The structure-specific endonuclease Ercc1–Xpf is required for targeted gene replacement in embryonic stem cells (original) (raw)
Abstract
The Ercc1–Xpf heterodimer, a highly conserved structure-specific endonuclease, functions in multiple DNA repair pathways that are pivotal for maintaining genome stability, including nucleotide excision repair, interstrand crosslink repair and homologous recombination. Ercc1–Xpf incises double-stranded DNA at double-strand/single-strand junctions, making it an ideal enzyme for processing DNA structures that contain partially unwound strands. Here we demonstrate that although Ercc1 is dispensable for recombination between sister chromatids, it is essential for targeted gene replacement in mouse embryonic stem cells. Surprisingly, the role of Ercc1–Xpf in gene targeting is distinct from its previously identified role in removing nonhomologous termini from recombination intermediates because it was required irrespective of whether the ends of the DNA targeting constructs were heterologous or homologous to the genomic locus. Our observations have implications for the mechanism of gene targeting in mammalian cells and define a new role for Ercc1–Xpf in mammalian homologous recombination. We propose a model for the mechanism of targeted gene replacement that invokes a role for Ercc1–Xpf in making the recipient genomic locus receptive for gene replacement.
Keywords: D-loops/double-strand breaks/heterology/homologous recombination
Introduction
hERCC1 was originally identified as the human gene that corrects hypersensitivity to UV irradiation in one of 11 Chinese hamster ovary (CHO) cell complementation groups (Westerveld et al., 1984), hence the name excision repair cross complementation group 1. The gene is highly conserved with homologs in mouse, mErcc1 (Selfridge et al., 1992), Saccharomyces cerevisiae, RAD10 (van Duin et al., 1986) and Schizosaccharomyces pombe, swi10 (Rodel et al., 1992). In cells, the Ercc1 protein exists in a tight complex with Xpf (Biggerstaff et al., 1993; van Vuuren et al., 1993). Heterodimerization is required to stabilize both proteins and for complementation of DNA repair defects. Xpf is also highly conserved (Sijbers et al., 1996), as is heterodimeric interaction. The Rad10–Rad1 protein complex was discovered to be an endonuclease (Tomkinson et al., 1993) that cleaves double-stranded (ds) DNA only on the strand with a 3′ single-strand (ss) tail (Bardwell et al., 1994). This substrate specificity has helped to define the role of Ercc1–Xpf in correcting UV sensitivity. The complex is responsible for making a cut in the damaged strand of duplex DNA 5′ to a lesion in nucleotide excision repair (NER) (Mu et al., 1995; Sijbers et al., 1996). In addition, the complex cleaves stem–loop and splayed arm structures within duplex DNA and only on the strand with the 3′ ss tail (de Laat et al., 1998a).
Although ERCC1 plays an essential role in NER, it is unable to complement the UV sensitivity of any of the eight complementation groups of fibroblast cell lines derived from NER-deficient xeroderma pigmentosum patients (van Duin et al., 1989). Nor has a viable mutation in hERCC1 been described. Furthermore, _Ercc1_–/– mice display a much more severe phenotype than other NER-deficient mouse models (McWhir et al., 1993; Weeda et al., 1997). These observations imply additional function(s) for Ercc1 outside NER. Hints as to this additional function(s) come from studies of mutant cell lines. Ercc1- and Xpf-deficient mammalian cells, unlike other NER-deficient cells, are sensitive to DNA crosslinking agents (Westerveld et al., 1984; Hoy et al., 1985). However, they are not ionizing radiation (IR)-sensitive. This is also true for rad10, rad1 (Moore, 1978) and swi10 mutants (Hang et al., 1996). These observations indicate a role for Ercc1–Xpf in NER-independent interstrand crosslink repair but not in double-strand break (DSB) repair.
In addition to involvement in NER and interstrand crosslink repair, there is evidence for a role of Ercc1–Xpf in recombination. For example in S.cerevisiae, mutations in rad1 or rad10 suppress mitotic recombination between direct or inverted repeats (Klein, 1988; Schiestl and Prakash, 1988, 1990; Prado and Aguilera, 1995). Furthermore, recombination between repeats yields different products in rad1 and rad10 compared with wild-type (wt) strains (Klein, 1988; Aguilera and Klein, 1989; Zehfus et al., 1990). Excisional products in which the sequence between the repeats is lost are less frequently recovered from the mutant strains. Similar results were obtained in Ercc1-deficient CHO cells (Sargent et al., 2000). The rad1 phenotype is not exaggerated by a mutation in rad10 (Schiestl and Prakash, 1990), implying that the role these proteins play in recombination is dependent upon protein heterodimer formation and therefore most likely its endonuclease function.
Intrachromosomal recombination between directly repeated sequences occurs most efficiently via single-strand annealing (SSA) (Paques and Haber, 1999). SSA requires a DSB between or within one of the repeats followed by exonuclease resection from both ends revealing complementary 3′ ss overhangs. The regions of homology within the two overhangs align and anneal, ultimately yielding a product in which one of the repeats and all intervening sequences are lost. The proposed role for Ercc1–Xpf in SSA is to remove nonhomologous termini from the 3′ ss overhangs after the overhangs have realigned. Indeed, Rad1 and Rad10 do cleave nonhomologous ss termini from the ends of a DSB to facilitate repair via homologous recombination (HR) in yeast (Fishman-Lobell and Haber, 1992; Ivanov and Haber, 1995; Prado and Aguilera, 1995; Paques and Haber, 1997), as do the hamster homologs in CHO cells (Sargent et al., 2000). Similarly, Ercc1 is required to remove nonhomologous tails from ends-in targeting constructs in CHO cells (Adair et al., 2000).
There is also mounting evidence that Ercc1–Xpf (and their homologs) participate in the processing of recombination intermediates in which the DNA substrates are not identical, but the sequence nonhomology is positioned between homologous domains rather than at an end. For example, gene conversion products resulting from recombination between direct repeats that differ by distally positioned deletions are not recovered in a rad1 strain (Klein, 1988). Similarly, in rad1 and rad10 strains, gene targeting using a linear construct is suppressed 40-fold (Schiestl and Prakash, 1988, 1990; Saparbaev et al., 1996). Furthermore, mutations in rad1 or rad10 suppress meiotic repair of loops created by patches of nonhomology between homologous chromosomes (Kirkpatrick and Petes, 1997; Kearney et al., 2001) as do mutations in the S.pombe homologs (Fleck et al., 1999). Finally, in Ercc1-deficient CHO cells intrachromosomal recombination between direct repeats separated by 600 bp of intervening sequence yields many more gene rearrangements compared with Ercc1-proficient cells (Sargent et al., 1997), in particular deletions beginning at the borders between the repeat and the intervening sequence.
