Nonmuscle Myosin Heavy Chain IIA Mutations Define a Spectrum of Autosomal Dominant Macrothrombocytopenias: May-Hegglin Anomaly and Fechtner, Sebastian, Epstein, and Alport-Like Syndromes (original) (raw)
Abstract
May-Hegglin anomaly (MHA) and Fechtner (FTNS) and Sebastian (SBS) syndromes are autosomal dominant platelet disorders that share macrothrombocytopenia and characteristic leukocyte inclusions. FTNS has the additional clinical features of nephritis, deafness, and cataracts. Previously, mutations in the nonmuscle myosin heavy chain 9 gene (MYH9), which encodes nonmuscle myosin heavy chain IIA (MYHIIA), were identified in all three disorders. The spectrum of mutations and the genotype-phenotype and structure-function relationships in a large cohort of affected individuals (_n_=27) has now been examined. Moreover, it is demonstrated that MYH9 mutations also result in two other FTNS-like macrothrombocytopenia syndromes: Epstein syndrome (EPS) and Alport syndrome with macrothrombocytopenia (APSM). In all five disorders, MYH9 mutations were identified in 20/27 (74%) affected individuals. Four mutations, R702C, D1424N, E1841K, and R1933X, were most frequent. R702C and R702H mutations were only associated with FTNS, EPS, or APSM, thus defining a region of MYHIIA critical in the combined pathogenesis of macrothrombocytopenia, nephritis, and deafness. The E1841K, D1424N, and R1933X coiled-coil domain mutations were common to both MHA and FTNS. Haplotype analysis using three novel microsatellite markers revealed that three E1841K carriers—one with MHA and two with FTNS—shared a common haplotype around the MYH9 gene, suggesting a common ancestor. The two new globular-head mutations, K371N and R702H, as well as the recently identified MYH9 mutation, R705H, which results in DFNA17, were modeled on the basis of X-ray crystallographic data. Altogether, our data suggest that MHA, SBS, FTNS, EPS, and APSM comprise a phenotypic spectrum of disorders, all caused by MYH9 mutations. On the basis of our genetic analyses, the name “MYHIIA syndrome” is proposed to encompass all of these disorders.
Introduction
Inherited giant-platelet disorders represent a group of rare disorders characterized by thrombocytopenia, large platelets, and variable bleeding symptoms (Mhawech and Saleem 2000). Given lack of reporting and misdiagnosis, the real prevalences of these disorders are most likely underestimated. The autosomal dominant disorders May-Hegglin anomaly (MHA [MIM 155100]; May 1909; Hegglin 1945), Fechtner syndrome (FTNS [MIM 153640]; Peterson et al. 1985), and Sebastian syndrome (SBS [MIM 605249]; Greinacher et al. 1990_b_) share the triad of thrombocytopenia, large platelets, and characteristic leukocyte inclusions (Döhle-like bodies) (table 1). MHA and SBS are differentiated by ultrastructural examination of leukocyte inclusions. These paracrystalline inclusions appear as highly parallel bodies in MHA but are smaller and less organized in SBS and FTNS. FTNS is distinguished by the additional clinical features of high-tone sensorineural deafness, cataracts, and nephritis (Peterson et al. 1985) (table 1). Two other FTNS-like macrothrombocytopenias of unknown genetic etiology have also been described. The first is a subset of autosomal dominant Alport syndrome (APS), but with platelet defects (APSM [MIM 153650]; Atkin et al. 1986) (table 1). The classic form of APS is an X-linked disorder of the glomerular basement membranes and is characterized by progressive renal failure, deafness, and ocular lesions (MIM 301050; Alport 1927). This form results from mutations in the COL4A5 gene, which encodes the type IV α-5 collagen chain (Barker et al. 1990). COL4A5 mutations have not been found in APSM-affected individuals. The second macrothrombocytopenia is Epstein syndrome (EPS [MIM 153650]), which has clinical similarities to FTNS and APSM, although cataracts and leukocyte inclusions have not been described (Epstein et al. 1972) (table 1).
Table 1.
Clinical and Morphological Features of the Five Autosomal Dominant Macrothrombocytopenias, MHA, SBS, FTNS, EPS, and APSM
| Clinical Featurea | |||||
|---|---|---|---|---|---|
| Disorder | MTCP | LeukocyteInclusions | Nephritis | High-Tone Sensorineural Deafness | Cataracts |
| MHA | + | + | − | − | − |
| SBS | + | + | − | − | − |
| EPS | + | − | + | + | − |
| FTNS | + | + | + | + | + |
| APSM | + | − | + | + | + |
Linkage-analysis studies in families with MHA, FTNS, SBS, and EPS had previously been performed and had localized the disease gene to chromosome 22q11-13 in all disorders (Martignetti et al. 2000; Toren et al. 2000). Subsequently, mutations in the nonmuscle myosin heavy chain 9 gene (MYH9), which lies in this region, were demonstrated in all three disorders: MHA, FTNS, and SBS (May-Hegglin/Fechtner Syndrome Consortium 2000). It has also been shown that a specific missense mutation in MYH9, R705H, results in nonsyndromic deafness, DFNA17, an autosomal dominant high-tone sensorineural deafness without platelet abnormalities (Lalwani et al. 2000).
MYH9 encodes nonmuscle myosin IIA (MYHIIA), which is expressed in many different tissues, including platelets, kidney (Simons et al. 1991), leukocytes (Toothaker et al. 1991), and cochlea (Lalwani et al. 2000). Myosins constitute a diverse superfamily, with, to date, 18 different classes. MYHIIA is classified as a class II conventional myosin. These are hexameric enzymes composed of two heavy chains and two pairs of light chains. Dimerization of two heavy chains results in a polar structure with two distinct regions. The amino terminus forms a globular head that binds to actin and ATP, has ATPase activity, and is required for motor activity. The C-terminal α-helical coiled-coil domain comprises the regulatory region (Harrington and Rodgers 1984). Interestingly, mutations in three unconventional myosins have previously been associated with deafness and inner ear hair-cell dysfunction, which is present in FTNS, EPS, and APSM. Mutations in the myosin VIIA gene can result in the Usher syndrome type IB (congenital deafness, vestibular areflexia, and progressive retinitis pigmentosa; Weil et al. 1995) and in recessive nonsyndromic deafness DFNB2 (Liu et al. 1997; Weil et al. 1997). DFNB3 is caused by mutations in the myosin XV gene (Wang et al. 1998). Finally, mutations in the myosin VI gene have been found to result in “_Snell’s waltzer_” deafness in mice (Avraham et al. 1995) and, most recently, in a nonsyndromic form of postlingual human deafness, DFNA22 (Melchionda et al. 2001).
