Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex (original) (raw)
Abstract
The RNA-binding protein Y14 binds preferentially to mRNAs produced by splicing and is a component of a multiprotein complex that assembles ∼20 nucleotides upstream of exon–exon junctions. This complex probably has important functions in post-splicing events including nuclear export and nonsense-mediated decay of mRNA. We show that Y14 binds to two previously reported components, Aly/REF and RNPS1, and to the mRNA export factor TAP. Moreover, we identified magoh, a human homolog of the Drosophila mago nashi gene product, as a novel component of the complex. Magoh binds avidly and directly to Y14 and TAP, but not to other known components of the complex, and is found in Y14-containing mRNPs in vivo. Importantly, magoh also binds to mRNAs produced by splicing upstream (∼20 nucleotides) of exon– exon junctions and its binding to mRNA persists after export. These experiments thus reveal specific protein–protein interactions among the proteins of the splicing-dependent mRNP complex and suggest an important role for the highly evolutionarily conserved magoh protein in this complex.
Keywords: exon–exon junction complex/magoh/mago nashi/splicing/Y14
Introduction
As nascent transcripts, pre-mRNAs become associated with abundant nuclear proteins collectively referred to as hnRNP proteins (Dreyfuss et al., 1993; Krecic and Swanson, 1999). Pre-mRNAs are converted to mRNAs by processing events that include addition of the cap structure, pre-mRNA splicing and polyadenylation. Following these nuclear processing events, mRNAs are exported from the nucleus to the cytoplasm, where they are translated into proteins. Several lines of evidence suggested that mRNA processing is linked to downstream events (reviewed in Shyu and Wilkinson, 2000). For example, the presence of an intron can enhance the efficiency of translation of some genes (Matsumoto et al., 1998). Furthermore, nonsense-mediated mRNA decay (NMD), a process that leads to degradation of mRNAs that contain premature stop codons (Li and Wilkinson, 1998; Nagy and Maquat, 1998; Culbertson, 1999; Czaplinski et al., 1999; Hentze and Kulozik, 1999; Hilleren and Parker, 1999), probably occurs in the cytoplasm and is linked to that pre-mRNA splicing (Carter et al., 1996; Zhang et al., 1998a,b). These and other observations suggested that pre-mRNA splicing alters the mRNA or endows it with proteins that are carried with the mRNA to the cytoplasm and required for NMD.
Recently, pre-mRNA splicing has been shown to alter the protein composition of mRNAs (Luo and Reed, 1999; Kataoka et al., 2000; Le Hir et al., 2000b). This change facilitates the nuclear export of spliced mRNAs compared with that of intronless mRNAs (Luo and Reed, 1999). Several proteins, including Aly/REF (Zhou et al., 2000), Y14 (Kataoka et al., 2000), RNPS1 (Mayeda et al., 1999), SRm160 (Blencowe et al., 1998; Le Hir et al., 2000a) and DEK (McGarvey et al., 2000), have been reported to bind preferentially to spliced mRNAs. These proteins have been shown to bind mRNAs in a position-specific manner ∼20 nucleotides upstream of exon–exon junctions in vitro (Le Hir et al., 2000a). Among these protein components, Aly/REF, Y14 and RNPS1 were shown to shuttle between the nucleus and the cytoplasm (Kataoka et al., 2000; Zhou et al., 2000; Lykke-Andersen et al., 2001). Aly/REF was reported to stimulate the export of both spliced and unspliced mRNAs (Zhou et al., 2000; Rodrigues et al., 2001). Importantly, the binding of Y14 has been shown to persist on newly exported spliced mRNA in the cytoplasm (Kataoka et al., 2000). Moreover, Y14 remains bound in the same position, 20 nucleotides upstream of exon–exon junctions, on mRNAs, whereas Aly/REF is dissociated from mRNAs after export (Kim et al., 2001b). Thus, Y14 may have a special role in communicating the position of exon–exon junctions of mRNAs to the cytoplasm.
To understand further the function of Y14, we studied its interactions with other components of the splicing-dependent exon–exon junction definition complex. Here we show that Y14 interacts with Aly/REF and RNPS1, but not with DEK in vitro. Y14 also interacts with the mRNA export factor TAP. We have also identified a novel component of the complex. This protein designated as magoh, a human homolog of the Drosophila mago nashi gene product (Zhao et al., 1998), interacts with Y14 directly. In Drosophila, mago nashi (Japanese for grandchildless) plays a role in axis formation during oogenesis (Boswell et al., 1991; Newmark and Boswell, 1994; Micklem et al., 1997; Newmark et al., 1997), but its precise function is unclear. Interestingly, magoh is a particularly avid interactor of TAP and, like the other components of the complex, it is associated with mRNAs produced by splicing 20 nucleotides upstream of exon– exon junctions.
Results
Y14 interacts with Aly/REF and RNPS1
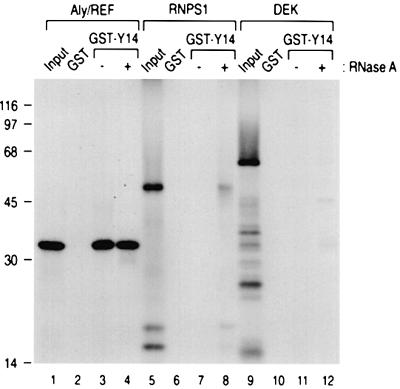
To investigate the protein–protein interaction between Y14 and other putative components of the splicing-dependent mRNP complexes, we carried out binding assays of [35S]methionine-labeled, in vitro translated candidate proteins to glutathione _S_-transferase (GST)– Y14. Of the known components of the complex, we could not test the binding of SRm160 to Y14 because SRm160 was not produced by in vitro translation (data not shown). Since several of these proteins are likely to be RNA-binding proteins, we also tested the binding activity after treatment with RNase A to avoid non-specific bridging through RNA. As shown in Figure 1, Y14 binds to Aly/REF, and RNase A treatment does not significantly affect the binding (lanes 3 and 4). This suggests that the binding of Aly/REF to Y14 is mediated by protein–protein interaction. This is consistent with previous results, which showed that Aly/REF can be co-immunoprecipitated with anti-Y14 antibodies from HeLa cell nucleoplasm (Kim et al., 2001b). RNPS1 showed weak binding to Y14, which was only detectable after RNase A digestion (Figure 1, lane 8). This indicates that these two proteins bind each other and suggests that RNPS1 binds to Y14 that is not bound to RNA. Under the same assay conditions, no binding of DEK to Y14 could be detected (Figure 1, lanes 9–12).