In the experiments described below, we discovered that Ercc1 is absolutely required for HR between gene replacement constructs and genomic loci in mouse embryonic stem (ES) cells. We tested linear constructs consisting of a selectable marker flanked by completely homologous arms and constructs including additional nonhomologous termini. This allowed us to determine whether the essential function of Ercc1 in gene replacement was removal of 3′ nonhomologous termini, similar to SSA, or processing of looped-out heteroduplex intermediates. Our data are consistent with the latter and have implications for the mechanism of targeted gene replacement in mammalian cells.
Results
Generation of mErcc1–/– and rescued ES cells
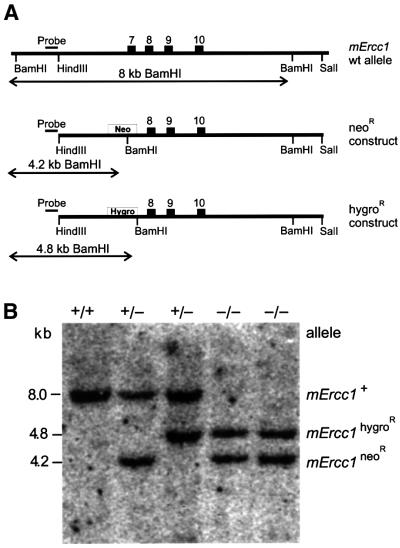
A clonal mErcc1+/– [neomycin-resistant (neoR)] ES cell line (Weeda et al., 1997) was transfected with a linearized mErcc1 targeting construct containing a hygromycin selectable marker (hygroR) with the goal of disrupting the second mErcc1 allele and thereby generating a nullizygous cell line (Figure 1A). More than 300 hygroR clones from multiple transfection experiments were analyzed by DNA blot analysis (Figure 1B). The absolute targeting efficiency of the mErcc1 locus was 16% as previously reported (Weeda et al., 1997). However, there was disequilibrium between targeting the wt and mutant alleles of the mErcc1+/– cells. In only two out of 48 targeted clones was the wt mErcc1 allele replaced by the hygroR construct. Thus, the frequency of generating _mErcc1_–/– cells was 12-fold lower than expected, suggesting selection against Ercc1-deficient ES cells. Indeed, the _mErcc1_–/– cells required daily passaging to maintain morphology and growth characteristics typical of wt ES cells.
Fig. 1. Targeting strategy for sequential inactivation of both mErcc1 alleles in mouse ES cells. (A) The top of the scheme depicts the 3′ end of the mErcc1 genomic locus with relevant restriction sites. Numbered boxes indicate exons. Beneath, the two targeting constructs are depicted. Exon 7 of mErcc1 was disrupted by insertion of either a neoR or hygroR marker containing a diagnostic _Bam_HI restriction site. ES cells were transfected first with the _mErcc1_neoR construct. Genomic DNA was isolated from resistant clones, digested with _Bam_HI and analyzed by DNA hybridization using a 5′ external probe (indicated above the constructs). The lengths of the expected genomic fragments are indicated with horizontal arrows below the constructs. mErcc1+/– cells were subsequently transfected with the _mErcc1_hygroR /em>construct and grown under hygromycin B selection. Positive clones were analyzed by DNA blotting. (B) An example of a DNA blot of _Bam_HI-digested genomic DNA isolated from ES clones and probed with the 5′ external probe is shown. The probe detects either an 8-kb fragment (wt allele), 4.8-kb fragment (hygroR mutant) or 4.2-kb allele (neoR mutant). Two independent double mutant clones were identified.
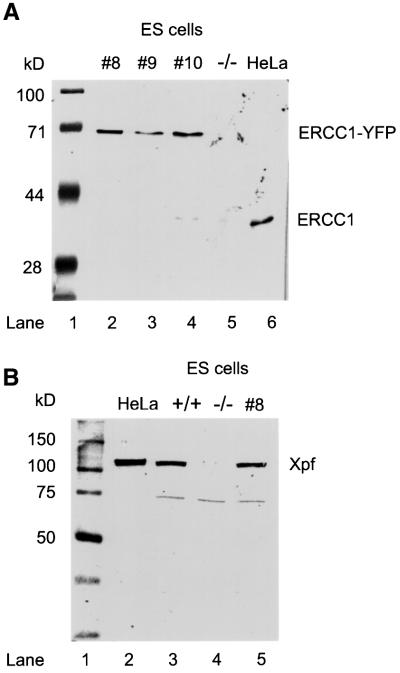
To verify that the phenotype of the _mErcc1_–/– cells was directly attributable to the loss of functional Ercc1, we generated rescued mutant lines. _mErcc1_–/– ES cells were transfected with a plasmid harboring the hERCC1 cDNA fused in-frame with a downstream sequence encoding yellow fluorescent protein (YFP). Transformants were screened for expression of ERCC1–YFP by fluorescence microscopy. In all of the clones screened, the fluorescent signal was low and restricted to the nucleus. These characteristics are inconsistent with expression of YFP alone. Immunoblot analysis of whole cell extracts (WCE) from the individual clones with α-hERCC1 antibody indicated expression of full-length ERCC1–YFP fusion protein but not ERCC1 by itself (Figure 2A). Furthermore, fusion protein levels were similar to levels of ERCC1 in HeLa cells, indicating physiological expression levels. The α-hERCC1 antibody does not recognize mouse Ercc1, therefore the absence of a protein signal in _mErcc1_–/– cell extract does not formally prove that there is no endogenous Ercc1 present in the mutant cell lines. However, immunoblot analysis with α-hXPF antibody, which does recognize the endogenous mouse protein, demonstrates markedly reduced levels of Xpf protein in the _mErcc1_–/– lines (Figure 2B). This is consistent with previous findings of destabilization of Xpf in the absence of Ercc1 (Sijbers et al., 1996; Houtsmuller et al., 1999; Gaillard and Wood, 2001). Xpf protein levels in the rescued mutant line were comparable to wt ES cells.
Fig. 2. Immunoblot analysis of wt, _mErcc1_–/– and rescued mutant ES cell lysates. (A) Immunoblot of WCE using α-hERCC1 polyclonal antibody. The antibody recognizes a 39 kDa band in the positive control (HeLa cells, lane 6). Lanes 2–4 contain lysates from three cell lines (#8, 9 and 10) expanded from individual MMC-resistant clones established by stable transfection of the _mErcc1_–/– ES cells with a construct containing the hERCC1 cDNA coupled in-frame with YFP. Expression of only the full-length fusion protein is seen as a single band with the predicted molecular mass of 66 kDa (39 kDa ERCC1 + 27 kDa YFP). Lane 5 contains a lysate from _mErcc1_–/– ES cells. (B) Immunoblot of WCE using α-hXPF polyclonal antibody. A band corresponding to XPF (115 kDa) is visible in the HeLa cell and wt ES cell lysates (lanes 2 and 3, respectively) but not in the _mErcc1_–/– mutant line (lane 4). Rescue of the mutant line with hERCC1–YFP restores the Xpf band to physiological levels (lane 5). The presence of a cross-reactive band in the ES cells (∼75 kDa) serves as a loading control.
