SMN interacts with a novel family of hnRNP and spliceosomal proteins (original) (raw)
Abstract
Spinal muscular atrophy (SMA) is a common neurodegenerative disease caused by deletion or loss-of-function mutations of the survival of motor neurons (SMN) protein. SMN is in a complex with several proteins, including Gemin2, Gemin3 and Gemin4, and it plays important roles in small nuclear ribonucleoprotein (snRNP) biogenesis and in pre-mRNA splicing. Here, we characterize three new hnRNP proteins, collectively referred to as hnRNP Qs, which are derived from alternative splicing of a single gene. The hnRNP Q proteins interact with SMN, and the most common SMN mutant found in SMA patients is defective in its interactions with them. We further demonstrate that hnRNP Qs are required for efficient pre-mRNA splicing in vitro. The hnRNP Q proteins may provide a molecular link between the SMN complex and splicing.
Keywords: hnRNP proteins/pre-mRNA splicing/spinal muscular atrophy/survival of motor neurons
Introduction
Spinal muscular atrophy (SMA) is a common, autosomal recessive neurodegenerative disease. The pathological hallmark of SMA is degeneration and loss of spinal cord α-motor neurons resulting in paralysis and muscular atrophy (reviewed in Melki, 1997). The survival of motor neurons gene (SMN) is the SMA determining gene (Lefebvre et al., 1995). The SMN gene is duplicated, giving rise to one centromeric (SMN2) and one telomeric (SMN1) copy on chromosome 5 at q13. SMA is caused by deletions or loss-of-function mutations of SMN1 (reviewed in Burghes, 1997). The SMN protein is ubiquitously expressed and its function is essential in all organisms studied, including mouse, Caenorhabditis elegans and Schizosaccharomyces pombe (Schrank et al., 1997; Miguel-Aliaga et al., 1999; Hannus et al., 2000; Owen et al., 2000; Paushkin et al., 2000). In humans, the amount of full-length SMN protein produced by SMN2 is sufficient for viability of all cells except motor neurons. Decreased protein levels of SMN correlate with phenotypic severity of SMA (Coovert et al., 1997; Lefebvre et al., 1997). Recently, several mouse models for SMA were described and all demonstrate that reduced SMN protein levels cause SMA (Frugier et al., 2000; Hsieh-Li et al., 2000; Jablonka et al., 2000; Monani et al., 2000). A general requirement of SMN for cell viability has recently been demonstrated (Wang and Dreyfuss, 2001). Thus, motor neurons are particularly sensitive to SMN reduction compared with other cells.
The SMN protein, itself oligomeric, is in a complex with three proteins known as Gemin2 (formerly SIP1), Gemin3 (a DEAD box RNA helicase) and Gemin4 (Liu et al., 1997; Charroux et al., 1999, 2000). In addition, SMN interacts directly with several protein substrates, including the core spliceosomal proteins (Sm proteins; Liu et al., 1997; Friesen and Dreyfuss, 2000). The SMN complex is found both in the cytoplasm and in the nucleus where it is concentrated in nuclear bodies known as gems (Liu and Dreyfuss, 1996). The Sm proteins are bound to all uridine-rich small nuclear RNAs (snRNAs) that function in pre-mRNA splicing (i.e. U1, U2, U4 and U5), with the exception of U6 snRNA. Sm proteins assemble on the Sm sites of snRNAs, in the cytoplasm, to form the small nuclear ribonucleoprotein particle (snRNP) core. Upon further processing, which includes hypermethylation of the 5′ cap of the snRNAs, the mature snRNPs are imported into the nucleus (reviewed in Mattaj, 1988; Mattaj et al., 1993). In addition to the common Sm proteins each snRNP is associated with specific proteins (reviewed in Will and Luhrmann, 1997). In the nucleus the stepwise assembly of spliceosomal snRNPs on pre-mRNAs, along with numerous other proteins, leads to the formation of the spliceosome (reviewed in Moore et al., 1993; Staley and Guthrie, 1998; Burge et al., 1999). SMN interacts directly with the arginine- and glycine-rich (RG) domains present in a subset of Sm proteins (Friesen and Dreyfuss, 2000) and functions in the biogenesis of spliceosomal snRNPs in the cytoplasm (Fischer et al., 1997). In the nucleus, SMN has a critical role in pre-mRNA splicing (Pellizzoni et al., 1998; Meister et al., 2000) and transcription (Pellizzoni et al., 2001). The molecular mechanism of SMN function in these processes is, however, unknown.
The hnRNP proteins are a family of ∼30 proteins that bind pre-mRNA and mRNA and play critical roles in all aspects of mRNA processing, including pre-mRNA splicing, mRNA transport and mRNA translation, stability and turnover (reviewed in Dreyfuss et al., 1993; Burd and Dreyfuss, 1994; Krecic and Swanson, 1999; Shyu and Wilkinson, 2000). Several members of the hnRNP family of proteins play important roles in regulating alternative pre-mRNA splicing. HnRNP A1, for example, can regulate 5′ splice site selection (Mayeda and Krainer, 1992). Neural-specific splicing of the c-src N1 exon is regulated by an intronic enhancer, which binds hnRNPs H and F (Chou et al., 1999) and a repressor element that binds the polypyrimidine tract binding protein (PTB/hnRNP I; Chan and Black, 1997). Interestingly, a dramatic remodeling of the PTB-containing hnRNP complex, prior to spliceosomal assembly on the c-src pre-mRNA has recently been demonstrated, strongly suggesting that regulated hnRNP complex remodeling may be critical for proper splice-site selection (Chou et al., 1999). A more general role for hnRNP F in splicing has also been reported (Gamberi et al., 1997).
Here we report the identification and functional characterization of a novel family of three hnRNP proteins (hnRNPs Q1, Q2 and Q3; collectively referred to as hnRNP Q proteins), which are produced by alternative splicing from a single gene and interact with SMN in vitro and in vivo. We show that the hnRNP Q proteins are novel components of the spliceosome and are required for efficient pre-mRNA splicing in vitro. The hnRNP Q proteins represent excellent candidates for mediating the nuclear function of the SMN complex, and they may provide a molecular link between SMN and the spliceosome.