Fig. 1. Y14 binds to Aly/REF and RNPS1, but not to DEK in vitro. His-Aly/REF, his-RNPS1 and his-DEK proteins, produced and labeled with [35S]methionine in vitro, were incubated with 5 µg each of GST or GST–Y14. After washing, bound proteins were eluted, separated on 12.5% SDS–PAGE gel and visualized by fluorography. The lanes marked Input contain 10% of the total protein used in each binding reaction. The position of molecular mass markers is shown on the left.
Y14 interacts avidly and specifically with magoh
To identify components of the Y14-containing complex, we carried out yeast two-hybrid screening on a HeLa cell cDNA library using the N-terminal 73 amino acids of Y14 as bait. Several positive clones were obtained, and one of these was identified as the human homolog of Drosophila mago nashi designated magoh (Zhao et al., 1998). Y14 has also been isolated from a yeast two-hybrid screening with magoh as a bait (Zhao et al., 2000). Therefore, Y14 and magoh have been identified as interactors in complementary yeast two-hybrid screens.
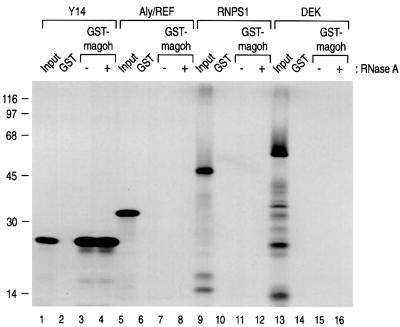
To confirm the specific binding of magoh to Y14, we carried out in vitro binding assays using recombinant GST–magoh. For these experiments, [35S]methionine-labeled Y14 was produced by transcription/translation in vitro. As shown in Figure 2, Y14 binds to GST–magoh, but not GST (lanes 2 and 3). Since Y14 has an RNA-binding motif (Kataoka et al., 2000) and mago has been implicated in RNA transport in oogenesis (Newmark and Boswell, 1994; Micklem et al., 1997; Newmark et al., 1997; Li et al., 2000), we also carried out binding experiments after digestion with RNase A. As shown in Figure 2, RNase A digestion did not affect the magoh–Y14 interaction (lane 4), indicating that magoh binds specifically and directly to Y14. We also tested the binding of magoh to other components of the post-splicing mRNP complex and found that, at least under these conditions, magoh binds only to Y14 and not to other known constituents of the complex (Figure 2).
Fig. 2. Magoh binds avidly and specifically to Y14 in vitro. In vitro translated and labeled proteins were produced and used for binding to GST or GST–magoh as described in Figure 1. The lanes marked Input contain 10% of the total protein used in each binding reaction.
Y14 and magoh interact with the mRNA export factor TAP
The mRNA export factor TAP is also present in the nuclear Y14 complex (Kataoka et al., 2000) and has been shown to interact with Aly/REF (Stutz et al., 2000; Rodrigues et al., 2001). We therefore tested whether TAP can bind to Y14 and magoh. TAP was produced by in vitro transcription/translation and incubated with immobilized GST fusion proteins. As reported previously (Stutz et al., 2000; Rodrigues et al., 2001), Aly/REF bound to TAP and RNase A has almost no effect on binding (Figure 3A). Similarly, Y14 bound to TAP and this binding was also unaffected by RNase A digestion (Figure 3A). Magoh also bound avidly to TAP under the same conditions (Figure 3A), and RNase A treatment had almost no effect on this interaction, suggesting that magoh and TAP interact directly. To verify this, we performed binding assays using recombinant magoh. As shown in Figure 3B, recombinant magoh binds to both GST–Y14 and TAP. RNase A treatment only slightly reduced magoh binding to Y14 and the binding of magoh to TAP was increased by RNase A digestion (Figure 3B). These results indicate that magoh interacts with Y14 and TAP.
Fig. 3. Y14 binds to TAP, and magoh binds to Y14 and TAP. (A) Aly/REF, Y14 and magoh bind to TAP in vitro. In vitro translated TAP was used for binding to 5 µg of the indicated GST fusion proteins. Ten percent of the total was loaded in the lane marked Input. (B) Magoh binds to Y14 and TAP in vitro. Two micrograms of bacterially expressed his-magoh protein were tested for binding to the indicated GST fusion proteins as described in Figure 1. Bound magoh was detected by western blotting with anti-T7 tag antibody (Novagen). The lane designated Input contains 20% of the total used for the binding reaction.