Characterization of mErcc1–/– ES cells
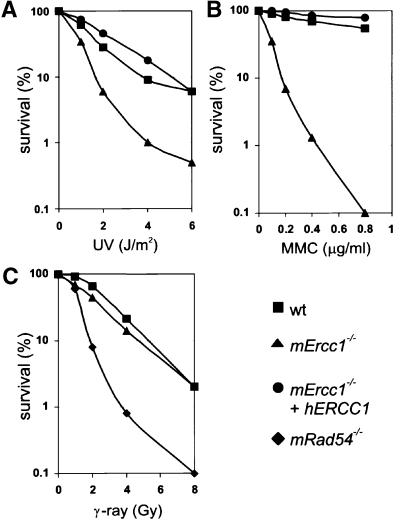
To characterize the _mErcc1_–/– cells, we examined their sensitivity to three types of DNA damaging agents: UV, mitomycin C (MMC) and IR. UV causes lesions that are substrates for NER, while MMC induces interstrand crosslinks and IR generates DSBs that in ES cells are efficiently repaired by HR (Dronkert et al., 2000). We observed that the _mErcc1_–/– cells are 2.7-fold more sensitive to UV than wt cells (Figure 3A). In fact, the Ercc1 mutant was equally as sensitive to UV irradiation as an _Xpa_–/– ES line (H.de Waard, unpublished data) consistent with a complete NER defect. The cell lines stably transfected with hERCC1 cDNA behaved identically to the parental wt line, demonstrating a complete restoration of NER. The _mErcc1_–/– cells were 10-fold more sensitive to low doses of MMC than the wt and rescued mutant cells (Figure 3B). These results were not crosslink agent-specific, as the _mErcc1_–/– ES cells were also 16-fold more sensitive to the crosslinking agent cisplatin and 12-fold more sensitive to 8-methoxypsoralen + UVA than the wt cells (data not shown). The _mErcc1_–/– cells were not sensitive to IR (Figure 3C). This was in contrast to the HR-defective _mRad54_–/– ES. Finally, to screen further for a role for Ercc1 in HR, the frequency of sister chromatid exchange (SCE) was measured in the different ES cell lines. The _mErcc1_–/– cells displayed the same number of SCEs as the parental wt line or the rescued mutant lines (data not shown), indicating that the absence of functional Ercc1 does not preclude HR. This was true whether spontaneous or MMC-induced SCEs were measured. Together, these data define the _mErcc1_–/– ES cells as NER-deficient, crosslink repair-deficient but DSB repair- and HR-proficient.
Fig. 3. Effect of UV, MMC and IR on wt and _mErcc1_–/– ES cells. (A) Clonogenic survival of wt (squares), _mErcc1_–/– (triangles) and a rescued mutant line (circles) after UV irradiation. The percent of irradiated cells that were able to form a colony compared with untreated cells is plotted against the dose of UV. (B) Clonogenic survival of wt, _mErcc1_–/– and a rescued mutant line after treatment with MMC for 1 h. (C) Clonogenic survival of ES cells after treatment with IR. Since the _mErcc1_–/– line was not hypersensitive to irradiation, a rescued line was not tested. An _mRad54_–/– ES line (diamonds) was used as a positive control for irradiation.
Measuring gene replacement frequency in mErcc1–/– ES cells
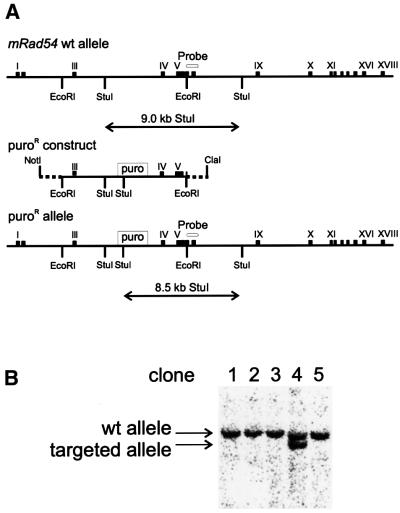
To explore the possibility of a role for Ercc1 in recombination between non-identical substrates, we examined the capacity of the mutant cells for gene targeting. In order to do this, the mutant, wt and rescued ES cells were transfected with a linear targeting construct consisting of a puromycin resistance (puroR) selectable marker flanked on both sides with ∼4 kb of sequence homologous to the mRad54 genomic locus (Figure 4). The construct was isolated either with or without 14–32 bp of heterologous sequences at each end.
Fig. 4. Strategy for quantitating gene replacement in wt and _mErcc1_–/– ES cells. (A) Top, the central portion of the mRad54 genomic locus is shown. Exons are depicted as numbered boxes. The targeting construct was generated by inserting a puroR expression cassette into intron 3. Once homologously recombined into the genome, the _Stu_I fragment encompassing this region is shortened from 9.0 to 8.5 kb. The targeting construct was isolated from a vector as either an _Eco_RI fragment (completely homologous to the mRad54 locus, with the exception of the selectable marker), or a _Cla_I–_Not_I fragment (which contains 32 bp of bacterial sequence on the 5′ and 14 bp on the 3′ end of the construct, indicated as dashed lines; not drawn to scale). Stable transformants were selected with puromycin. (B) Genomic DNA was isolated from individual resistant clones and analyzed by DNA hybridization after _Stu_I digestion. Targeting was detected by the appearance of an 8.5-kb DNA fragment using a 3′ external probe from exons 7 and 8 of mRad54 cDNA.
In the wt cell line, the construct with the nonhomologous termini targeted the mRad54 locus with 30% efficiency (Table I). In stark contrast, transfection of the same construct into _mErcc1_–/– cells did not yield a single targeting event in >225 puroR clones. This represents a >78-fold drop in targeting efficiency. In the _mErcc1_–/– ES line rescued with a tagged hERCC1 cDNA, the targeting efficiency returned to wt levels. Unexpectedly, the complete absence of targeting events in the _mErcc1_–/– ES cells was also observed when the ends of the transfected linear construct were completely homologous to the genomic locus. Thus, these data define a new role for Ercc1 in gene targeting in mammalian cells that is distinct from the previously identified role in removing nonhomologous termini from recombinational intermediates.