In the present study, we performed molecular analyses of 27 previously unstudied families with MHA, SBS, FTNS, EPS, and APSM, defined the common genetic etiology of these five autosomal dominant macrothrombocytopenias, and analyzed structure-function and genotype-phenotype characteristics of detected MYH9 mutations. Our results, combined with the mutation data of previous studies (Kelley et al. 2000; Lalwani et al. 2000; May-Hegglin/Fechtner Syndrome Consortium 2000 Kunishima et al. 2001), suggest that all six syndromes represent one class of disorder with phenotypic variability. Therefore, the name “MYHIIA syndrome” is proposed to encompass these disorders.
Subjects and Methods
Individuals and Families
After informed consent and institutional review board approval from the corresponding institutions were obtained, blood samples were obtained from 27 individuals with diagnoses of MHA, SBS, FTNS, EPS, and APSM (table 2) and, where possible, from relevant family members. Clinical diagnoses were provided by referring physicians. Urinalysis, ophthalmologic examination, and hearing tests were performed to determine the clinical features of affected individuals. Leukocyte inclusions were characterized by electron microscopy. Genomic DNA was isolated from whole blood (Nicolaides and Stoeckert 1990).
Table 2.
Clinical and Morphological Features of 27 Individuals with a Diagnosis of MHA, FTNS, SBS, EPS, or APSM[Note]
| Clinical Featureb | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient IDNumber | Disordera | Ethnic Origin and Country of Origin | Familial orSporadic | MTCP | Döhle-like bodies | Nephritis | Deafness | Cataracts | Reference for Clinical Detailsc |
| 1 | SBS | European, Germany | Familial | + | + | − | − | − | Greinacher et al. 1990_a_ |
| 2 | SBS | European, Germany | Familial | + | + | − | − | − | Toren et al. 2000 |
| 3 | FTNS | European, Germany | ND | + | + | + | + | + | … |
| 4 | SBS | European, Germany | Familial | + | + | − | − | − | … |
| 5 | MHA/SBS | European, Germany | Familial | + | + | − | − | − | … |
| 6 | FTNS | European, U.S.A. | Familial | + | + | − | + | − | … |
| 7 | FTNS | African American, U.S.A. | Familial | + | ND | + | + | + | … |
| 8 | MHA | European, U.S.A. | ND | + | + | − | − | − | … |
| 9 | FTNS | European, U.S.A. | Familial | + | ND | + | + | − | … |
| 10 | FTNS | European, U.S.A. | Familial | + | + | − | + | − | … |
| 11 | MHA/SBS | European, U.S.A. | Familial | + | + | − | − | − | … |
| 12 | FTNS | Iraqi Jewish, Israel | Familial | + | + | + | + | + | Toren et al. 1999 |
| 13 | FTNS | European, U.S.A. | Familial | + | + | + | + | + | Peterson et al. 1985 |
| 14 | FTNS | European, Italy | Familial | + | + | + | + | + | Rocca et al. 1993 |
| 15 | APSM | European, U.S.A. | Familial | + | ND | + | + | − | … |
| 16 | EPS | European, U.S.A. | Familial | + | − | + | + | − | Epstein et al. 1972 |
| 17 | APSM | European, U.S.A. | Sporadic | + | ND | + | − | − | … |
| 18 | FTNS | African American, U.S.A. | Sporadic | + | − | + | + | − | Moxey-Mims et al. 1999 |
| 19 | APSM | European, U.S.A. | Sporadic | + | ND | + | + | − | … |
| 20 | APSM | European, U.K. | Sporadic | + | − | + | + | + | … |
| 21 | FTNS | European, Italy | Familial | + | + | + | + | − | Velasco et al. 2000 |
| 22 | EPS | European, U.S.A. | Familial | + | − | + | + | − | Epstein et al. 1972 |
| 23 | FTNS | European, Belgium | Familial | + | + | + | + | + | … |
| 24 | EPS | European, Belgium | Familial | + | − | + | + | − | Nurden and Nurden 1996 |
| 25 | FTNS | European, Belgium | Familial | + | + | − | + | − | … |
| 26 | MHA/SBS | European, Australia | Familial | + | + | − | − | − | … |
| 27 | FTNS | Chinese, Australia | Familial | + | + | + | − | − | Colville et al. 2000 |
Mutation Analysis
Primers were designed to amplify all 40 coding exons (exons 1–40), their intron/exon boundaries, and regulatory regions (Kawamoto 1994; Beohar and Kawamoto 1998; human MYH9 GenBank accession number 3135984) (fig. 1 and table 2). PCR amplifications were performed in a 25-μl volume with 10 ng of genomic DNA, 20 μM of each primer, 200 μM dNTPs, 10 mM Tris pH 8.8, 50 mM KCl, 1.5 mM MgCl2, and 0.8 U of Ampli_Taq_ GOLD DNA polymerase (Perkin Elmer). PCR cycle conditions included an initial denaturation of 95°C for 10 min, followed by 40 cycles of 95°C for 1 min, 53°C–61°C (table 3) for 1 min, 72°C for 1 min, and a final 5-min extension at 72°C. Amplification of the promoter (see fig. 1) required 5% dimethyl sulfoxide, whereas the exon 31–32 amplimer required 0.4 mM dNTP and an extension time of 2.5 min.
Figure 1.
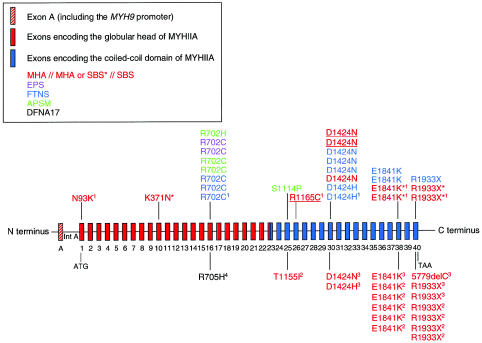
Schematic representation of MYH9 genomic structure (not drawn to scale) and the spectrum of mutations identified in the MYH9 gene. The 40 coding exons and the promoter (noncoding exon A) are represented by vertical colored bars. Intron A (Int A) contains enhancer elements. Mutations shown above the genomic structure have been identified in this cohort or in our previous study (May-Hegglin/Fechtner Syndrome Consortium 2000, indicated by a superscript “1”). Mutations shown below the genomic structure have been reported by other groups (Kelly et al. 2000 [“2”]; Kunishima et al. 2001 [“3”]; Lalwani et al. 2000 [“4”]). Clinical disorders are color-coded as shown in the key. Red mutations indicate the purely hematological disorders. Within this group, individuals with MHA are labeled in red, those with SBS are in red and underlined, and those in whom a diagnosis of MHA or SBS could not been differentiated are marked with an asterisk (*).