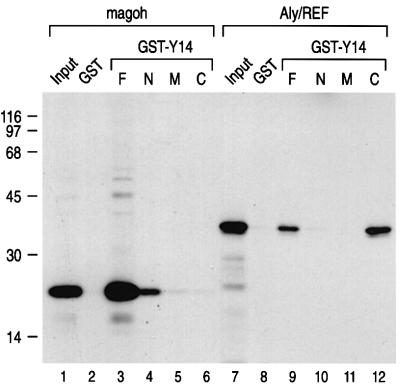
To map the domains of Y14 that mediate its binding to magoh, several Y14 deletion mutants were constructed. Y14 is a 174-amino-acid protein with a central RNP motif RNA-binding domain (RBD) (Kataoka et al., 2000). Since the N-terminus of Y14 was used for the yeast two-hybrid screening, this portion was expected to interact with magoh. We therefore fused GST with amino acids 1–73, 74–139 and 140–174 of Y14, and designated those proteins GST–N, –M and –C, respectively. The GST–M protein contains the RBD of Y14. As expected, magoh bound predominantly to the N-terminus of Y14 (Figure 4). However, binding of magoh to the N-terminus of Y14 was considerably weaker than that of full-length Y14, suggesting that other sequences of Y14 contribute to the binding or to the presentation of the N-terminus for efficient binding. We also found that the C-terminus of Y14 binds to Aly/REF (Figure 4). These results suggest that Y14 may have the capacity to bind magoh and Aly/REF at the same time, perhaps through different domains.
Fig. 4. Magoh and Aly/REF bind to different domains of Y14. Magoh and Aly/REF proteins were produced and used for in vitro binding experiments as described in Figure 1. GST fused to Y14 amino acids 1–174, 1–73, 74–139 and 140–174 is designated GST–F, –N, –M and –C, respectively. The input lanes contain 10% of the total used for the binding reaction.
Magoh is associated with nuclear Y14-containing complexes in vivo
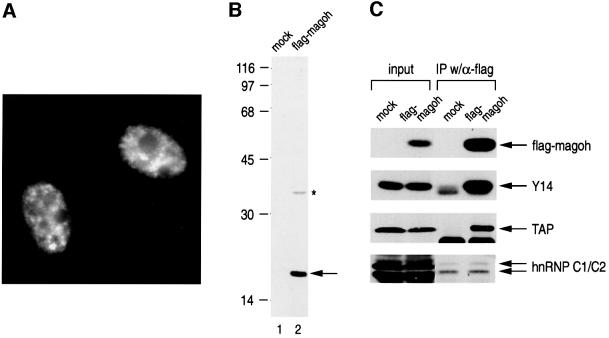
The results described above suggested that magoh is in the nuclear Y14-containing complexes. To test this directly, we transiently expressed flag-tagged magoh by transient transfection in HEK 293T cells and carried out immunoprecipitations. However, we first examined the localization of flag-magoh in HeLa cells. As shown in Figure 5A, flag-magoh is localized in the nucleoplasm and shows a pattern similar to that observed for Y14 (Kataoka et al., 2000). Next, we checked the expression of flag-magoh in HEK293T cells by western blotting with anti-flag-tag monoclonal antibodies. This showed predominantly a 17 kDa protein and one additional larger polypeptide (Figure 5B). The identity of the larger product is not known, but it may be a glycosylated form of magoh, since magoh has two candidate sites for glycosylation and in vitro translation reaction produces larger glycosylated products in addition to the 17 kDa one (Zhao et al., 1998).
Fig. 5. Magoh is in nuclear Y14 complexes. (A) Magoh is mainly localized in the nucleoplasm in HeLa cells. Flag-magoh cDNA was transfected into HeLa cells, which were subsequently immunostained using anti-flag-tag antibody (M2, Sigma) as described in Materials and methods. (B) Flag-magoh is expressed as a 17 kDa protein in HeLa cells. The expression of flag-magoh was tested by western blotting using anti-flag antibody with vector-transfected (mock) and flag-magoh cDNA-transfected HEK293T cell extracts. (C) Nuclear magoh is associated with Y14 and TAP. Immuno precipitation with anti-flag antibody was carried out from the nucleoplasm of HEK293T cells transfected either with flag-vector (mock) or flag-magoh cDNA (flag-magoh). Co-immunoprecipitated proteins were analyzed by western blotting to detect the indicated proteins. Two percent of total input is shown on the left.
To determine whether magoh is in the nuclear Y14-containing complexes, immunoprecipitations from the nucleoplasm of cells transiently expressing flag-magoh were carried out, and co-immunoprecipitated proteins were resolved by SDS–PAGE and analyzed by western blotting with several different antibodies. Y14 and TAP were co-immunoprecipitated efficiently with flag-tagged magoh (Figure 5C). In contrast, only small amounts of the abundant hnRNP C proteins were co-immunoprecipitated (Figure 5C). These results show that magoh is in nuclear complexes that also contain Y14 and TAP.
