The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site (original) (raw)
Abstract
Cell attachment and membrane penetration are functions of the rotavirus outer capsid spike protein, VP4. An activating tryptic cleavage of VP4 produces the N-terminal fragment, VP8*, which is the viral hemagglutinin and an important target of neutralizing antibodies. We have determined, by X-ray crystallography, the atomic structure of the VP8* core bound to sialic acid and, by NMR spectroscopy, the structure of the unliganded VP8* core. The domain has the β-sandwich fold of the galectins, a family of sugar binding proteins. The surface corresponding to the galectin carbohydrate binding site is blocked, and rotavirus VP8* instead binds sialic acid in a shallow groove between its two β-sheets. There appears to be a small induced fit on binding. The residues that contact sialic acid are conserved in sialic acid-dependent rotavirus strains. Neutralization escape mutations are widely distributed over the VP8* surface and cluster in four epitopes. From the fit of the VP8* core into the virion spikes, we propose that VP4 arose from the insertion of a host carbohydrate binding domain into a viral membrane interaction protein.
Keywords: galectin/hemagglutinin/rotavirus/sialic acid/structure
Introduction
Cell entry by non-enveloped viruses is still a poorly understood process. The Reoviridae have an especially challenging cell entry problem. Their transcriptionally active units are large icosahedral particles (Figure 1), which must be delivered intact across a membrane and into the cytoplasm in order to initiate viral gene expression. The transcriptionally active cores of the rotaviruses, orthoreoviruses and orbiviruses have related overall architectures, reflecting similar strategies of genome transcription, replication and packaging. In contrast, their outermost layers, responsible for cell entry, are quite different from each other. (Structures of the Reoviridae are reviewed in Harrison et al., 1996.) This structural diversity reflects the diverse virus–host interactions of the Reoviridae. Different genera within the family replicate in the intestinal epithelium (rotaviruses), spread to the central nervous system (some orthoreoviruses) or infect both insect and mammalian hosts (orbiviruses).
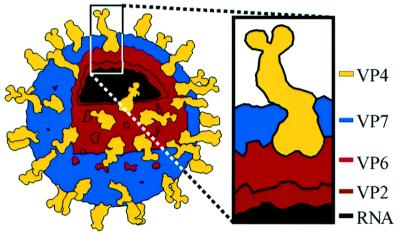
Fig. 1. The rotavirus triple-layered virion. VP4 and VP7 make up the outer capsid, which constitutes the entry apparatus. The RNA, VP2 and VP6 are the major structural components of the double-layered particle, which is the transcriptionally active core. The line drawing is based on an electron cryomicroscopy-based reconstruction (Yeager et al., 1990).
Understanding cell entry is particularly important in the case of rotavirus, because of its clinical and economic impact. Dehydrating diarrhea caused by rotavirus infection is responsible for ∼6% of all deaths of children under the age of 5 years, worldwide (World Health Organization, 1999). Since the withdrawal of a rotavirus vaccine due to a temporal association between immunization and intestinal intussusception (Centers for Disease Control, 1999), it is once more a major childhood illness for which no licensed vaccine is available. Rotavirus is also an important veterinary pathogen, causing disease in calves, sheep, swine and poultry. The process of cell entry by rotavirus is a promising target for therapeutic and preventative interventions against rotavirus diarrhea.
The rotavirus outer capsid consists of the coat glycoprotein, VP7, and the spike protein, VP4 (Figure 1). The virion must be activated by trypsin for efficient entry (Estes et al., 1981). Trypsin cleaves VP4 into an N-terminal fragment, VP8*, the viral hemagglutinin (Fiore et al., 1991), and a C-terminal fragment, VP5*, which permeabilizes membranes (Dowling et al., 2000). Electron cryomicroscopy of treated and untreated virions demonstrates that trypsinization confers icosahedral ordering on the VP4 spikes (Crawford et al., 2001). The biochemical correlate of this activation-associated conformational change is a protease-induced dimerization of the VP5* region of VP4 (Dormitzer et al., 2001).
In some strains of rotavirus, VP8* binding to a neuraminidase-sensitive sialic acid moiety constitutes the initial interaction with the target host cell and restricts productive entry to the apical membrane (Fiore et al., 1991; Ciarlet et al., 2001). However, most rotavirus strains, including a large majority of those that infect humans, do not require a neuraminidase-sensitive sialic acid moiety for entry (Ciarlet et al., 2002). The isolation of a sialic acid binding rotavirus mutant that is not inhibited by neuraminidase treatment of cells and the documentation of cell type-specific sialic acid dependence (Mendez et al., 1996; Ludert et al., 1998) complicate the simple dichotomy between sialic acid-dependent and -independent strains. These findings may reflect the binding of some rotavirus strains to neuraminidase-insensitive internal sialic acid moieties on glycolipids (Guo et al., 1999; Delorme et al., 2001) or to modified sialic acids that are resistant to neuraminidase treatment. It is probable that both sialic acid-dependent and -independent rotavirus strains also bind a second cellular receptor, possibly an integrin (Guerrero et al., 2000; Hewish et al., 2000).
The majority of neutralizing monoclonal antibodies (mAbs) that recognize VP4 of hemagglutinating rotavirus strains select mutations in VP8* (Burns et al., 1988; Mackow et al., 1988; Giammarioli et al., 1996). Several of these mAbs block cell attachment (Ruggeri and Greenberg, 1991). In contrast, the majority of neutralizing mAbs that recognize VP4 of sialic acid-independent human rotavirus strains select mutations in the VP5* fragment (Kobayashi et al., 1990; Padilla-Noriega et al., 1995; Kirkwood et al., 1996). As VP8*-specific neutralizing antibodies show limited cross-neutralization between rotavirus strains (Shaw et al., 1986; Burns et al., 1988), VP8* is the main determinant of rotavirus ‘P’ serotype. VP8* may also have intracellular functions in virus replication, as it has been shown to activate cell signaling pathways upon binding to tumor necrosis factor receptor-associated factors (TRAFs; LaMonica et al., 2001).
To probe the process of cell entry by rotavirus, we have determined, by nuclear magnetic resonance (NMR), solution structures of the rhesus rotavirus (RRV) sialic acid binding domain and, by X-ray diffraction, the crystal structure of this domain complexed with sialic acid.
Results and discussion
Biochemical strategy
A previous biochemical analysis of purified, recombinant RRV VP4 demonstrated that the VP8* region of VP4 contains a compact and homogeneous protease-resistant core from residues A46 to R231 (Dormitzer et al., 2001). This core contains all mapped antigenic sites on VP8* (references in Table IV), the hypervariable region (residues T72–C203) and the hemagglutination region (residues V93–I208; Fuentes-Panana et al., 1995). Because the VP8* core is monomeric (Dormitzer et al., 2001), purified preparations do not hemagglutinate (our unpublished data). Escherichia coli expression of a construct equivalent to this core (EcVP846–231) yields sufficient quantities of protein for structural analysis. EcVP846–231 is highly soluble (to >65 mg/ml) and monodisperse (polydispersity 9.5% for a 3 mg/ml solution by dynamic laser light scattering). Although EcVP846–231 did not crystallize, it produced good NMR spectra.
The NMR spectra showed that the N-terminal 17 residues and the C-terminal five residues of EcVP846–231 are disordered (not shown). To truncate these N- and C-terminal ‘tails’, which might prevent crystallization, a new construct that incorporated only residues E62–L224 was designed. Mass spectrometry and N-terminal sequencing revealed that the resulting product, EcVP862–224 was heterogeneous, retaining an N-terminal leader of 8–16 residues, probably due to steric interference with the intended trypsin cleavage of the glutathione-_S_-transferase (GST) tag. Nevertheless, VP862–224 crystallized in the presence, but not in the absence, of a sialoside, forming tetragonal crystals with rounded edges. The sialoside, 2-_O_-methyl-α-d-_N_-acetyl neuraminic acid, is the simplest sialoside that bound the VP8* core efficiently in an NMR analysis (P.R.Dormitzer, Z.-Y.J.Sun, O.Blixt, J.C.Paulson, G.Wagner and S.C.Harrison, in preparation).
Structure determinations
The X-ray crystal structure of EcVP862–224 in complex with sialoside was determined to 1.4 Å resolution by single isomorphous replacement (Table I). The final structure has an _R_free of 18.1% and mean thermal parameters of 14.5 for the protein and 9.6 for the sialoside. The reported structure includes residues L65–L224 of VP4, the sialoside, 190 ordered water molecules, one sulfate ion, one glycerol molecule and 22 amino acids with alternative conformations. A Ramachandran plot (not shown) contains 89.2% of the non-proline or glycine residues within the most favored regions and the remaining 10.8% within additionally allowed regions.
