Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein (original) (raw)
Abstract
Mass spectrometry was used to identify novel proteins associated with the human 17S U2 snRNP and one of its stable subunits, SF3b. Several additional proteins were identified, demonstrating that 17S U2 snRNPs are significantly more complex than previously thought. Two of the newly identified proteins, namely the DEAD-box proteins SF3b125 and hPrp5 (a homologue of Saccharomyces cerevisiae Prp5p) were characterized further. Immunodepletion experiments with HeLa nuclear extract indicated that hPrp5p plays an important role in pre-mRNA splicing, acting during or prior to prespliceosome assembly. The SF3b-associated protein SF3b125 dissociates at the time of 17S U2 formation, raising the interesting possibility that it might facilitate the assembly of the 17S U2 snRNP. Finally, immunofluorescence/FISH studies revealed a differential subnuclear distribution of U2 snRNA, hPrp5p and SF3b125, which were enriched in Cajal bodies, versus SF3b155 and SF3a120, which were not; a model for 17S U2 snRNP assembly based on these findings is presented. Taken together, these studies provide new insight into the composition of the 17S U2 snRNP and the potential function of several of its proteins.
Keywords: DEAD-box/pre-mRNA splicing/SF3b/U2 snRNP
Introduction
Spliceosomes, multi-component enzymes that catalyse pre-mRNA splicing, form step-wise by the ordered interaction of UsnRNPs and non-snRNP proteins with short conserved regions of the pre-mRNA at the 5′ and 3′ splice sites and branch site (for reviews, see Reed and Palandjian, 1997; Burge et al., 1999). Spliceosome assembly is initiated by the interaction of the U1 snRNP with the 5′ splice site, forming the E complex. The latter also contains the 17S U2 snRNP, which at this stage associates via a non-base pairing interaction (Das et al., 2000). In a subsequent ATP-dependent step, the U2 snRNA base pairs with the branch site of the pre-mRNA leading to stable association of the U2 snRNP and formation of the so-called A complex or prespliceosome. Finally, the U4/U6.U5 tri-snRNP complex binds, generating the B complex, and after a major conformational change, the C complex is formed. After splicing catalysis, the spliceosome dissociates into its snRNP subunits, which take part in ensuing rounds of splicing. Pre-mRNA splicing is thus a highly dynamic process in which the splicing machinery must be repeatedly assembled and disassembled. During spliceosome assembly, a complex network of dynamic RNA–RNA and RNA–protein interactions is formed. Many of the conformational changes in this network appear to be catalysed by spliceosomal proteins that are members of the DExH/D-box family of NTPases and RNA unwindases/RNPases; at least eight DExH/D-box proteins that act at various stages of the splicing process have been identified (for a review, see Staley and Guthrie, 1998).
The U1, U2, U5 and U4/U6 snRNPs are the main subunits of the U2-dependent spliceosome. They consist of one or two snRNA molecules complexed with seven common proteins (B/B′, D3, D2, D1, E, F and G), plus additional particle-specific proteins (Will and Lührmann, 1997). Of the major snRNPs, the 17S U2 snRNP is the least well-characterized in terms of its protein composition. To date, at least 12 specific proteins have been identified in purified, human 17S U2 snRNPs (Behrens et al., 1993; Will et al., 2001). These include the stably associated U2-A′ and U2-B′′ polypeptides, and the heteromeric protein complexes SF3a and SF3b, which contain subunits of 60, 66 and 120 kDa (reviewed in Krämer, 1996), or 14, 49, 130, 145 and 155 kDa, respectively (Das et al., 1999; Krämer et al., 1999; Will et al., 2001; and references therein). Biochemically purified HeLa 17S U2 snRNPs contain at least two additional proteins of 35 and 92 kDa, whose identity has not yet been established (Behrens et al., 1993). Recent studies indicate that SPF30/SMNrp is also a 17S U2-associated protein (Meister et al., 2001; Rappsilber et al., 2001).
At moderate salt concentrations (>250 mM), the 17S U2 snRNP dissociates into a 12S snRNP particle (containing A′/B″ and the Sm proteins), and the heteromeric complexes SF3a and SF3b (Behrens et al., 1993; Brosi et al., 1993b). The latter are highly stable protein complexes that remain intact even at high ionic strengths (Brosi et al., 1993b; Das et al., 1999; Will et al., 2001). Both were originally biochemically purified as non-snRNP complexes and shown to be essential for prespliceosome formation (Brosi et al., 1993b). The formation of the 17S U2 snRNP in vitro involves the sequential interaction of SF3b with the 12S U2 snRNP (forming an intermediate 15S particle) followed by SF3a (Brosi et al., 1993a; Krämer et al., 1999). At present, little is known about proteins facilitating the formation of the 17S U2 snRNP. In Saccharomyces cerevisiae, the protein Cus2p appears to help fold the U2 snRNA into a conformation that allows the subsequent association of SF3b and SF3a (Yan et al., 1998). The human homologue of Cus2p, Tat-SF1, has been proposed to act in a similar fashion (Yan et al., 1998; Perriman and Ares, 2000), but direct evidence that it facilitates U2 snRNP assembly is lacking.
Within the cell, formation of the 17S U2 particle from the core U2 snRNP (i.e. U2 snRNA and Sm proteins) appears to be a nuclear event. However, the subnuclear site(s) of 17S U2 snRNP assembly is presently not clear. After their assembly in the cytoplasm, core U2 snRNPs are transported to the nucleus where they interact with U2-specific proteins (for a review, see Will and Lührmann, 2001). Recent studies indicate that the newly assembled core snRNPs first accumulate in Cajal bodies (CBs) and subsequently in so-called nuclear speckles (Sleeman and Lamond, 1999a). The latter contain high concentrations of splicing factors and appear to serve as storage and/or assembly sites for spliceosomal components (Sleeman and Lamond, 1999b and references therein). In addition to being present in nuclear speckles, both U2 snRNA and U2-B′′, and thus presumably 12S U2 snRNPs, are enriched in CBs (Wu et al., 1991; Carmo-Fonseca et al., 1992; Matera and Ward, 1993). More recent studies demonstrated that SF3b155 accumulates in nuclear speckles, but not in CBs; SF3a66 was also detected in speckles, but whether it is enriched in CBs was not investigated (Schmidt-Zachmann et al., 1998). Thus, relatively little is currently known about the subnuclear distribution of the various U2 snRNP components, in particular those specifically found in the 17S particle.
The U2 snRNP plays an important role during the early stages of splicing complex formation and, ultimately the U2 snRNA, together with the U6 snRNA, is thought to form one of the active centers of the spliceosome (reviewed in Burge et al., 1999). In addition to its RNA moiety, U2 snRNP proteins also contribute to U2 function. SF3 proteins, for example, play essential roles during prespliceosome assembly, tethering the U2 snRNP to the pre-mRNA and contributing to branch site recognition (for a review, see Will and Lührmann, 1997). All known SF3a and SF3b subunits, with the exception of SF3b130 can be cross-linked to the pre-mRNA in the vicinity of the branch site (Gozani et al., 1996). While most SF3 subunits cross-link to a 20 nucleotide region upstream of the branch site, SF3b155 cross-links are found both 5′ and 3′ of the branch site, and the SF3b subunit, p14/SF3b14a, cross-links to the branch site adenosine, the nucleophile for the first transesterification step of splicing (Gozani et al., 1996; Will et al., 2001). Thus, due to their apparent close proximity to the branch adenosine, several SF3b components are likely to be core components of the spliceosome. The functional importance of SF3b is further underscored by the fact that it is also present in the U11/U12 snRNP, a subunit of the minor, U12-dependent spliceosome (Will et al., 1999).