Table I. Frequency of gene replacement events in ES cells of the indicated genotypes.
| ES cell genotype | Targeted locus: | mRad54 | mRad54 | mRb | mCsb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Construct ends: | non-homologous | homologous | non-homologous | non-homologous | ||||||
| wt | 30.9%a | (30/97) | 25.7% | (37/144) | 20.0%e,f | (19/95) | 17.9%e | (10/56) | ||
| _mErcc1_–/– | <0.4%b | (0/227) | <0.4%b | (0/269) | <0.5%b | (0/212) | <0.6%b | (0/160) | ||
| _mErcc1_–/– + hERCC1 | 14.7%c,d | (21/143) | 26.1%c,d | (36/138) | n.d. | n.d. | ||||
| _mRad54_–/– | 1.6% | (4/249) | 2.1% | (6/284) | 3.3%e | (2/61) | 4.3%e | (3/69) |
To exclude the possibility that the abrogation of gene targeting in _mErcc1_–/– ES cells was construct related, we also tested other linear ‘ends-out’ constructs that target the murine retinoblastoma (mRb) or Cockayne syndrome B (mCsb) locus (Table I). With these additional constructs, we again never observed a single event of gene replacement (0/212 puroR clones and 0/160 puroR clones, respectively) in the Ercc1-deficient cells, although the targeting efficiency was 18–20% in wt ES cells. Nor was the abrogation of gene targeting peculiar to the particular Ercc1–/– cell line studied, as examination of a second Ercc1–/– and two additional rescued mutant lines yielded results in accordance with Table I (0/165 puroR clones in the mutant cell line; 7/38 and 6/30 puroR clones in the rescued line). By way of comparison, we also determined the targeting frequency of these same constructs in an HR-deficient _mRad54_–/– ES cell line. In the _mRad54_–/– cells, targeted integration was reduced by 5- to 15-fold, but not abolished. The difference in the level of suppression of gene replacement between the _mErcc1_–/– and _mRad54_–/– ES cells was statistically significant (p <0.001).
Discussion
Generation of mErcc1–/– and rescued ES cell lines
Both alleles of the mErcc1 locus were disrupted in ES cells via sequential rounds of gene targeting (Figure 1). In the second round of targeting, disruption of the remaining wt Ercc1 allele occurs 12-fold less frequently than re-targeting of the previously replaced allele, suggesting selection against Ercc1-deficient ES cells, as previously reported in primary mouse embryonic fibroblasts (MEFs) (Weeda et al., 1997). A rescued mutant line (_mErcc1_–/– cells stably transfected with _hERCC1_–YFP cDNA) was also generated for use as a positive control during phenotypic analysis of the _mErcc1_–/– ES cells. The heterodimeric partner of Ercc1, Xpf, is not detected in _mErcc1_–/– ES cells by immunoblotting, but is detected in the rescued line (Figure 2B). This result demonstrates that in ES cells, like in CHO cells (Sijbers et al., 1996), the absence of Ercc1 destabilizes Xpf. This result also provides evidence for the absence of functional Ercc1 protein in the _mErcc1_–/– cells. The mErcc1 nullizygous cells are as sensitive to UV irradiation (Figure 3A) as a complete NER-deficient ES cell line (_Xpa_–/– ES cells; H.de Waard, unpublished results). This level of sensitivity corresponds to that of immortalized MEFs derived from a knock-out mouse and to a CHO mutant line (Melton et al., 1998). The mutant ES cells are also extremely sensitive to the crosslinking agent MMC (Figure 3B), much more so than are NER- or HR-deficient ES cells (Essers et al., 1997). This is consistent with the hypothesis that DNA interstrand crosslinks are repaired via a mechanism that can involve both NER and HR (Sladek et al., 1989) and that Ercc1 is involved in both contributory pathways (Dronkert and Kanaar, 2001). CHO Ercc1 mutants are as sensitive to MMC as the ES mutant cells (Westerveld et al., 1984). However, primary (Weeda et al., 1997) and immortalized _Ercc1_–/– MEFs (Melton et al., 1998) are less sensitive, possibly reflecting differential utilization of the various recombinational repair pathways by the different cell types.
Role of Ercc1 in homologous recombination
Since SCE and repair of DSBs can occur via HR (Paques and Haber, 1999; Sonoda et al., 1999), assays to measure either are used to screen for novel genes involved in recombination. The _mErcc1_–/– ES cells are not sensitive to DSBs induced by IR (Figure 3C), similar to rad10 yeast (Moore, 1978) and _Ercc1_–/– CHO cells (Darroudi and Natarajan, 1985). Thus, Ercc1 is not essential for DSB repair in any organism yet tested. Furthermore, the wt and _mErcc1_–/– ES cells display identical frequencies of spontaneous and MMC-induced SCEs (data not shown). Thus, HR appears not to be suppressed in Ercc1-deficient mammalian cells. Our findings are consistent with the demonstrated HR-proficiency of Ercc1–/– CHO cells (Nairn et al., 1991; Sargent et al., 1997) and immortalized MEFs (Melton et al., 1998). Together, these data support the notion that if Ercc1 does play a role in HR, it is limited to a selected subset of recombinational events.
Indeed, Ercc1 is absolutely essential for a particular class of HR in ES cells: gene replacement. We screened a total of 868 puroR clones derived from transfection of _mErcc1_–/– ES cells with constructs carrying a puroR selectable marker and targeting three different genomic loci, yet we never observed a single gene replacement event (Table I). In total, these data reflect a >200-fold drop in targeting efficiency in the Ercc1-deficient cells compared with wt ES cells. Stable transfection of the Ercc1-deficient cells with a tagged hERCC1 cDNA corrects the targeting efficiencies to wt levels. Surprisingly, failure to homologously recombine targeting constructs in the _mErcc1_–/– cells is not limited to constructs carrying heterologous terminal sequences. This provides the first evidence that in mammalian cells, Ercc1 is required for HR between non-identical DNA substrates in a capacity other than removing nonhomologous termini.
The data obtained with the _mErcc1_–/– ES cells can be contrasted with the results obtained with _mRad54_–/– ES cells. Rad54-deficient cells are sensitive to IR (Figure 3 and Essers et al., 1997) and have a reduced capacity for spontaneous and DNA damage-induced SCE (Dronkert et al., 2000), yet gene replacement is not abrogated in _mRad54_–/– ES cells (Table I and Essers et al., 1997). This serves to highlight the differential roles of the proteins in HR; whereas Rad54 facilitates the general mechanism of HR, Ercc1 has a limited, yet essential function in a subset of recombinational events.
The finding that Ercc1 plays a role in gene replacement in ES cells is consistent with observations made in S.cerevisiae, the first organism for which targeted gene replacement was developed (Orr-Weaver et al., 1981; Rothstein, 1983). Replacement is suppressed 40-fold in rad1 or rad10 deletion strains compared with wt strains (Schiestl and Prakash, 1988, 1990; Saparbaev et al., 1996). In contrast, however, the role of Ercc1 in mammalian cells appears to be essential to gene replacement, whereas in yeast there is either another protein that can substitute for Rad1–Rad10 or there is an additional Rad10-independent mechanism of gene replacement.
A model for Ercc1-dependent gene replacement
Based on our findings in ES cells, we propose a model for the mechanism of gene replacement that incorporates an essential role for the endonuclease function of Ercc1–Xpf (Figure 5). Currently, at least three models exist for the mechanism of gene replacement using ends-out targeting constructs: double cross-over, SSA and break-induced replication (Paques and Haber, 1999). We implement the double cross-over mechanism as the basis of our model (Szostak et al., 1983). Although our data are in no way inconsistent with the other mechanisms, we found them more difficult to reconcile with the incorporation of large stretches of heterologous DNA into a mammalian genome.