Table I. Crystallographic data and refinement statistics.
| Data collection statistics | ||
|---|---|---|
| Crystal | Native | Selenomethionine |
| Resolution limit (Å) | 1.40 | 1.75 |
| Unique reflectionsa | 31 002 (1993) | 16 220 (1030) |
| Redundancya | 8.25 (5.71) | 8.39 (4.71) |
| Completenessa (%) | 99.5 (98.3) | 99.7 (97.1) |
| I/σa | 37.4 (5.51) | 36.0 (4.73) |
| _R_syma,b (%) | 5.0 (25.1) | 5.5 (25.4) |
| Phasing statistics | ||
| Number of sites | 3 | |
| _R_isoc (%) | 7.3 | |
| Phasing powerd | ||
| centric | 1.540 | |
| acentric | 1.885 | |
| _R_cullise (%) | ||
| centric | 0.568 | |
| acentric | 0.597 | |
| Figure of merit | ||
| centric | 0.631 | |
| acentric | 0.375 | |
| Refinement statistics | ||
| Protein atoms | 1273 | |
| Sialoside atoms | 22 | |
| Water molecules | 190 | |
| Glycerol molecules | 1 | |
| Sulfate ions | 1 | |
| Amino acids with alternative conformations | 22 | |
| R.m.s. bond lengths (Å) | 0.0149 | |
| R.m.s. bond angles (°) | 1.858 | |
| R.m.s. main chain B (Å2) | 1.294 | |
| Resolution range (Å) | 25–1.4 (all data) | |
| _R_-factorf | 0.169 | |
| Free _R_-factorf | 0.181 |
An ensemble of NMR solution structures of residues L65–L224 of EcVP846–231 in the absence of ligand was determined (Table II). The final set of ligand-free solution structures was obtained after the NMR spectra were systematically reviewed for evidence of differences from the liganded crystal structure. This review resulted in only minor local changes from a preliminary set of structures obtained prior to determining the crystal structure. The final calculations included 11.1 nuclear Overhauser effect (NOE) constraints per residue. The 20 structures with the lowest violations of NOE distance constraints were selected from 25 randomly annealed structures. These structures contain no violations of NOE distance constraints >0.15 Å and no violations of TALOS-derived dihedral angle constraints >5°.
Table II. NMR data and refinement statistics.
| NOE distance restraints | |
|---|---|
| intraresidue | 338 |
| sequential | 519 |
| medium range (2≤|i – j | ≤4) |
| long range (|i – _j | _>4) |
| total | 1793 |
| Hydrogen bonds | 84 |
| Dihedral angle restraints | 116 |
| Ramachandran plot | |
| most favored region (%) | 72.4 |
| additionally allowed region (%) | 24.9 |
| generously allowed region (%) | 2.0 |
| disallowed region (%) | 0.8 |
| <R.m.s.d.> from ideal geometry | |
| bonds (Å) | 0.0011 ± 0.0002 |
| angles (°) | 0.2660 ± 0.0118 |
| impropers (°) | 0.1177 ± 0.0170 |
| <R.m.s.d.> from mean structure, all residues | |
| backbone (Å) | 0.779 ± 0.122 |
| side chain, non-hydrogen (Å) | 1.242 ± 0.138 |
| <R.m.s.d.> from mean structure, selected residuesa | |
| backbone (Å) | 0.513 ± 0.062 |
| side chain, non-hydrogen (Å) | 1.048 ± 0.130 |
Structure description
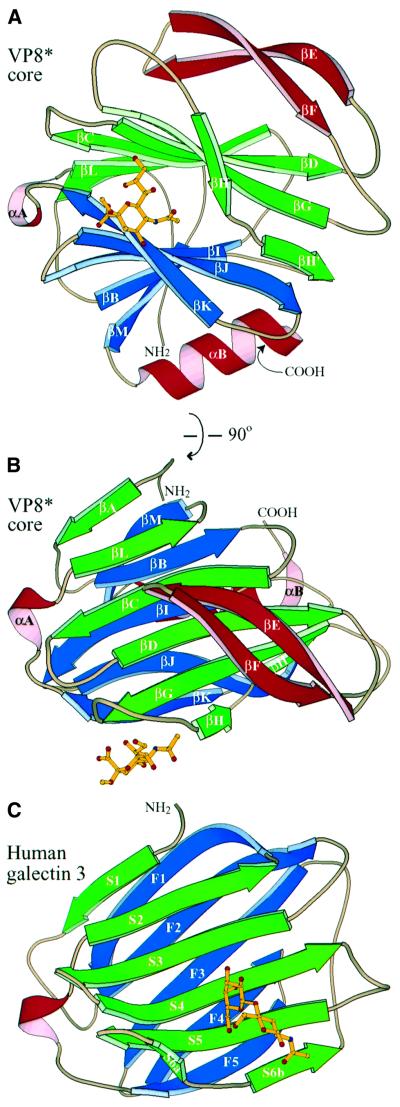
The NMR and X-ray analyses revealed the same basic protein structure. The rotavirus VP8* core is a single, compactly folded, globular domain with dimensions of 36.6 × 37.7 Å (height by width in Figure 2A) by 28.3 Å (height in Figure 2B), as measured on a Cα trace. Although there is a 2-fold rotational symmetry axis in space group _P_41212, the 2-fold crystal contact (centered around residues A89, E109, P110, W138 and K163) does not suggest a stable dimeric interaction. The NMR spectra contain no NOE cross-peaks that would come from a dimer in solution. The VP8* core contains two cysteines (C203 and C216), but they do not form a disulfide bond in the folded structure. Prolines 68 and 182 are in the cis configuration. In addition, there is electron density for two alternative positions of the P157 carbonyl, indicating that the crystals contain a mixture of molecules with the G156–P157 peptide bond in either the cis or trans configuration.
Fig. 2. Basic structural features and galectin homology. (A) Ribbon diagram of the rotavirus sialic acid binding domain, as viewed along arrow 1 and in panel B of Figure 7. The C-terminus is hidden behind the last turn of αB. The sialoside is depicted by balls and sticks. (B) Ribbon diagram of the rotavirus sialic acid binding domain, as viewed along arrow 2 and in panel C of Figure 7. This view is rotated 90° about a horizontal axis relative to the view in (A). (C) Ribbon diagram of human galectin 3, based on PDB file 1A3K (Seetharaman et al., 1998). The coloring matches that of the rotavirus structures. The view is equivalent to that in (B). _N_-acetyl-lactosamine is depicted with balls and sticks. The central β-sandwich (including αA) of the rotavirus sialic acid binding domain has an r.m.s.d. of 2.9 Å from human galectin-3 for 89 corresponding Cα pairs and a similarly good fit to crystallographic models of galectins-1, -2, -7 and -10 (PDB files 1SLC, 1HLC, 5GAL and 1LCL).
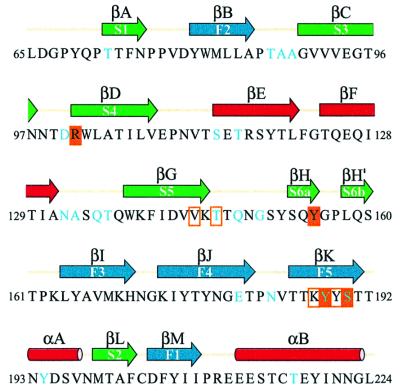
The central structural feature of the domain is an 11-stranded anti-parallel β-sandwich (Figure 2A and B), formed from a five-stranded β-sheet (in blue, strands βM, βB, βI, βJ and βK) and a six-stranded β-sheet with an interrupted top strand (in green, strands βA, βL, βC, βD, βG and βH/H′). Figure 3 shows the alignment of the secondary structure with the amino acid sequence. The two β-sheets are joined by five short inter-sheet loops as well as by a brief stretch of parallel β-structure between strand βH′ of the six-stranded sheet and βJ of the five-stranded sheet (Figure 2A). The cleft between the β-sheets is broad near the carbohydrate binding site but narrows toward the bridging parallel β-strands. The cleft is filled by a dense core of hydrophobic side chains contributed by all strands of the sheets except for βH and βH′. The α-carbons of the N-terminal (L65) and C-terminal (L224) residues are only 10 Å apart. Thus, the VP8* core arises from a narrow attachment to the remainder of the VP4 spike. Indeed, the side chains of L65 and L224 contribute to the same hydrophobic core.
Fig. 3. Sequence of the rotavirus sialic acid binding domain, showing secondary structure assignments as arrows (β-strands) or rods (α-helices). The β-strand and α-helix designations for the VP8* core are shown above the arrows and rods; the equivalent β-strands in galectins (Seetharaman et al., 1998) are indicated within the arrows. The colors of the arrows and rods match the colors of the strands and helices in Figure 2. Cyan lettering in the amino acid sequence indicates neutralizing antibody escape mutations (references in Table IV). Solid orange boxes indicate residues that make hydrogen bonds with the sialoside. Outline orange boxes indicate residues that make van der Waals contacts but not hydrogen bonds with the sialoside.
The domain contains three other structural elements (colored red in Figures 2A and B, and 3). First, the inter-sheet loop connecting strands βK and βL contains a short α-helix (αA). Secondly, a longer α-helix (αB) at the C-terminus packs against strands βM, βB, βI and βJ of the five-stranded β-sheet. Thirdly, an extended β-ribbon (strands βE and βF) arises from the loop between strands βD and βG and packs against strands βC, βD, βG, βH and βH′ of the six-stranded β-sheet.
The tight fold of the β-sandwich, the cross-bracing of the β-sheets by the β-ribbon and the C-terminal α-helix, the short loops between strands and the dense hydrophobic cores between the major structural elements suggest a rigid structure that is unlikely to undergo major rearrangements during cell entry. The compact structure accounts for the protease resistance and stability of the VP8* core, which shows no evidence of degradation or denaturation after storage for months at 4°C in the absence of protease inhibitors (not shown). This physical resistance may be an adaptation to the harsh conditions in the gut and the external environment.
Similarity to galectins
The galectins (or S-type lectins) are a diverse group of animal lectins that bind β-galactoside-containing oligosaccharides (reviewed in Leffler, 1997). The β-sandwich of the rotavirus sialic acid binding domain and the galectin carbohydrate binding domain have the same fold (Figure 2B and C). In Figure 3, equivalent β-strands of the rotavirus and galectin domains are indicated by labels above and within the arrows, respectively. Despite their three-dimensional congruence, the VP8* core and the galectins have no significant sequence similarity (9% sequence identity with human galectin-3 in structurally equivalent residues).