Magoh binds spliced mRNAs at ∼20 nucleotides upstream of exon–exon junctions
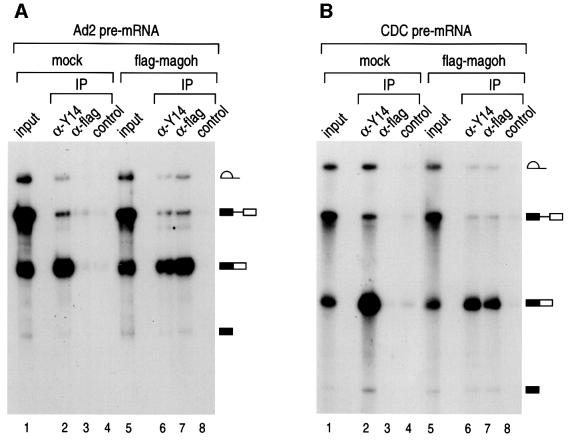
We have previously shown that the anti-Y14 antibody 4C4 preferentially immunoprecipitates mRNAs produced by splicing in vitro and in vivo (Kataoka et al., 2000). Since magoh interacts with Y14 and associates with the nuclear Y14 complex (Figure 5C), it was expected that it would also be associated with spliced mRNA. To examine this, we wished to use the strategy we have previously employed to map the position of Y14 on mRNAs which involve RNase H digestion and immunoprecipitation. However, because anti-magoh antibodies were not available, we devised the following experimental scheme. Splicing of radiolabeled adenovirus-derived pre-mRNA (Ad2) was carried out in vitro using extracts prepared either from flag vector-transfected (mock) or flag-magoh-transfected HEK293T cells. Immunoprecipitation of splicing products was then performed using anti-flag antibodies and the products were analyzed by denaturing PAGE. The flag-magoh in HEK293T cell extracts had no effect on pre-mRNA splicing (Figure 6A, lanes 1 and 5). As shown previously, anti-Y14 antibody, 4C4, immunoprecipitated predominantly spliced mRNA from both extracts (Figure 6; Kataoka et al., 2000). As shown in Figure 6A, anti-flag antibodies immunoprecipitated predominantly spliced mRNA from flag-magoh-containing extracts, but not from mock-transfected extracts (lanes 3 and 7). We conclude that magoh, which specifically interacts with Y14, is preferentially associated with spliced mRNA. To determine whether this is unique to Ad2 mRNA, the same experiment was carried out with a different pre-mRNA, the one derived from chicken delta crystallin (CDC). Similar to the case for Ad2, anti-flag antibodies immunoprecipitated efficiently only the spliced mRNA from flag-magoh-transfected extracts but not from mock-transfected extracts (Figure 6B). Therefore, magoh binds to mRNAs that are produced by splicing.
Fig. 6. Magoh is preferentially associated with mRNA produced by pre-mRNA splicing in vitro. (A) Immunoprecipitation was performed from in vitro splicing reaction with 32P-labeled Ad2 pre-mRNA. For in vitro splicing, HeLa cell nuclear extracts were mixed with extracts from HEK293T cells that had been transfected either with flag-vector (mock) or with flag-magoh cDNA (flag-magoh). RNAs immunoprecipitated with anti-Y14 (α-Y14), anti-flag (α-flag) and non-immune antibody SP2/0 (control) were analyzed on a 10% polyacrylamide denaturing gel. A schematic representation of RNAs is shown on the right. The antibodies against flag-tag (M2), Y14 (4C4) and control non-immune antibody (SP2/0) were used for this assay. The input lane contains 10% of the total. (B) RNA immunoprecipitation was carried out as described in (A) with CDC pre-mRNA.
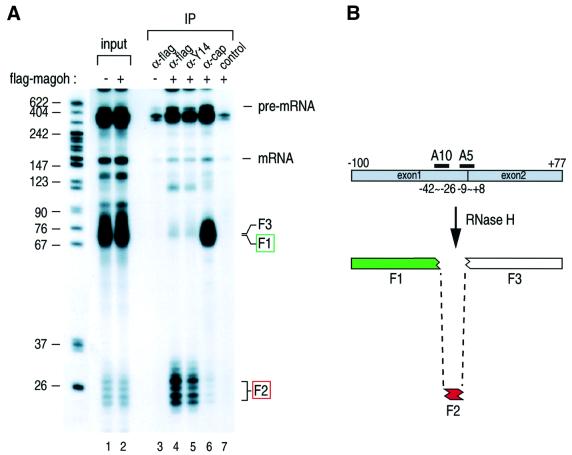
To determine the specific position of magoh on the spliced mRNAs, we used the RNase H digestion and immunoprecipitation methods as described previously (Le Hir et al., 2000a; Kim et al., 2001b). Sequence-specific cleavage of Ad2 mRNA with two oligos, A5 and A10, generates three fragments, as illustrated in Figure 7. The shortest fragment, F2, contains the region 20 nucleotides upstream of the exon–exon junctions. As reported previously, the anti-Y14 antibody (4C4) immunoprecipitated the shortest F2 fragment from the in vitro splicing reaction, while the anti-cap structure antibody, H20, precipitated the 5′ cap structure-containing F1 fragment (Figure 7). Similarly, the anti-flag antibody immunoprecipitated the F2 fragment from extracts containing flag-magoh (lane 4), but not from mock-transfected extracts (lane 3). A similar result was obtained by performing the same experiments starting with CDC pre-mRNA (data not shown). These results indicate that magoh, like Y14, is bound, ∼20 nucleotides upstream of exon–exon junctions to mRNAs produced by splicing.
Fig. 7. Magoh is bound to spliced mRNA ∼20 nucleotides upstream of the exon–exon junction. (A) In vitro splicing with 32P-labeled Ad2 pre-mRNA was performed as described in Figure 6. After splicing, RNAs were subjected to RNase H digestion with 7 µM each A5 and A10 oligonucleotides, followed by immunoprecipitation with anti-flag (α-flag), anti-Y14 (α-Y14) and non-immune antibody SP2/0 (control). Immunoprecipitated RNA fragments were recovered and analyzed on 10% polyacrylamide denaturing gel. Lanes for input contain 10% of total material. The position of RNA size markers is shown in nucleotides on the left. (B) The positions of oligonucleotides and predicted RNA fragments from the spliced RNase H-cleaved Ad2 mRNA. The red box shows the fragment associated with magoh. The green box indicates the 5′-most fragment.