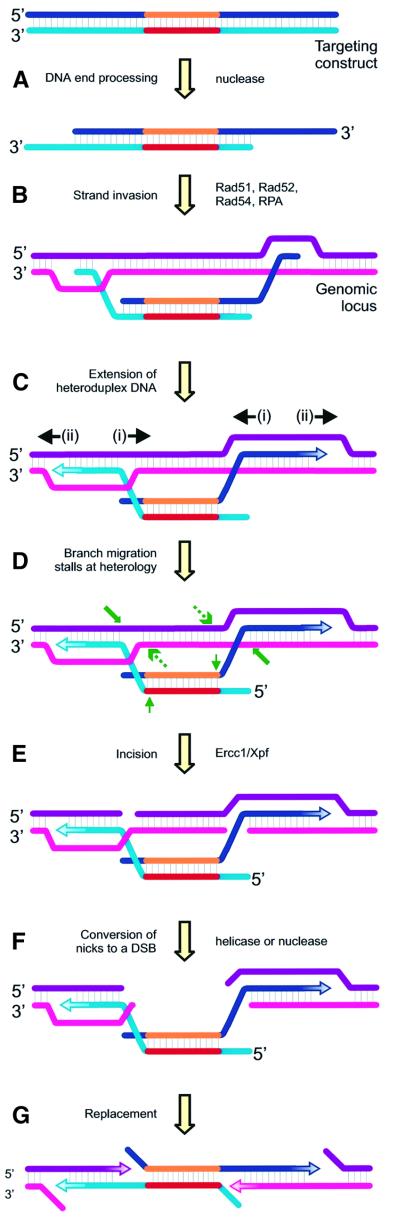
Fig. 5. Model for Ercc1–Xpf-dependent gene replacement. The two strands of a linear targeting construct or donor DNA are depicted in shades of blue. The central portion of the donor DNA (shown in reds) indicates a stretch of sequence without homology to the recipient DNA. The domain of nonhomology is typically a selectable marker gene. The two strands of genomic or recipient DNA are pictured in shades of purple. New DNA synthesis is indicated by arrows with a gradient-fill pattern. (A) Exonucleolytic processing of the donor to reveal 3′ ss ends. (B) Strand invasion by both 3′ ss ends of the donor DNA into homologous sequences of the recipient DNA to create two D-loops. (C) Expansion of the D-loops proximally by branch migration (i), and possibly also distally by DNA replication using the invading donor strand as a primer and the recipient strand as a template (ii). (D) Branch migration continues until the region of nonhomology in the donor DNA blocks further heteroduplex formation. Substrates for the structure-specific Ercc1–Xpf endonuclease are indicated by arrows. The slanted green arrow indicates the predicted site of primary Ercc1–Xpf cleavage. The dashed green arrow indicates an alternative cleavage site that can also result in gene replacement. The thin green arrow indicates a third potential substrate for Ercc1–Xpf that is nonproductive because it cannot result in gene replacement. (E) Ercc1–Xpf incises the stalled intermediate at characteristic ds/ss junctions just 5′ to a 3′ ss domain (at bold arrows shown in D). (F) Conversion of the nicks in the recipient DNA to a DSB. The break is stabilized by the heteroduplex DNA holding the two genomic ends in proximity. (G) Gene replacement is achieved by nonconservative insertion of the nonhomologous sequence into the DSB either with or without polymerization from the new 3′ ends in the recipient DNA by Ercc1–Xpf incision. Resolution requires the activities of a flap endonuclease and DNA ligase.
Our proposed model is applicable to the replacement of a continuous stretch of genomic DNA (recipient) of indeterminate length with a large stretch of nonhomologous sequence (donor DNA; typically a selectable marker). The novel feature of our model is that Ercc1 is required for initiating what becomes, in effect, a DSB in the recipient DNA. We propose that cleavage by the Ercc1–Xpf endonuclease is required for resolution of the heteroduplex intermediate formed between the donor and recipient DNA when branch migration becomes stalled at the point of sequence nonhomology. Accurate localization of the endonuclease cleavage is directed by the junctions between heterology and homology.
Our model proceeds as follows (Figure 5). First, both processed 3′ ends of the linear donor DNA identify homologous sequences in the recipient DNA with the aid of the general HR machinery, including Rad51, Rad52, Rad54 and the ss DNA binding protein RPA (Paques and Haber, 1999). Strand invasion at each end creates two D-loops (Figure 5B). Observations that are consistent with the hypothesis that gene replacement is initiated via invasion by both ends of the donor DNA include: (i) the frequency and the fidelity of gene replacement increase dramatically if the donor DNA is linear and both ends are homologous to the recipient DNA (Berinstein et al., 1992; Thomas et al., 1992; Negritto et al., 1997); and (ii) the length of nonhomologous donor sequence does not affect targeting efficiency (Surosky and Tye, 1985; Mansour et al., 1990; Deng et al., 1993; Li and Baker, 2000).
Secondly, the extent of the heteroduplex DNA is maximized (Li and Baker, 2000). This occurs by branch migration in an inward direction via pairing of homologous sequences between the donor and recipient DNA (Figure 5Ci) and perhaps also outwardly by leading strand DNA synthesis from the 3′ invading ends using the donor as a template (Figure 5Cii). D-loop expansion continues until heterology between the donor and recipient DNA sequences is reached, at which point branch migration is stalled and the intermediate stabilized (Negritto et al., 1997; Belotserkovskii et al., 1999; Constantinou et al., 2001).
Thirdly, we propose that nicking by the structure-specific endonuclease Ercc1–Xpf is required to initiate resolution of the stalled intermediate. The substrate specificity of the endonuclease dictates the sites of incision which will occur in ds DNA just 5′ to a 3′ ss domain (de Laat et al., 1998a). Within the proposed intermediate, cleavage of both strands of the recipient DNA favors the outcome of gene replacement (possible cut sites indicated with green arrows in Figure 5D). In NER, it has been established that RPA binds the undamaged strand and by virtue of its polarity, directs ERCC1–XPF cleavage exclusively to the opposite (damaged) strand (de Laat et al., 1998b). In recombination, it is likely that RPA, bound to the invading ends of the donor DNA, similarly directs Ercc1–Xpf to cleave the strand with the opposite polarity (cleavage sites indicated by bold green arrows in Figure 5D). However, based on the currently available data, we cannot exclude the possibility that cleavage instead, or in addition, occurs immediately 5′ to both of the D-loops (dashed green arrows in Figure 5D). Nevertheless, the net result is cleavage of both strands of the recipient genomic locus, making it receptive for productive gene replacement. It should also be noted that based on the substrate specificity of the Ercc1–Xpf endonuclease alone, the looped-out nonhomologous sequence of the donor DNA could also be a substrate (cleavage sites indicated with thin green arrows in Figure 5D). If, however, cleavage of the donor DNA occurs, the homologous arms are removed, eliminating the possibility of homologous gene replacement. In this case, the recipient DNA is not affected, therefore this outcome cannot be detected. The heteroduplex intermediate we propose as a substrate for Ercc1–Xpf (and therefore the concomitant role for the enzyme in HR) is fundamentally different from the role that Rad10–Rad1 plays in recombination between DNA substrates that differ by small mismatches (Nicholson et al., 2000) because small patches of heterology are unlikely to stall branch migration.