To better understand the structure of the U2 snRNP and its function during splicing, we immunoaffinity-purified native HeLa 17S U2 snRNPs and the U2-associated complex SF3b, and determined their protein compositions using mass spectrometry. We identified several additional U2 proteins, including the DExH/D-box proteins hPrp43p and a homologue of yeast Prp5p (hPrp5p). In subsequent studies, we provide evidence that the latter protein functions in pre-mRNA splicing. Experimental observations additionally suggest that the DEAD-box protein SF3b125 may be involved in the assembly of the 17S U2 snRNP. Finally, we performed subcellular localization studies with antibodies against several 17S U2 proteins to learn more about the potential site of 17S U2 assembly within the nucleus.
Results
Identification of novel SF3b proteins
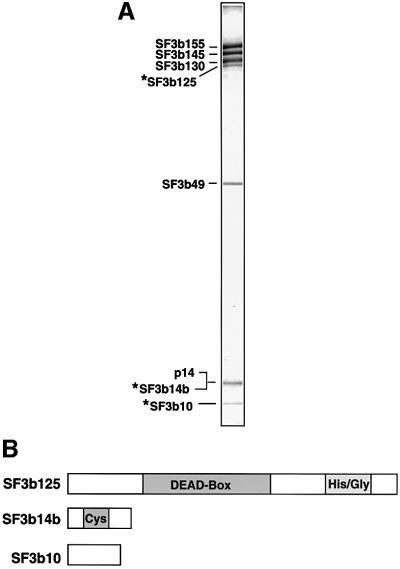
During our characterization of the branch site-interacting protein, p14, we described the isolation at 600 mM KCl of highly purified SF3b complexes using antibodies against SF3b155 (Will et al., 2001). Immunoblotting analyses of these complexes revealed the presence of a previously unknown SF3b subunit, namely p14/SF3b14a. To identify additional SF3b components, we have now analysed these complexes using matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS). In addition to all known SF3b proteins (i.e. SF3b155, SF3b145, SF3b130, SF3b49 and p14/SF3b14a), three novel SF3b components, with apparent molecular masses of 10, 14 and 125 kDa (which we designate SF3b10, SF3b14b and SF3b125, respectively), were identified (Figure 1A).
Fig. 1. Identification of novel SF3b proteins. (A) Protein composition of affinity-purified SF3b. Proteins were separated by SDS–PAGE on a 10/13% polyacrylamide gel and stained with Coomassie Blue. The identity of the bands, as determined by MS, is shown on the left. (B) Domain structure of SF3b10, SF3b14b and SF3b125. Abbreviations are as follows: DEAD-box, helicase domain characteristic of members of DExH/D box family; Cys, cysteine-rich; His/Gly, histidine/glycine-rich.
Multiple cDNAs encoding SF3b10 and SF3b14b were found in the human EST database. The putative proteins encoded by the longest SF3b10 and SF3b14b ESTs were 86 and 110 amino acids long, with a predicted molecular weight of 10.1 and 12.4 kDa, respectively. Significantly, the in vitro translation product of the identified cDNAs co-migrated with the corresponding endogenous protein, confirming that they both encode full-length proteins (data not shown). Apparent orthologues of both proteins were identified in a wide range of organisms. SF3b10 (NCBI accession No. NP_112577), which contains no motifs indicative of function (Figure 1B), is evolutionarily conserved, exhibiting 83 or 60% identity between humans and Drosophila melanogaster or Arabidopsis thaliana, respectively, or 50% between humans and Schizo saccharomyces pombe. SF3b14b is cysteine-rich, and in S.cerevisiae, belongs to a subfamily of zinc finger proteins termed zinc/binuclear cluster proteins exemplified by the transcriptional regulator Gal4p (Akache et al., 2001). The human protein (accession No. Q9UH06) exhibits 96 and 94% identity with its apparent orthologues from D.melanogaster and A.thaliana, respectively, and 55% identity with that of S.cerevisiae. This exceptionally high degree of conservation is consistent with the idea that SF3b14b, like other SF3b subunits, plays an important role in splicing. Indeed, gene deletion studies originally demonstrated that the S.cerevisiae SF3b14b homologue (YPR094W) is required for yeast viability (Winzeler et al., 1999); however, more recent studies indicate that YPR094W is not an essential gene (Akache et al., 2001).
SF3b125 is a member of the DExH/D-box family of RNA unwindases/RNPases
A database search using the peptide mass information obtained from the band designated SF3b125 identified a putative RNA helicase of unknown function (accession No. AAH15505), 819 amino acids long with a predicted molecular weight of 90 kDa. SF3b125 contains all of the sequence motifs (i.e. motifs I–VI) characteristic of members of the DEAD-box family of RNA unwindases/RNPases (data not shown). Database searches revealed potential orthologues of similar length in several organisms, including D.melanogaster, Caenorhabditis elegans and A.thaliana, but no obvious candidate homologue in S.cerevisiae. Homologous regions are found not only in their RNA helicase domains, but also in regions both directly upstream and downstream of this domain. However, as the N- and C-terminal-most domains of the human protein are distinct from those of the D.melanogaster, C.elegans and A.thaliana proteins, it is presently not clear whether these proteins are true orthologues.
Immunoaffinity purification of native 17S U2 snRNPs
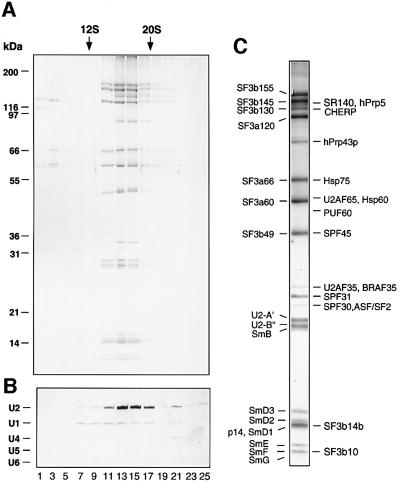
We next investigated whether the novel SF3b proteins are also present in U2 snRNPs. To obtain highly purified 17S U2 particles, we performed immunoaffinity chromatography with nuclear extract and anti-peptide antibodies against SF3a66. Bound snRNPs were eluted with an excess of SF3a66 peptide and the eluate subsequently fractionated on a glycerol gradient (Figure 2). Protein and RNA were isolated from gradient fractions and analysed by SDS–PAGE (Figure 2A) or denaturing PAGE (Figure 2B), respectively. As shown by the presence of U2 snRNA and several known subunits of SF3a/b, eluted U2 snRNPs peaked in the 17S region of the gradient (i.e. fractions 13–15). Significantly, these fractions contained almost exclusively U2 snRNPs, with only low levels of U1 snRNA detectable.