Magoh is associated with spliced mRNA in the cytoplasm
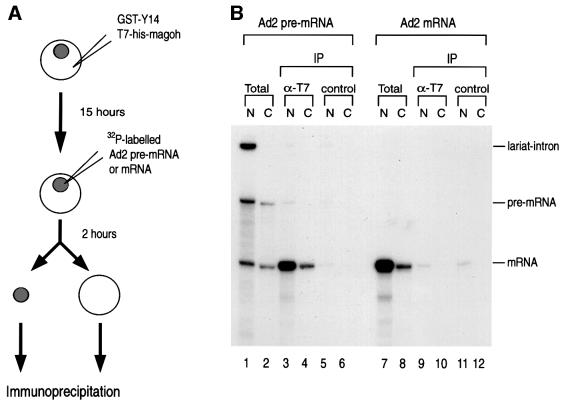
The in vitro splicing experiments described above indicated that magoh is a novel component of the exon–exon junction complex (Figure 7). Of the known components of the complex, so far only the binding of Y14 has been shown to persist on newly exported mRNAs in the cytoplasm (Kataoka et al., 2000; Kim et al., 2001b). To examine whether magoh is found on mRNAs in the cytoplasm, we performed microinjection of radiolabeled pre-mRNA into the nuclei of Xenopus oocytes in order to employ the same immunoprecipitation strategy that we have described previously for Y14 and Aly (Kim et al., 2001b). Since a specific antibody against the Xenopus mago protein is not available, we used tagged bacterially expressed recombinant proteins. In preliminary experiments we found that cytoplasmically injected GST–magoh remained in the cytoplasm and could not be imported into the nucleus (data not shown). This is consistent with a recent report that Y14 and magoh are imported into the nucleus as a complex by importin 13, while magoh itself does not bind to importin 13 (Mingot et al., 2001). We therefore carried out injections of his-tagged magoh together with GST–Y14 into the cytoplasm of oocytes and incubated them for an additional 15 h to allow the Y14–magoh complex to be imported into the oocyte nuclei (Figure 8A). Nuclear injection of Ad2 pre-mRNA or mRNA was then performed and the oocytes were incubated for an additional 2 h followed by immunoprecipitation, as shown in Figure 8B. The his-tagged magoh protein also has a T7 tag, and therefore anti-T7 tag antibody was used to immunoprecipitate injected his- magoh. As shown in Figure 8B, the anti-T7 tag antibody efficiently and preferentially immunoprecipitated spliced mRNA from the nuclear fraction (lane 3). In contrast, anti-T7 tag antibody did not precipitate intronless mRNA (lane 9). These results demonstrate that cytoplasmically injected magoh is imported to the nucleus and binds to mRNA that is produced by splicing. Importantly, the anti-T7 tag antibody also immunoprecipitated spliced mRNA that was exported to the cytoplasm (lane 4). However, intronless mRNA produced by transcription in vitro was not immunoprecipitated by anti-T7 tag antibody from the cytoplasm, indicating that if any his-magoh remained in the cytoplasm it could not bind mRNA there (lane 10). These results strongly suggest that magoh is a nucleocytoplasmic shuttling protein, which, like Y14, binds to mRNA produced by pre-mRNA splicing in the nucleus, stays on it during export and remains associated with newly exported mRNA in the cytoplasm.
Fig. 8. Magoh is preferentially associated with mRNA produced by pre-mRNA splicing both in the nucleus and the cytoplasm. (A) Schematic representation of the experimental protocol including injection, cell fractionation and immunoprecipitation. Recombinant proteins were first injected into the cytoplasm of Xenopus oocytes. After 15 h incubation, 32P-labeled Ad2 pre-mRNA or mRNA was injected into the nuclei of the same oocytes. Oocytes were then incubated for an additional 2 h before dissection, to allow pre-mRNA splicing and mRNA export. (B) Immunoprecipitations and RNA analysis were carried out following nuclear injection of 32P-labeled RNA from either the nuclear (N) or the cytoplasmic (C) fraction. Anti-T7 (Novagen) and control antibodies (SP2/0) were used for immunoprecipitation (IP). The lanes marked Total contain 5% of input materials.
Discussion
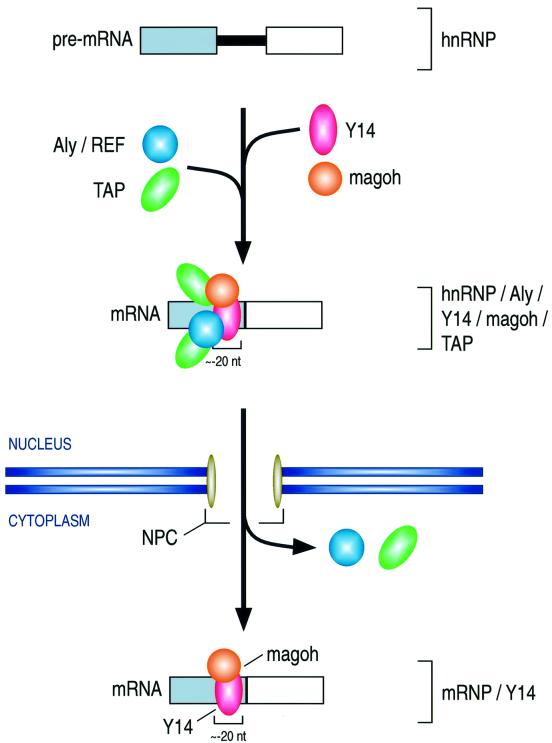
We describe here a novel component of the splicing-dependent complex that assembles near exon–exon junctions. This protein, magoh, is the human homolog of the Drosophila mago protein (Zhao et al., 1998), the product of the mago nashi gene. The amino acid sequence of magoh is extremely highly conserved among divergent organisms (Newmark and Boswell, 1994; Li et al., 2000), suggesting that it has a fundamental role in cellular function. Indeed, both null mutation in flies and disruption of mRNA by RNAi in Caenorhabditis elegans show embryonic lethality, indicating that mago is essential for viability (Boswell et al., 1991; Micklem et al., 1997; Li et al., 2000). Magoh binds Y14 avidly and specifically, but it does not appear to interact with other known components of the exon–exon junction complex. Also of note is the very efficient binding of magoh to mRNA export factor TAP, which appears to be much more avid than that of Aly/REF and Y14. Although the binding of magoh to the splicing-dependent complex is position specific and, like Y14, could serve to indicate the sites of exon–exon junctions, the specific function of magoh is unknown. The particularly avid binding of magoh to TAP and Y14 suggests that it has an important influence on the interaction of these factors with the spliced mRNAs. It may, therefore, serve as a specific adaptor for Y14 and TAP (Figure 9), and may thus enhance the export of spliced mRNA.