The C-terminal α-helix (αB), the bridging parallel β-structure and the β-ribbon distinguish the VP8* core from the galectins. The loop between galectin strands S4 and S5 (Figure 2C), which has been extended into the β-ribbon in the VP8* core (Figure 2B), varies in length among galectins and is a determinant of galectin carbohydrate specificity (Lobsanov and Rini, 1997). In VP8*, the β-ribbon neatly blocks the surface corresponding to the galectin carbohydrate binding site. Many of the VP8* residues that anchor the β-ribbon to the six-stranded β-sheet (residues V91, W102, A104, I106, Y152 and Q154) are in positions equivalent to galectin carbohydrate binding residues (human galectin-3 residues R144, H158, N160, R162, E184 and R186; Seetharaman et al., 1998).
There are several similarities between the processing, localization, and function of the VP8* sialic acid binding domain and the galectins. Both are lectins. Both are found in the cytoplasm and on the cell surface (Leffler, 1997; Nejmeddine et al., 2000). Galectins-3, -4, -6 and -9 reside in the mammalian intestinal epithelium (Hughes, 1997), the site of rotavirus replication. Galectins-1 and -3 undergo non-signal, non-Golgi-dependent translocation across the plasma membrane (Hughes, 1997). VP4 is also translocated across the plasma membrane by a non-signal, non-Golgi-dependent mechanism (Nejmeddine et al., 2000). Both galectins and VP8* modulate cell signaling (Leffler, 1997; LaMonica et al., 2001). It remains to be determined whether these similarities reflect common functions mediated by common structures.
Description of the sialoside binding site
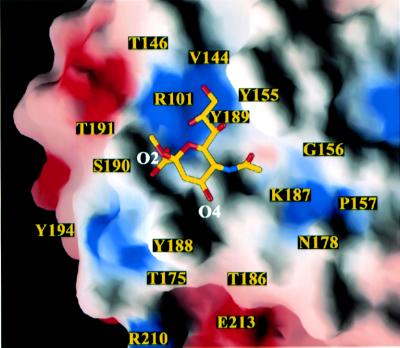
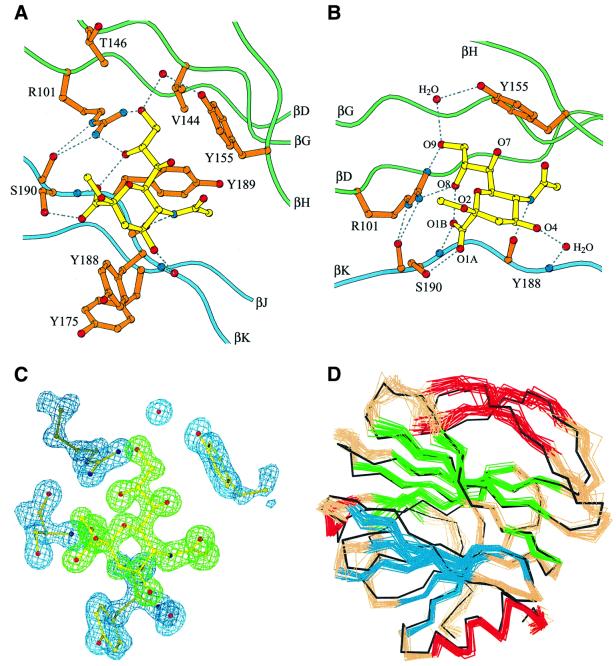
The VP8* sialic acid binding site lies above the cleft between the two β-sheets (Figure 2A and B). It is shifted not only from the galectin carbohydrate binding site (Figure 2C), but also from the binding sites of the legume lectins (Sharma and Surolia, 1997) and the pentraxins (Emsley et al., 1994), which have more distantly related β-sandwich folds. On a surface representation of the VP8* core, the sialoside binding site appears to be an open-ended, shallow groove (Figure 4). The Y188 and S190 side chains form one rim of the groove; the Y155 aromatic ring forms the opposite rim; and the R101, V144, K187 and Y189 side chains form the floor (Figures 4 and 5A). Residues Y155, Y188 and S190 were identified previously as likely to be involved in sialic acid binding on the basis of alanine-scanning mutagenesis and neutralization escape mutant studies (Giammarioli et al., 1996; Isa et al., 1997).
Fig. 4. Surface representation of the sialic acid binding site. Surfaces with positive electrostatic potential are colored blue; surfaces with negative potential are colored red. The amino acids discussed in the text are labeled. As viewed along arrow 1 and in panel B of Figure 7.
Fig. 5. Details of sialoside binding by the VP8* core. (A) Ball and stick diagram of the sialoside binding site. Dotted lines indicate hydrogen bonds. As viewed along arrow 1 and in panel B of Figure 7. (B) Ball and stick diagram showing the hydrogen bonds anchoring the sialoside: two bonds from the R101 side chain guanidinium (atoms NH1 and NH2) to the sialoside glycerol side chain (atoms O8 and O9); one from the Y155 side chain hydroxyl to the sialoside glycerol (atom O9) via a water bridge; one from the S190 side chain hydroxyl to the sialoside carboxylate (atom O1A); one from the S190 main chain amide to the sialoside carboxylate (atom O1B); one from the Y188 main chain carbonyl to the sialoside acetamido nitrogen; and one from the Y188 main chain amide to the sialoside O4 hydroxyl via a water bridge. (C) Electron density of the sialoside, R101, Y155, Y188, S190 and two water molecules. The simulated annealing omit map was calculated in CNS with a phasing model that excludes the sialoside and the depicted amino acid residues and water molecules. The map is contoured at 1.2σ. The sialoside is colored green. The amino acid residues and water molecules are colored blue. Same view as in (A). (D) Superposition of the Cα trace of the crystal structure on a set of 20 NMR solution structures. The NMR Cα traces are colored as in Figure 2. The crystal Cα trace is black. Same view as in (A) and (C).
The electron density of the bound sialoside is very well defined (Figure 5C), allowing unambiguous assignment of the contacts that it makes with VP8* residues. The sialoside is anchored to VP8* by seven hydrogen bonds (Figure 5B). An internal hydrogen bond between the sialoside carboxylate (atom O1B) and glycerol side chain (atom O8) fixes its conformation. The sialoside makes van der Waals contacts with the side chains of residues V144, T146, Y155, K187 (not shown, see below), Y188 and Y189 (Figure 5A).
R101 has a critical role in binding the sialoside, making two hydrogen bonds to the sialoside glycerol side chain. The R101 side chain is buttressed in position by hydrogen bonds from its guanidinium group (atoms Nε and NH2) to the S190 main chain carbonyl (Figure 5A and B) and by van der Waals contacts between its side chain and the T191 side chain (not shown). Y188 is stabilized in its carbohydrate binding position by a stacking interaction with Y175 from the adjacent strand (Figure 5A). Although the sialoside also makes a crystal contact with a symmetry-related VP8* core, a sialic acid moiety in an oligosaccharide side chain could not bind at the alternative site due to steric interference from proximal sugar residues.
The interactions between VP8* and sialic acid resemble those in other viral hemagglutinins. Hydrogen bonds from amino acid side chain hydroxyl and guanidinium groups, hydrogen bonds from backbone amides and carbonyls, water-mediated contacts and van der Waals contacts with aromatics rings have been noted in sialic acid binding by influenza hemagglutinin (HA), polyomavirus VP1 and Newcastle disease virus hemagglutinin–neuraminidase (Weis et al., 1988; Stehle and Harrison, 1997; Crennell et al., 2000).
Variability of the sialoside binding residues
The key sialic acid binding residues are strongly conserved among rotavirus strains (P genotypes 1, 2, 3 and 7) that are known to bind sialic acid (Table III, top five rows). Among these strains, the conservation of residues R101, Y189 and S190 is absolute; residues Y155 and Y188 undergo only conservative mutations to histidine. In a number of sialic acid-independent or non-hemagglutinating strains (from P genotypes 4, 6, 8, 11 and 19) the key sialic acid binding residue R101 is replaced by residues with hydrophobic side chains, precluding the formation of hydrogen bonds to the glycerol group of a sialoside (Table III, bottom eight rows). In these strains, substitutions of hydrophilic residues for Y155, Y188 and Y189 further disrupt the binding site. Thus, structural data correlate well with functional data, although not all strains with conserved sialic acid binding residues are sialic acid dependent (e.g. strain LP14; Table III, row 7).