Fig. 2. Identification of novel U2-associated proteins. 17S U2 snRNPs were immunoaffinity purified from HeLa nuclear extract and subjected to 10–30% glycerol gradient centrifugation. Distribution of (A) protein and (B) snRNA across the gradient. Proteins were separated by SDS–PAGE and visualized as in Figure 1. RNA was fractionated by denaturing PAGE and stained with silver. The peak positions of 12S U1 and 20S U5 snRNPs, run in parallel, are indicated at the top. (C) Protein composition of the peak 17S U2 snRNP gradient fraction. Proteins were fractionated as in (A) and identified by MS.
Identification of 17S U2 snRNP proteins by mass spectrometry
Proteins in the peak 17S U2 gradient fraction were analysed by MALDI MS or additionally by liquid chromatography-coupled tandem mass spectrometry (LC-MSMS). Purified 17S U2 snRNPs contained all known U2 proteins, including all previously characterized SF3a/b subunits, U2-A′, U2-B″, SPF30/SMNrp and the Sm proteins (Figure 2C; Table I). In contrast, despite small amounts of U1 in the 17S U2 snRNP fractions, no U1 snRNP proteins were detected, demonstrating the relative purity of our preparation and underscoring the significance of those proteins identified. Fifteen additional proteins that previously eluded detection by biochemical and/or immunological methods were also identified (Behrens et al., 1993). These included SPF31, SPF45 and CHERP, proteins previously detected in a mixture of spliceosomal complexes (Neubauer et al., 1998), as well as SF3b10 and SF3b14, which co-migrated with SmF and SmD1, respectively. SF3b125, in contrast, was not detected, suggesting that it may dissociate upon integration of SF3b into the U2 snRNP. Surprisingly, nearly stoichiometric amounts of the human homologue of the yeast DEAH-box protein Prp43p, which plays a role in spliceosome disassembly in yeast (Arenas and Abelson, 1997), were also found. Sub stoichiometric amounts of the splicing factors SF2/ASF, PUF60, U2AF35 and U2AF65 (as shown by their staining intensity; Figure 2C) were detected. Likewise, low levels of the heat shock proteins Hsp75 and Hsp60, and BRAF35, a 35 kDa DNA-binding protein that interacts with the breast cancer-associated protein BRCA2 (Marmorstein et al., 2001) were also observed; the functional significance of their co-purification with U2 is presently not clear.
Table I. 17S U2 snRNP-associated proteins.
| Protein group | MW (apparent; kDa) | Accession No. | Features |
|---|---|---|---|
| Known proteins | |||
| SF3b155 | 160 | NP_036565 | HEAT repeats |
| SF3b145 | 150 | NP_006833 | SAP, Pro-rich, Glu-rich |
| SF3b130 | 130 | NP_036558 | CPSF A |
| SF3a120 | 120 | NP_005868 | Pro-rich, 2 SURP, Gln-rich, Glu-rich, UBQ |
| SF3a66 | 66 | NP_009096 | C2H2-zinc finger, Pro-rich |
| SF3a60 | 60 | NP_006793 | Glu-rich, C2H2-zinc finger, SAP |
| SF3b49 | 50 | NP_005841 | 2 RRMs, His-rich, Pro-rich |
| SPF30 | 31 | AAC64086 | Tudor domain |
| U2-A′ | 30 | P09661 | LRR |
| U2-B′′ | 29 | P08579 | 2 RRMs |
| SmB/B′ | 28 | NP_003082 | Sm |
| SmD3 | 17 | NP_004166 | Sm |
| SmD2 | 16 | NP_004588 | Sm |
| SmD1 | 15 | NP_008869 | Sm |
| p14 | 15 | AAK94041 | RRM |
| SmE | 10 | NP_003085 | Sm |
| SmF | 9 | Q15356 | Sm |
| SmG | 8 | NP_003087 | Sm |
| Novel proteins | |||
| hPrp5p | 140 | NP_055644 | SR-domain, Lys-rich, Glu-rich, DEAD-box |
| SR140 | 140 | BAA20790 | RRM, Pro-rich, SWAP, Glu-rich, SR-domain |
| CHERP | 130 | NP_006378 | SWAP, Gln-rich, Pro-rich, SR-domain, G-patch |
| hPrp43 | 90 | O43143 | Arg-rich, His-rich, DEAH-box |
| Hsp75 | 66 | NP_057376 | |
| PUF60 | 63 | NP_055096 | 3 RRMs |
| U2AF65 | 61 | NP_009210 | SR-domain, 3 RRMs |
| Hsp60 | 61 | P10809 | |
| SPF45 | 50 | NP_006441 | G-patch, RRM |
| U2AF35 | 35 | NP_006749 | C3H1-zinc finger, RRM, SR-domain, Gly-rich |
| BRAF35 | 35 | AAG01174 | HMG |
| SPF31 | 33 | NP_055095 | DnaJ domain |
| SF2/ASF | 31 | Q07955 | 2 RRMs, Gly-rich, SR-domain |
| SF3b14b | 15 | Q9UH06 | Cys-rich |
| SF3b10 | 9 | NP_112577 |
The human homologue of yeast Prp5p is present in 17S U2 snRNPs
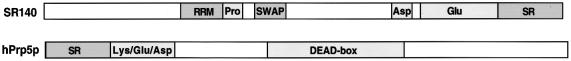
Database searches using peptide masses obtained from a band with an apparent molecular weight of 140 kDa revealed the presence of two proteins of unknown function. The first of these (which we designate SR140; accession No. BAA20790) contains a domain structure characteristic of classical SR proteins, exhibiting an N-terminal RNA recognition motif (RRM) and C-terminal domain rich in SR dipeptides (Figure 3). In addition, SR140 contains a so-called SWAP/SURP domain that is found in a number of pre-mRNA splicing factors (Denhez and Lafyatis, 1994). Potential SR140 orthologues with a similar domain structure were detected in D.melanogaster, C.elegans and A.thaliana and shared 61, 54 and 51% similarity, respectively, with the human protein.
Fig. 3. Domain structure of the newly identified U2 proteins, hPrp5p and SR140. Abbreviations are as follows: SR, domain rich in arginine-serine dipeptides; RRM, RNA recognition motif; SWAP, suppressor-of-white-apricot domain; Lys/Glu/Asp, lysine/glutamic acid/aspartic acid-rich; Pro, proline-rich.
A second protein in this band (accession No. NP_055644) was identified as a DEAD-box protein of unknown function. Interestingly, it contains not only all of the sequence motifs characteristic of RNA helicases, but also an N-terminal domain rich in SR dipeptides (Figure 3). The overall domain structure is thus reminiscent of several mammalian DExH/D-box proteins known to be involved in pre-mRNA splicing (Staley and Guthrie, 1998). A search for homologous proteins identified several potential orthologues in diverse organisms such as C.elegans, A.thaliana and S.pombe, whose sequence similarities with the human protein ranged from 52 to 62%. The two S.cerevisiae proteins exhibiting the highest degree of homology were Dbp2p (41% identity) and Prp5p (34% identity), a DEAD-box protein required for prespliceosome formation in yeast (Dalbadie-McFarland and Abelson, 1990; Ruby et al., 1993). Despite its lower homology score, several observations suggest that Prp5p is the yeast orthologue of this 17S U2 protein. The region of homology between the human protein and Dbp2p is limited to their helicase domains and Dbp2p shares much greater homology with the human p68 DEAD-box protein (55% identity). In contrast, Prp5p and the human 17S U2 protein also share regions of homology outside of their helicase domains, including blocks near their C-terminus (see Supplementary data available at The EMBO Journal Online for alignment) and, moreover, are more similar in length [1031 amino acids versus 849 (Prp5p) and 546 (Dbp2p)]. Although Prp5p lacks a classical SR domain, many charged residues and a few SR dipeptides are found in its N-terminal domain. Furthermore, Prp5p interacts genetically with several U2 components (Ruby et al., 1993; Wells and Ares, 1994) and was recently shown to physically associate with the yeast U2 snRNP (Abu Dayyeh et al., 2002). Significantly, we demonstrate that the human Prp5p homologue (hPrp5p) is associated with HeLa 17S U2 snRNPs and provide evidence that it, like Prp5p, is required for prespliceosome formation (see below). Based on these criteria, we propose that the newly identified 17S U2 snRNP protein is the human orthologue of Prp5p.