Fig. 9. Schematic representation of the formation and transport of the Y14 and magoh complex. See text for details.
The amino acid sequence of magoh does not show significant similarity to other proteins, nor does it contain any recognizable motifs that predict specific functions. Magoh does, however, contain a sequence predicted to have a very high propensity to form a coiled-coil structure, encompassing amino acids 50–80 (Coils, ISREC, Switzerland; Lupas et al., 1991). Whether this sequence in fact engages in coiled-coil interactions and, if so, whether these are homotypic or heterotypic is not known. We note, however, that the coiled domain prediction program suggests that amino acids 180–380 of TAP are also likely to form a coiled-coil structure. This domain of TAP confers its binding activity to viral constitutive transport element (CTE) RNA (Braun et al., 1999; Herold et al., 2000). Interestingly, RNase A treatment enhances the binding of magoh to TAP (Figure 5, lanes 6 and 7). It is therefore possible that the magoh-binding domain of TAP is in the N-terminal portion of the protein, which confers its RNA- and substrate-binding activity (Braun et al., 1999; Herold et al., 2000).
We further showed that magoh remains associated with spliced mRNA after export to the cytoplasm (Figure 8B). Transiently expressed flag-tagged magoh is localized mainly to the nucleus in HeLa cells (Figure 5A) and magoh first binds to spliced mRNA in the nucleus (Figure 8B). We conclude that magoh can shuttle between the nucleus and the cytoplasm, although direct experiments to confirm this by use of heterokaryon assays (Piñol-Roma and Dreyfuss, 1992; Michael et al., 1995) could not be carried out because the required fusions of large reporters to magoh interfere with its nuclear import (data not shown). It has been suggested that magoh plays important roles in mRNA functions in the cytoplasm, since point mutations in Drosophila mago cause several deficiencies in cytoplasmic events in oocytes. For example, mago mutations cause disruption of the anterior–posterior transport of oskar and gurken mRNAs (Micklem et al., 1997; Newmark et al., 1997). These mutations also cause mislocalization of the oocyte nucleus to the posterior region (Micklem et al., 1997). These findings strongly suggest that magoh is involved in mRNA localization in the cytoplasm through interaction with the cytoskeleton. It is likely that the cytoskeleton has an important role in the cytoplasmic sorting and localization of mRNAs (reviewed in Oleynikov and Singer, 1998; Jansen, 1999, 2001). We have previously shown that the binding of Y14 persists on newly exported mRNAs in the cytoplasm (Kataoka et al., 2000) and Y14 remains bound to the same position after export (Kim et al., 2001b). If magoh is also present in the cytoplasmic Y14 complex, magoh and Y14 may serve as adaptors for the interaction of mRNAs with microtubule-based cytoskeletons.
Another possible function for magoh present in the Y14 complex is in NMD. Mutations that cause premature termination can be deleterious, since they may produce C-terminal truncated proteins, often resulting in severe human diseases (Frischmeyer and Dietz, 1999). NMD is a process that helps avoid accumulation of mutant proteins by degrading the mRNAs that bear such mutations. Although degradation of mRNA through the NMD pathway is thought to occur in the cytoplasm, pre-mRNA splicing has an important role in NMD (Carter et al., 1996; Zhang et al., 1998a,b). Therefore, the exon–exon junction complex is an excellent candidate to mediate transfer of molecular memory of splicing and the position of introns on mRNAs to the cytoplasm. By tethering various components of the exon–exon junction complex downstream of stop codon, RNPS1 was found to recruit activity that mediates NMD (Lykke-Andersen et al., 2001). Y14 was also shown to have a moderate activity in that system (Lykke-Andersen et al., 2001). The human proteins involved in NMD were identified and designated as hUpf proteins (Lykke-Andersen et al., 2000; Mendell et al., 2000; Serin et al., 2001). Recently, we have demonstrated that hUpf3 interacts with Y14 and is a component of the exon–exon junction complex (Kim et al., 2001a). Since both Y14 and magoh binding persist on the mRNA in the cytoplasm (Figure 8B; Kataoka et al., 2000; Kim et al., 2001b), it appears likely that Y14 and magoh together communicate the position of exon–exon junctions on mRNAs to the cytoplasm and play a role in NMD. The specific roles of these proteins in NMD remain to be investigated.
We have also studied here the protein–protein interaction between Y14 and other components of the splicing-dependent exon–exon junction complex. Y14 interacts with Aly/REF, which has a role in nuclear export of mRNAs (Zhou et al., 2000; Rodrigues et al., 2001). Since RNase A treatment did not affect its binding and the domain on Y14 which binds to Aly/REF is outside of the RBD, the interaction between these two proteins is a protein–protein interaction. These results are consistent with previously reported co-immunoprecipitation of Aly/REF with Y14 from the nucleoplasm even in the presence of RNase A (Kim et al., 2001b). It is also likely that Y14 and Aly/REF interact with each other in the exon–exon junction complex (Figure 9). Interestingly, Y14 interacts with another component of this complex, RNPS1, and this is only detected after treatment with RNase A. This suggests that this is a protein–protein interaction, but that these proteins may not interact with each other when they are bound to mRNAs. We could not detect binding of Y14 to DEK, or find any other evidence for conclusions that DEK is a component of the complex. It will be important to determine how and when the proteins of the exon–exon junction complex are recruited to mRNAs.