Table III. Sequence diversity of sialoside binding residues.
| Straina | Host | P typeb | HAc | Sialic acid dependenced | 101 | 155 | 188 | 189 | 190 |
|---|---|---|---|---|---|---|---|---|---|
| RRV | simian | 5B[3] | + | + | R | Y | Y | Y | S |
| NCDV-Lin | bovine | 6[1] | + | + | R | H | Y | Y | S |
| SA11 cl3 | simian | 5B[2] | + | + | R | Y | H | Y | S |
| CU-1 | canine | 5A[3] | + | + | R | Y | Y | Y | S |
| OSU | porcine | 9[7] | + | + | R | H | Y | Y | S |
| L338 | equine | 12[18] | + | +/–e | R | Y | Y | Y | S |
| Lp14 | ovine | [15] | NDf | – | R | Y | Y | Y | S |
| FRV-1 | feline | [9] | – | ND | R | Y | Y | Y | L |
| K8 | human | 3A[9] | – | – | R | Y | Y | Y | L |
| ALA | lapine | 11[14] | ND | – | R | Y | Y | Y | L |
| HAL1166 | human | 11[14] | ND | – | R | Y | Y | Y | L |
| EC | murine | 10[16] | ND | +/–e | R | Y | Y | Y | A |
| Ty-1 | avian | [17] | ND | – | R | M | Y | Y | S |
| EHP | murine | [20] | – | +/–g | R | Y | Y | Y | T |
| UK | bovine | 7[5] | – | –h | R | Y | G | Y | F |
| 69M | human | 4[10] | – | – | R | H | G | Y | P |
| H-2 | equine | 4[12] | – | – | R | Y | G | Y | V |
| RV-5 | human | 1B[4] | – | ND | F | R | S | S | N |
| Gott | porcine | 2B[6] | ND | – | V | Q | Y | S | T |
| ST3 | human | 2A[6] | – | – | I | K | Y | S | S |
| Wa | human | 1A[8] | – | – | F | R | S | S | S |
| B233 | bovine | 8[11] | – | – | F | R | T | Y | S |
| 116E | human | 8[11] | ND | ND | L | R | T | Y | F |
| PRV 4F | porcine | [19] | ND | – | V | K | Y | S | T |
| Mc323 | human | [19] | ND | ND | V | K | Y | S | T |
Several animal and human strains (e.g. from genotypes 5, 9, 14, 16, 17 and 20) have retained some, but not all, of the sialic acid binding residues (Table III). Many of these strains display neuraminidase-insensitive cell entry or fail to hemagglutinate human type O red blood cells. Their sequences suggest the loss of only a single hydrogen bond to sialic acid. These strains may bind sialosides that have been modified or that are presented in different contexts. Supporting this possibility, the neuraminidase-resistant, non-hemagglutinating, bovine strain UK (P genotype 5) binds sialic acid-containing gangliosides, but with different specificity from that of the sialic acid-dependent strains SA11 and NCDV (Mochizuki and Nakagomi, 1995; Delorme et al., 2001). Similarly, the non-hemagglutinating, murine strain EHP (P genotype 20) is neuraminidase resistant when tested on monkey kidney-derived (MA104) cells (Ciarlet et al., 2002), but neuraminidase sensitive when tested on human colonic epithelium-derived (CaCo-2) cells (Ludert et al., 1996). Therefore, the variability in the sialic acid binding residues of VP8* adds evidence that different rotavirus strains employ a variety of initial cell attachment strategies.
Implications of the structure for binding specificity
The methyl substituent of the sialoside at the O2 position, which takes the place of the penultimate residue of a glycoprotein’s oligosaccharide side chain, faces the mouth of the groove adjacent to S190 and projects away from the protein into solvent (Figures 4, 5A and B, and 6C). Thus, VP8* may bind sialosides with a variety of oligo saccharide linkages proximal to the sialic acid moiety. Indeed, synthetic thiosialosides with either α(2,6) or α(2,4) linkages to galactose inhibit rotavirus strains NCDV and SA11 at equivalent concentrations (Kiefel et al., 1996), and α(2,3)- and α(2,6)-sialyllactose inhibit rotavirus strain OSU at equivalent concentrations (Rolsma et al., 1998).
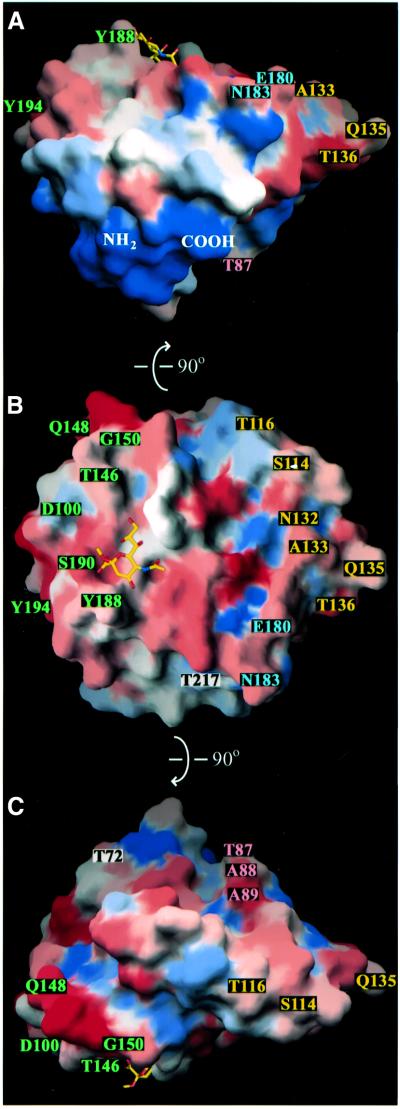
Fig. 6. Surface representations of the rotavirus VP8* core, colored according to the variability between the rotavirus strains listed in Table III. Blue represents the most conserved surfaces and red represents the most variable surfaces. Labeled amino acids indicate neutralization escape mutations (Table IV). Labels colored by epitope: 8-1, green; 8-2, blue; 8-3, yellow; 8-4, pink; and not assigned, black. (A) As viewed along arrow 3 of Figure 7. (B) As viewed along arrow 1 and in panel B of Figure 7. (C) As viewed along arrow 2 and in panel C of Figure 7. (A) and (C) are rotated 90° in either direction around the horizontal axis relative to (B), as indicated by arrows on the figure.
The sialoside O4 faces an opening at the other end of the groove, adjacent to Y188, and also projects out into the solvent (Figures 4, 5A and B, and 6A). This may allow VP8* to bind internal sialic acids linked at O4 to distal sugar residues in glycolipid moieties, as suggested by binding of rotavirus strain SA11, NCDV and UK triple-layered particles to gangliosides with such internal sialic acids (Delorme et al., 2001). In the crystals, which were frozen in 20% glycerol, the VP8* core binds a glycerol molecule. The glycerol makes hydrogen bonds with residues T186, R210 and E213 and contacts the aromatic ring of Y175 (Figure 4, glycerol not modeled). A sugar linked to the O4 position of the sialoside could potentially make similar contacts.
Several studies indicate that rotavirus preferentially binds modified sialosides: the loss of the inhibition of SA11 by bovine salivary mucin that has been treated with specific neuraminidases suggests a preference for 7-_O_-acetylated sialic acid (Willoughby and Yolken, 1990); enhanced inhibition of strain NCDV by an acetylated synthetic thiosialoside suggests a preference for 9-_O_-acetylated sialic acid (Kiefel et al., 1996); and the interactions of strains OSU, SA11 and NCDV with specific gangliosides suggest a preference for _N_-glycolyl neuraminic acid (Rolsma et al., 1998; Delorme et al., 2001). The crystal structure demonstrates that modification is not required for sialoside binding by RRV VP8*. As O7 points out of the sialic acid binding groove (Figure 5B) however, 7-_O_-acetylation could be accommodated, with the added methyl group interacting with an aromatic ring at position 155. The O9 position is more constrained (Figures 4 and 5B), but an acetyl group might be accommodated projecting away from the floor of the groove.
The glycolyl group of an _N_-glycolyl sialoside (which has a hydroxyl in place of one hydrogen on the acetamido-methyl group) would be adjacent to residues that have multiple conformations in the VP8* core bound to an _N_-acetyl sialoside (Figure 4): the K187 side chain has ambiguous electron density beyond Cβ; the G156–P157 peptide bond takes on both cis and trans conformations; and N178 has two alternative side chain conformations. This area varies among sialic acid binding strains: K187 is replaced by glycine in other sialic acid-dependent rotaviruses, and its replacement by arginine preserves sialic acid binding, but eliminates dependence upon sialic acid for entry (Mendez et al., 1996; Ludert et al., 1998). Although the structure does not suggest an obvious explanation for the latter observation, enhanced binding of an _N_-glycolyl sialoside could be explained by a potential hydrogen bond between the hydroxyl of an _N_-glycolyl substituent and the G156 amide (not shown).
Binding-induced structural changes in the VP8* core
Comparison of the liganded crystal and unliganded NMR structures shows no evidence for a major conformational rearrangement induced by sialic acid binding (Figure 5D). The backbone trace of the crystal structure superimposes on the mean NMR structure with a root mean squared deviation (r.m.s.d.) of 1.34 Å. For comparison, within the suite of 20 accepted NMR structures, the backbone r.m.s.d. is 0.78 Å.
There is, however, evidence that sialoside binding causes local changes in the VP8* core. Specifically, the cleft above which the sialoside binds is slightly narrowed in the crystal structure relative to the NMR structure (Figure 5D). In the triple resonance NMR spectra, the S190 backbone peaks are missing, and the T191 backbone peaks have two alternative amide proton chemical shifts, suggesting flexibility in the absence of ligand (and preventing accurate T2 relaxation measurements). In the crystal structure, S190 and T191 have well-defined electron density (S190 electron density shown in Figure 5C) and low thermal parameters (7.3 for S190 atom N, and 8.7 for T191 atom N), indicating that their conformation is stabilized in the presence of ligand. The network of hydrogen bonds between the sialoside glycerol and carboxylate groups and residues R101, Y155 and S190 (Figure 5B) is probably responsible for this stabilization. These findings suggest a binding-induced fit of the sialoside and its recognition site.