Once the incisions are made by Ercc1–Xpf, there are a number of means by which the nicked intermediate might be resolved in order to achieve gene replacement. First, depending on the distance between the nicks, the DNA may be unwound either spontaneously or enzymatically by a helicase. Secondly, a 5′- to -3′ exonuclease may remove the intervening sequence by chewing back from the nicks. Thirdly, another round of endonucleolytic cleavage immediately 5′ to the D-loops may remove the intervening sequence. This site is also a potential substrate for Ercc1–Xpf, as indicated above. Regardless of the mechanism, the net result must be removal of the recipient DNA between the nicks, thereby creating a frank DSB in the chromosome (Figure 5F). After removal of the sequence between the Ercc1–Xpf-generated nicks, incorporation of the nonhomologous donor sequences into the genomic locus can be completed by enzymes involved in DNA replication (Figure 5G). A portion of the donor sequences may be replicated by leading strand synthesis initiated from the new 3′ termini of the recipient DNA (Bardwell et al., 1994). The activities of a flap endonuclease and DNA ligase are required to restore continuity to both DNA strands of the replaced genomic locus.
In addition to the fact that creating incisions in the recipient DNA greatly facilitates conceptual understanding of how exogenous sequences can be incorporated into the genome, there is experimental evidence for this event. In mammalian cells, including ES cells, the introduction of a DSB in a chromosome stimulates targeting by exogenous DNA by several orders of magnitude (Rouet et al., 1994; Smih et al., 1995; Richardson et al., 1999). In meiotic recombination in yeast, rad1 or rad10 strains fail to repair nonhomologous genomic insertions, as reflected by an increase in post-meiotic segregation compared with wt strains (Kirkpatrick and Petes, 1997; Kearney et al., 2001). However, one class of segregation events is suppressed in the mutant strains: 2:6 conversions (the conversion of a wt allele to a mutant allele containing the inserted loop). This is consistent with the Rad1–Rad10 endonuclease mediating loop repair via cleavage of the recipient strand to allow integration of the nonhomologous sequence, similar to the mechanism proposed above for gene replacement in ES cells. Finally, precedence for a D-loop endonuclease that stimulates gene replacement exists in phages. Rap (for recombination adept with plasmid) is a phage protein that is a functional homolog of the Escherichia coli RusA Holliday junction resolvase. Rap preferentially cleaves branched structures and 3′-tailed D-loops identically to the proposed mechanism for Ercc1 (Sharples et al., 1998).
Although the role we propose for Ercc1–Xpf in gene replacement is to initiate the formation of a DSB in the genome, there are several aspects of our model that contribute to the preservation of genomic integrity. First, the trigger for Ercc1–Xpf cleavage is an unresolved heteroduplex structure, which if left unresolved could itself trigger undesirable consequences via DNA and/or RNA polymerase blockage. Secondly, since the proposed cleavage is internal to two stretches of heteroduplex DNA, the break is stabilized and well-poised for rapid rejoining of the correct ends. This is consistent with the observation that there is a minimum amount of homology required to favor gene replacement in ES cells (Hasty et al., 1991; Deng and Capecchi, 1992; Thomas et al., 1992), which may be indicative of the amount of homology required to stabilize the proposed intermediate. Thirdly, the structure specificity of the endonuclease dictates that cuts are only made within ds sequences immediately adjacent to ds/ss borders (Bardwell et al., 1994; de Laat et al., 1998a). This minimizes the loss of sequence information from the chromosome. Similarly, sequence information is not lost from the targeting construct, which is supported by the observation that gene replacement is highly accurate even in the absence of gain-of-function selection (Zheng et al., 1991; Li and Baker, 2000).
In conclusion, we have demonstrated that Ercc1 is essential for gene replacement in ES cells. This is true even if the replacement construct is completely homologous to the genomic target, with the exception of a centrally positioned selectable marker. Thus, the role for Ercc1 that we discovered is distinct from its previously established role in removing 3′ overhangs of nonhomologous sequence from DSBs, SSA intermediates and gene insertion constructs. Considering the substrate specificity of the Ercc1–Xpf endonuclease, the most likely function of Ercc1 in gene replacement is nicking of heteroduplex intermediates within D-loops. Therefore, in addition to defining a new role for Ercc1 in HR and providing evidence for a new substrate of the Ercc1–Xpf endonuclease, these data help to elucidate the mechanism of gene replacement in mammalian cells.
Materials and methods
Generation of mErcc1–/– ES cells
mErcc1 targeting constructs and the production of Ercc1+/– murine ES cells were as previously described (Weeda et al., 1997). Briefly, a 12-kb _Hin_dIII/_Sal_I genomic DNA fragment from a 129/CCE genomic library including the last four exons (exons 7–10) of mErcc1 was subcloned into pBR322. A unique _Cla_I restriction site was created in exon 7 allowing disruption of the exon by introducing a neoR expression cassette into the site. The neoR construct was transfected into wt ES cells, G418-resistant colonies isolated and targeted clones identified by DNA blotting (Weeda et al., 1997). To target the second Ercc1 allele in the mErcc1+/– ES line, the targeting construct was modified by replacing the neoR marker with a hygroR cassette. A linker containing a diagnostic _Bam_HI site was included 3′ to the selectable marker to facilitate identification of targeted clones. The construct was electroporated into the mErcc1+/– ES cells and selected for with hygromycin. An _mErcc1_-specific probe that hybridizes to sequences outside (5′) of the targeting constructs was used to distinguish between wt, neoR and hygroR alleles in DNA blot analysis of _Bam_HI-digested genomic DNA isolated from hygroR clones. Positive clones were further characterized using probes against the resistance markers and sequences external to the 3′ homologous arm of the constructs.
Rescue of mErcc1–/– ES cells with hERCC1 cDNA
The cDNA encoding hERCC1 was cloned into N-pEYFP (Promega) as an N-terminal in-frame fusion with eYFP. The fusion cDNA was subsequently cloned into a phosphoglycerate kinase promoter–poly(A) expression cassette (Essers et al., 1997). The vector was electroporated into the _mErcc1_–/– ES cells either alone or with a second plasmid carrying a puromycin expression cassette. Since the parental knock-out line is sensitive to crosslinking agents (see Results) selection of stably transformed cells was achieved by adding 5.0 × 10–8 M MMC to the media or in the case of cotransfection with puromycin (1 µg/ml). Resistant clones were screened for the presence of fluorescence by microscopy. Expression of the full-length fusion protein was confirmed by immunoblotting of 10 µg of WCE using a polyclonal α-hERCC1 antibody (van Vuuren et al., 1995).
DNA damage sensitivities and sister chromatid exchange
Sensitivity to various DNA damages was measured as colony-forming ability of the different ES cell lines after exposure to various agents as described (Essers et al., 1997). All data points were measured in triplicate. The cloning efficiency of untreated cells was typically 20–30%. SCEs were measured as described previously (Dronkert et al., 2000). More than 50 metaphase spreads were analyzed for each data point.