Results
hnRNP Qs are a novel family of hnRNP proteins
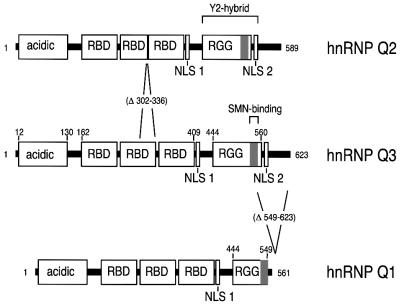
Using SMN as bait in a yeast two-hybrid screen we isolated four clones encoding a 122-amino-acid protein fragment that is identical to a portion of a 623-amino-acid protein of unknown function, named NSAP1, for NS1-associated protein 1 or Gry-rbp (for glycine- and tyrosine-rich RNA binding protein). NSAP1 was isolated previously by a yeast two-hybrid screen on a HeLa cDNA library using as bait a portion of NS1, a protein from minute virus of mice (Harris et al., 1999). However, in vitro or in vivo (mammalian cells) interaction between NS1 and NSAP1 could not be demonstrated (Harris et al., 1999). Gry-rbp is listed as a sequence-only entry in the DDBJ/EMBL/GenBank database (accession No. AF037448). Database searches revealed the presence of several expressed sequence tags (ESTs) coding for two additional, alternatively spliced forms of NSAP1. All three NSAP1 isoforms were cloned and sequenced and were assigned as novel hnRNP proteins: Q1/Q2/Q3 (see below; sequences deposited in DDBJ/EMBL/GenBank under accession Nos AY034483, AY034482 and AY034481, respectively). The primary structure is presented in Figure 1 and we shall collectively refer to them as the hnRNP Q proteins. Database searches revealed that the gene encoding the hnRNP Qs is located on chromosome 6 (Unigene Cluster Hs.155489). hnRNP Q3 is composed of an ∼140 N-terminal acidic domain followed by three consecutive RNP motifs (also known as RNA binding domains, RBD), a putative nuclear localization signal (NLS), an unusually large ∼120-amino-acid arginine- and glycine-rich domain (RGG box), a second putative NLS and an ∼40-amino-acid C-terminal domain rich in glutamine and asparagine residues (Figure 1). The sequence of hnRNP Q3 is 83% identical to that of hnRNP R, a recently identified hnRNP protein (Hassfeld et al., 1998; alignment not shown). The protein products of the yeast two-hybrid clones correspond to amino acids 427–549 of hnRNP Q3, which represent most of the RGG box (Figure 1). Two smaller isoforms appear to be derived by alternative splicing of hnRNP Q3: hnRNP Q2 lacks 34 amino acids from the second RBD (amino acids 302–336), and hnRNP Q1 lacks the last 74 amino acids from the C-terminus of hnRNP Q3, which have been replaced by the sequence VKGVEAGPDLLQ (Figure 1).
Fig. 1. Schematic representation of hnRNP Q isoforms. Acidic, protein domain rich in acidic amino acids; RBD, RNA binding domain; RGG, arginine-glycine-glycine-rich domain; NLS, nuclear localization signal. Numbers refer to amino acids (aa). hnRNP Q2 lacks aa 302–336 (Δ302–336) from the second RBD of hnRNP Q3; hnRNP Q1 lacks aa 549–623 (Δ549–623) from hnRNP Q3 and contains the unique C-terminal sequence VKGVEAGPDLLQ. Y2-hybrid clones: mapping of protein products of the clones isolated from the yeast dihybrid screen (see text). Gray area represents the SMN binding domain (aa 518–549 of hnRNP Q3).
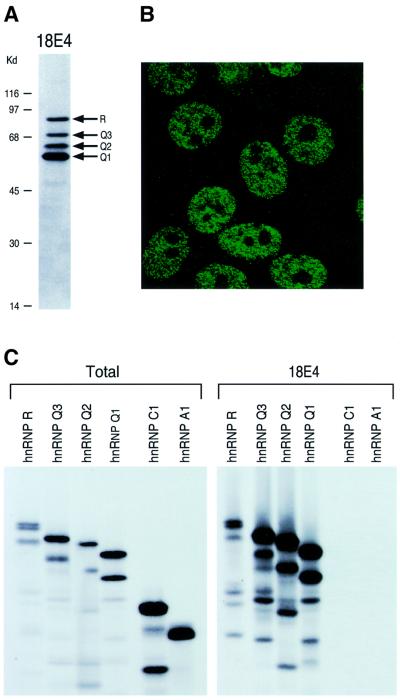
To further characterize the hnRNP Qs we produced monoclonal antibodies against bacterially expressed, recombinant hnRNP Q3, and one of the hybridomas, designated 18E4, was selected for further studies. Monoclonal antibody (mAb) 18E4 recognizes four bands on a western blot of total HeLa cell lysate corresponding to molecular masses of ∼80, ∼70, ∼60 and ∼55 kDa (Figure 2A). The observation of an ∼80 kDa band was unexpected, as hnRNP Q has three isoforms of ∼70– 55 kDa. The ∼80 kDa reactive band was subsequently identified as hnRNP R, which migrates at ∼80 kDa on SDS–PAGE (Hassfeld et al., 1998) and is highly similar (∼83%) to the hnRNP Qs. Indirect immunofluorescence laser confocal microscopy on HeLa cells using 18E4 showed that the hnRNP Q proteins are localized to the nucleoplasm displaying a punctate staining pattern (Figure 2B). We also performed immunoprecipitations using 18E4 with in vitro translated hnRNP R and the three isoforms of hnRNP Q under very stringent conditions (1% Empigen). mAb 18E4 immunoprecipitated hnRNP R and all three hnRNP Q isoforms but not other hnRNP proteins tested (Figure 2C).
Fig. 2. Properties of monoclonal antibody 18E4. (A) 18E4 recognizes four bands corresponding to hnRNPs Q1, Q2, Q3 and R (indicated), on the western blot of total HeLa cell lysate. Molecular weight markers (kDa) are shown on the left. (B) Laser confocal image of indirect immunofluorescence on HeLa cells using 18E4. (C) Immunoprecipi tations of the indicated proteins, produced by in vitro translation and labeled with [35S]methionine, were performed with monoclonal antibody 18E4 under stringent conditions (1% Empigen); immuno precipitated proteins were resolved by SDS–PAGE and detected by fluorography. Lanes marked total contain 10% of input fraction used for immunoprecipitations. The doublets seen in some in vitro translated polypeptides are due to aberrant translation initiation.
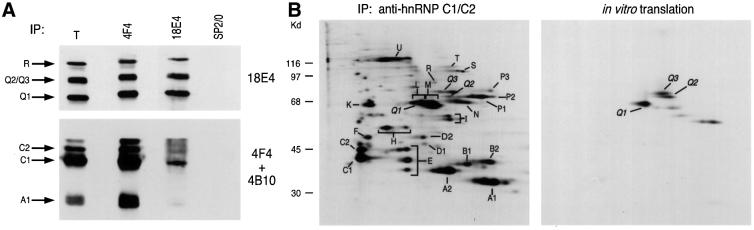
Since the SMN-interacting proteins isolated from the yeast two-hybrid screen were highly similar to hnRNP R, we expected that they too are hnRNP proteins. To investigate this possibility we performed immunoprecipitations using 4F4, a monoclonal antibody that recognizes hnRNP C1/C2 and specifically immunoprecipitates the hnRNP complexes (Choi and Dreyfuss, 1984a,b; Pinol-Roma et al., 1988), or with 18E4 (against hnRNP Qs/R), or SP2/0, as a negative control. The immunoprecipitates were then analyzed by western blotting with mAbs 18E4, 4F4 and 4B10 (against hnRNP A1). As seen in Figure 3A, hnRNPs Q1, Q2, Q3 and R are present in the 4F4 immunoprecipitates, demonstrating that they are indeed components of hnRNP complexes. However, hnRNP A1 and C do not co-immunoprecipitate with the hnRNP Q and R proteins, suggesting that hnRNP Qs form subcomplexes, distinct from the general hnRNP complexes obtained by immunoprecipitations using the anti-hnRNP C1/C2 antibody 4F4 (Pinol-Roma et al., 1988). We further assigned the three hnRNP Q isoforms as specific spots on the two-dimensional (2D) map of hnRNP complexes (Pinol-Roma et al., 1988). To do this, hnRNP complexes were first immunopurified using 4F4 from [35S]methionine-labeled HeLa cells and resolved by 2D non-equilibrium pH gradient gel electrophoresis (2D-NEPHGE). In parallel, 2D-NEPHGE was carried out on in vitro translated products from the three isoforms of hnRNP Q (the ∼70, ∼60 and ∼55 kDa bands seen on the western blot; Figure 2A) and hnRNP R, either alone or in various combinations, and these were compared with the pattern of migration of hnRNP complexes. A representative result is shown in Figure 3B. This analysis revealed that the 623-amino-acid isoform corresponds to a spot previously designated as hnRNP Q (Pinol-Roma et al., 1988) and that we now rename hnRNP Q3. The 589-amino-acid isoform is slightly more basic, migrating just below hnRNP Q3 and we name it hnRNP Q2. The 561-amino-acid isoform co-migrates with the spot corresponding to hnRNP L, a previously cloned hnRNP (Pinol-Roma et al., 1989), and we name this isoform hnRNP Q1. Despite the co-migration of hnRNP Q3 and hnRNP L (both proteins have the same predicted isoelectric point, pi, of 7.42), the amino acid sequences of the two proteins are different. The faster migrating spots seen in Figure 3B represent shorter protein products, most probably due to aberrant internal translation initiation from the template mRNA used in the in vitro translation reactions. These in vitro produced polypeptides are also seen as faster migrating bands on the one dimension SDS–PAGE shown in Figures 2C and 4A.