hPrp5p associates predominantly with U2 snRNPs
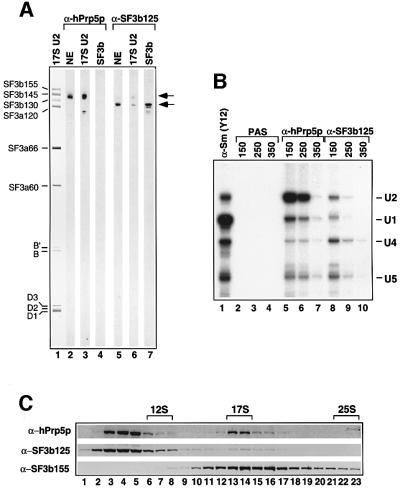
To further characterize hPrp5p and SF3b125, we raised antibodies against each of these proteins. Both anti-hPrp5p and anti-SF3b125 affinity-purified antibodies reacted specifically with a protein of the expected molecular weight in nuclear extract (Figure 4A, lanes 2 and 5). Consistent with our MS data, immunoblotting detected hPrp5p in 17S U2 snRNPs, but not in affinity-purified SF3b complexes (Figure 4A, lanes 3 and 4). Furthermore, anti-hPrp5p antibodies precipitated predominantly U2 snRNPs and only a very low level of U1, U4 and U5 from nuclear extract at moderate salt concentrations (150–250 mM NaCl; Figure 4B, compare lanes 2 and 3 with 5 and 6). U2 precipitation was nearly abolished at higher salt concentrations, indicating that hPrp5p dissociates from the U2 snRNP under these conditions (Figure 4B, lane 7). To determine whether hPrp5p is predominantly free or snRNP-associated at 150 mM salt, nuclear extract was fractionated on a 10–30% glycerol gradient and the migration behaviour of hPrp5, as well as SF3b155 and SF3b125, determined by immunoblotting. The majority of hPrp5p was detected in fractions 3–5, with a second peak observed in fractions 13 and 14 (Figure 4C, upper panel). The latter correspond to the ∼17S region of the gradient as shown by the distribution of the spliceosomal snRNAs (data not shown); the vast majority of SF3b155 also peaked in this region (lower panel). Thus, ∼15% of hPrp5p in nuclear extract is U2-associated.
Fig. 4. hPrp5p, but not SF3b125, is associated with 17S U2 snRNPs. (A) Proteins from nuclear extract (lanes 2 and 5), purified 17S U2 snRNPs (lanes 1, 3 and 6), or purified SF3b (lanes 4 and 7) were stained with Ponceau S (lane 1), affinity-purified anti-hPrp5p (lanes 2–4) or anti-SF3b125 (lanes 5–7) antibodies. The identities of the major 17S U2 proteins are shown on the left, and the positions of hPrp5p and SF3b125 are indicated on the right. (B) Immuno precipitations were performed with nuclear extract at 150–350 mM NaCl (as indicated) and PAS alone (lanes 2–4) or PAS-bound with anti-hPrp5p (lanes 5–7), anti-SF3b125 (lanes 8–10) or anti-Sm protein (Y12) antibodies (lane 1). Precipitated RNAs were labelled with [32P]pCp and analysed by denaturing PAGE. The identity of the snRNAs is indicated on the right. (C) Nuclear extract was fractionated on a 10–30% glycerol gradient, and proteins were subsequently separated by SDS–PAGE, blotted to nitrocellulose and probed with antibodies against hPrp5p (top), SF3b125 (middle) or SF3b155 (bottom). The 12S U1, 17S U2 and 25S U4/U6.U5 snRNP peaks are indicated at the top.
Immunoblotting clearly detected SF3b125 in purified SF3b complexes, but only a very low level in 17S U2 snRNPs (Figure 4A, compare lanes 6 and 7). Note that the blot strips contain approximately twice as many moles of 17S U2 snRNP as SF3b. Anti-SF3b125 antibodies precipitated only low levels of U1, U2, U4 and U5 snRNPs from nuclear extract at low salt concentrations (Figure 4B, lanes 8–10). Consistent with an association with U2 snRNPs, a very small fraction of SF3b125 was found in the 17S region of the glycerol gradient (Figure 4C, middle panel). However, the majority of SF3b125 migrated at the top of the gradient, presumably as free protein or as part of small protein complexes.
hPrp5p and SF3b125 are present in CBs and nuclear speckles
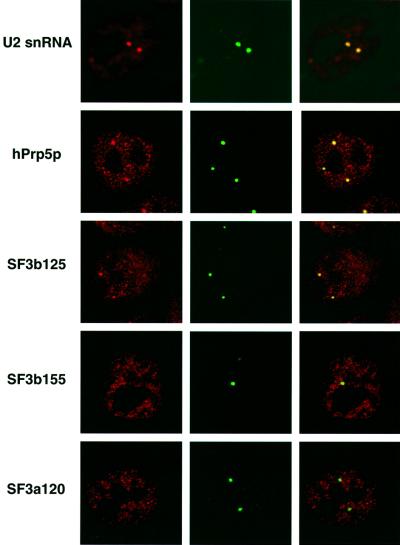
To learn more about the subcellular distribution of components of the 17S U2 snRNP, and potentially gain insight into the subnuclear site of 17S U2 assembly, we performed immunofluoresence studies with HeLa cells and affinity-purified antibodies directed against hPrp5p, SF3b125, SF3b155 or SF3a120, or FISH with a fluorescently-labelled oligonucleotide complementary to the U2 snRNA (Figure 5). Cells were counterstained with anti-coilin antibodies to mark the site of CBs. Nucleoplasmic staining with several intensely stained foci amid diffuse, less intense speckles was observed with the anti-U2 oligonucleotide (Figure 5, left panel). Consistent with previous observations (Carmo-Fonseca et al., 1992; Matera and Ward, 1993), these foci co-localized with structures stained by anti-coilin antibodies, as shown by their yellow colour in the confocal overlay (right panel), demonstrating that U2 snRNA is highly enriched in CBs. Each of the affinity-purified antibodies yielded a nucleoplasmic, speckled staining pattern, characteristic of splicing factors (left panels). In addition, hPrp5p and SF3b125, but not SF3b155 or SF3a120, were enriched in foci that co-localized with CBs (right panels). Thus, the subcellular distribution of hPrp5p and SF3b125 is consistent with their designation as splicing factors. In addition, the apparent low levels of SF3b155 and SF3a120 in CBs suggests that 17S U2 snRNPs do not accumulate in CBs and possibly assemble elsewhere in the nucleus.