Table 3.
Oligonucleotide Sequences, PCR Annealing Temperature, and DHPLC Conditions for Each MYH9 Amplicon
| Oligonucleotide Sequence (5′→3′) | DHPLC Conditions(starting–final %of solution B[temperature in °C])b | |||||
|---|---|---|---|---|---|---|
| MYH9 Amplicona | Sense | Antisense | Amplimer Size(bp) | Annealing Temperature(°C) | 1 | 2 |
| Prom | CAG TGG GTG TAG CAG GAA GG | GCG ATG AAG GTG CCA ACT A | 395 | 61 | 59–65 (68) | 59–65 (69) |
| Enh | CTT AGC CTC CCT GAG CCT CT | CAA AGC CAA AGG GAA ACT CA | 211 | 55 | 54–60 (61) | 54–60 (62) |
| Ex1 | TCC TTC TCC TCC CCG CTT AG | TCC TTC AAG CCC CCT TCT CA | 500 | 55 | 59–65 (63) | 60–66 (61) |
| Ex2 | CAT CAG GGA GTG CCT TCA CA | ATC ACC AGC CAC TAG ATC AA | 400 | 55 | 60–66 (57) | 60–66 (58) |
| Ex3 | CAT CTG TGA CAC TGT GCT CC | GGT TTC AGT AGG AGA CCT CAA G | 210 | 55 | 54–60 (58) | 54–60 (59) |
| Ex4 | ACA CGT TGG GTC CTT CAC G | GTG CTC TTC CTC CAT CAT CCC | 284 | 57 | 58–64 (62) | 58–64 (63) |
| Ex5 | AAA CCT CTG TTC CTG GCA CG | ATA GCA CCG AGT CTG AAC CG | 340 | 55 | 57–63 (62) | 57–63 (64) |
| Ex6 | TGG CCT GGT ACA TGG GTC | AGT GCT GGA ATC AGG AGG CA | 302 | 55 | 58–64 (59) | 58–64 (58) |
| Ex7 | CAT GCC ACT GCA CTC CAA C | TCT TCA GGT GCT CTC CAG C | 290 | 55 | 55–63 (57)c | 55–63 (58)c |
| Ex8 | ACG TCA CTG GCT ACT CTG CG | CCA CAC TCG ACC ATA GGA GG | 320 | 55 | 58–64 (61) | 58–64 (62) |
| Ex9 | TGG CTT GAG GTG CCT CTG AT | ATT TCC GCA AGA CCT TCC CTC | 270 | 52 | 55–61 (62) | 55–61 (61) |
| Ex10 | TTG CTC ATT CAC CCC GTG G | CCT CAA CTG TGC TGC TGC A | 329 | 55 | 54–60 (62) | 57–63 (59) |
| Ex11 | TTA CTG GGG CAT AGG GTA TGA GG | GGA TAA GGC AAC CAA CAG GC | 287 | 55 | 56–62 (61) | 56–62 (63) |
| Ex12 | TGG GTG GAT AAA GGG AAG GC | AAC CAA CAC AGA GCT GAG GT | 430 | 55 | 59–65 (61) | 59–65 (62) |
| Ex13 | TCT GTG GGA TTC AGG GGA TT | ACT GGG TGA GTC ATT GTG CA | 420 | 55 | 55–61 (62) | 55–61 (63) |
| Ex14 | CTT AGG GGT GGA AAT GCT GGA | ACT GTG GAG GTG GGA AGA TGA | 324 | 55 | 55–61 (60) | 55–61 (62) |
| Ex15 | GGT CCT GTT GTT TCA TTC TGT CTC C | GAG AAA CGA CTG AAG GCT CTG TG | 445 | 55 | 60–66 (62) | 60–66 (63) |
| Ex16 | GTT CCC TGA CTA TTC TCC GAC TG | CAC CTC TGG GAC TCA CTG CA | 256 | 55 | 53–59 (65) | 53–59 (66) |
| Ex17 | CCT TGT CTC ATT CAG CCG AAG A | GCA TCC ACC GAC CAC TGA T | 362 | 55 | 56–62 (61) | 58–64 (57) |
| Ex18 | CCT TCC CAG CAT CCT GTT G | GAA CTG CCC GAT TCT ACT CCT C | 697 | 55 | 61–67 (62) | 63–69 (60) |
| Ex19 | AGC TTG AGG ACA AGA CCA GG | AGC CAG GTA TGT ATG GTG GTG | 254 | 55 | 56–62 (62) | 56–62 (63) |
| Ex20 | TTC CAG CCG AGC ATG TCT CT | CCT GGA CTG AGC CTG CAC TG | 210 | 55 | 56–62 (61) | 56–62 (63) |
| Ex21 | GGC TCT CCA GAT GAA AGC TAC TT | AAA GGG AAC ACC TCT CAC TG | 496 | 55 | 59–65 (62) | 59–65 (63) |
| Ex22 | TGG GCT CAC AGA CCT TGC TA | CAG AAG AGA CAG GAA GCA GC | 370 | 55 | 59–65 (64) | 59–65 (65) |
| Ex23 | GTA CCT CGC TGT TTC AGG GG | AGT GCT GTA GTG TGA CCC AG | 450 | 55 | 61–67 (59) | 61–67 (58) |
| Ex24 | GAG ACA GAA CCC ATG GCA CT | AGT GCC GAG AAC TAG GGC CAG | 305 | 55 | 57–63 (64) | 57–63 (65) |
| Ex25 | TGT CCT GCA AAC TCT GCT CC | TCC ATG TCT CCA AGC CAA GG | 350 | 55 | 54–60 (62) | 54–60 (61) |
| Ex26 | GAA AAG CTG CCT GGA GTG CC | CAG GAC TGG TTT GGA TTC TG | 341 | 55 | 56–64 (63)c | 56–64 (62)c |
| Ex27 | GGG TCC AGT GAT GAT AGA CC | CAG AGA GCA CAC ATG CAC CT | 370 | 55 | 61–67 (61) | 58–64 (63) |
| Ex28 | TTG TGA CTC AGG TCC AGC TTT | TGC GTG GAC ACA GAG GCC T | 260 | 55 | 55–61 (63) | 55–61 (64) |
| Ex29 | CTA AAT CAG CAG GAC CAG CT | CCT TGA GAG CAC TGA TGT GGG | 416 | 55 | 58–64 (64) | 60–66 (62) |
| Ex30 | (GGC)3TGA GCA GGT GCC ATC TCG Gd | TCA ACA AGC CAG AGC CTG AG | 454 | 59 | 54–60 (65) | 54–60 (66) |
| Ex31 | TCC CCA GGG AGC TTA GGC | CCG ACC CTC TGT GAT GAC CC | 361 | 59 | 58–64 (63) | 58–64 (62) |
| Ex32 | CCT GAC TTG GGC TCT CTG GG | AGA GAA CAG AAG CCT GCG TG | 309 | 55 | 57–63 (63) | 59–65 (64) |
| Ex31/32 | TCC CCA GGG AGC TTA GGC | ACC CCG AGA CAG GAG GCT A | 805 | 55 | 63–69 (62) | 63–69 (63) |
| Ex33 | CTG AGT TCA GAG CTA GGG CA | TGT AGT TGG CTC AGT CGG GT | 290 | 55 | 56–62 (63) | 56–62 (62) |
| Ex34 | GCA TTG AGT GGA GCA CCA GC | GGA GCC CCG CTA TGA AAC GG | 412 | 55 | 60–66 (62) | 60–66 (61) |