Fit to an electron cryomicroscopy reconstruction
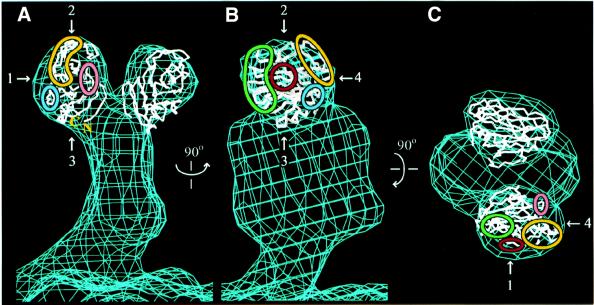
The sialic acid binding domain fits the size and shape of the heads of the rotavirus spike, as seen in an electron cryomicroscopy-based reconstruction (Yeager et al., 1990; Figure 7). Like the VP8* core, the heads make no dimeric contacts. The placement of the crystal structure in Figure 7 buries the highly conserved region near the N- and C-termini (see below) and exposes the sialic acid binding site and the widely distributed neutralizing antibody escape mutations. This positioning is also consistent with an image reconstruction of trypsinized particles decorated with mAb 7A12 (Tihova et al., 2001). A precise orientation of the sialic acid binding domain within the head can not be determined, however, because of its globular shape and the 26 Å resolution of the map.
Fig. 7. Placement of the Cα trace of the sialic acid binding domain within an electron cryomicroscopy-based reconstruction of the VP4 spike on trypsinized RRV virions. The map, contoured at 0.8σ, is courtesy of Drs Kelly Dryden and Mark Yeager (Yeager et al., 1990). (A) View along arrow 4. (B) View along arrow 1. (C) View along arrow 2. The view in (B) is rotated 90° about the vertical axis relative to the view in (A). The view in (C) is rotated 90° about the horizontal axis relative to the view in (B). Green outline, epitope 8-1; blue outline, epitope 8-2; yellow outline, epitope 8-3; pink outline, epitope 8-4; red outline, sialoside binding site.
The identification of residues L65–L224 as the ‘heads’ of the VP4 spike indicates that the first 64 residues of VP8* form a part of the ‘body’, interacting with VP5*. It is, therefore, likely that non-covalent interactions of these N-terminal residues (and possibly P225–R231) with VP5* link authentic VP8* to the virion after trypsin activation. This placement also indicates that the trypsin activation region (R231–R247) must be near the junction of the head and the body. Antibody decoration experiments locate the membrane interaction motif of VP5* at the top of the body, near this junction (Prasad et al., 1990; Tihova et al., 2001). This placement suggests that trypsin cleavage may permit an unmasking of the membrane interaction motif. As the N- and C-termini of the VP8* core are separated by only 10 Å (Figure 6A) and are located in a prominence with a strong negative charge (not indicated), the connection between head and body may be flexible.
Antigenic surfaces
The antigenic topography of VP8* has been investigated extensively because antibodies against VP8* can both neutralize virus (Shaw et al., 1986; Burns et al., 1988; Padilla-Noriega et al., 1995) and protect experimental animals from disease (Matsui et al., 1989). Neutralization escape mutant analyses have provided sequence-specific data that allow correlation of antigenic maps of the VP8* core with its structure (Figures 3, 6 and 7, and Table IV). None of the 20 mutated residues listed in Table IV is located in the center of a β-sheet (Figure 3). Five escape mutations map to residues with evidence for significant flexibility (residues Q135, Q148, G150, E180 and S190), but the other 15 residues selected are relatively inflexible.
Table IV. Neutralization escape mutations.
| Residuea | Mutation | Epitopeb | Strainc | Antibody | Commentd | References |
|---|---|---|---|---|---|---|
| 72 | T to I | NA | ST3 | HS6 | neutralizes human strain | Padilla-Noriega et al. (1995) |
| 87 | T to A | 8–4 | RRV | M11 | Mackow et al. (1988) | |
| 88 | T to L | 8–4 | RRV | A1 | Mackow et al. (1988) | |
| 89 | A to P | 8–4 | RRV | A15 | Mackow et al. (1988) | |
| 100 | D to N | 8–1 | RRV | 1A9 | blocks binding | Mackow et al. (1988); Ruggeri and Greenberg (1991) |
| 114 | S to F | 8–3 | RRV | 5D9 | Mackow et al. (1988) | |
| 116 | E to D | 8–3 | C486 | 2E8 | Lee et al. (1998) | |
| 132 | Y to D | 8–3 | RRV | 3G6 | IgA | Giammarioli et al. (1996) |
| 133 | A to N | 8–3 | RRV | 2A10 | IgA | Giammarioli et al. (1996) |
| 133 | A to V | 8–3 | RRV | 3F12 | IgA | Giammarioli et al. (1996) |
| 135 | Q to K | 8–3 | RRV | 4B7 | IgA | Giammarioli et al. (1996) |
| 135 | Q to R | 8–3 | RRV | 4C2 | IgA | Giammarioli et al. (1996) |
| 136 | D to N | 8–3 | SA11 cl3 | 9F6 | Zhou et al. (1994) | |
| 146 | Q to R, K, or Y | 8–1 | SA11 4f | hyperimmunee | Gorziglia et al. (1990) | |
| 148 | Q to R | 8–1 | RRV | 2B12 | IgA | Giammarioli et al. (1996) |
| 148 | Q to R | 8–1 | RV-5 | RV5:2 | neutralizes human strain | Kirkwood et al. (1996) |
| 148 | Q to R | 8–1 | RRV | M14 | IgM, blocks binding, passively protects mice | Mackow et al. (1988); Ruggeri and Greenberg (1991); Matsui et al. (1989) |
| 150 | G to E | 8–1 | RRV | 5C4 | V150 is neuraminidase insensitive | Mackow et al. (1988); Ludert et al. (1998) |
| 180 | Q to R | 8–2 | SA11 cl3 | 7G6 | destabilizes virus | Zhou et al. (1994) |
| 183 | N to D | 8–2 | SA11 cl3 | 10G6 | destabilizes virus | Zhou et al. (1994) |
| 188 | Y to F | 8–1 | RRV | 7A12 | blocks binding | Mackow et al. (1988); Ruggeri and Greenberg (1991) |
| 190 | S to L | 8–1 | RRV | 4B6 | IgA; v4B6 does not HA | Giammarioli et al. (1996) |
| 194 | Y to C | 8–1 | RRV | 954/23 | blocks binding; v194 does not HA | Zhou et al. (1994); Ruggeri and Greenberg (1991) |
| 217 | E to K | NA | ST3 | HS11 | neutralizes human strain | Padilla-Noriega et al. (1995) |
Antibody competition experiments and escape mutant analyses indicate that VP8* contains several interrelated epitopes recognized by neutralizing antibodies (Shaw et al., 1986; Burns et al., 1988; Mackow et al., 1988; Zhou et al., 1994; Giammarioli et al., 1996). Neutralization escape mutations against hemagglutinating strains are widely distributed across the accessible surface of the VP8* core, but do show clustering (Figures 6 and 7). We call the clusters epitopes 8-1, 8-2, 8-3 and 8-4 (Figures 6 and 7 and Table IV). Antibody competition experiments and analysis of the cross-resistance of variants, while reflecting this clustering, also show some competition between antibodies located in different epitopes. For example, mAb 954/23, which selects a mutation in epitope 8-1 at residue 194, competes for binding with mAbs that select mutations in epitopes 8-2 and 8-3 at residues 136, 180 and 183 (Shaw et al., 1986; Burns et al., 1988).
Epitope 8-1 (green in Figures 6 and 7) is located near the bound sialic acid and the positions of proximal sugar residues in an oligosaccharide side chain. Some antibodies that select mutations at residues in this epitope interfere with early entry events. For example, mAbs that select escape mutations at residues 100, 148 and 188 block binding to cells (Ruggeri and Greenberg, 1991); a S190 to L escape mutant no longer hemagglutinates (Giammarioli et al., 1996); and infection by a G150 to D escape mutant is no longer sensitive to neuraminidase digestion of cells (Ludert et al., 1998).
Epitope 8–2 (blue in Figures 6 and 7) is defined by mutations selected at residues E180 and N183 by mAbs that destabilize the outer capsid of the virion (Zhou et al., 1994). The residues from G179 to V184, which include the βJ–βK loop, make key contacts (Figures 2A and 3), participating in the parallel β-strands (βJ and βH′) that link the five-stranded and six-stranded β-sheets and forming hydrogen bonds with Q137 in the βF–βG loop, T161 in the βH′–βI loop and N221 near the C-terminus of αB. Antibody binding may disrupt these interactions, and transmission of the resulting distortions to the remainder of the spike through the domain’s C-terminus may result in the observed outer capsid destabilization.
Residues in epitope 8-3 (yellow in Figures 6 and 7) are located in the β-ribbon and the loops that connect the β-ribbon to the six-stranded β-sheet (Figures 2A and 3). A number of IgA monoclonals map to this epitope (Giammarioli et al., 1996). Although some escape mutations in epitope 8-3 are located close to those in epitope 8-2, antibodies that select these mutations do not destabilize the outer capsid (Zhou et al., 1994), and their mechanism of neutralization has not been determined.
Epitope 8-4 (pink in Figures 6 and 7) consists of three adjacent residues (Mackow et al., 1988) on the βB–βC loop, which connects the two β-sheets (Figures 2B and 3). It is predicted to lie on an accessible surface at the edge of the cleft between the two heads of the spike. A mechanism of neutralization has not been determined for mAbs mapping to this epitope.