Quantitation of gene targeting events
Cloning of a 9-kb _Eco_RI mRad54 genomic fragment (encompassing exons 3–8) into a bacterial vector was as described previously (Essers et al., 1997). In an attempt to achieve high targeting efficiency but not necessarily disruption of Rad54 protein expression, a puroR expression cassette was inserted into a unique _Nco_I site in intron 3 of the genomic fragment. When homologously recombined into the locus, the construct converts the 9.0-kb genomic _Stu_I fragment to an 8.5-kb fragment, permitting diagnosis of targeting. The targeting construct was isolated from the vector as a linear _Eco_RI fragment with flanking arms completely homologous to the chromosomal mRad54 target. Alternatively, the construct was isolated from the vector using a _Cla_I–_Not_I double digest, leaving 32 bp of vector sequence on the 5′ end and 14 bp on the 3′ end of the linear fragment, effectively creating short termini without homology to the mRad54 locus. The linear fragments were electroporated into one wt, two Ercc1–/– and three rescued mutant cell lines, as described above. Since all of the rescued mutant lines were phenotypically indistinguishable regardless of how they were generated (data not shown), only lines selected by MMC were used in this assay so that selection of targeted clones using the last selectable marker, puromycin, was possible. Resistant clones were picked and expanded, then lysed and genomic DNA was isolated. DNA preparations were digested with _Stu_I and analyzed by DNA blotting using a 3′ external probe recognizing exons 7 and 8. Targeting frequencies are reported as the total number of HR events divided by the number of homologous plus nonhomologous integration events. The χ2 test was used to evaluate the significance of the differences between the observed HR frequencies.
Acknowledgments
Acknowledgements
We thank H.de Waard, H.M.Kearney and T.D.Petes for communication of unpublished results and helpful discussion. This research was supported by the Netherlands Organization for Scientific Research (NWO), the Dutch Cancer Society (KWF), the Human Frontier Science Program Organization (grant RGP0298/2001-M) and the Association for International Cancer Research. G.W. was a fellow of The Netherlands Royal Academy of Arts and Sciences. L.J.N. was supported by the National Science Foundation (Grant INT-9900963; 6-1999 to 1-2000) and the American Cancer Society (Grant PF-99-142; 1-2000 to present).
References
- Adair G.M., Rolig,R.L., Moore-Faver,D., Zabelshansky,M., Wilson,J.H. and Nairn,R.S. (2000) Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J., 19, 5552–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A. and Klein,H.L. (1989) Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics, 123, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell A.J., Bardwell,L., Tomkinson,A.E. and Friedberg,E.C. (1994) Specific cleavage of model recombination and repair intermediates by the yeast Rad1–Rad10 DNA endonuclease. Science, 265, 2082–2084. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii B.P., Reddy,G. and Zarling,D.A. (1999) DNA hybrids stabilized by heterologies. Biochemistry, 38, 10785–10792. [DOI] [PubMed] [Google Scholar]
- Berinstein N., Pennell,N., Ottaway,C. and Shulman,M.J. (1992) Gene replacement with one-sided homologous recombination. Mol. Cell. Biol., 12, 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggerstaff M., Szymkowski,D.E. and Wood,R.D. (1993) Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J., 12, 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A., Davies,A.A. and West,S.C. (2001) Branch migration and Holliday junction resolution catalyzed from mammalian cells. Cell, 104, 259–268. [DOI] [PubMed] [Google Scholar]
- Darroudi F. and Natarajan,A.T. (1985) Cytological characterization of repair-deficient CHO cell line 43–3B. I. Induction of chromosomal aberrations and sister-chromatid exchanges by UV and its modulation with 3-aminobenzamide. Mutat. Res., 149, 239–247. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Appeldoorn,E., Jaspers,N.G. and Hoeijmakers,J.H.J. (1998a) DNA structural elements required for ERCC1–XPF endonuclease activity. J. Biol. Chem., 273, 7835–7842. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Appeldoorn,E., Sugasawa,K., Weterings,E., Jaspers,N.G. and Hoeijmakers,J.H.J. (1998b) DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev., 12, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C. and Capecchi,M.R. (1992) Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol., 12, 3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Thomas,K.R. and Capecchi,M.R. (1993) Location of crossovers during gene targeting with insertion and replacement vectors. Mol. Cell. Biol., 13, 2134–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert M.L.G. and Kanaar,R. (2001) Repair of DNA interstrand cross-links. Mutat. Res., 486, 217–247. [DOI] [PubMed] [Google Scholar]
- Dronkert M.L.G., Beverloo,H.B., Johnson,R.D., Hoeijmakers,J.H.J., Jasin,M. and Kanaar,R. (2000) Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol., 20, 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H.J. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J. and Haber,J.E. (1992) Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science, 258, 480–484. [DOI] [PubMed] [Google Scholar]
- Fleck O., Lehmann,E., Schar,P. and Kohli,J. (1999) Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nature Genet., 21, 314–317. [DOI] [PubMed] [Google Scholar]
- Gaillard P.H. and Wood,R.D. (2001) Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res., 29, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang H., Hager,D.N., Goriparthi,L., Hopkins,K.M., Shih,H. and Lieberman,H.B. (1996) Schizosaccharomyces pombe rad23 is allelic with swi10, a mating-type switching/radioresistance gene that shares sequence homology with human and mouse ERCC1. Gene, 170, 113–117. [DOI] [PubMed] [Google Scholar]
- Hasty P., Rivera-Perez,J. and Bradley,A. (1991) The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol., 11, 5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers,S., Nigg,A.L., Hoogstraten,D., Hoeijmakers,J.H.J. and Vermeulen,W. (1999) Action of DNA repair endonuclease ERCC1/XPF in living cells. Science, 284, 958–961. [DOI] [PubMed] [Google Scholar]
- Hoy C.A., Thompson,L.H., Mopney,C.L. and Salazar,E.P. (1985) Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res., 45, 1737–1743. [PubMed] [Google Scholar]
- Ivanov E.L. and Haber,J.E. (1995) RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney H.M., Kirkpatrick,D.T., Gerton,J.L. and Petes,T.D. (2001) Meiotic recombination involving heterozygous large insertions in S. cerevisiae: formation and repair of large, unpaired DNA loops. Genetics, 158, 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D.T. and Petes,T.D. (1997) Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature, 387, 929–931. [DOI] [PubMed] [Google Scholar]
- Klein H.L. (1988) Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics, 120, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. and Baker,M.D. (2000) Mechanisms involved in targeted gene replacement in mammalian cells. Genetics, 156, 809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S.L., Thomas,K.R., Deng,C. and Capecchi,M.R. (1990) Introduction of a lacZ reporter gene into the mouse int-2 locus by homologous recombination. Proc. Natl Acad. Sci. USA, 87, 7688–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhir J., Selfridge,J., Harrison,D.J., Squires,S. and Melton,D.W. (1993) Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nature Genet., 5, 217–223. [DOI] [PubMed] [Google Scholar]