| Ex35/36 | GGG AAG GAT GGT CTT GTG GG | AGG CCA GCT CTG CCG TGG TG | 420 | 55 | 60–66 (63) | 60–66 (64) |
| Ex37 | CCC AAG GGT TTG GTG GGA TC | CTG GTT GTG GCC CAG ATT TG | 403 | 55 | 60–66 (62) | 60–66 (61) |
| Ex38 | CCG GAC CCT CTG AAG GAG G | CCC TGC CTG TCA CCC CAT CT | 229 | 52 | 55–61 (64) | 55–61 (65) |
| Ex39 | AGA TGG GGT GAC AGG CAG GG | GCC AGA AGG GGC AGG GAT TG | 401 | 59 | 60–66 (62) | 60–66 (63) |
| Ex40 | GAG TGG TCC TGT CTA GCT CAG | GGA GGC TGT GGT GTC TGT CT | 346 | 57 | 57–63 (62) | 55–61 (64) |
Mutation screening was performed using the Transgenomic Wave denaturing high-performance liquid chromatography (DHPLC) system (Transgenomic). All amplicons were analyzed at two melting temperatures (table 3). Detected heteroduplexes were subsequently sequenced. PCR samples for sequencing were purified (Qiagen) and were sequenced in both directions using ABI BigDye terminator sequencing (Applied Biosystems) on an ABI 3700 DNA Sequencer. Data were analyzed using ABI Sequencing Analysis 3.3 (Applied Biosystems) and Sequencher 3.11 (Gene Codes Corp.) software programs.
DNA sequencing of two independent amplification products confirmed mutations. To rule out polymorphisms, genomic DNA isolated from 94 unrelated control subjects were screened for the presence of the identified mutations, using DHPLC followed by sequencing if any heteroduplex pattern was observed.
Haplotype Analysis
Haplotype analysis was performed to determine the ancestry of certain identified mutations. Four microsatellite markers from the Marshfield Medical Research Foundation and three novel dinucleotide markers, D22S1745, D22S1746, and D22S1747, were analyzed (fig. 2). Novel markers were generated. Tandem repeats flanking the MYH9 gene were identified using the Tandem Repeats Finder program (Benson 1999; Tandem Repeats Finder Web site). The sense and antisense primers (5′→3′), respectively, of the new dinucleotide markers are as follows: D22S1745, CACACACATTTTCTTCATCCAC and CTGGATGAAGTTGGACACC; D22S1746, TTCCCACAGAACCATTCC and CTACTTGAGAATGAGCCACC; and D22S1747, CCTGGAGAGGTCTGAGTATT and GGTCAGGATAAACAGTGGAG. Markers were amplified by PCR in multiplexes, on the basis of their expected sizes and fluorescent end-labels, and then were electrophoretically separated on a 4.2% denaturing polyacrylamide gel, on an ABI 377 DNA Sequencer. Data were analyzed using the ABI Genescan 3.11 and Genotyper 2.5 (Applied Biosystems) software packages. Haplotypes were then determined according to the pedigrees.
Figure 2.
A, Chromosome location of MYH9 and of the microsatellite markers used in the haplotype analysis. Drawing is not to scale. Physical distances are indicated. Heterozygosity (Htz) values for each marker are shown, as determined from the Marshfield Medical Research Foundation human genetic map or by the amplification of DNA from 95 healthy white control individuals (data not shown). B, Haplotype analysis of markers flanking the MYH9 locus in three E1841K carriers. Shared haplotype is indicated in boldface, and genotypes where phase was determined are underlined. M = E1841K mutation. Individual A was reported in our previous study as “individual 3” (May-Hegglin/Fechtner Syndrome Consortium 2000).
Molecular Modeling
The globular-head mutations were modeled onto the X-ray crystallographic structure of the chick smooth-muscle myosin motor domain (Dominguez et al. 1998), obtained from the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics (ID 1BR2). The atomic coordinates of this structure served as a template for the identified human MYHIIA mutations. All of the amino acids and potentially interacting residues in the chick smooth-muscle myosin were conserved and were identical with respect to the human MYHIIA sequence (fig. 3), and they are numbered according to this sequence. All in silico substitutions for the mutations, as well as model building, were calculated and rendered with the computer software suites QUANTA, INSIGHT II (Molecular Simulations), or O (Jones et al. 1991), for molecular visualization. Since no detailed high-resolution structural information is available for the coiled-coil domain, mutations in this region were modeled using helical wheel diagrams (Schiffer and Edmundson 1967).