Although a number of neutralizing mAbs map to VP5* from sialic acid-independent human rotavirus strains (Taniguchi et al., 1988; Kobayashi et al., 1990; Padilla-Noriega et al., 1995), only three neutralizing mAbs have been mapped to VP8* of such strains. One such VP8*-specific mAb selects a mutation at residue 148 in epitope 8-1 (Kirkwood et al., 1996), demonstrating that interference with sialic acid binding is not the sole mechanism of neutralization for epitope 8-1 mAbs. The other two mAbs select mutations at residues 72 and 217 (black lettering in Figure 6; Padilla-Noriega et al., 1995), which are located outside the known neutralization epitopes of sialic acid-dependent strains and are remote from the sialic acid binding site. The paucity of neutralizing mAbs against VP8* of sialic acid-independent strains and the separate locations of these two escape mutations suggest that there are substantially different roles for VP8* from sialic acid-independent and -dependent strains in early entry events.
Overall surface variability
VP8* surface residues are highly variable among P genotypes (Figure 6), probably reflecting selection pressure for diversity by the host immune response. The residues around the N- and C-termini form the only surface that is highly conserved (Figure 6A). As this surface contains the point of attachment to the remainder of VP4 (Figure 7), much of it is probably buried on the complete spike. This surface variability poses a challenge for using the VP8* core as an immunogen and as a target for structure-based drug design: although most human disease is caused by P genotypes 4 and 8 (Gentsch et al., 1996), a number of other P genotypes also infect humans (Griffin et al., 2000), raising the possibility of the early emergence of strains that escape neutralization or resist antivirals.
Evolutionary implications
The galectin-like fold of the rotavirus VP8* core does not resemble the folds of reovirus σ3, µ1 or the σ1 knob (Olland et al., 2001; Chappell et al., 2002; Liemann et al., 2002). The reovirus σ1 sialic acid binding region is predicted to have an unrelated β-spiral fold (Chappell et al., 2002). This suggests independent evolution of the rotavirus and reovirus sialic acid binding domains.
The similarity of the rotavirus sialic acid binding domain to the galectin carbohydrate binding domain could result from either convergent evolution or common ancestry. As VP8* and the galectins bind carbohydrates on different surfaces of the β-sandwich (Figure 2), it is unlikely that carbohydrate binding drove convergence to a common fold. It is particularly difficult to explain the precision with which the VP8* β-ribbon blocks the galectin carbohydrate binding site (Figure 2B and C) on the basis of convergent evolution. Rather, we propose that rotavirus VP4 arose by the insertion of a host-derived, galectin-like carbohydrate binding domain into an ancestral membrane interaction protein. Subsequently, VP4 acquired a new carbohydrate binding site and specificity, and the insertion of the β-ribbon blocked the galectin carbohydrate binding site. These molecular events were probably accompanied by significant changes in viral tropism and pathogenicity.
Other viruses have modified a membrane interaction protein by inserting a carbohydrate binding domain. Structural data indicate that influenza HA arose from the insertion of a sialic acid binding domain into an ancestral membrane interaction domain (Zhang et al., 1999). The NADC-1 strain of porcine adenovirus has an enlarged fiber head due to the insertion of a sequence that has strong similarity to the galectin carbohydrate binding domain (Kleiboeker, 1995).
Sequence alignments (not shown) of VP4 from rotavirus serogroups A, B and C (DDBJ/EMBL/GenBank accession numbers M91434, AF184084, U03556, AF323981, AF323980 and those listed in Table III) show that the acquisition of the galectin-like domain preceded the divergence of the serogroups. Subsequent to this split, variability arose in the VP8* core of the group A rotaviruses (Figure 6). The correlation of sequence diversity in the carbohydrate binding site with functional diversity in carbohydrate binding (Table III) indicates that rotavirus is capable of considerable flexibility in its initial interaction with its host cell, the mature enterocyte. This diversity is probably driven by intense selection pressure from the host immune system and possibly by developmental and species-specific differences in host carbohydrate receptors.
Subsequent to cell attachment, the outer layer of rotavirus must accomplish membrane penetration and controlled disassembly. We have crystallized the VP5* core and VP7 (Dormitzer et al., 2000; our unpublished data), which are responsible for these functions. High-resolution structures of these proteins will give additional insights into the evolution and mechanisms of rotavirus cell entry. Structural comparisons between the rotavirus entry apparatus and its counterparts in the other Reoviridae will reveal the variety of strategies that these viruses have evolved to solve their common entry problem.
Materials and methods
Molecular biology
DNA oligonucleotide primers were synthesized and plasmid DNA was sequenced by the Howard Hughes Medical Institute Biopolymer Facility (Boston). DNA and amino acid sequences were manipulated using the Lasergene suite of sequence analysis software (DNAstar), and sequences were aligned using Clustal X (Thompson et al., 1997).
Constructs
To construct plasmid pGex-VP846–231, the nucleotide sequence encoding residues A46 to R231 of RRV VP4 was amplified by PCR from plasmid pRRV4, which contains a clone of RRV gene segment 4 described previously (Mackow et al., 1988; Dormitzer et al., 2001). The amplified sequence was subcloned into pGex 4T-1 (Amersham-Pharmacia Biotech) to create an in-frame fusion downstream of GST. The same strategy was used to construct plasmid pGex-VP862–224 except that the nucleotide sequence encoding residues E62–L224 was amplified.
Protein expression and purification
Escherichia coli strain BL21 DE3, transformed with the plasmids described above, was grown at 37°C to an _A_600 of 0.6 in Luria–Bertani (LB) medium supplemented with 100 µg/ml of ampicillin. The cultures were then incubated at 25°C and, after 1 h, were induced with 1 mM isopropyl-β-d-thiogalactopyranoside. Cells were harvested by pelleting 4 h after induction and frozen. For production of selenomethionine-substituted protein, M9 minimal medium containing selenomethionine in place of methionine was used. For production of isotopically labeled protein, cells were grown in M9 minimal medium, modified by the substitution of [15N]NH4Cl, [13C]glucose and/or D2O for the equivalent unlabeled compounds. To produce protein selectively labeled at specific residue types, E.coli strain DL39, an auxotroph for amino acid synthesis, was grown in M9 supplemented with a 15N-labeled amino acid (valine, leucine, isoleucine, phenylalanine or tyrosine) and with other unlabeled amino acids. Culture times were optimized for each labeling regimen.
Frozen cell pellets were thawed in 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA (TNE), supplemented with 1% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride (PMSF). The suspension was sonicated and centrifuged at 235 400 g for 2 h. The supernatant was passed over a glutathione–Sepharose column (Amersham-Pharmacia Biotech), which was then washed with 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM CaCl2 (TNC) and digested with 5 µg/ml TPCK-treated trypsin (Worthington Biochemical) for 2 h at room temperature. The cleaved protein was eluted with TNC, the eluate was passed over benzamidine–Sepharose (Amersham-Pharmacia Biotech), and 1 mM PMSF and 2.5 mM benzamidine were added. The protein was then concentrated by ultrafiltration using a Centricon 10 unit and subjected to size exclu sion chromatography over a Superdex 200 Hi-Load 16/60 column (Amersham-Pharmacia Biotech) equilibrated in 20 mM NaPO4 pH 7.0, 100 mM NaCl (for NMR studies) or TNE (for crystallographic studies), using an FPLC system (Amersham-Pharmacia Biotech). The concentration of pooled fractions was estimated by _A_280. During purification of selenomethionine-substituted protein, 5–10 mM dithiothreitol (DTT) was included in all buffers. The purified proteins were analyzed by MALDI time-of-flight mass spectrometry and N-terminal sequencing (carried out by the Tufts Core Protein Chemistry Facility) and by dynamic laser light scattering, using a DynaPro 801 instrument (Protein Solutions, Inc.).
NMR data collection
NMR spectra were obtained at 500 MHz (Varian Unity), 600 MHz (Bruker Avance) and 750 MHz (Varian INOVA). EcVP846–231 samples were dialyzed against 20 mM NaPO4 pH 7.0, 10 mM NaCl, 0.02% sodium azide and concentrated to ∼21 mg/ml. Aqueous solutions were made 10% in D2O for spectrometer field-locking. Backbone and side chain resonance assignments were made as described previously (Matsuo et al., 1997). Distance constraints were obtained from a three-dimensional 15N-NOESY-HSQC spectrum and a two-dimensional NOESY spectrum. A three-dimensional 13C-NOESY-HSQC experiment was used to confirm some ambiguous assignments in the two-dimensional NOESY. Relaxation data were obtained by 15N-1H HSQC experiments with the T2 time progressively increased from 0 to 164 ms.
NMR data processing and structure calculation
NMR data were processed by PROSA (Guntert et al., 1992). Peak assignments were made using XEASY (Bartels et al., 1995). Dihedral angle constraints based on chemical shifts were derived using TALOS (Cornilescu et al., 1999). Integrated peak volumes were converted into distance constraints using DYANA (Guntert et al., 1997). Hydrogen bond constraints were introduced based on characteristic NOE patterns for α-helices and β-sheets, on slow exchange amide protons identified from D2O buffer exchange experiments, and on the proximity and orientation of potential hydrogen bond partners in annealed structures. Structures were calculated by simulated annealing using CNS (Brunger et al., 1998). Annealed structures were analyzed for proper stereochemistry (Table II) using AQUA and Procheck-NMR (Laskowski et al., 1996).