Materials and methods
Plasmid construction and protein overexpression
The bait plasmid for the yeast two-hybrid screening was made by inserting the N-terminus of Y14 cDNA (amino acids 1–73) between the _Eco_RI and _Xho_I sites of LexA plasmid (Clontech). Full-length magoh cDNA was PCR amplified from a human HeLa cDNA (Clontech) with Pfu polymerase (Stratagene). To make the flag-magoh plasmid, this cDNA was inserted between the _Hin_dIII and _Xho_I sites of pFLAG-CMV-4 (Sigma). To construct GST– and his-magoh plasmids, the full-length cDNA was cloned into the _Eco_RI and _Xho_I sites of pGEX5X-1 (Amersham-Pharmacia) and pET28a (Novagen), respectively. To construct his-DEK, his-Aly/REF, those full-length cDNAs were amplified by PCR from Human HeLa Marathon-Ready cDNA (Clontech). His-Y14 was produced as described elsewhere (Kataoka et al., 2000). For production of GST–Y14 protein for in vitro binding experiments, we cloned the full-length Y14 cDNA into _Bam_HI- and _Hin_dIII-cleaved pET-GSTII (Nakielny et al., 1999). For protein injection experiments, the full-length Y14 cDNA was inserted between _Bam_HI and _Xho_I sites of plasmid pGV66 (a gift from Dr Van Duyne, University of Pennsylvania). Proteins were overexpressed in Escherichia coli BL21(DE3) codonplus cells (Stratagene). Induction was carried out at 17°C overnight. Proteins were purified according to the manufacturer’s recommendation.
Yeast two-hybrid screening
The HeLa MATCHMAKER LexA cDNA library, yeast strains and cloning vectors were obtained from Clontech. All library screening and yeast manipulations were carried out as recommended by the manufacturer. Saccharomyces cerevisiae strain EGY48 was transformed simultaneously with pLexA-Y14 and the HeLa cell cDNA library. Transformants (1 × 106) were plated onto 20 plates (150 mm) of X-gal-synthetic medium lacking histidine, uracil, tryptophan and leucine, and blue colonies were isolated. Library plasmids were isolated from these yeast cells and sequenced.
In vitro protein binding experiments
In vitro protein binding experiments were carried out essentially as described previously (Kataoka et al., 1999). Briefly, 5 µg of GST fusion proteins were immobilized on glutathione beads. In vitro translated proteins were prepared by using a TNT-coupled wheat germ extract system (Promega). Approximately 2 × 105 c.p.m. of in vitro translated products were added to the beads in the binding buffer (50 mM Tris–HCl pH 7.4, 200 mM NaCl, 2 mM EDTA, 0.1% NP-40) and mixed at 4°C for 1 h. After binding, the beads were washed five times, and binders were eluted with sample buffer and analyzed by SDS–PAGE. For direct binding experiments, 2 µg of his-magoh proteins were used per binding reaction and bound proteins were detected by western blotting with anti-T7 tag antibody (Novagen). For RNase treatment, RNase A (USB) was added to a final concentration of 2 µg/ml.
Cell culture, transfection and cell fractionation
HeLa and HEK293 cell culture was performed as described previously (Siomi et al., 1997). For transfection, a Calphos™ mammalian transfection kit (Clontech) was used according to the manufacturer’s recommendation. Subcellular fractionation of HEK293T cells was carried out as described previously (Siomi et al., 1997).
Immunoprecipitation, western blotting and immunofluorescence microscopy
Flag-magoh complex was immunoprecipitated from either vector alone or flag-magoh cDNA-transfected human HEK293T cell nucleoplasm for 30 min at 4°C with the Flag antibodies on beads (Sigma). After extensive washing, the bound fractions were eluted by boiling in SDS–PAGE sample buffer and analyzed by SDS–PAGE. Western blotting was performed as described previously (Siomi et al., 1997). For detection of TAP and Y14, the monoclonal antibodies 53H8 (L.Abel, V.N.Kim and G.Dreyfuss, unpublished data), 1F12 (Kim et al., 2001b) were used, and the flag tag was detected with antibody M2 (Sigma). Immunofluorescence microscopy was carried out with M2 antibody (1:200 dilution) as described previously (Siomi et al., 1997).
In vitro splicing, RNase H digestion and immunoprecipitation
Template preparation and in vitro transcription were performed as described previously (Kataoka et al., 2000). The in vitro splicing reactions in combination with HEK293T cell extracts were performed as described previously (Ohno and Shimura, 1996) with slight modifications. To disrupt HEK293T cells, sonication was used instead of the glass beads method. Briefly, a typical 40 µl splicing reaction contained 4 µl of HeLa nuclear extract and 20 µl of HEK293T whole-cell extract, as well as 4 µl of 10× SP buffer (Pellizzoni et al., 1998). RNase H digestion with A5 and A10 oligos (Kim et al., 2001b) and immunoprecipitation were performed as described previously (Hanamura et al., 1998). The anti-monomethylguanosine cap structure antibody H20 (Bochnig et al., 1987) was a kind gift from Dr Reinhard Lührmann.