Fig. 5. Subcellular localization of hPrp5p and SF3b125, and other U2 components, in HeLa cells. Fluorescence microscopy was performed with HeLa cells stained with anti-coilin antibodies and affinity-purified antibodies against either SF3b155, SF3a120, SF3b125 or hPrp5p, or a fluorescently-labelled oligonucleotide complementary to the U2 snRNA, as indicated on the left. Left panels show the staining patterns of the U2 components alone, middle panels anti-coilin staining, and right panels show the confocal overlay of both.
hPrp5p plays a role in splicing prior to, or during, A complex formation
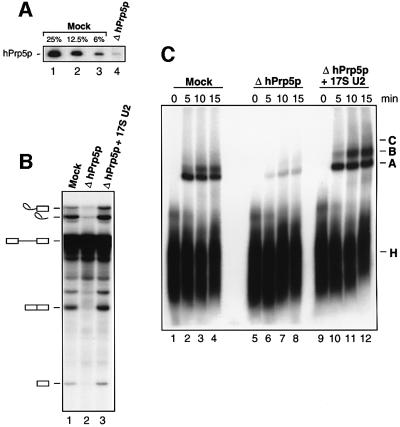
To determine whether hPrp5p and SF3b125 function in splicing, we attempted to deplete nuclear extract of these proteins with anti-hPrp5p or anti-SF3b125 antibodies. Immunodepletion was performed at high salt concentrations (600 mM) in order to prevent the co-depletion of U2 snRNPs and other splicing factors. Under these conditions, 17S U2 snRNPs dissociate, but normally reassemble during the subsequent dialysis step. While attempts to remove the majority of SF3b125 were unsuccessful, >97% of hPrp5p, as shown by immunoblotting, could be depleted (Figure 6A). Importantly, no detectable decrease in U2 snRNA (or any other spliceosomal snRNA) was observed in the hPrp5p-depleted (ΔhPrp5p) extract (data not shown). In vitro splicing was subsequently performed with a 32P-labelled adenovirus pre-mRNA substrate. Splicing activity was significantly reduced in the ΔhPrp5p versus mock-depleted extract, but could be restored by addition of very small amounts (∼5% of the endogenous U2) of immunoaffinity-purified 17S U2 snRNP (Figure 6B, lanes 1–3). To determine at what step of splicing hPrp5p acts, we analysed splicing complex formation by native PAGE. Relative to the mock-depleted extract, a clear decrease in the formation of A, B and C complexes was observed with the ΔhPrp5p extract (Figure 6C, compare lanes 1–4 with 5–8); complex formation was fully restored by the addition of 17S U2 snRNP (lanes 9–12). Thus, like its yeast counterpart, hPrp5p appears to function in splicing either prior to or during A complex formation.
Fig. 6. Immunodepletion of hPrp5p from HeLa nuclear extract leads to inhibition of both splicing and splicing complex formation. (A) Immunoblot of mock-depleted extract (lanes 1–3) or hPrp5p- depleted extract (lane 4) probed with affinity-purified anti-hPrp5p antibodies. For quantitative purposes, 25, 12.5 and 6% of the mock- depleted extract versus 100% of the depleted extract are compared. (B) In vitro splicing of 32P-labelled MINX pre-mRNA in mock- depleted (lane 1), hPrp5p-depleted (lane 2) or hPrp5p-depleted nuclear extract complemented with 17S U2 snRNPs (lane 3). RNA was analysed on a 7 M urea–14% polyacrylamide gel and visualized by autoradiography. The positions of the pre-mRNA and splicing intermediates/products are indicated on the left. (C) Spliceosome assembly in mock-depleted (lanes 1–4), hPrp5p-depleted (lanes 5–8) and hPrp5p-depleted nuclear extract complemented with 17S U2 snRNPs (lanes 9–12) was analysed on a native gel at the indicated times. The positions of the H, A, B and C complexes are indicated.
To determine whether 17S U2 assembly was blocked in ΔhPrp5p extracts, mock and depleted extracts were fractionated on glycerol gradients and the distribution of the U2 snRNA analysed by denaturing PAGE. No significant difference in the level of 17S U2 formed in mock versus ΔhPrp5p extract was observed (data not shown), suggesting that, during the dialysis step following immunodepletion, most 17S U2 proteins reassociate efficiently even in the absence of hPrp5p.
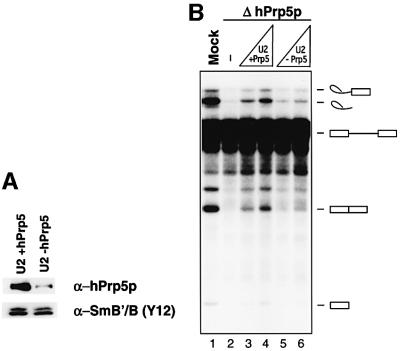
As it was not possible to isolate recombinant or purified hPrp5p, add-back experiments with hPrp5p alone could not be performed. However, to provide additional evidence that the block in splicing was due solely to the absence of hPrp5p, we incubated immunoaffinity-purified 17S U2 snRNPs with PAS alone (mock) or PAS containing anti-hPrp5p antibodies in order to remove the subpopulation (maximally 15–20%) of 17S U2 snRNPs containing hPrp5p. Indeed, immunoblotting of mock-depleted and hPrp5p-depleted 17S U2 snRNPs revealed an ∼10-fold reduction in hPrp5p levels in the latter particles (upper panel); in contrast similar levels of Sm B/B′, a core U2 component, were detected (lower panel). Importantly, a loss of other 17S U2 snRNP components was not observed as analysed by SDS–PAGE (data not shown). Sig nificantly, splicing activity was more efficiently restored to the ΔhPrp5p extract by the mock-depleted versus hPrp5p-depleted 17S U2 snRNPs (Figure 7B, compare lanes 3 and 4 with 5 and 6). These results thus additionally support the conclusion that hPrp5p plays an essential role in splicing.
Fig. 7. Splicing in hPrp5p-depleted extracts is less efficiently restored by 17S U2 snRNPs with reduced hPrp5p levels. (A) Immunoblots of mock-depleted (+hPrp5, left lane) or hPrp5p-depleted 17S U2 snRNPs (–hPrp5, right lane) probed with anti-hPrp5p or anti-Sm (Y12) antibodies. (B) Splicing in mock-depleted (lane 1), hPrp5p-depleted (lane 2) or hPrp5p-depleted nuclear extract complemented with 32.5 or 65 fmol of 17S U2 snRNPs with wild type (lanes 3–4) or reduced levels of hPrp5p (lanes 5–6). RNA was analysed as described in Figure 6.
Discussion
The human 17S U2 snRNP is more complex than previously thought
Immunoaffinity chromatography has allowed the preparative isolation of highly purified, native 17S U2 snRNPs and SF3b complexes, which are ideally suited for both biochemical and functional studies. Subsequent MS analyses resulted in the identification of several additional U2-associated polypeptides, including the previously uncharacterized 35 and 92 kDa proteins, which likely correspond to SPF31 and hPrp43, respectively (Figure 2). Aside from the advantage in identifying proteins afforded by MS, this increase in 17S U2 complexity may, in part, be due to the new isolation procedure which, while highly specific, is less harsh than earlier methods employed (Behrens et al., 1993). Alternatively, many of the newly identified proteins may have previously escaped detection either due to their co-migration on SDS– polyacrylamide gels with other U2 snRNP proteins (e.g. CHERP, SPF45, SF3b14 and SF3b10) or potentially due to their relatively low abundance. Whether 17S U2 proteins that are present in substoichiometric amounts were partially lost during the isolation procedure or are normally present in only a subpopulation of U2 snRNPs is presently not clear.