Crystallization
2-_O_-methyl-α-d-_N_-acetyl neuraminic acid was obtained from Sigma Chemical Co. Crystals were grown by hanging drop vapor diffusion. A sample solution containing 17.6 mg/ml EcVP862–224, 52 mM sialoside, 5.6 mM Tris–HCl pH 8.0, 14 mM NaPO4 pH 7.0, 35 mM NaCl, 0.3 mM EDTA, 0.02% sodium azide and 0.1 mM benzamidine was mixed with an equal volume of a well solution containing 1.60–1.75 M ammonium sulfate, 2.3–2.5% PEG 400 and 100 mM PIPES pH 6.5. Crystals formed at 30°C over the course of 3 days to 3 weeks, reaching maximal dimensions of 170 × 170 × 340 µm. For crystallization of seleno methionine-labeled protein, 5–10 mM DTT was included in the crystallization solutions. Crystals were frozen by immersion in liquid nitrogen after a brief soak in 15.8 mM sialoside, 1.8 M ammonium sulfate, 2.6% PEG 400, 100 mM PIPES pH 6.5, 0.02% sodium azide, 0.1 mM benzamidine and 20% glycerol.
X-ray diffraction data collection and processing
The crystals belong to space group _P_41212 and have unit cell parameters of a = b = 48.03 Å and c = 130.40 Å. There is one protein–sialoside complex per asymmetric unit, predicting a solvent content of 25.2%. X-ray diffraction data were obtained at Advanced Photon Source beamline 14C, using a Quantum-4 detector. Data collection statistics are shown in Table I. Complete data sets were obtained at a wavelength of 1 Å from one native and one selenomethionine-substituted frozen crystal. The native crystal diffracted X-rays to an interplanar spacing of <1.4 Å (the limit of the collection area on the detector). The data sets were integrated and scaled using HKL 2000 (HKL Research, Inc.).
X-ray crystallographic structure determination and refinement
Phasing and refinement statistics are summarized in Table I. Initial phases were obtained by single isomorphous replacement using a selenomethionine-substituted protein crystal. Native and derivative data sets were scaled to each other using SCALEIT from the CCP4 suite (CCP4, 1994). Subsequent calculations were performed using CNS (Brunger et al., 1998). Three selenium sites were located and refined based on isomorphous Patterson maps. The electron density map calculated using the resulting phase information was improved by solvent flipping. All but one residue of the final peptide backbone, most side chains and the sialoside could be unambiguously traced in the resulting map. The structure was refined by successive rounds of simulated annealing, energy minimization and _B_-factor refinement alternating with rebuilding in O (Jones et al., 1991). Five percent of the reflections were set aside for the calculation of _R_free. The structure was analyzed for proper stereochemistry using PROCHECK (Laskowski et al., 1993).
Structure analysis
Structure-based sequence alignment, r.m.s.d. calculation and structural similarity database searching were performed using LSQKAB in CCP4 (CCP4, 1994), MOLMOL (Koradi et al., 1996) and DALI (Holm and Sander, 1993). For the initial search for structures similar to the VP8* core, the entire crystal structure was used as a query model. Subsequent searches used residues T72–V108 and K139–I207 of the structure. Amino acid variability was calculated by AMAS (Livingstone and Barton, 1993).
Illustration
Figures were produced using Molscript (Figures 2 and 5A, B and D; Kraulis, 1991), Grasp (Figures 4 and 6; Nicholls et al., 1991), Spock (Figure 5C; Christopher, 1997), O (Figure 7; Jones et al., 1991), Photoshop 6.0 (Figures 1, 4, 6 and 7; Adobe Systems, Inc.) and Illustrator 8.0 (all figures; Adobe Systems, Inc.).
Coordinates
The coordinates of the crystal structure of the VP8* core in complex with sialoside and the structure factor amplitudes have been deposited in the Protein Data Bank (PDB) (http://www.rcsb.org) with the identification code 1KQR. The coordinates of the NMR structures of the VP8* core without ligand and the chemical shifts of assigned atoms have been deposited in the PDB with the identification code 1KRI.
Acknowledgments
Acknowledgements
We thank Marina Babyonyshev for her skillful technical assistance; Mark Yeager and Kelly Dryden for the electron cryomicroscopy map of VP4; Yorgo Modis for collecting X-ray diffraction data; Emelia Boiadgieva for molecular cloning assistance; Harry Greenberg for scientific advice and materials; Ulu Unligil and Stephen Litster for computational expertise; Don Wiley, Thilo Stehle, Andrea Carfi, Stephen DeWall, Katya Heldwein, Mykol Larvie, Susanne Liemann and Erik Vogan for helpful and stimulating discussions; Gary Navrotski for assistance with data collection at the Advanced Photon Source; and Mary Estes and Max Ciarlet for a careful reading of the manuscript. This work was supported by National Institutes of Health grants K08 AI 01496 to P.R.D., GM 47467 to G.W. and CA 13202 to S.C.H. S.C.H. is an investigator in the Howard Hughes Medical Institute. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38.
References
- Bartels C., Xia,T.-H., Billeter,M., Guntert,P. and Wuthrich,K. (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR, 5, 1–10. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burns J.W., Greenberg,H.B., Shaw,R.D. and Estes,M.K. (1988) Functional and topographical analyses of epitopes on the hemagglutinin (VP4) of the simian rotavirus SA11. J. Virol., 62, 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control (1999) Withdrawal of rotavirus vaccine recommendation. MMWR Morb. Mortal. Wkly Rep., 48, 1007. [PubMed] [Google Scholar]
- Chappell J.D., Prota,A.E., Dermody,T.S. and Stehle,T. (2002) Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J., 21, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher J.A. (1997) Spock. Center for Macromolecular Design, College Station, TX.
- Ciarlet M., Crawford,S.E. and Estes,M.K. (2001) Differential infection of polarized epithelial cell lines by sialic acid-dependent and sialic acid-independent rotavirus strains. J. Virol., 75, 11834–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlet M., Ludert,J.E., Iturriza-Gomara,M., Liprandi,F., Gray,J.J., Desselberger,U. and Estes,M.K. (2002) The initial interaction of rotavirus strains with _N-_acetyl-neuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Crawford S.E., Mukherjee,S.K., Estes,M.K., Lawton,J.A., Shaw,A.L., Ramig,R.F. and Prasad,B.V. (2001) Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol., 75, 6052–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crennell S., Takimoto,T., Portner,A. and Taylor,G. (2000) Crystal structure of the multifunctional paramyxovirus hemagglutinin– neuraminidase. Nature Struct. Biol., 7, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Delorme C., Brussow,H., Sidoti,J., Roche,N., Karlsson,K.A., Neeser,J.R. and Teneberg,S. (2001) Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol., 75, 2276–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer P.R., Greenberg,H.B. and Harrison,S.C. (2000) Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virology, 277, 420–428. [DOI] [PubMed] [Google Scholar]
- Dormitzer P.R., Greenberg,H.B. and Harrison,S.C. (2001) Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol., 75, 7339–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling W., Denisova,E., LaMonica,R. and Mackow,E.R. (2000) Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol., 74, 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley J., White,H.E., O’Hara,B.P., Oliva,G., Srinivasan,N., Tickle,I.J., Blundell,T.L., Pepys,M.B. and Wood,S.P. (1994) Structure of pentameric human serum amyloid P component. Nature, 367, 338–345. [DOI] [PubMed] [Google Scholar]
- Estes M.K., Graham,D.Y. and Mason,B.B. (1981) Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol., 39, 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore L., Greenberg,H.B. and Mackow,E.R. (1991) The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology, 181, 553–563. [DOI] [PubMed] [Google Scholar]
- Fuentes-Panana E.M., Lopez,S., Gorziglia,M. and Arias,C.F. (1995) Mapping the hemagglutination domain of rotaviruses. J. Virol., 69, 2629–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch J.R., Woods,P.A., Ramachandran,M., Das,B.K., Leite,J.P., Alfieri,A., Kumar,R., Bhan,M.K. and Glass,R.I. (1996) Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis., 174, S30–S36. [DOI] [PubMed] [Google Scholar]
- Giammarioli A.M., Mackow,E.R., Fiore,L., Greenberg,H.B. and Ruggeri,F.M. (1996) Production and characterization of murine IgA monoclonal antibodies to the surface antigens of rhesus rotavirus. Virology, 225, 97–110. [DOI] [PubMed] [Google Scholar]
- Gorziglia M., Larralde,G. and Ward,R.L. (1990) Neutralization epitopes on rotavirus SA11 4fM outer capsid proteins. J. Virol., 64, 4534–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.D., Kirkwood,C.D., Parashar,U.D., Woods,P.A., Bresee,J.S., Glass,R.I. and Gentsch,J.R. (2000) Surveillance of rotavirus strains in the United States: identification of unusual strains. J. Clin. Microbiol., 38, 2784–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C.A., Mendez,E., Zarate,S., Isa,P., Lopez,S. and Arias,C.F. (2000) Integrin αvβ3 mediates rotavirus cell entry. Proc. Natl Acad. Sci. USA, 97, 14644–14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntert P., Dotsch,V., Wider,G. and Wuthrich,K. (1992) Processing of multi-dimensional NMR data with the new software PROSA. J. Biomol. NMR, 2, 619–629. [Google Scholar]
- Guntert P., Mumenthaler,C. and Wuthrich,K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol., 273, 283–298. [DOI] [PubMed] [Google Scholar]
- Guo C.T., Nakagomi,O., Mochizuki,M., Ishida,H., Kiso,M., Ohta,Y., Suzuki,T., Miyamoto,D., Hidari,K.I. and Suzuki,Y. (1999) Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J. Biochem., 126, 683–688. [DOI] [PubMed] [Google Scholar]
- Harrison S.C., Skehel ,J.J and Wiley,D.C. (1996) Virus structure. In Fields,B.N., Knipe,D.M., Howley,P.M., Chanock,R.M., Melnick,J.L, Monath,T.P, Roizman,B. and Strauss,S.E (eds), Field’s Virology. Lippincott-Raven, Philadelphia, PA, pp. 59–99.
