31, 8406–8414. [DOI] [PubMed] [Google Scholar]
- Christiansen J., Kofod,M. and Nielsen,F.C. (1994) A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res., 22, 5709–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E., Voeller,D., Koeller,D.M., Drake,J.C., Takimoto,C.H., Maley,G.F., Maley,F. and Allegra,C.J. (1993) Identification of an RNA binding site for human thymidylate synthase. Proc. Natl Acad. Sci. USA, 90, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery T.A., Harris,J.B., Willems,P.J., Oostra,B.A., Irwin,S.A., Weiler,I.J. and Greenough,W.T. (1997) Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl Acad. Sci. USA, 94, 5401–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin F., Bouillon,M., Fortin,A., Morin,S., Rousseau,F. and Khandjian,E.W. (1997) The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet., 6, 1465–1472. [DOI] [PubMed] [Google Scholar]
- Devys D., Lutz,Y., Rouyer,N., Bellocq,J.P. and Mandel,J.L. (1993) The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nature Genet., 4, 335–340. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Eberhart D.E., Malter,H.E., Feng,Y. and Warren,S.T. (1996) The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet., 5, 1083–1091. [DOI] [PubMed] [Google Scholar]
- Ercikan-Abali E.A., Banerjee,D., Waltham,M.C., Skacel,N., Scotto,K.W. and Bertino,J.R. (1997) Dihydrofolate reductase protein inhibits its own translation by binding to dihydrofolate reductase mRNA sequences within the coding region. Biochemistry, 36, 12317–12322. [DOI] [PubMed] [Google Scholar]
- Feng Y., Absher,D., Eberhart,D.E., Brown,V., Malter,H.E. and Warren,S.T. (1997) FMRP associates with polyribosomes as an mRNP and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell, 1, 109–118. [DOI] [PubMed] [Google Scholar]
- Fry M. and Loeb,L.A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L. and Benchimol,S. (1997) Participation of the human p53 3′UTR in translational repression and activation following γ-irradiation. EMBO J., 16, 4117–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K., Parsons,J.T., Beard,J.W., Beard,D. and Green,M. (1970) Mechanism of carcinogenesis by RNA tumor viruses. 3. Formation of RNA, DNA complex and duplex DNA molecules by the DNA polymerase(s) of avian myeloblastosis virus. Proc. Natl Acad. Sci. USA, 67, 1432–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T.J., Rice,P.M., Thompson,J.D. and Heringa,J. (1993) KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem. Sci., 18, 331–333. [DOI] [PubMed] [Google Scholar]
- Gray N.K. and Hentze,M.W. (1994) Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J., 13, 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman R.J. (1996) Fragile X Syndrome: Diagnosis, Treatment and Research. Johns Hopkins University Press, Baltimore, MD.
- Hamilton B.J., Burns,C.M., Nichols,R.C. and Rigby,W.F. (1997) Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription and phosphorylation. J. Biol. Chem., 272, 28732–28741. [DOI] [PubMed] [Google Scholar]
- Imbert G., Feng,Y., Nelson,D., Warren,S.T. and Mandel,J.-L. (1998) FMR1 and mutations in fragile X syndrome: molecular biology, biochemistry and genetics. In Warren,S.T. and Wells,R.D. (eds), Genetic Instabilities and Hereditary Neurological Diseases. Academic Press, pp. 27–53.
- Jin P. and Warren,S.T. (2000) Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet., 9, 901–908. [DOI] [PubMed] [Google Scholar]
- Jing N., Marchand,C., Liu,J., Mitra,R., Hogan,M.E. and Pommier,Y. (2000) Mechanism of inhibition of HIV-1 integrase by G-tetrad-forming oligonucleotides in vitro. J. Biol. Chem., 275, 21460–21467. [DOI] [PubMed] [Google Scholar]
- Kang C., Zhang,X., Ratliff,R., Moyzis,R. and Rich,A. (1992) Crystal structure of four-stranded Oxytricha telomeric DNA. Nature, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- Katahira M., Moriyama,K., Kanagawa,M., Saeki,J., Kim,M.H., Nagaoka,M., Ide,M., Uesugi,S. and Kono,T. (1995) RNA quadruplex containing G and A. Nucleic Acids Symp. Ser., 34, 197–8. [PubMed] [Google Scholar]
- Kim J., Cheong,C. and Moore,P.B. (1991) Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature, 351, 331–332. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA, 83, 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1989) Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol., 9, 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B., Ostareck,D., Keidel,E.M., Ostareck-Lederer,A. and Fischer,U. (2001) Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet., 10, 329–338. [DOI] [PubMed] [Google Scholar]
- Laughlan G., Murchie,A.I., Norman,D.G., Moore,M.H., Moody,P.C., Lilley,D.M. and Luisi,B. (1994) The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science, 265, 520–524. [DOI] [PubMed] [Google Scholar]