Figure 3.
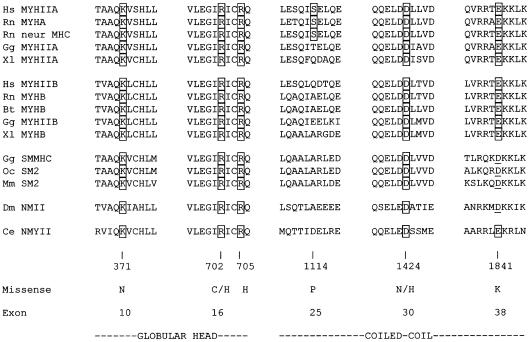
Clustal W alignment of the human MYHIIA amino acid sequence with the amino acid sequences of nonmuscle and smooth-muscle myosins, using the BLASTP program from NCBI. The predicted affected amino acids for the identified missense mutations are shown in boxes. Conserved amino acid changes at the positions of the mutated residues are underlined. Abbreviations and GenBank accession numbers are as follows: Hs MYHIIA, Homo sapiens MYHIIA (accession number P35579); Rn MYHA, Rattus norvegicus nonmuscle myosin heavy chain A (accession number AAA74950); Rn neur MHC, R. norvegicus neuronal myosin heavy chain (accession number S21801); Gg MYHIIA, Gallus gallus MYHIIA (accession number AAA48974); Xl MYHIIA, Xenopus laevis MYHIIA (accession number AAC83556); Hs MYHIIB, H. sapiens MYHIIB (accession number AAA99177); Rn MYHB, R. norvegicus nonmuscle myosin heavy chain B (accession number AAF61445); Bt MYHB, Bos taurus MYHB (accession number BAA36494); Gg MYHIIB, G. gallus MYHIIB (accession number AAA48988); Xl MYHB, X. laevis MYHIIB (accession number AAA49915); Gg SMMHC, G. gallus SMMHC (accession number P10587); Oc SM2, Oryctolagus cuniculus smooth-muscle myosin 2 (accession number P35748); Mm SM2, Mus musculus SM2 (accession number JC5421); Dm NMII, Drosophila melanogaster nonmuscle myosin heavy chain II (accession number AAB09049); and Ce NMYII, Caenorhabditis elegans nonmuscle myosin heavy chain II (accession number AAA83339).
Results
Spectrum of MYH9 Mutations
The 40 MYH9 coding exons (exons 1–40), the promoter found in exon A, and a 120-bp region of intron A containing enhancer elements (fig. 1) were screened in 27 individuals with a diagnosis of MHA (_n_=4), SBS (_n_=3), FTNS (_n_=13), EPS (_n_=3), or APSM (_n_=4), using DHPLC followed by DNA sequencing.
MYH9 mutations were identified for the first time in EPS (R702C) and APSM (R702C, R705H, and S1114P). In total, MYH9 mutations were identified in 20 affected individuals (table 4). The location of the mutations is represented schematically in figure 1. Eight different single-nucleotide substitutions were identified, including three that were previously undescribed: 1119G→C (K371N) in exon 10, 2105G→A (R702H) in exon 16, and 3340T→C (S1114P) in exon 25 (table 4). Three of the eight mutations were located in the globular head, whereas the remaining five were present in the coiled-coil domain (table 4; fig. 1). Four mutations were frequently observed in this cohort: R702C in exon 16, D1424N in exon 30, E1841K in exon 38, and R1933X in exon 40. The missense mutation R702C was identified in individuals diagnosed with FTNS, EPS, and APSM, whereas the D1424N, E1841K, and R1933X mutations were found in individuals with FTNS, MHA, and SBS (fig. 1).
Table 4.
MYH9 Mutations Identified in the Cohort of 27 Individuals with a Diagnosis of MHA, FTNS, SBS, EPS, or APSM, as Indicated
| Patient IDNumbera | Disorder | Ethnic Origin andCountry of Origin | Exon | Nucleotide Substitution (Codon Change) | ProteinAlteration | MYHIIADomain |
|---|---|---|---|---|---|---|
| 1 | SBS | European, Germany | 30 | 4270G→A (GAC→AAC) | D1424N | Coiled coil |
| 2 | SBS | European, Germany | 30 | 4270G→A (GAC→AAC) | D1424N | Coiled coil |
| 3 | FTNS | European, Germany | 16 | 2104C→T (CGT→TGT) | R702C | Globular head |
| 5 | MHA/SBS | European, Germany | 10 | 1119G→C (AAG→AAC) | K371Nb | Globular head |
| 8 | MHA | European, U.S.A. | 30 | 4270G→A (GAC→AAC) | D1424N | Coiled coil |
| 9 | FTNS | European, U.S.A. | 30 | 4270G→A (GAC→AAC) | D1424N | Coiled coil |
| 10 | FTNS | European, U.S.A. | 30 | 4270G→C (GAC→CAC) | D1424H | Coiled coil |
| 13 | FTNS | European, U.S.A. | 30 | 4270G→A (GAC→AAC) | D1424N | Coiled coil |
| 14 | FTNS | European, Italy | 38 | 5521G→A (GAG→AAG) | E1841K | Coiled coil |
| 15 | APMS | European, U.S.A. | 16 | 2105G→A (CGT→CAT) | R702Hb | Globular head |
| 16 | EPS | European, U.S.A. | 16 | 2104C→T (CGT→TGT) | R702C | Globular head |
| 17 | APSM | European, U.S.A. | 25 | 3340T→C (TCT→CCT) | S1114Pb | Coiled coil |
| 18 | FTNS | African American, U.S.A. | 16 | 2104C→T (CGT→TGT) | R702C | Globular head |
| 19 | APSM | European, U.S.A. | 16 | 2104C→T (CGT→TGT) | R702C | Globular head |
| 20 | APSM | European, U.K. | 16 | 2104C→T (CGT→TGT) | R702C | Globular head |
| 21 | FTNS | European, Spain | 38 | 5521G→A (GAG→AAG) | E1841K | Coiled coil |
| 22 | EPS | European, U.S.A. | 16 | 2104C→T (CGT→TGT) | R702C | Globular head |
| 23 | FTNS | European, U.S.A. | 30 | 4270G→A (GAC→AAC) | D1424N | Coiled coil |
| 26 | MHA/SBS | European, Australia | 40 | 5797C→T (CGA→TGA) | R1933X | Coiled coil |
| 27 | FTNS | Chinese, Australia | 40 | 5797C→T (CGA→TGA) | R1933X | Coiled coil |
Several lines of evidence indicated that these mutations were pathological. First, where family members were available, each mutation cosegregated with the respective phenotype. In cases in which the mutation was not present in either parent, it was therefore consistent with a de novo mutation. Second, each of the sequence changes was absent in 94 unaffected, unrelated control individuals. Finally, amino acid sequence-alignment analysis of 15 smooth-muscle and nonmuscle myosins (Sellers 1999, 2000) revealed conservation of the substituted residues (fig. 3) (May-Hegglin/Fechtner Syndrome Consortium 2000).