The presence of several of the newly identified proteins in U2 snRNPs is not entirely unexpected. Both subunits of U2AF and the U2AF65-related protein PUF60 facilitate the stable association of U2 with the pre-mRNA during A complex formation (Zamore and Green, 1989; Page-McCaw et al., 1999). Although these proteins can bind the pre-mRNA independently of U2, they nonetheless likely possess affinity for one or more components of the U2 snRNP. Indeed, both U2AF65 and U2AF35 bind directly SF3b155, a core component of the 17S U2 snRNP, in vitro (Gozani et al., 1998). The association of hPrp5p with U2 snRNPs is consistent with studies in S.cerevisiae demonstrating (i) both genetic and physical interactions between Prp5p and the U2 snRNP (Ruby et al., 1993; Wells and Ares, 1994; Abu Dayyeh et al., 2002) and (ii) a link between Prp5p and a structural rearrangement in the U2 snRNP (O’Day et al., 1996). The presence of SF3b14b and SF3b10 in U2 is consistent with the fact that all previously characterized SF3b subunits are also found in 17S U2 snRNPs. Finally, SPF31, SPF45 and CHERP were previously found in spliceosomal complexes (Neubauer et al., 1998), and thus their association with a spliceosomal snRNP is not surprising. In addition, SPF45 has recently been shown to mediate 3′ splice site selection during the alternative splicing of the Drosophila sex-lethal pre-mRNA, a result consistent with its association with U2 snRNPs (Lallena et al., 2002).
A role for hPrp43 at an early stage of splicing?
The identification of the DEAH-box protein hPrp43 in 17S U2 snRNPs, on the other hand, was somewhat surprising. In yeast, Prp43p facilitates release of excised intron product from post-splicing complexes and thus acts at a late stage of the splicing process (Arenas and Abelson, 1997). The presence of hPrp43 in U2 snRNPs suggests that, at least in mammals, this putative RNA unwindase/RNPase potentially functions at a very early stage of spliceosome assembly or even facilitates 17S U2 formation. Alternatively, hPrp43 may associate with the pre-mRNA as part of the 17S U2 snRNP, but remain inactive until after splicing catalysis, when it is first activated by an unknown mechanism. Its association with U2 snRNPs further suggests that one or more U2 components may be the target upon which hPrp43 ultimately acts or may be responsible for its activation either during post-splicing complex formation or potentially at an earlier step of splicing. It will thus be particularly interesting to determine hPrp43’s interaction partner(s) within the 17S U2 snRNP.
hPrp5p potentially acts prior to, or during, prespliceosome assembly
We have identified a human homologue of the S.cerevisiae, DEAD-box protein Prp5p, and shown that it is an integral component of the 17S U2 snRNP. In S.cerevisiae, Prp5p is required for prespliceosome formation and appears to catalyse a conformational change in the yeast U2 particle that leads to exposure of its branch site-interacting region (Ruby et al., 1993; O’Day et al., 1996). Immunodepletion studies suggested that hPrp5p is also required for prespliceosome formation in higher eukaryotes and, thus, that it is a is true orthologue of the yeast protein; a clear reduction in A complex formation was observed in nuclear extracts where the vast majority of hPrp5p had been removed (Figure 6). In contrast, subsequent to hPrp5p depletion, 17S U2 snRNP assembly appeared, for the most part, to be uneffected (data not shown). However, the method employed to assay 17S U2 formation (i.e. gradient centrifugation) would not reveal minor structural changes in the U2 snRNP or the absence of a only few proteins, and thus it is possible that the 17S U2 snRNPs formed in ΔhPrp5p extracts are functionally compromised. Unfortunately, attempts to overexpress hPrp5p in Escherichia coli or to isolate endogenous hPrp5p via immunoaffinity techniques were not successful and thus complementation studies with hPrp5p alone could not be performed. However, add-back experiments with purified 17S U2 snRNPs support the conclusion that hPrp5p alone (and not a co-depleted splicing factor) is responsible for the block in A complex formation in ΔhPrp5p extracts. That is, very low amounts of 17S U2 snRNPs fully restored splicing and complex formation to ΔhPrp5p extracts, consistent with idea that an enzymatic activity was lacking. Furthermore, 17S U2 snRNPs with reduced levels of hPrp5p, but no apparent loss of other U2 proteins (at least as judged by SDS–PAGE analysis), were less active in restoring splicing (Figure 7).
Interestingly, only substoichiometric amounts of hPrp5p were found in purified 17S U2 snRNPs (Figure 2). Assuming that large amounts have not dissociated during purification, hPrp5p may normally be associated with only a subpopulation of U2. Thus, it may interact in a dynamic manner, associating at some point during 17S U2 assembly and potentially dissociating, for example, after catalysing a conformational change in the U2 particle or upon U2 interaction with the pre-mRNA. Nonetheless, our data demonstrate that hPrp5p physically associates with one or more components of the U2 snRNP. As the interaction partners of hPrp5p are potential targets or activators of this enzyme, their identification will be of prime importance to ultimately understand how hPrp5p acts at the molecular level. Interestingly, hPrp5p contains an N-terminal SR domain that is lacking in S.cerevisiae Prp5p and its binding behaviour may thus differ from that of the yeast protein, with other SR proteins in particular serving as potential interaction partners. hPrp5p may also be involved in functionally important intermolecular interactions within the 17S U2 and/or prespliceosome that are not related to its enzymatic activity. Indeed, studies in yeast have shown a requirement for Prp5p even in the absence of ATP (Perriman and Ares, 2000).
A role for SF3b125 in 17S U2 assembly?
Three novel proteins were identified in purified SF3b, two of which, SF3b10 and SF3b14b, were also found in the 17S U2 snRNP. Considering the established functions of known SF3b subunits (see Introduction), it is likely that SF3b10 and SF3b14b also facilitate the interaction of U2 with the branch site. Indeed, both proteins are highly conserved across eukaryotes, consistent with the idea that they play an essential role in splicing. Intriguingly, a third novel protein, the DEAD-box protein SF3b125, was present substoichiometrically in SF3b, but nearly absent from 17S U2 snRNPs. In this respect, SF3b125 appears to be the only SF3b component that is largely lost upon 17S U2 snRNP formation. As the major portion of SF3b that we isolate is generated by dissociation of 17S U2 snRNPs at high salt concentrations, it is not surprising that only a subpopulation of purified SF3b complexes contain this putative helicase/RNPase. Our results suggest that SF3b125 dissociates either at the time of SF3b association with U2 or at a subsequent step of 17S U2 assembly. Thus, it could conceivably act at this time as an RNPase, facilitating association of SF3b by catalysing a conformational change in the 12S U2 particle. Alternatively, it could act after SF3b has bound and enable SF3a or other proteins to interact with the 15S U2 snRNP. As with hPrp5p, the future identification of interaction partners of SF3b125 may help shed light on its presumed function during U2 biogenesis.