- Hewish M.J., Takada,Y. and Coulson,B.S. (2000) Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol., 74, 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hughes R.C. (1997) The galectin family of mammalian carbohydrate-binding molecules. Biochem. Soc. Trans., 25, 1194–1198. [DOI] [PubMed] [Google Scholar]
- Isa P., Lopez,S., Segovia,L. and Arias,C.F. (1997) Functional and structural analysis of the sialic acid-binding domain of rotaviruses. J. Virol., 71, 6749–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kiefel M.J., Beisner,B., Bennett,S., Holmes,I.D. and von Itzstein,M. (1996) Synthesis and biological evaluation of _N_-acetylneuraminic acid-based rotavirus inhibitors. J. Med. Chem., 39, 1314–1320. [DOI] [PubMed] [Google Scholar]
- Kirkwood C.D., Bishop,R.F. and Coulson,B.S. (1996) Human rotavirus VP4 contains strain-specific, serotype-specific and cross-reactive neutralization sites. Arch. Virol., 141, 587–600. [DOI] [PubMed] [Google Scholar]
- Kleiboeker S.B. (1995) Sequence analysis of the fiber genomic region of a porcine adenovirus predicts a novel fiber protein. Virus Res., 39, 299–309. [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Taniguchi,K. and Urasawa,S. (1990) Identification of operationally overlapping and independent cross-reactive neutralization regions on human rotavirus VP4. J. Gen. Virol., 71, 2615–2623. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wuthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics, 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kraulis J. (1991) MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- LaMonica R., Kocer,S.S., Nazarova,J., Dowling,W., Geimonen,E., Shaw,R.D. and Mackow,E.R. (2001) VP4 differentially regulates TRAF2 signaling, disengaging JNK activation while directing NF-κB to effect rotavirus-specific cellular responses. J. Biol. Chem., 276, 19889–19896. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the sterochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Lee J., Babiuk,L.A. and Yoo,D. (1998) A neutralizing monoclonal antibody to bovine rotavirus VP8 neutralizes rotavirus infection without inhibiting virus attachment to MA-104 cells. Can. J. Vet. Res., 62, 63–67. [PMC free article] [PubMed] [Google Scholar]
- Leffler H. (1997) Introduction to galectins. Trends Glycosci. Glycotech., 9, 9–19. [Google Scholar]
- Liemann S., Chandran,K., Baker,T.S, Nibert,M. and Harrison,S.C. (2002) Structure of the membrane penetration protein, µ1, in complex with its protector protein, σ3. Cell, 108, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone C.D. and Barton,G.J. (1993) Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci., 9, 745–756. [DOI] [PubMed] [Google Scholar]
- Lobsanov Y.D. and Rini,J.M. (1997) Galectin structure. Trends Glycosci. Glycotech., 9, 145–154. [Google Scholar]
- Ludert J.E., Feng,N., Yu,J.H., Broome,R.L., Hoshino,Y. and Greenberg,H.B. (1996) Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol., 70, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludert J.E., Mason,B.B., Angel,J., Tang,B., Hoshino,Y., Feng,N., Vo,P.T., Mackow,E.M., Ruggeri,F.M. and Greenberg,H.B. (1998) Identification of mutations in the rotavirus protein VP4 that alter sialic-acid-dependent infection. J. Gen. Virol., 79, 725–729. [DOI] [PubMed] [Google Scholar]
- Mackow E.R., Shaw,R.D., Matsui,S.M., Vo,P.T., Dang,M.N. and Greenberg,H.B. (1988) The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc. Natl Acad. Sci. USA, 85, 645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui S.M., Offit,P.A., Vo,P.T., Mackow,E.R., Benfield,D.A., Shaw,R.D., Padilla-Noriega,L. and Greenberg,H.B. (1989) Passive protection against rotavirus-induced diarrhea by monoclonal antibodies to the heterotypic neutralization domain of VP7 and the VP8 fragment of VP4. J. Clin. Microbiol., 27, 780–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H., Li,H., McGuire,A.M., Fletcher,C.M., Gingras,A.C., Sonenberg,N. and Wagner,G. (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nature Struct. Biol., 4, 717–724. [DOI] [PubMed] [Google Scholar]
- Mendez E., Arias,C.F. and Lopez,S. (1996) Interactions between the two surface proteins of rotavirus may alter the receptor-binding specificity of the virus. J. Virol., 70, 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki M. and Nakagomi,O. (1995) Haemagglutination by rotaviruses in relation to VP4 genotypes. Res. Virol., 146, 371–374. [DOI] [PubMed] [Google Scholar]
- Nejmeddine M., Trugnan,G., Sapin,C., Kohli,E., Svensson,L., Lopez,S. and Cohen,J. (2000) Rotavirus spike protein VP4 is present at the plasma membrane and is associated with microtubules in infected cells. J. Virol., 74, 3313–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Taniguchi,K., Torres,A., Hoshino,Y., Green,K., Kapikian,A.Z., Chanock,R.M. and Gorziglia,M. (1988) Comparative analysis of the VP3 gene of divergent strains of the rotaviruses simian SA11 and bovine Nebraska calf diarrhea virus. J. Virol., 62, 4022–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olland A.M., Jane-Valbuena,J., Schiff,L.A., Nibert,M.L. and Harrison,S.C. (2001) Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J., 20, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Noriega L., Dunn,S.J., Lopez,S., Greenberg,H.B. and Arias,C.F. (1995) Identification of two independent neutralization domains on the VP4 trypsin cleavage products VP5* and VP8* of human rotavirus ST3. Virology, 206, 148–154. [DOI] [PubMed] [Google Scholar]
- Prasad B.V., Burns,J.W., Marietta,E., Estes,M.K. and Chiu,W. (1990) Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature, 343, 476–479. [DOI] [PubMed] [Google Scholar]
- Rolsma M.D., Kuhlenschmidt,T.B., Gelberg,H.B. and Kuhlenschmidt,M.S. (1998) Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol., 72, 9079–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri F.M. and Greenberg,H.B. (1991) Antibodies to the trypsin cleavage peptide VP8 neutralize rotavirus by inhibiting binding of virions to target cells in culture. J. Virol., 65, 2211–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman J., Kanigsberg,A., Slaaby,R., Leffler,H., Barondes,S.H. and Rini,J.M. (1998) X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-Å resolution. J. Biol. Chem., 273, 13047–13052. [DOI] [PubMed] [Google Scholar]
- Sharma V. and Surolia,A. (1997) Analyses of carbohydrate recognition by legume lectins: size of the combining site loops and their primary specificity. J. Mol. Biol., 267, 433–445. [DOI] [PubMed] [Google Scholar]
- Shaw R.D., Vo,P.T., Offit,P.A., Coulson,B.S. and Greenberg,H.B. (1986) Antigenic mapping of the surface proteins of rhesus rotavirus. Virology, 155, 434–451. [DOI] [PubMed] [Google Scholar]
- Spence L., Fauvel,M., Petro,R. and Babiuk,L.A. (1978) Comparison of rotavirus strains by hemagglutination inhibition. Can. J. Microbiol., 24, 353–362. [DOI] [PubMed] [Google Scholar]
- Stehle T. and Harrison,S.C. (1997) High-resolution structure of a polyomavirus VP1–oligosaccharide complex: implications for assembly and receptor binding. EMBO J., 16, 5139–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K., Maloy,W.L., Nishikawa,K., Green,K.Y., Hoshino,Y., Urasawa,S., Kapikian,A.Z., Chanock,R.M. and Gorziglia,M. (1988) Identification of cross-reactive and serotype 2-specific neutralization epitopes on VP3 of human rotavirus. J. Virol., 62, 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tihova A., Dryden,K.A., Bellamy,A.R., Greenberg,H.B. and Yeager,M. (2001) Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: Implications for cell entry. J. Mol. Biol., 314, 985–992. [DOI] [PubMed] [Google Scholar]
- Weis W., Brown,J.H., Cusack,S., Paulson,J.C., Skehel,J.J. and Wiley,D.C. (1988) Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature, 333, 426–431. [DOI] [PubMed] [Google Scholar]
- Willoughby R.E. and Yolken,R.H. (1990) SA11 rotavirus is specifically inhibited by an acetylated sialic acid. J. Infect. Dis., 161, 116–119. [DOI] [PubMed] [Google Scholar]
- World Health Organization (1999) Rotavirus vaccines. Wkly Epidemiol. Rec., 74, 33–38. [PubMed] [Google Scholar]
- Yeager M., Dryden,K.A., Olson,N.H., Greenberg,H.B. and Baker,T.S. (1990) Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J. Cell Biol., 110, 2133–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Rosenthal,P.B., Formanowski,F., Fitz,W., Wong,C.H., Meier-Ewert,H., Skehel,J.J. and Wiley,D.C. (1999) X-ray crystallographic determination of the structure of the influenza C virus haemagglutinin–esterase-fusion glycoprotein. Acta Crystallogr. D, 55, 945–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.J., Burns,J.W., Morita,Y., Tanaka,T. and Estes,M.K. (1994) Localization of rotavirus VP4 neutralization epitopes involved in antibody-induced conformational changes of virus structure. J. Virol., 68, 3955–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]