Evolutionary History of Mutations
To investigate the possible evolution of the mutations, haplotype analysis was performed on the region flanking the MYH9 locus, using seven microsatellite markers. Four markers previously used for the localization of the gene were used (Martignetti et al. 2000): D22S1142, D22S1173, D22S283, and D22S272 (fig. 2). Three novel markers, D22S1745, D22S1746, and D22S1747 (Genome Database), were generated against tandem repeat sequences in close proximity to the MYH9 locus (fig. 2). D22S1745 and D22S1746 lie ∼4.2 kb and ∼9.1 kb centromeric (3′) to the MYH9 gene, respectively, and D22S1747 is positioned ∼3.9 kb telomeric (5′) to the MYH9 gene (fig. 2). Genomic DNA from >100 white individuals was amplified for each marker. The heterozygosity values for these novel markers were .36, .58, and .46, respectively, and the number of observed alleles was 6, 13, and 6, respectively (see the Genome Database Web site). Using these seven markers, haplotypes were determined for seven R702C carriers, two families with D1424N, and three E1841K carriers (two families and one individual) from this cohort (table 4) and our previous study (May-Hegglin/Fechtner Syndrome Consortium 2000). Phase was determined, where possible, in two families with the D1424N mutation (FTNS individuals 9 and 23) and in two E1841K carriers: individual 21 (with FTNS) in this cohort and one individual (with MHA/SBS) from our previous study (May-Hegglin/Fechtner Syndrome Consortium 2000). The phase was indeterminable in the third E1841K carrier (individual 14), and a possible disease haplotype was constructed from the alleles present at each genotype. A common haplotype was demonstrated for three E1841K carriers (fig 2). No common haplotype was demonstrated for the R702C or D1424N carriers (data not shown).
Molecular Modeling
The MYHIIA globular-head mutations K371N and R702H, first described in this cohort, as well as the R705H mutation identified in nonsyndromic deafness, DFNA17 (Lalwani et al. 2000), were modeled on the X-ray crystallographic structure of chick smooth-muscle myosin (fig. 4) (Dominguez et al. 1998). Each of the specific residues is invariant within known nonmuscle myosins and the chick smooth-muscle myosin heavy chain (SMMHC) (fig. 3).
Figure 4.
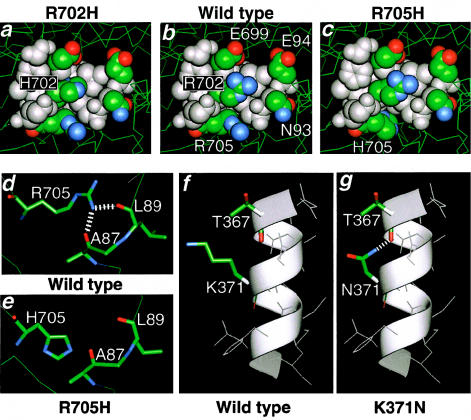
A space-filling representation of the wild-type chick smooth-muscle myosin (b) compared with the mutated residues R702H (a) and R705H (c); This 4.5-Å radial sphere was derived from the X-ray crystallographic structure (Dominguez et al. 1998), as described in the Subjects and Methods section. d, Possible interactions of wild type. e, Ablation of these interactions in the R705H mutation. f, Alpha helical conformation of wild type. g, Potential stabilization of mutation K371N.
The R702H and R705H mutations lie within or adjacent to the “SH1-SH2” helix of the globular-head domain, proximal to the highly reactive “SH1” cysteine that is responsible for the power transduction state. On the basis of predictions from the wild-type chick SMMHC crystallographic structure, the δ-guanido groups of the two wild-type arginines supply positive charge density within the overall negatively charged cleft (fig. 4_b_). The introduction of a histidine residue at position 702 (fig. 4_a_ and 4_c_) would reduce the positive charge density. The wild-type arginine at position 705 can form potential stabilizing interactions with either L89 or A87 (fig. 4_d_). The mutation to H705 disrupts the potential bonding pattern, preventing these stabilizing interactions (fig. 4_e_).
K371 resides in the middle of an α-helix (fig. 4_f_). The potentially reactive ε-amino group of the wild-type lysine residue is oriented perpendicular to the axis of the helix, facing towards the solvent and completely exposed, with no apparent interactions with other portions of the protein (fig. 4_f_). The K371N mutation results in the potential creation of a bond between the amide nitrogen of the asparagine and the carbonyl oxygen of the T367 residue at the n-4 position in the α-helix (fig. 4_g_). Thus, this mutation could result in the abnormal stabilization of the region.
Helical wheel representations (Schiffer and Edmundson 1967) were used to assess the possible altered interactions resulting from the S1114P and D1424N mutations in the coiled-coil domain (data not shown). Although the serine residue is only conserved in the myosin IIA subclass, a change to proline would be expected to have a major effect on the α-helical secondary structure, since it is a classical α-helix “breaker” and thus could alter coiled-coil interactions. The other amino acids found at this position within the myosin family are compatible with the maintenance of an α-helical conformation. Most notably, the conserved α-helical domain, ∼1,100 amino acids in length, contains no other prolines. The D1424N mutation involves the change to the amide form of the wild-type residue. Although there is no drastic change in the side group conformation, a loss of a potentially favorable coulombic interaction occurs in a structurally critical position, as suggested by conservation of this residue (May-Hegglin/Fechtner Syndrome Consortium 2000) (fig. 3).
Discussion
In the present study, the spectrum of MYH9 mutations and the genotype-phenotype relationship in 27 individuals diagnosed with MHA, SBS, FTNS, and two FTNS-like disorders—EPS and APSM—were investigated. The combination of previously identified mutations, MYH9 mutations reported here, and haplotype data indicates that MHA, SBS, FTNS, APSM, and EPS represent a class of allelic disorders with variable phenotypic diversity. This spectrum of disorders is collectively being termed “MYHIIA syndrome.”