Fig. 3. (A) hnRNP Qs and R are part of hnRNP complexes and form distinct hnRNP subcomplexes. Immunoprecipitations (IP) were performed from total HeLa cell lysates with antibodies against hnRNP C1/C2 (4F4), hnRNPs Q1, Q2, Q3 and R (18E4) or SP2/0, as negative control. The immuno precipitates were analyzed by western blotting with monoclonal antibodies 18E4, 4F4 and 4B10 (against hnRNP A1). Total (T) shows ∼3% of the input used for IPs. (B) Assignment of hnRNP Q isoforms as hnRNPs Q1, Q2 and Q3 on the 2D map of hnRNP complexes. hnRNP complexes were immunopurified with monoclonal antibody 4F4 against hnRNP C1/C2 (IP, anti-hnRNP C1/C2) from [35S]methionine-labeled HeLa cells and were analyzed by 2D non-equilibrium pH gradient gel electrophoresis (2D-NEPHGE). The first dimension is NEPHGE and the second dimension is SDS–PAGE (11.5% polyacrylamide). Molecular weight markers (kDa) are shown on the left. In vitro translated and [35S]methionine-labeled hnRNP Q isoforms (in vitro translation) were combined and similarly analyzed on a separate gel under identical conditions; hnRNP proteins are labeled.
Fig. 4. Interaction of hnRNP Qs with SMN in vitro and in vivo; identification of a minimal SMN binding domain and interaction of hnRNP R with hnRNP Q3. (A) Schematic representation of hnRNP Q1 wild type and deletion mutants used in binding assays and amino acid sequence of minimal SMN binding domain. Numbers refer to amino acids. Gray area represents the RGG box. Amino acids are shown in single letter code; RG dipeptides and RGG sequences are highlighted. (B) Indicated proteins were produced by in vitro translation, labeled with [35S]methionine and incubated with recombinant GST–SMN; bound proteins were analyzed by SDS–PAGE and fluorography. (C) In vitro translated and [35S]methionine-labeled SMN was incubated with recombinant GST fused to the RGG domain of hnRNP Q1 [GST–Q1(RGG)] or with GST; bound protein was analyzed as above. Lanes marked total contain 10% of input fraction used for bindings. (D) hnRNPs Q1, Q2, Q3 and R associate with SMN in vivo. Immunoprecipitations (IP) were performed from total HeLa cell lysates with antibodies against SMN (α-exon7) or SP2/0 as negative control. The immunoprecipitates were analyzed by western blotting with monoclonal antibody 18E4 against hnRNPs Q1, Q2 Q3 and R. Total (T) shows ∼3% of the input used for IPs. (E) hnRNP R protein was produced by in vitro translation, labeled with [35S]methionine, and incubated with recombinant GST–hnRNP Q3; bound protein was analyzed as above; total refers to 10% of input fraction used for binding. The doublets seen in some in vitro translated polypeptides are due to aberrant translation initiation.
The hnRNP Q proteins interact with SMN in vitro and in vivo
To validate the yeast two-hybrid results we carried out in vitro binding experiments. For this assay recombinant glutathione _S_-tranferase (GST)–SMN fusion protein or GST was immobilized on glutathione–Sepharose beads and incubated with [35S]methionine-labeled proteins produced by in vitro transcription and translation in rabbit reticulocyte lysate. To exclude nucleic acid-mediated interactions, all in vitro translated products were treated with DNase I and RNase A prior to bindings. As shown in Figure 4B, all hnRNP Q isoforms, but not hnRNP R, C1 or A1, bind to GST–SMN. None of the proteins showed binding to GST. Purified, bacterially produced recombinant hnRNP Q3 also binds to recombinant GST–SMN, indicating that the interaction between the two proteins is direct (data not shown).
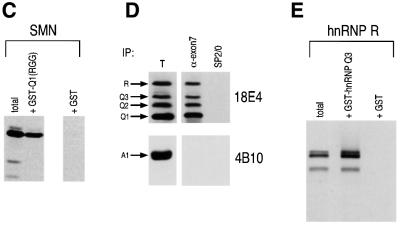
SMN interacts directly with the RG domains of Sm proteins (Friesen and Dreyfuss, 2000) and with the RGG box of RNA helicase A (RHA; Pellizzoni et al., 2001). Similarly, the RGG box of hnRNP Qs, like the RGG box of hnRNP U (Liu and Dreyfuss, 1996), interacts with SMN in the yeast two-hybrid system. To confirm this finding and to determine whether the RGG box of hnRNP Q is necessary and sufficient for binding to SMN, in vitro binding experiments were performed using immobilized GST– SMN with either wild-type in vitro translated hnRNP Q1 or three deletion mutants of hnRNP Q1 that lack either the entire RGG box [hnRNP Q1(1–410)] or portions of it [hnRNP Q1(1–480) and hnRNP Q1(1–517)], as illustrated in schematic form in Figure 4A. As shown in Figure 4B, none of the deletion mutants of hnRNP Q1 interacts with SMN. In a parallel experiment in vitro translated SMN bound efficiently to a fusion protein of GST with the RGG box of hnRNP Q1 (GST–Q1RGG) but not to GST alone, demonstrating that the RGG box of hnRNP Qs is sufficient for interaction with SMN (Figure 4C). In summary, the hnRNP Q proteins bind SMN via their RGG domains, and amino acids 518–549 of hnRNP Q1 are required for this interaction. The amino acid sequence of this minimal SMN-binding domain is shown in Figure 4A. Interest ingly, this sequence contains mostly RG dipeptides. This is similar to the requirement for RG-rich sequences for SMN binding to the Sm proteins (Friesen and Dreyfuss, 2000).