- Lee J.S. (1990) The stability of polypurine tetraplexes in the presence of mono- and divalent cations. Nucleic Acids Res., 18, 6057–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang,Y., Ku,L., Wilkinson,K.D., Warren,S.T. and Feng,Y. (2001) The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res., 29, 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Parsels,L.A., Voeller,D.M., Allegra,C.J., Maley,G.F., Maley,F. and Chu,E. (2000) Characterization of a _cis_-acting regulatory element in the protein coding region of thymidylate synthase mRNA. Nucleic Acids Res., 28, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lee,A. and Gilbert,W. (1995) Gene disruption of a G4-DNA-dependent nuclease in yeast leads to cellular senescence and telomere shortening. Proc. Natl Acad. Sci. USA, 92, 6002–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A.W., Bogdarina,I., Schroeder,E., Taylor,I.A. and Kneale,G.G. (2000) Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures. J. Mol. Biol., 301, 575–584. [DOI] [PubMed] [Google Scholar]
- Pitulle C., Dorsch,M., Kazda,J., Wolters,J. and Stackebrandt,E. (1992) Phylogeny of rapidly growing members of the genus Mycobacterium. Int. J. Syst. Bacteriol., 42, 337–343. [DOI] [PubMed] [Google Scholar]
- Rangan A., Fedoroff,O.Y. and Hurley,L.H. (2001) Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem., 276, 4640–4646. [DOI] [PubMed] [Google Scholar]
- Romby P. et al. (1996) The expression of E.coli threonyl-tRNA synthetase is regulated at the translational level by symmetrical operator–repressor interactions. EMBO J., 15, 5976–5987. [PMC free article] [PubMed] [Google Scholar]
- Schenck A., Bardoni,B., Moro,A., Bagni,C. and Mandel,J.-L. (2001) A highly conserved protein family interacting with the fragile X mental retardation protein (FNRP) and displaying selective interactions with FNRP-related proteins FXR1P and FXR2P. Proc. Natl Acad. Sci. USA, 98, 8844–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper W., Meinsma,D., Holthuizen,P.E. and Sussenbach,J.S. (1995) Long-range RNA interaction of two sequence elements required for endonucleolytic cleavage of human insulin-like growth factor II mRNAs. Mol. Cell. Biol., 15, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D. and Gilbert,W. (1990) A sodium–potassium switch in the formation of four-stranded G4-DNA. Nature, 344, 410–414. [DOI] [PubMed] [Google Scholar]
- Simonsson T., Pecinka,P. and Kubista,M. (1998) DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res., 26, 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Choi,M., Siomi,M.C., Nussbaum,R.L. and Dreyfuss,G. (1994) Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell, 77, 33–39. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Zhang,Y., Siomi,H. and Dreyfuss,G. (1996) Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol., 16, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittler A., Devys,D., Weber,C. and Mandel,J.L. (1996) Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum. Mol. Genet., 5, 95–102. [DOI] [PubMed] [Google Scholar]
- Smith F.W. and Feigon,J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- Sundquist W.I. and Heaphy,S. (1993) Evidence for interstrand quadruplex formation in the dimerization of human immuno deficiency virus 1 genomic RNA. Proc. Natl Acad. Sci. USA, 90, 3393–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist W.I. and Klug,A. (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- Sung Y.J., Conti,J., Currie,J.R., Brown,W.T. and Denman,R.B. (2000) RNAs that interact with the fragile X syndrome RNA binding protein FMRP. Biochem. Biophys. Res. Commun., 275, 973–980. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Ovchinnikov,L.P., Dreyfuss,G. and Sonenberg,N. (1996) General RNA binding proteins render translation cap dependent. EMBO J., 15, 7147–7155. [PMC free article] [PubMed] [Google Scholar]
- Tamanini F. et al. (1999) Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum. Mol. Genet., 8, 863–869. [DOI] [PubMed] [Google Scholar]
- Wan L., Dockendorff,T.C., Jongens,T.A. and Dreyfuss,G. (2000) Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell. Biol., 20, 8536–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler I.J. et al. (1997) Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl Acad. Sci. USA, 94, 5395–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- Williamson J.R., Raghuraman,M.K. and Cech,T.R. (1989) Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell, 59, 871–880. [DOI] [PubMed] [Google Scholar]
- Zahler A.M., Williamson,J.R., Cech,T.R. and Prescott,D.M. (1991) Inhibition of telomerase by G-quartet DNA structures. Nature, 350, 718–720. [DOI] [PubMed] [Google Scholar]
- Zhang Y., O’Connor,J.P., Siomi,M.C., Srinivasan,S., Dutra,A., Nussbaum,R.L. and Dreyfuss,G. (1995) The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J., 14, 5358–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]