Mutation Analysis
Eight different mutations in the MYH9 gene were identified in this new group of affected families, including three that were previously undescribed: K371N, R702H, and S1114P. The R702C mutation, previously identified in patients with FTNS, has now also been identified in patients with EPS and APSM, suggesting that MYH9 mutations can also be the cause of these two disorders. Moreover, the clinical phenotype of FTNS, APSM, and EPS, as well as our genetic data, suggest that all three conditions are, in fact, the same disease entity.
Our data reveal the presence of frequent mutations within this spectrum of clinically and morphologically distinguishable macrothrombocytopenias. This is more apparent when our data are combined with all reported mutations (fig. 1; Kelley et al. 2000; May-Hegglin/Fechtner Syndrome Consortium 2000; Kunishima et al. 2001). The mutation-detection rate for MYHIIA syndrome was 74% (20/27). If each phenotypic variation is taken into consideration, the mutation-detection rates were as follows: MHA/SBS, 71% (5/7); FTNS (including APSM), 76% (13/17); and EPS, 67% (2/3).
Genotype-Phenotype Correlations
The clinical phenotype of individuals sharing the same mutation can be variable, as is demonstrated by individuals with the D1424N, E1841K, and R1933X mutations. First, the D1424N mutation in exon 30, previously described in only one patient with MHA (Kunishima et al. 2001), was detected in an additional patient with MHA (individual 8), and now in individuals with FTNS (individuals 9, 13, and 23) and SBS (individuals 1 and 2). Second, the E1841K mutation in exon 38, previously described in patients with MHA and MHA/SBS (Kelley et al. 2000; May-Hegglin/Fechtner Syndrome Consortium 2000), has now been found in two patients with FTNS, individuals 14 and 21. Third, the exon 40 missense mutation, R1933X, previously found in a large Italian family with MHA and originally used for linkage of this trait (Martignetti et al. 2000; May-Hegglin/Fechtner Syndrome Consortium 2000), has now been identified in an additional seven patients with MHA: individual 26 of the present study, four individuals reported by Kelley et al. (2000), and two individuals reported by Kunishima et al. (2001), as well as in a patient with FTNS (individual 27 of the present study). Thus, all three mutations have been found in patients with FTNS and MHA/SBS.
Finally, the R702C mutation in exon 16 has now been shown to occur in a total of eight individuals with a diagnosis of FTNS, EPS, or APSM. Since these three disorders share the clinical features of macrothrombocytopenia, deafness, and nephritis, the genetics suggest that all three disorders may be classified as one syndrome with mild phenotypic variation, including the presence/absence of presenile cataracts. Moreover, a different mutation at the same codon, R702H, was identified in individual 15, who was also diagnosed with APSM. In marked contrast, no mutations have been found in this codon in the purely hematological disorders, MHA and SBS (_n_=7). Therefore, substitutions at codon 702 are consistently associated with the manifestation of nephritis and deafness in addition to macrothrombocytopenia, indicating that the preservation of codon 702 is critical for the maintenance of MYHIIA function in these affected organs.
A different mutation located in the same region, R705H, was recently identified in a family with nonsyndromic deafness, DFNA17 (Lalwani et al. 2000). Interestingly no platelet abnormalities, leukocyte inclusions, nephritis, or cataracts were observed in any of the family members (Lalwani et al. 2000). Thus, the mutations in these two codons, R702C, R702H, and R705H, appear to define a common region associated with the pathogenesis of deafness, whereas codon 702 seems specifically involved in the molecular pathology of macrothrombocytopenia and nephritis.
Structure-Function Relationships
Previously, six MYHIIA mutations (May-Hegglin/Fechtner Syndrome Consortium 2000) with structure-function correlates were revealed, according to structural data derived from the molecular modeling of the chick smooth-muscle X-ray crystallographic structure (Dominguez et al. 1998) for the N-terminal globular-head mutations and from helical wheel analysis for the carboxy-terminal coiled-domain α-helical mutations. The R702C mutation occurs in the “SH1-SH2” helix that links the interface of the converter domain with the amino terminus of the myosin head. There are two highly reactive cysteine residues at either end of this “SH1-SH2” helix, C694 and C704, which are critical to the conformational changes of the globular-head domain accompanying specific states of power transduction in the myosin-ATPase system (Harrington and Rodgers 1984). Additionally, this region has been implicated in the “swinging lever arm” hypothesis to explain the structural basis of motility (reviewed by Houdusse and Sweeney [2001]).
Two additional mutations lying in this region of MYHIIA—R702H (individual 15) and R705H (Lalwani et al. 2000)—were analyzed. The R702H and R705H mutations share several common structural features. Both residues are present in α-helices of the “SH1-SH2” region (Dominguez et al. 1998) and involve the alteration of basic amino acid residues, which are among the most energetically favorable and stabilizing side chains in α-helices. Judging from amino acid substitutions on a well-characterized synthetic peptide that forms an α-helical polypeptide (O’Neil and DeGrado 1990), the mutations to histidine at both the R702 and R705 positions would result in an energy difference of 0.62 kcal/mole in the α-helix, thus destabilizing the secondary structure. Myosin cross-linking studies have biochemically confirmed these potential interactions and the close proximity of the residues within the domain depicted in figure 4_a_ and c (Lu and Wong 1989). Lastly, modifications in the region of the two cysteines defining the functionality of this helical region have been shown to have severe biochemical ramifications, by altering nucleotide exchange, ATPase activity, and actin sliding activity (Tiepold et al. 2000). In addition to the destabilizing forces of the R→H mutations within this helical region, the R705H mutation involves the disruption of potential bonds with A87 or L90, resulting in a lower propensity for stability of this secondary structure motif (fig. 4_e_).
The S1114P mutation present in individual 17 is expected to result in a highly unstable protein. Energy calculations using a synthetic peptide show that the replacement of a serine with a proline greatly destabilizes the helix, by >2.65 kcal/mol (O’Neil and DeGrado 1990). It is interesting to note that, in accord with the structural constraints imposed by proline, no proline residues are present in the wild-type MYHIIA coiled-coil domain. This ∼1,100–amino acid sequence stretches from the end of the head domain (P836) to within 33 amino acids of the carboxy terminus (P1927, P1931, and P1958).