To determine whether SMN and hnRNP Qs are in a complex in vivo, immunoprecipitations were performed from HeLa cell lysates, using anti-exon 7 antiserum, a highly specific polyclonal serum that recognizes an epitope located in the last 16 amino acids of human SMN, encoded by exon 7 of the SMN gene (Pellizzoni et al., 1998). The immunoprecipitated proteins were resolved by SDS–PAGE, blotted and probed with the anti-hnRNP Qs monoclonal antibody 18E4 or with 4B10 against hnRNP A1. As seen in Figure 4D, hnRNP R and hnRNP Qs, but not hnRNP A1, are co-immunoprecipitated with SMN, suggesting that they are associated in vivo. Although hnRNP R does not bind directly to SMN, it interacts with hnRNP Q1 (Figure 4E) and this interaction may explain the co-immunoprecipitation of hnRNP R with anti-SMN antibodies. Alternatively, hnRNP R may interact with one of the other proteins of the SMN complex or it may also interact with SMN in vivo but not in vitro.
The most prevalent SMN mutant found in SMA patients is defective in its interactions with the hnRNP Q proteins
To define more precisely the domain of SMN that is required for binding to the hnRNP Q proteins, we tested the binding of in vitro translated SMN deletion mutants to recombinant GST–hnRNP Q3 fusion protein. All of the SMN deletion mutants used for these experiments contained an N-terminal myc tag. As shown in Figure 5, an N-terminal deletion of SMN lacking the first 91 amino acids (SMNΔ91) bound to hnRNP Q3 as efficiently as wild-type SMN does. In contrast, deletion of the last 26 amino acids of SMN (SMNΔY/G), which encompasses the YG box, or deletion of the last 16 amino acids of SMN (SMNEx7), which corresponds to the most prevalent form found in SMA patients, were unable to bind to GST–hnRNP Q3. We conclude that the last 16 amino acids of SMN are required for binding to hnRNP Q1 and, more importantly, that the most common SMN mutant found in SMA patients is deficient in its capacity to interact with the hnRNP Q proteins.
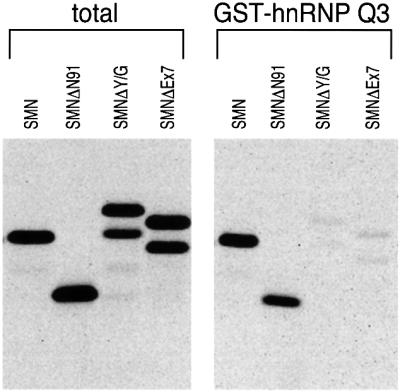
Fig. 5. The C-terminal domain of SMN is required for binding to hnRNP Q1, and SMNΔEx7, the most prevalent mutant found in SMA patients, shows defective interaction with hnRNP Q1. In vitro translated and [35S]methionine-labeled wild-type SMN and myc-tagged, N-terminal (SMNΔN91) or C-terminal (SMNΔY/G and SMNΔEx7) SMN deletions were incubated with recombinant GST–hnRNP Q1. Bound proteins were resolved by SDS–PAGE and detected by fluorography. The slower migration of SMNΔY/G is due to its anomalous electrophoretic behavior on SDS–PAGE gels. The doublets seen in the C-terminal myc-tagged SMN deletion proteins are due to the utilization of two translation initiation sites, one contributed from the myc epitope and the other from the first codon of SMN.
The hnRNP Q proteins are components of the spliceosome
Recently, a comprehensive survey of spliceosome-associated proteins by nanoelectrospray mass spectrometry identified several novel components (Neubauer et al., 1998). One of the major ones was hnRNP Q3 (then known as Gry-rbp; Neubauer et al., 1998), suggesting that hnRNP Qs may have a role in pre-mRNA splicing. To investigate this possibility we examined the possible role of hnRNP Qs in pre-mRNA splicing in an in vitro system. Three model pre-mRNAs were used, two of which contain a single intron: Ad2 derived from Adenovirus 2 major late transcription unit, and CDC, derived from chicken δ-crystallin gene (Kataoka et al., 2000), and a more complex pre-mRNA containing two introns (CDC-2, derived from chicken δ-crystallin gene; Ohno et al., 1987; Kataoka et al., 2000).
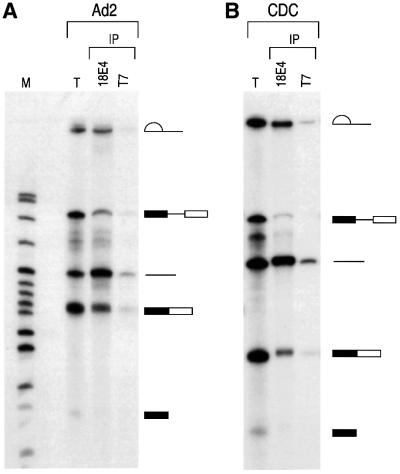
We first wished to determine whether hnRNP Qs are stably associated with pre-mRNA. For this we incubated 32P-labeled Ad2, CDC or CDC-2 pre-mRNA with HeLa nuclear extract under splicing conditions and performed immunoprecipitations using mAb 18E4. As shown in Figure 6, 18E4 efficiently immunoprecipitated intron-containing RNA intermediates, mRNA and pre-mRNA from in vitro splicing reactions of Ad2 or CDC-2 pre-mRNAs. When CDC pre-mRNA was used, the anti-hnRNP Qs antibody 18E4 preferentially immunoprecipitated intron-containing RNA intermediates and mRNA. As a negative control, immunoprecipitations with an equal amount of anti-T7 antibody, an unrelated monoclonal antibody, did not immunoprecipitate a significant amount of any of the splicing products or pre-mRNA. We also used the anti-hnRNP C proteins antibody 4F4 in immunoprecipitation experiments from in vitro splicing reactions using CDC-2 pre-mRNA. As shown in Figure 6C, 4F4 immunoprecipitated very efficiently the pre-mRNA and intron-containing RNA intermediates but not mature mRNA. These results demonstrate that the hnRNP Q proteins are associated with pre-mRNA and with intron-containing RNA intermediates, extending the observation that they are novel spliceosomal proteins.
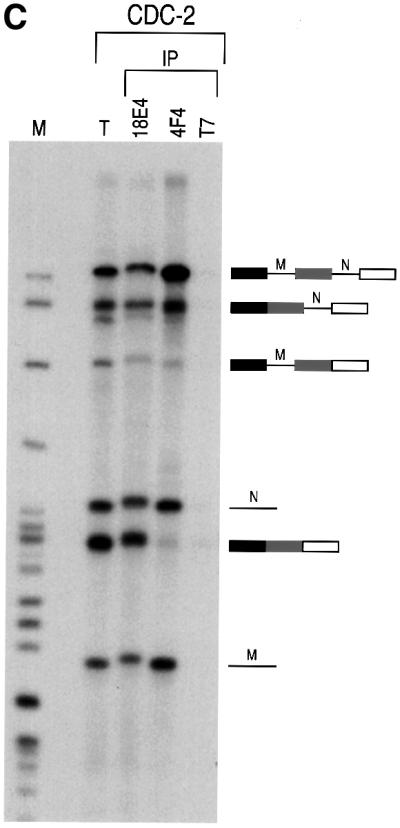
Fig. 6. hnRNP Qs associate with pre-mRNA, intron-containing RNA intermediates and mRNA. 32P-labeled Ad2 (A), CDC (B) or CDC-2 (C) pre-mRNAs were incubated with HeLa cell nuclear extracts under splicing conditions. Following immunoprecipitation with anti-hnRNP Qs and R antibody (18E4), anti-hnRNP C1/C2 antibody (4F4) and anti-T7 tag antibody (T7), as negative control, retained RNA was analyzed on 10% (A and B) or 6% (C) denaturing polyacrylamide gels. Lanes marked T (total) contain 5% of input RNA. Lanes marked M contain 32P-labeled pBR322/_Msp_I marker. RNA products are shown on the right in schematic form. Exons are represented as boxes and introns as lines or lariats. Designation of products and splicing intermediates of CDC-2 pre-mRNA (C) is based on previous characterization (Ohno et al., 1987).