Insight into the assembly of the 17S U2 snRNP within the nucleus
Cellular localization studies (Figure 5) revealed a differential subnuclear distribution of the U2 snRNA versus core 17S U2 proteins (e.g. SF3b155 and SF3a120). Consistent with previous observations, FISH demonstrated that the U2 snRNA is highly enriched in CBs. As U2-B′′ is also known to be enriched in CBs (Wu et al., 1991; Carmo-Fonseca et al., 1992) large amounts of 12S U2 snRNPs appear to be present in these subnuclear structures. Consistent with previous studies (Schmidt-Zachmann et al., 1998), SF3b155 and SF3a120, on the other hand, were detected almost exclusively outside of CBs, either in nuclear speckles or in the nucleoplasm. Immunoblotting studies showed that the majority of SF3b155 in nuclear extract is present in 17S U2 snRNPs or higher molecular weight complexes (Figure 4C). Therefore, the observed subnuclear distribution of SF3b155, and presumably also SF3a120, most likely reflects that of assembled 17S U2 snRNPs, as opposed to free SF3b155 or SF3b complexes. Our results thus suggest that little or no 17S U2 accumulates in CBs. The simplest interpretation of these results is that 17S U2 snRNP assembly does not occur in CBs. However, it is also conceivable that the association of SF3a and SF3b with the 12S U2 snRNP occurs in CBs, but that the resultant 17S U2 particles rapidly move to other subnuclear sites. Additional studies, e.g. with fluorescently labelled SF3b or SF3a complexes, which track the movement of 12S versus 17S U2 components are needed to clarify this point.
The subcellular distribution of both hPrp5p and SF3b125 is consistent with their designation as snRNP/splicing factors; antibodies against either protein show a predominantly nucleoplasmic staining with a characteristic speckled pattern (Figure 5). However, in contrast to SF3b155 and SF3a120, an enrichment of both DEAD-box proteins in CBs was observed. Thus, CBs appear to contain an excess of hPrp5p and SF3b125 over 17S U2 snRNPs. Consistent with this, a major portion of both hPrp5p and SF3b125 is not associated with snRNPs in nuclear extract (Figure 4). A number of open questions concerning the transport of these factors to and within the nucleus remain. For example, where does SF3b125 first associate with SF3b in the cell and at what site does it dissociate from SF3b/17S U2 snRNPs? Based on the apparent excess of this putative helicase over other SF3b components in CBs, it is tempting to speculate that SF3b125 may dissociate within these subnuclear structures concomitant with the formation of 17S U2 snRNPs. However, answers to these and many other questions await further experimentation.
Future studies
The data presented here provide insight into the complexity of the 17S U2 snRNP and reveal an important role for hPrp5p in splicing. Additional experiments are clearly required to establish whether any of the other newly identified U2 proteins contribute to U2 snRNP biogenesis and/or function. The availability of highly purified, native 17S U2 snRNPs should allow the future investigation of the U2/pre-mRNA branch site interaction in a purely in vitro system. Such studies may reveal in more detail the role of U2 proteins (and other spliceosomal proteins) in this functionally important interaction. In addition, immunoaffinity-purified SF3b and 17S U2 particles are well suited for ultrastructural investigation, for example via electron microscopy (EM). The more complete inventory of components of both SF3b and the 17S U2 snRNP presented here should also aid in the elucidation of their molecular architecture by EM.
Materials and methods
Immunological techniques
To generate antibodies against SF3a66, hPrp5p and SF3b125, rabbits were immunized with the ovalbumin-coupled peptides CMLRPPLPSEGPGNIP, CLQNSYQPTNKGRYKVL and MAEVEDQAARDMKRLEC, respectively. Anti-SF3a66 antibodies reacted specifically with SF3a66 on immunoblots (data not shown). Antibodies were affinity purified on a SulfoLink column (Pierce) containing the respective peptide. For immunoaffinity purification/immunodepletion, affinity-purified antibodies were covalently coupled to protein A–Sepharose beads with dimethylpimelimidate. For immunoblotting, proteins from nuclear extract, affinity-purified 17S U2 snRNPs/SF3b, or nuclear extract separated on a 10–30% glycerol gradient, as described below for 17S U2 snRNPs, were fractionated by SDS–PAGE, transferred electrophoretically to nitrocellulose and immunostained using an ECL-Detection Kit (Amersham). Immunoprecipitations were performed with HeLa nuclear extract (Dignam et al., 1983) and precipitated RNAs were detected by 3′ end-labelling with [32P]pCp (Will et al., 2001).
Purification of SF3b and 17S U2 snRNPs
SF3b was immunoaffinity purified from nuclear extract and separated on a 15–35% glycerol gradient (Will et al., 2001). To isolate 17S U2 snRNPs, nuclear extract was dialysed against G150 buffer (20 mM HEPES pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 5% glycerol, 0.5 mM DTE, 0.5 mM PMSF) for 4 h, diluted 1:1 with G150 buffer and loaded onto an anti-SF3a66 column. The column was washed with 50 vols of G150 buffer and complexes were eluted by incubating three times for 30 min with 0.1 M SF3a66 peptide in G150 buffer. Eluted snRNPs were fractionated on a 10–30% glycerol gradient containing G150 buffer, by centrifuging at 75 000 g for 17 h at 4°C. Gradients were fractionated manually from top to bottom into 150 µl aliquots. RNA and protein were recovered and analysed on 7 M urea–10% polyacrylamide gels (RNA) or by 10/13% SDS–PAGE (proteins).
Mass spectrometry
Proteins separated by SDS–PAGE were excised from the gel and prepared as described at http://www.narrador.embl-heidelberg.de. Peptide mass fingerprinting was performed under standard conditions with a Bruker-Reflex I (Bruker Daltronics, Bremen, Germany). Proteins were identified in the NCBI non-redundant (nr) database using Mascot® (Matrix Science, London, UK) as a search engine. For LC-MSMS analysis, samples were extracted (see above website), dissolved in 10% formic acid and subjected to MS analysis using a Q-TOF (Micromass, Manchester, UK) coupled to an HPLC system. The fragment ion series generated were used to search the NCBI nr database as described above.
Immunofluorescence microscopy
HeLa cells were immunostained with affinity-purified rabbit antibodies against SF3b155 (Will et al., 2001), SF3a120 (Will et al., 2001), hPrp5p or SF3b125, and mouse anti-coilin antibodies, and detected with a mixture of Texas Red-conjugated anti-rabbit and Alexa 488-conjugated anti-mouse secondary antibodies as described by Fabrizio et al. (1997). FISH was performed with a Cy3-labelled oligonucleotide complementary to nucleotides 4–44 of the human U2 snRNA as described previously (Taneja et al., 1992), using 15% formamide for hybridization and washing. Fluoresence microscopy was carried out with a Leica SP2 confocal laser scanning microscope.