- Melton D.W., Ketchen,A.M., Nunez,F., Bonatti-Abbondandolo,S., Abbondandolo,A., Squires,S. and Johnson,R.T. (1998) Cells from ERCC1-deficient mice show increased genome instability and a reduced frequency of S-phase-dependent illegitimate chromosome exchange but a normal frequency of homologous recombination. J. Cell Sci., 111, 395–404. [DOI] [PubMed] [Google Scholar]
- Moore C.W. (1978) Responses of radiation-sensitive mutants of Saccharomyces cerevisiae to the lethal effects of bleomycin. Mutat. Res., 51, 165–180. [DOI] [PubMed] [Google Scholar]
- Mu D., Park,C.-H., Matsunaga,T., Hsu,D.S., Reardon,J.T. and Sancar,A. (1995) Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem., 270, 2415–2418. [DOI] [PubMed] [Google Scholar]
- Nairn R.S., Adair,G.M., Christmann,C.B. and Humphrey,R.M. (1991) Ultraviolet stimulation of intermolecular homologous recombination in Chinese hamster ovary cells. Mol. Carcinog., 4, 519–526. [DOI] [PubMed] [Google Scholar]
- Negritto M.T., Wu,X., Kuo,T., Chu,S. and Bailis,A.M. (1997) Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol. Cell. Biol., 17, 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson A., Hendrix,M., Jinks-Robertson,S. and Crouse,G.F. (2000) Regulation of mitotic homeologous recombination in yeast. Functions of mismatch repair and nucleotide excision repair genes. Genetics, 154, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Szostak,J.W. and Rothstein,R.J. (1981) Yeast transformation: a model system for the study of recombination. Proc. Natl Acad. Sci. USA, 78, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1997) Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F. and Aguilera,A. (1995) Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10 and RAD52 genes. Genetics, 139, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C., Moynahan,M.E. and Jasin,M. (1999) Homologous recombination between heterologs during repair of a double-strand break. Suppression of translocations in normal cells. Ann. N. Y. Acad. Sci., 886, 183–186. [DOI] [PubMed] [Google Scholar]
- Rodel C., Kirchhoff,S. and Schmidt,H. (1992) The protein sequence and some intron positions are conserved between the switching gene swi10 of Schizosaccharomyces pombe and the human excision repair gene ERCC1. Nucleic Acids Res., 20, 6347–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R.J. (1983) One-step gene disruption in yeast. Methods Enzymol., 101, 202–211. [DOI] [PubMed] [Google Scholar]
- Rouet P., Smih,F. and Jasin,M. (1994) Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol., 14, 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M., Prakash,L. and Prakash,S. (1996) Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1–RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics, 142, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent R.G., Rolig,R.L., Kilburn,A.E., Adair,G.M., Wilson,J.H. and Nairn,R.S. (1997) Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl Acad. Sci. USA, 94, 13122–13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent R.G., Meservy,J.L., Perkins,B.D., Kilburn,A.E., Intody,Z., Adair,G.M., Nairn,R.S. and Wilson,J.H. (2000) Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res., 28, 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R.H. and Prakash,S. (1988) RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol. Cell. Biol., 8, 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R.H. and Prakash,S. (1990) RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol., 10, 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selfridge J., Pow,A.M., McWhir,J., Magin,T.M. and Melton,D.W. (1992) Gene targeting using a mouse HPRT minigene/HPRT-deficient embryonic stem cell system: inactivation of mouse ERCC-1 gene. Somat. Cell Mol. Genet., 18, 325–336. [DOI] [PubMed] [Google Scholar]
- Sharples G.J., Corbett,L.M. and Graham,I.R. (1998) λ Rap protein is a structure-specific endonuclease involved in phage recombination. Proc. Natl Acad. Sci. USA, 95, 13507–13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbers A.M. et al. (1996) Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell, 86, 811–822. [DOI] [PubMed] [Google Scholar]
- Sladek F., Munn,M., Rupp,W. and Howard-Flanders,P. (1989) In vitro repair of psoralen-DNA cross-links by RecA, UvrABC and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem., 264, 6755–6765. [PubMed] [Google Scholar]
- Smih F., Rouet,P., Romanienko,P.J. and Jasin,M. (1995) Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res., 23, 5012–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky R.T. and Tye,B.K. (1985) Construction of telocentric chromosomes in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 82, 2106–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver,T.L., Rothstein,R.J. and Stahl,F.W. (1983) The double-strand-break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Thomas K.R., Deng,C. and Capecchi,M.R. (1992) High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol. Cell. Biol., 12, 2919–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson A.E., Bardwell,A.J., Bardwell,L., Tappe,N.J. and Friedberg,E.C. (1993) Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature, 362, 860–862. [DOI] [PubMed] [Google Scholar]
- van Duin M., de Wit,J., Odijk,H., Westerveld,A., Yasui,A., Koken,M.H.M., Hoeijmakers,J.H.J. and Bootsma,D. (1986) Molecular characterization of the human excision repair gene ERCC-1: cDNA cloning and amino acid homology with the yeast DNA repair gene RAD10. Cell, 44, 913–923. [DOI] [PubMed] [Google Scholar]
- van Duin M. et al. (1989) The cloned human DNA excision repair gene ERCC-1 fails to correct xeroderma pigmentosum complementation groups A through I. Mutat. Res., 217, 83–92. [DOI] [PubMed] [Google Scholar]
- van Vuuren A.J., Appeldoorn,E., Odijk,H., Yasui,A., Jaspers,N.G.J., Bootsma,D. and Hoeijmakers,J.H.J. (1993) Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO J., 12, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vuuren A.J., Appeldoorn,E., Odijk,H., Humbert,S., Moncollin,V., Eker,A.P.M., Jaspers,N.G.J., Egly,J.M. and Hoeijmakers,J.H.J. (1995) Partial characterization of the DNA repair protein complex, containing the ERCC1, ERCC4, ERCC11 and XPF correcting activities. Mutat. Res., 337, 25–39. [DOI] [PubMed] [Google Scholar]
- Weeda G. et al. (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol., 7, 427–439. [DOI] [PubMed] [Google Scholar]
- Westerveld A., Hoeijmakers,J.H.J., van Duin,M., de Wit,J., Odijk,H., Pastink,A., Wood,R.D. and Bootsma,D. (1984) Molecular cloning of a human DNA repair gene. Nature, 310, 425–429. [DOI] [PubMed] [Google Scholar]
- Zehfus B.R., McWilliams,A.D., Lin,Y.-H., Hoekstra,M.F. and Keil,R.L. (1990) Genetic control of RNA polymerase I-stimulated recombination in yeast. Genetics, 126, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Hast,P., Brenneman,M.A., Grompe,M., Gibbs,R.A., Wilson,J.H. and Bradley,A. (1991) Fidelity of targeted recombination in human fibroblasts and murine embryonic stem cells. Proc. Natl Acad. Sci. USA, 88, 8067–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]