The hnRNP Q proteins are required for efficient pre-mRNA splicing
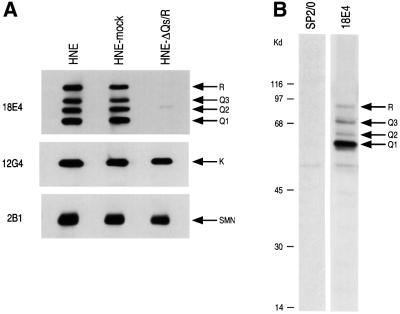
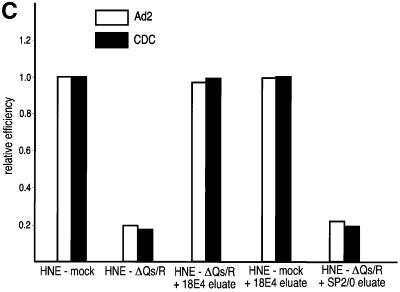
We next asked whether the hnRNP Q proteins have a function in pre-mRNA splicing. To address this, HeLa nuclear extracts were incubated with mAb 18E4 to immunodeplete the hnRNP Q and R proteins. To increase the likelihood of specificity of the depletion, immunodepletions were carried out at 700 mM KCl. The depleted nuclear extracts were subsequently dialyzed in splicing buffer (buffer D). As a negative control, mock depletion of nuclear extracts was carried out with the non-immune control antibody SP2/0 under identical conditions. The extent of depletion was monitored on equal aliquots of non-depleted, mock-depleted and hnRNP Qs- and R-depleted nuclear extracts by western blotting using various antibodies. As shown in Figure 7A, hnRNP Qs and hnRNP R were specifically depleted only from the nuclear extracts that were incubated with mAb 18E4 while the amounts of other proteins such as hnRNP K or SMN were not affected. The mock- and hnRNP Qs- and R-depleted nuclear extracts were then used for in vitro splicing reactions using 32P-labeled Ad2 and CDC as template pre-mRNAs. Equal amounts of extract, pre-mRNA and incubation time (1 h) were used for all experiments. As shown in Figure 8, splicing efficiency was significantly impaired for both pre-mRNAs in 18E4-immunodepleted, compared with mock-depleted extracts. The amount of mRNA produced in hnRNP Qs- and R-depleted extracts was considerably reduced with a concomitant increase in intron-containing RNA intermediates. These findings suggest that there is an overall decrease in splicing efficiency rather than a specific block of one of the two steps of pre-mRNA splicing.
Fig. 7. (A) Specific immunodepletion of hnRNPs Q1, Q2, Q3 and R from HeLa nuclear extracts (HNE). An equivalent amount of native (HNE), mock-depleted with SP2/0 (HNE-mock), or depleted with monoclonal antibody 18E4 (HNE-ΔQs/R) extracts, was separated by SDS–PAGE followed by western blotting with the indicated antibodies. HnRNPs Q1, Q2, Q3 and R are specifically depleted in the nuclear extracts that had been treated with 18E4 (HNE-ΔQs/R); levels of other hnRNP proteins (hnRNP K) or SMN remained essentially unaltered. (B) Purification of hnRNP Qs and R with immunoaffinity chromato graphy. Total HeLa cell lysate was passed through a column containing either covalently coupled 18E4 (against hnRNPs Q1, Q2, Q3 and R) or SP2/0 (negative control). After extensive washings, bound proteins were eluted with low pH, immediately neutralized and dialyzed in buffer D (not containing glycerol). Aliquots from the eluates (18E4 and SP2/0) were separated by SDS–PAGE and visualized by silver staining. The lane labeled 18E4 contains 1 µg of total protein. hnRNPs Q1, Q2, Q3 and R are present only in the 18E4 eluate.
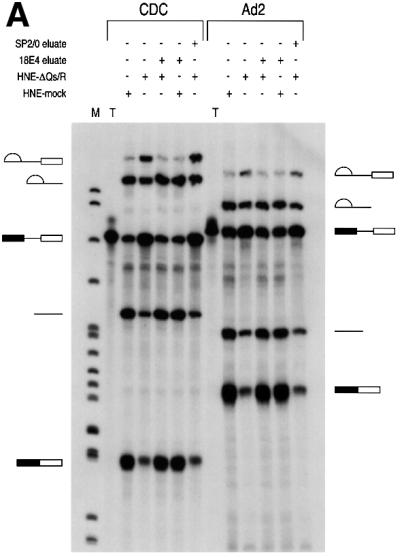
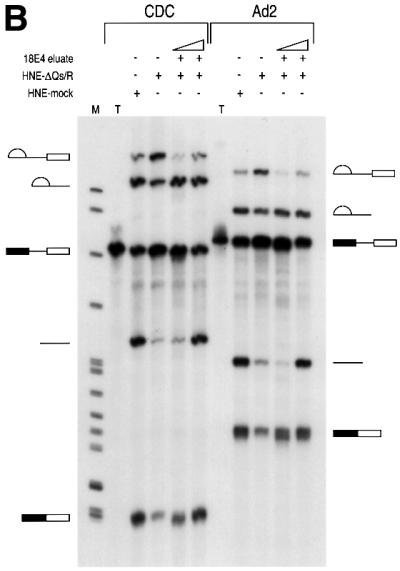
Fig. 8. hnRNP Qs are required for efficient pre-mRNA splicing. In vitro splicing reactions using 32P-labeled CDC and Ad2 pre-mRNAs were performed using HeLa nuclear extracts that had been depleted of hnRNP Q1, Q2, Q3 and R (HNE-ΔQs/R) or that had been mock depleted (HNE-mock). 25% of input pre-mRNA used in splicing reactions (but not subjected to splicing) is also shown (T). In (A), ∼400 ng of total protein of immunopurified hnRNP Q1, Q2, Q3 and R obtained from the 18E4-containing column (18E4 eluate), or an equal volume of eluate from the SP2/0-containing column (SP2/0 eluate), was added to nuclear extracts as indicated, prior to splicing. In (B), either ∼100 or ∼400 ng of total protein of immunopurified hnRNP Q1, Q2, Q3 and R (18E4 eluate) were added to nuclear extracts as indicated. After splicing, RNA products were analyzed on 6% denaturing polyacrylamide gels. Lanes marked M contain 32P-labeled pBR322/_Msp_I marker. The schematic RNA products are shown on the left for CDC pre-mRNA and on the right for Ad2 pre-mRNA. Exons are represented as boxes and introns as lines or lariats. (C) Quantitation of the data shown in (A). The relative efficiency of splicing in the indicated extracts was compared with that of the mock-depleted extracts (HNE-mock), which was arbitrarily set to 1.0. White bars represent data from splicing reactions using Ad2 pre-mRNA and black bars represent data from CDC pre-mRNA. The quantitative analysis was performed as indicated (Materials and methods).