Immunodepletion and in vitro splicing
For immunodepletion, nuclear extract in D buffer (Dignam et al., 1983) was first adjusted to 600 mM KCl. Two hundred microlitres of PAS (for mock-depletion) or PAS coupled with affinity-purified anti-hPrp5p antibodies were incubated for 1.5 h at 4°C with 1 ml of blocking buffer (phosphate-buffered saline pH 8.0, containing 0.1 µg/µl tRNA, 0.5 µg/µl bovine serum albumin and 0.1 µg/µl glycogen). After washing three times with D buffer containing 600 mM KCl, 100 µl of PAS (± antibody) was added twice to 200 µl of extract and incubated for 2 h at 4°C with end-over-end rotation each time. The extract was dialysed for 5 h at 4°C against D buffer containing 60 mM KCl. For complementation, ∼65 fmol of immunoaffinity-purified 17S U2 snRNP (equivalent to 5% of the U2 present in the extract used for splicing) were added prior to initiation of the splicing reaction. 17S U2 ± hPrp5p was generated by incubating 150 µl immunoaffinity-purified 17S U2 snRNPs in G150 buffer (∼6.5 pmol) with 100 µl of pre-blocked PAS (± anti-hPrp5p antibodies) for 3 h at 4°C with end-over-end rotation. In vitro splicing was carried out with 32P-labelled MINX pre-mRNA according to Will et al. (2001). Splicing complexes were analysed on gels containing 3.5% acrylamide and 0.5% agarose (Behrens et al., 1993).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank G.Heyne and M.Raabe for excellent technical assistance, and P.Kempkes, T.Conrad, B.Hildebrandt and H.Kohansel for preparing nuclear extract/UsnRNPs. We are grateful to C.Schneider for help with immunoprecipitations, D.Ingelfinger for initial cellular localization studies with YFP-labelled SF3b125, N.Watkins and P.Fabrizio for helpful comments on the manuscript, and the RZPD Deutsches Ressourcenzentrum für Genomforschung for providing EST clones. This work was supported by grants from the Deutsche Forschungs gemeinschaft (SFB 523/A8) and the BMBF (031U215B) to R.L., and a grant from the BMBF (031U215A) to M.W.
References
- Abu Dayyeh B.K, Quan,T.K., Castro,M. and Ruby,S.W. (2002) Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J. Biol. Chem., 277, 20221–20233. [DOI] [PubMed] [Google Scholar]
- Akache B., Wu,K. and Turcotte,B. (2001) Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res., 29, 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J.E. and Abelson,J.N. (1997) Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl Acad. Sci. USA, 94, 11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S.E., Tyc,K., Kastner,B., Reichelt,J. and Lührmann,R. (1993) Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell. Biol., 13, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R., Groning,K., Behrens,S.E., Lührmann,R. and Krämer,A. (1993a) Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science, 262, 102–105. [DOI] [PubMed] [Google Scholar]
- Brosi R., Hauri,H.P. and Krämer,A. (1993b) Separation of splicing factor SF3 into two components and purification of SF3a activity. J. Biol. Chem., 268, 17640–17646. [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Carmo-Fonseca M., Pepperkok,R., Carvalho,M.T. and Lamond,A.I. (1992) Transcription-dependent colocalization of the U1, U2, U4/U6 and U5 snRNPs in coiled bodies. J. Cell Biol., 117, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbadie-McFarland G. and Abelson,J. (1990) PRP5: a helicase-like protein required for mRNA splicing in yeast. Proc. Natl Acad. Sci. USA, 87, 4236–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B.K., Xia,L., Palandjian,L., Gozani,O., Chyung,Y. and Reed,R. (1999) Characterisation of a protein complex containing spliceosomal proteins SAPs 49, 130, 145 and 155. Mol. Cell. Biol., 19, 6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., Zhou,Z. and Reed,R. (2000) Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell, 5, 779–787. [DOI] [PubMed] [Google Scholar]
- Denhez F. and Lafyatis,R. (1994) Conservation of regulated alternative splicing and identification of functional domains in vertebrate homologs to the Drosophila splicing regulator, suppressor-of-white-apricot. J. Biol. Chem., 269, 16170–16179. [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Laggerbauer,B., Lauber,J., Lane,W.S. and Lührmann,R. (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J., 16, 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Feld,R. and Reed,R. (1996) Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev., 10, 233–243. [DOI] [PubMed] [Google Scholar]
- Gozani O., Potashkin,J. and Reed,R. (1998) A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol., 18, 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer A., Gruter,P., Groning,K. and Kastner,B. (1999) Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol., 145, 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena M.J., Chalmers,K.J., Llamazares,S., Lamond,A.I. and Valcárcel,J. (2002) Splicing regulation at the second catalytic step by sex-lethal involves 3′ splice site recognition by SPF45. Cell, 109, 285–296. [DOI] [PubMed] [Google Scholar]
- Marmorstein L.Y., Kinev,A.V., Chan,G.K., Bochar,D.A., Beniya,H., Epstein,J.A., Yen,T.J. and Shiekhattar,R. (2001) A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell, 104, 247–257. [DOI] [PubMed] [Google Scholar]
- Matera A.G. and Ward,D.C. (1993) Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol., 121, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Hannus,S., Plottner,O., Baars,T., Hartmann,E., Fakan,S., Laggerbauer,B. and Fischer,U. (2001) SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J., 20, 2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterisation of the multi-protein spliceosome complex. Nat. Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- O’Day C.L., Dalbadie-McFarland,G. and Abelson,J. (1996) The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J. Biol. Chem., 271, 33261–33267. [DOI] [PubMed] [Google Scholar]
- Page-McCaw P.S., Amonlirdviman,K. and Sharp,P.A. (1999) PUF60: a novel U2AF65-related splicing activity. RNA, 5, 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R. and Ares,M.,Jr (2000) ATP can be dispensable for prespliceosome formation in yeast. Genes Dev., 14, 97–107. [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ajuh,P., Lamond,A.I. and Mann,M. (2001) SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J. Biol. Chem., 276, 31142–31150. [DOI] [PubMed] [Google Scholar]
- Reed R. and Palandjian,L. (1997) Spliceosome assembly. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. IRL Press, Oxford, UK, pp. 103–129.
- Ruby S.W., Chang,T.H. and Abelson,J. (1993) Four yeast spliceosomal proteins (PRP5, PRP9, PRP11 and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev., 7, 1909–1925. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann M.S., Knecht,S. and Krämer,A. (1998) Molecular characterisation of a novel, widespread nuclear protein that colocalizes with spliceosome components. Mol. Biol. Cell, 9, 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999a) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999b) Nuclear organization of pre-mRNA splicing factors. Curr. Opin. Cell Biol., 11, 372–377. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Taneja K.L., Lifshitz,L.M., Fay,F.S. and Singer,R.H. (1992) Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J. Cell Biol., 119, 1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E. and Ares,M.,Jr (1994) Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 6337–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- Will C.L., Schneider,C., Reed,R. and Lührmann,R. (1999) Identification of both shared and distinct proteins in the major and minor spliceosomes. Science, 284, 2003–2005. [DOI] [PubMed] [Google Scholar]
- Will C.L., Schneider,C., MacMillan,A.M., Katopodis,N.F., Neubauer,G., Wilm,M., Lührmann,R. and Query,C.C. (2001) A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J., 20, 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterisation of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Wu Z.A., Murphy,C., Callan,H.G. and Gall,J.G. (1991) Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres and snurposomes. J. Cell Biol., 113, 465–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Perriman,R., Igel,H., Howe,K.J., Neville,M. and Ares,M.,Jr (1998) CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol., 18, 5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D. and Green,M.R. (1989) Identification, purification and biochemical characterisation of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl Acad. Sci. USA, 86, 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]