Xenopus oocyte microinjections
Injections were carried out as described previously (Kataoka et al., 2000). Briefly, co-purified GST–Y14 and T7-his-magoh proteins were injected into the cytoplasm of Xenopus oocytes. After 15 h incubation, in vitro transcribed RNAs were injected into the nuclei of oocytes. Oocytes were incubated at 17°C for 2 h to allow pre-mRNA splicing and nuclear export of mRNA to take place, and were manually dissected to the nucleus and the cytoplasm. Each fraction, nucleus and cytoplasm, was used for immunoprecipitation as described (Kataoka et al., 2000).
Acknowledgments
Acknowledgements
We thank Drs Gregory Van Duyne, Akila Mayeda and Reinhard Lührmann for the plasmid, RNPS1 cDNA and anti-cap antibodies, respectively. We are grateful to our laboratory members, especially Drs Westley Friesen, Livio Pellizzoni, Josée Dostie, Lili Wan and Jennifer Baccon, for comments on this manuscript. We are also grateful to Gina Daly for secretarial assistance. This work was supported by grants from the National Institute of Health and the Human Frontier Science Program Organization. G.D. is an Investigator of the Howard Hughes Medical Institute.
References
- Blencowe B.J., Issner,R., Nickerson,J.A. and Sharp,P.A. (1998) A coactivator of pre-mRNA splicing. Genes Dev., 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochnig P., Reuter,R., Bringmann,P. and Lührmann,R. (1987) A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur. J. Biochem., 168, 461–467. [DOI] [PubMed] [Google Scholar]
- Boswell R.E., Prout,M.E. and Steichen,J.C. (1991) Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development, 113, 373–384. [DOI] [PubMed] [Google Scholar]
- Braun I.C., Rohrbach,E., Schmitt,C. and Izaurralde,E. (1999) TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J., 18, 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.S., Li,S. and Wilkinson,M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Culbertson M.R. (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Gonzalez,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis,M.J., Piñol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A. and Dietz,H.C. (1999) Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet., 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- Hanamura A., Cáceres,J.F., Mayeda,A., Franza,B.R.,Jr and Krainer,A.R. (1998) Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA, 4, 430–444. [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Herold A., Suyama,M., Rodrigues,J.P., Braun,I.C., Kutay,U., Carmo-Fonseca,M., Bork,P. and Izaurralde,E. (2000) TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol., 20, 8996–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.P. (1999) RNA–cytoskeletal associations. FASEB J., 13, 455–466. [PubMed] [Google Scholar]
- Jansen R.P. (2001) mRNA localization: message on the move. Nature Rev. Mol. Cell. Biol., 2, 247–256. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Bachorik,J.L. and Dreyfuss,G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol., 145, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Yong,J., Kim,V.N., Velazquez,F., Perkinson,R.A., Wang,F. and Dreyfuss,G. (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell, 6, 673–682. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Kataoka,N. and Dreyfuss,G. (2001a) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science, 293, 1832–1836. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Yong,J., Kataoka,N., Abel,L., Diem,M.D. and Dreyfuss,G. (2001b) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J., 20, 2062–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000a) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Moore,M.J. and Maquat,L.E. (2000b) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Li W., Boswell,R. and Wood,W.B. (2000) mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol., 218, 172–182. [DOI] [PubMed] [Google Scholar]
- Luo M.J. and Reed,R. (1999) Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA, 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Wassarman,K.M. and Wolffe,A.P. (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J., 17, 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Badolato,J., Kobayashi,R., Zhang,M.Q., Gardiner,E.M. and Krainer,A.R. (1999) Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J., 18, 4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T. et al. (2000) The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon–product complexes. J. Cell Biol., 150, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., Medghalchi,S.M., Lake,R.G., Noensie,E.N. and Dietz,H.C. (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol., 20, 8944–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W.M., Choi,M. and Dreyfuss,G. (1995) A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell, 83, 415–422. [DOI] [PubMed] [Google Scholar]
- Micklem D.R., Dasgupta,R., Elliott,H., Gergely,F., Davidson,C., Brand,A., Gonzalez-Reyes,A. and St Johnston,D. (1997) The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol., 7, 468–478. [DOI] [PubMed] [Google Scholar]
- Mingot J.M., Kostka,S., Kraft,R., Hartmann,E. and Görlich,D. (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J., 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Shaikh,S., Burke,B. and Dreyfuss,G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J., 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark P.A. and Boswell,R.E. (1994) The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development, 120, 1303–1313. [DOI] [PubMed] [Google Scholar]
- Newmark P.A., Mohr,S.E., Gong,L. and Boswell,R.E. (1997) mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development, 124, 3197–3207. [DOI] [PubMed] [Google Scholar]
- Ohno M. and Shimura,Y. (1996) A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev., 10, 997–1007. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y. and Singer,R.H. (1998) RNA localization: different zipcodes, same postman? Trends Cell Biol., 8, 381–383. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka,N., Charroux,B. and Dreyfuss,G. (1998) A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell, 95, 615–624. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode,M., Gatfield,D., Blencowe,B., Carmo-Fonseca,M. and Izaurralde,E. (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA, 98, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G., Gersappe,A., Black,J.D., Aronoff,R. and Maquat,L.E. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol., 21, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Eder,P.S., Kataoka,N., Wan,L., Liu,Q. and Dreyfuss,G. (1997) Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol., 138, 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Seraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y., LaDuca,J.P. and Maquat,L.E. (1998a) At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998b) Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.F., Colaizzo-Anas,T., Nowak,N.J., Shows,T.B., Elliott,R.W. and Aplan,P.D. (1998) The mammalian homologue of mago nashi encodes a serum-inducible protein. Genomics, 47, 319–322. [DOI] [PubMed] [Google Scholar]
- Zhao X.F., Nowak,N.J., Shows,T.B. and Aplan,P.D. (2000) MAGOH interacts with a novel RNA-binding protein. Genomics, 63, 145–148. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Luo,M.J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]