To further assess the possible role of the hnRNP Q proteins in pre-mRNA splicing, we initially attempted to restore the splicing inhibition seen with hnRNP Qs- and R-depleted extracts by adding recombinant, bacterially produced hnRNP Q3. However, addition of hnRNP Q3 alone did not restore splicing. This suggested that either the bacterially produced protein is not active or that other hnRNP Q isoforms or hnRNP R are required for efficient splicing. To address this, all hnRNP Q isoforms and hnRNP R were immunopurified from HeLa extracts using 18E4 cross-linked to an agarose matrix. After extensive washings the captured proteins were eluted with low pH, immediately neutralized, dialyzed in splicing buffer and used for reconstitution experiments. As a negative control, eluate was produced in exactly the same manner, using non-immune antibody (SP2/0). To determine the purity and concentration of the eluates, aliquots were subjected to SDS–PAGE followed by silver staining (Figure 7B). Only the eluate from the 18E4-immunoaffinity column contained the three isoforms of hnRNP Q and hnRNP R. Western blots using mAb 18E4 confirmed the presence of the hnRNP Qs and hnRNP R only in the eluate from the 18E4-immunoaffinity column (data not shown).
We next added equal amounts of immunopurified hnRNP Qs and hnRNP R (∼400 ng per splicing reaction) to either 18E4-depleted or mock-depleted extracts, incubated for 10 min on ice and performed splicing reactions using Ad2 and CDC pre-mRNAs. As shown in Figure 8A, addition of hnRNP Qs and hnRNP R purified from HeLa cells completely restored the splicing defect of 18E4-depleted extracts but had no effect on the mock-depleted extracts. As expected, the SP2/0 eluate was unable to restore splicing when added to 18E4-depleted extracts. Quantitative analysis of the data confirms the reduction in splicing efficiency in hnRNP Qs/R-depleted extracts and its restoration upon addition of immunopurified hnRNP Qs/R (Figure 8C). A titration experiment was also performed by adding either 100 or 400 ng of purified hnRNP Qs and hnRNP R to 18E4-depleted extracts (Figure 8B). Judged by the production of mRNA, even a small amount of purified hnRNP Qs and hnRNP R was able to substantially increase the efficiency of splicing for both Ad2 and CDC pre-mRNAs. In summary, these findings demonstrate that hnRNP Qs and possibly hnRNP R are required for efficient pre-mRNA splicing in vitro. However, we can not exclude the possibility that another factor(s) that is tightly associated with the immunopurified hnRNP Qs and R may be responsible for the reconstitution of splicing upon addition to hnRNP Qs- and R-depleted extracts. The purity of the immunopurified hnRNP Qs and R (Figure 7B) makes this possibility rather unlikely but a definitive answer will require native recombinant hnRNP Qs and R produced in a heterologous eukaryotic expression system.
Discussion
We report here the characterization of a novel family of three hnRNP proteins derived by alternative splicing of a single gene. Based on the 2D map of the major hnRNP proteins (Pinol-Roma et al., 1988), we assign them as hnRNPs Q1, Q2 and Q3 and refer to them collectively as the hnRNP Q proteins. The hnRNP Q proteins are highly similar to hnRNP R, a recently identified hnRNP protein (Hassfeld et al., 1998). Two independent lines of evidence demonstrate that the hnRNP Qs are bona fide components of the spliceosome. First, the mAb 18E4, which specifically recognizes the hnRNP Qs, immunoprecipitates both pre-mRNA and splicing intermediates (intron-containing RNAs) from in vitro splicing reactions. This reflects a general association rather than a transcript-specific one, as the same pattern of immunoprecipitation is observed with three different pre-mRNAs. Secondly, hnRNP Qs (then known as Gry-rbp) have been found to be components of purified functional spliceosomes (Neubauer et al., 1998). The presence of three different hnRNP Q isoforms all highly similar to hnRNP R suggests that hnRNP Qs and hnRNP R may have different functions. In the analysis of the spliceosomal proteins on 2D gels by mass spectrometry, five different spots were identified as Gry-rbp, implying that all hnRNP Q isoforms and possibly post-translationally modified forms of hnRNP Qs are associated with functional spliceosomes. In that same analysis hnRNP R was not identified. The absence of hnRNP R from purified spliceosomes suggests that hnRNP R is not a stable or consistent component of the splicing machinery (Neubauer et al., 1998).
Interestingly, mRNA produced from splicing is also immunoprecipitated with 18E4, indicating that hnRNP Qs remain associated with mature mRNA after splicing (Figure 6). In contrast, the abundant hnRNP C proteins, which are also detected in spliceosomes (Neubauer et al., 1998), do not co-immunoprecipitate mRNA from the same in vitro splicing reactions although they efficiently immunoprecipitate pre-mRNA and intron-containing RNA intermediates (Figure 6C). This demonstrates that distinct hnRNP proteins, such as hnRNP Qs, associate with mRNA produced from splicing, while others, such as hnRNP C, do not, and suggests a possible role of hnRNP Qs in the function of mRNP complexes produced after splicing. This latter possibility is strengthened by the recent finding that NSAP1 (which is hnRNP Q3) is part of a cytoplasmic multiprotein complex that binds to the major determinant of instability (mCRD) of the c-fos proto-oncogene mRNA and regulates its stability and translatability (Grosset et al., 2000). Further experiments are required to elucidate the putative role of hnRNP Qs in post-splicing events, such as mRNA export and translation. It is also interesting to note that pre-mRNA is less efficiently immunoprecipitated than intron-containing RNA intermediates or mRNA with mAb 18E4 (Figure 6). This is particularly evident with CDC pre-mRNA. Similarly, the amount of CDC-2 pre-mRNA that is immunoprecipitated by mAb 18E4 is much less than that which is immunoprecipitated with 4F4, while the efficiency of immunoprecipitation of intron-containing RNA intermediates is not substantially different (Figure 6C). Taken together, these findings suggest that hnRNP Qs are either more accessible to mAb 18E4, or are associated preferentially with the products of splicing (both intron-containing RNA intermediates and mRNA) than with the pre-mRNA. We conclude that hnRNP Qs, and possibly hnRNP R, have a general role in splicing as they are required for efficient pre-mRNA splicing in vitro. The molecular mechanism of hnRNP Qs function remains to be identified, but is most likely mediated by direct interactions of hnRNP Qs with pre-mRNA.
The interaction of SMN with hnRNP Qs expands the repertoire of SMN targets and extends previous observations that indicated an important role for the SMN complex in pre-mRNA splicing (Pellizzoni et al., 1998). It was previously shown that a mutant of SMN lacking the first 27 amino acids (SMNΔN27) has a dominant-negative effect on pre-mRNA splicing (Pellizzoni et al., 1998), although how the function of SMN in splicing is mediated has not been clear. The function of SMN in splicing may simply be secondary to the known requirement of the SMN complex in spliceosomal snRNP biogenesis; in the absence of a functional SMN complex the snRNPs may be improperly assembled, leading to splicing impairment. Although this is likely, we also consider a direct role of the SMN complex in splicing. Indeed, addition of anti-SMN antibodies to nuclear extracts during in vitro splicing reactions inhibits splicing (Pellizzoni et al., 1998; Meister et al., 2000). Antibodies to SMN, Gemin2, Gemin3 and Gemin4 do not immunoprecipitate pre-mRNA or splicing intermediates from in vitro splicing reactions, suggesting that the SMN complex is not stably associated with the splicing machinery (our unpublished data). More likely, the SMN complex interacts transiently with components of the splicing machinery and perhaps only prior to their assembly into the spliceosome. The hnRNP Q proteins, because they are components of the spliceosome and interact with SMN, may provide a direct molecular link between SMN and pre-mRNA splicing and perhaps also other RNA processing events.
SMN interacts with an RGG box present in hnRNP Qs. This is consistent with the observation that SMN binds to RG-rich domains found in other proteins, including the Sm proteins (Friesen and Dreyfuss, 2000), and to RHA (Pellizzoni et al., 2001). SMN, however, does not interact indiscriminately with all RGG box-containing proteins as it does not bind hnRNP R or hnRNP A1 in vitro. Moreover, only a small portion of the RGG box of hnRNP Qs (comprising amino acids 518–549 of hnRNP Q1) is required for SMN interaction. It seems likely that specific RG-rich domains, sequences that are found mainly in RNA binding proteins, constitute a common molecular motif of SMN protein targets. The participation of the SMN complex in diverse RNA processing events is likely to be mediated by direct binding of the SMN complex to its many target proteins: Sm proteins (Liu et al., 1997; Pellizzoni et al., 1999; Friesen and Dreyfuss, 2000), RHA (Pellizzoni et al., 2001) and hnRNP Qs (this study). The general theme that emerges is that the SMN complex has a general role in the assembly of many RNP complexes. If a general RNP assembly/restructuring role is the modus operandi of the SMN complex, it can be anticipated that the SMN complex participates in additional pathways in which RNPs function. In this vein, we note that SMN also interacts with fibrillarin (Liu and Dreyfuss, 1996), an RG-domain protein that is found in small nucleolar RNPs (snoRNPs) and functions in ribosome biogenesis.
Finally, of particular interest is the fact that SMN lacking the last 16 amino acids (SMNΔEx7), the most common mutant form found in SMA patients, is defective in its interactions with the hnRNP Q proteins. Similarly, SMNΔEx7 is unable to bind to Sm proteins (Pellizzoni et al., 1999; Friesen and Dreyfuss, 2000) and to RHA (Pellizzoni et al., 2001). This highlights the importance of the C-terminal domain of SMN for proper target binding. In addition, these results strongly suggest that impaired interactions between SMN patient mutants and its targets (including Sm proteins, hnRNP Qs and RHA), which most probably result in defects in mRNA biogenesis, are likely to play a role in SMA pathogenesis.
Materials and methods
Cloning of hnRNP Qs and construction of plasmids and recombinant proteins
A yeast two-hybrid screen of a HeLa cDNA library, using SMN as bait, was performed as previously described (Liu and Dreyfuss, 1996) and led to the isolation of four identical clones encoding a 122-amino-acid protein. Database searches revealed that this was a fragment from Gry-rbp, a protein of unknown function deposited in DDBJ/EMBL/GenBank. Further searches revealed the presence of ESTs coding for two additional alternatively spliced isoforms (see text and Figure 1). All three full-length isoforms and hnRNP R were cloned by RT–PCR from HeLa cell mRNA, sequenced, and subcloned in the following vectors: pcDNA3 (for in vitro translations); pGEX-6P-2 (for production of recombinant proteins fused to GST); and pTYB2 (for production of untagged recombinant proteins). Plasmids coding for hnRNP Q1 lacking the RGG box or portions thereof, and for GST fused to the RGG box of hnRNP Q, were generated by PCR. The SMN constructs have been described previously (Pellizzoni et al., 1999; Friesen and Dreyfuss, 2000).
Antibodies
mAb 18E4 against hnRNP Qs was prepared by immunizing mice with recombinant untagged hnRNP Q protein. Other antibodies used were mAbs: 2B1 and rabbit polyclonal α-exon7, against SMN (Liu and Dreyfuss, 1996; Pellizzoni et al., 1998); mAb 4F4 against hnRNP C1/C2 and mAb 4B10 against hnRNP A1 (Choi and Dreyfuss, 1984a,b); mAb 12G4 against hnRNP K (Matunis et al., 1992); and mAb against T7 tag (Novagen).
Immunostaining, immunoblotting and immunoprecipitations
Ascites fluid from mAb 18E4 was used at 1:1000 dilution for indirect immunofluorescence of HeLa cells and samples were visualized by laser confocal microscopy. The same dilution was used for western blottings with all mAbs. Immunoprecipitations were performed from total cell lysate of HeLa cells as previously described (Pinol-Roma et al., 1988).
In vitro protein-binding assay
This was performed as previously described (Charroux et al., 1999). To exclude nucleic acid-mediated interactions, all in vitro translated products were treated with DNase I (0.5 U/µl) and RNase A (100 µg/ml) for 15 min at 30°C, prior to bindings.
Splicing assays
In vitro splicing reactions and immunoprecipitations were performed as described (Kataoka et al., 2000). Immunodepletion of hnRNP Qs and R from HeLa nuclear extracts was performed as previously described (Chua and Reed, 1999). Briefly, 40 µl of 18E4 ascites were coupled to 100 µl bed volume of protein G–agarose (Gibco) and used to deplete 200 µl of nuclear extract, whose salt concentration had been raised to 700 mM KCl. Three sequential depletions (1 h each) were performed at 4°C with constant rotation. After depletion the extracts were dialyzed in buffer D (Dignam et al., 1983). For hnRNP Qs and hnRNP R purification, mAb 18E4 was covalently coupled to Affi-Gel Hz (Bio-Rad), according to the manufacturer’s instructions. Total HeLa cell lysate (prepared in a buffer containing 20 mM Tris–HCl pH 7.4, 250 mM NaCl and 0.1% Triton X-100) was passed three times through a column containing the coupled 18E4 beads. After extensive washings with washing buffer (20 mM Tris–HCl pH 7.4, 500 mM NaCl and 0.5% Triton X-100), captured proteins were eluted with 100 mM glycine pH 2.5 and immediately neutralized in a buffer containing 1 M Tris pH 9.0. The eluted proteins were dialyzed in buffer D (without glycerol), concentrated, and used immediately for reconstitution experiments. Splicing gels were scanned, the efficiency was quantitated and results presented in a graphical format as previously described (Graveley et al., 1998). Briefly, percentages of splicing were calculated by the equation %splicing = P/(P + S), where P is the final reaction product (mRNA) and S is the sum of the precursor (pre-mRNA) and splicing intermediates.
Protein gel electrophoresis
Protein samples were electrophoresed on 11.5% SDS–PAGE gels. Two-dimensional NEPHGE was carried out as previously described (Pinol-Roma et al., 1988). The first dimension was separated by using pH 3–10 ampholine gradients for 4 h at 400 V, and the second dimension was by 11.5% SDS–PAGE.
Cell culture and labeling
This was performed as previously described (Pinol-Roma et al., 1988).
Acknowledgments
Acknowledgements
We are grateful to members of our laboratory, especially Westley Friesen, Livio Pellizzoni, Sergey Paushkin, Jin Wang, Jennifer Baccon, Severine Massenet and Amelie Gubitz for discussions, insight and critical reading of the manuscript. This work was supported by NIH grants to G.D. and Z.M. (K08 NS02199). G.D. is an investigator of the Howard Hughes Medical Institute.
References
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNA by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Burghes A.H.M. (1997) When is a deletion not a deletion? When it is converted. Am. J. Hum. Genet., 61, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.C. and Black,D.L. (1997) The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol., 17, 4667–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni,L., Perkinson,A.R., Shevchenko,A., Mann,M. and Dreyfuss,G. (1999) Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol., 147, 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni,L., Perkinson,R.A., Yong,J., Shevchenko,A., Mann,M. and Dreyfuss,G. (2000) Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol., 148, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.D. and Dreyfuss,G. (1984a) Isolation of the heterogeneous nuclear RNA–ribonucleoprotein complex (hnRNP): a unique supra molecular assembly. Proc. Natl Acad. Sci. USA, 81, 7471–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.D. and Dreyfuss,G. (1984b) Monoclonal antibody characteriz ation of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol., 99, 1997–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.Y., Rooke,N., Turck,C.W. and Black,D.L. (1999) hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol., 19, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K. and Reed,R. (1999) Human step II splicing factor hSlu7 functions in restructuring the spliceosome between the catalytic steps of splicing. Genes Dev., 13, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert D.D. et al. (1997) The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet., 6, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Fischer U., Liu,Q. and Dreyfuss,G. (1997) The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell, 90, 1023–1029. [DOI] [PubMed] [Google Scholar]
- Friesen W.J. and Dreyfuss,G. (2000) Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN). J. Biol. Chem., 275, 26370–26375. [DOI] [PubMed] [Google Scholar]
- Frugier T., Tiziano,F.D., Cifuentes-Diaz,C., Miniou,P., Roblot,N., Dierich,A., Le Meur,M. and Melki,J. (2000) Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet., 9, 849–858. [DOI] [PubMed] [Google Scholar]
- Gamberi C., Izaurralde,E., Beisel,C. and Mattaj,I.W. (1997) Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol. Cell. Biol., 17, 2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R., Hertel,K.J. and Maniatis,T. (1998) A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J., 17, 6747–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosset C., Chen,C.Y., Xu,N., Sonenberg,N., Jacquemin-Sablon,H. and Shyu,A.B. (2000) A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell, 103, 29–40. [DOI] [PubMed] [Google Scholar]
- Hannus S., Buhler,D., Romano,M., Seraphin,B. and Fischer,U. (2000) The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet., 9, 663–674. [DOI] [PubMed] [Google Scholar]
- Harris C.E., Boden,R.A. and Astell,C.R. (1999) A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol., 73, 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassfeld W., Chan,E.K.L., Mathison,D.A., Portman,D., Dreyfuss,G., Steiner,G. and Tan,E.M. (1998) Molecular definition of heterogeneous nuclear ribonucleoprotein R (hnRNP R) using autoimmune antibody: immunological relationship with hnRNP P. Nucleic Acids Res., 26, 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li H.M., Chang,J.G., Jong,Y.J., Wu,M.H., Wang,N.M., Tsai,C.H. and Li,H. (2000) A mouse model for spinal muscular atrophy. Nature Genet., 24, 66–70. [DOI] [PubMed] [Google Scholar]
- Jablonka S., Schrank,B., Kralewski,M., Rossoll,W. and Sendtner,M. (2000) Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: an animal model for spinal muscular atrophy type III. Hum. Mol. Genet., 9, 341–346. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Yong,J., Kim,V.N., Velazquez,F., Perkinson,R.A., Wang,F. and Dreyfuss,G. (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell, 6, 673–682. [DOI] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Lefebvre S. et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burlet,P., Liu,Q., Bertrandy,S., Clermont,O., Munnich,A., Dreyfuss,G. and Melki,J. (1997) Correlation between severity and SMN protein level in spinal muscular atrophy. Nature Genet., 16, 265–269. [DOI] [PubMed] [Google Scholar]
- Liu Q. and Dreyfuss,G. (1996) A novel nuclear structure containing the survival of motor neurons protein. EMBO J., 15, 3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fischer,U., Wang,F. and Dreyfuss,G. (1997) The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell, 90, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. (1988) U snRNP assembly and transport. In Birnstiel,M. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer Verlag, New York, NY, pp. 100–114.
- Mattaj I.W., Boelens,W., Izaurralde,E., Jarmolowski,A. and Kambach,C. (1993) Nucleocytoplasmic transport and snRNP assembly. Mol. Biol. Rep., 18, 79–83. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Michael,W.M. and Dreyfuss,G. (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol., 12, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- Meister G., Buhler,D., Laggerbauer,B., Zobawa,M., Lottspeich,F. and Fischer,U. (2000) Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet., 9, 1977–1986. [DOI] [PubMed] [Google Scholar]
- Melki J. (1997) Spinal muscular atrophy. Curr. Opin. Neurol., 10, 381–385. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I., Culetto,E., Walker,D.S., Baylis,H.A., Sattelle,D.B. and Davies,K.E. (1999) The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum. Mol. Genet., 8, 2133–2143. [DOI] [PubMed] [Google Scholar]
- Monani U.R. et al. (2000) The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn–/– mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet., 9, 333–339. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Query,C.C. and Sharp,P.A. (1993) Splicing of precursors to mRNA by the spliceosome. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–358.
- Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- Ohno M., Sakamoto,H. and Shimura,Y. (1987) Preferential excision of the 5′ proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc. Natl Acad. Sci. USA, 84, 5187–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen N., Doe,C.L., Mellor,J. and Davies,K.E. (2000) Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum. Mol. Genet., 9, 675–684. [DOI] [PubMed] [Google Scholar]
- Paushkin S., Charroux,B., Abel,L., Perkinson,R.A., Pellizzoni,L. and Dreyfuss,G. (2000) The survival motor neuron protein of Schizosacharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J. Biol. Chem., 275, 23841–23846. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka,N., Charroux,B. and Dreyfuss,G. (1998) A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell, 95, 615–624. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Charroux,B. and Dreyfuss,G. (1999) SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl Acad. Sci. USA, 96, 11167–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Charroux,B., Rappsilber,J., Mann,M. and Dreyfuss,G. (2001) A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol., 152, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S., Choi,Y.D., Matunis,M.J. and Dreyfuss,G. (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev., 2, 215–227. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Swanson,M.S., Gall,J.G. and Dreyfuss,G. (1989) A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell Biol., 109, 2575–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B., Gotz,R., Gunnersen,J.M., Ure,J.M., Toyka,K.V., Smith,A.G. and Sendtner,M. (1997) Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl Acad. Sci. USA, 94, 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Wang J. and Dreyfuss,G. (2001) A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J. Biol. Chem., 276, 9599–9605. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Luhrmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]