Unexpectedly, and in contrast to the aforementioned changes, the previously undescribed globular-head mutation K371N in individual 5 (fig. 4_g_) results in an apparent stabilization of the region, by the creation of a bond between the amide nitrogen of the asparagine with the carbonyl oxygen of the T367 residue at the n-4 position in this exposed surface α-helix.
Three of the five missense mutations present in the coiled-coil domain, D1424H, E1841K, and R1933X, were described in the first study (May-Hegglin/Fechtner Syndrome Consortium 2000). These coiled-coil domain mutations cannot be directly modeled onto homologous structures that are solved to atomic resolution by current structural biological approaches. The common D1424N mutation resides in a region of the α-helical coiled coil, producing a higher-ordered left-handed superhelix. This is an energetically feasible way of stabilizing the long helix at the carboxy terminus. Electrostatic interactions of residues surrounding the hydrophobic interface of the coiled coils provide further stabilization of the chains. Although the change of an aspartic acid to its corresponding nonionizable amide asparagine is a relatively conservative alteration, with respect to the possible interacting surface areas and three-dimensional space-filling conformation, the examination of this mutation through use of a helical wheel diagram indicates a potential destabilizing effect. A net change in the charge density (−1 to 0) may destabilize the interaction, with concomitant deleterious effects upon the normal protein dimerization and assembly in that portion of the α-helical coiled coil.
Haplotype Analysis
The haplotype analysis indicated a common haplotype at a minimal distance of 2.3 Mb surrounding the MYH9 locus (D221746 to D22S272) in the three E1841K carriers. Similar analyses suggested multiple origins of the R702C and D1424N mutations. One of the three E1841K carriers had a clinical diagnosis of MHA/SBS (May-Hegglin/Fechtner Syndrome Consortium 2000), whereas two had FTNS (individuals 14 and 21). These two individuals had nephritis and deafness, although only individual 14 had cataracts. Further clinical and historical investigation of the family with MHA/SBS confirmed the absence of nephritis, deafness, and ocular abnormalities. The allele frequencies of the four markers that form the shared haplotype were determined in a white control population or were taken from the CEPH database. Given the frequency of the shared alleles, the probability of three unrelated individuals sharing this haplotype was calculated to be <1 in 2×106. Therefore, these three families may share a common ancestor. Thus, it is hypothesized that MHA/SBS and FTNS are phenotypic variations of a single syndrome. This could explain why intrafamilial phenotypic variation occurs in these disorders, as observed by us and others (Peterson et al. 1985; Rocca et al. 1993).
Conclusion
Mutations were not identified in all affected individuals (7/27). The absence of a mutation may be explained both at the clinical and molecular level. Macrothrombocytopenia is a genetically heterogeneous disorder (Mhawech and Saleem 2000). In addition, there is a normal distribution of platelet size, ranging from very small to giant (Bain 1985). The combination of macrothrombocytopenia, leukocyte inclusions, and the additional phenotypic features of nephritis, deafness, and cataracts are unique to FTNS, EPS (no cataracts), or APSM and thus are more “readily diagnosed” as a single syndrome entity. MHA and SBS are diagnosed on the basis of the presence of macrothrombocytopenia and Döhle-like bodies, as well as from the pattern of inheritance. Döhle-like bodies are generally visible only if a blood smear is stained with May-Grünwald Giemsa stain in a time-dependent manner (Greinacher et al. 1990_a,_ 1992). Ultrastructural features of these inclusions are only detectable by electron microscopy, and this preparation is also time sensitive. Thus, the diagnosis may either be missed or incorrectly applied, depending on the availability of appropriate testing facilities.
At the molecular level, a mutation in MYH9 may not have been identified for different reasons. First, because of technical limitations: DHPLC has been shown to be a highly sensitive mutation-detection technique (Choy et al. 1999), but, as with any screening strategy, mutations may still be missed. Second, the mutation may be present outside the analyzed regions—for example, in a noncoding region that may have a regulatory function. Third, the mutation may be a deletion or duplication on the order of a single exon, which would be undetected by DHPLC and DNA sequencing, or the mutation could be a major deletion or duplication that would only be detected by northern or Southern analysis. Finally, since MYH9 mutations were identified in 74% of individuals, genetic heterogeneity cannot be excluded. Mutations may be present in the associated myosin light chains, in another nonmuscle myosin, or in a protein that forms complexes with MYHIIA.
In summary, MYH9 mutations in patients with EPS and APSM, as well as additional MYH9 mutations in patients with MHA, FTNS, and SBS, have been identified. These mutations occurred in conserved positions and critical regions in both the globular-head and the coiled-coil domains of MYHIIA and are therefore predicted to result in altered assembly, dimerization, or stability of the quaternary myosin complex. Whether the molecular pathogenesis of these disorders arises from a dominant-negative effect caused by the formation of normal-defective myosin dimers or by haploinsufficiency is currently under investigation. The causes of the phenotypic variation also remain to be delineated. For example, a gene-modifier effect (reviewed by Nadeau [2001]) could be present. A second possibility is the involvement of a regulatory factor that affects the differential expression of MYH9 in the affected tissues. Altogether, the molecular and clinical data indicate that these six disorders—MHA, SBS, FTNS, EPS, APSM, and DFNA17—represent variants of a single syndrome, MYHIIA syndrome, with a broad phenotypic spectrum ranging from a lack of symptoms, mild bleeding tendencies, or high-tone sensorineural deafness to severe macrothrombocytopenia with deafness, presenile cataracts, and nephritis, resulting in end-stage renal failure.
Acknowledgments
The authors thank the families and individuals for participation in this study, as well as their referring physicians and scientists: N. Bizzaro, Simon Davies, M. Espinosa, Barbara Greenbaum, Jeffrey Hord, Naomi Luban, Jenny Kim, Eric Legius, Gert Matthijs, Alan Nurden, Chris Van Geet, Lionel van Maldergen, and D. Velasco. We also thank Maria Ramirez and Dan Musat, for their technical assistance, and I. Visiers, for discussion. This work represents partial fulfillment of the Ph.D. requirements for A.T., at the Sackler Faculty of Medicine, Tel Aviv University. K.E.H. was funded, in part, by the Charles H. Revson Foundation; A.C.B. was funded, in part, by Spanish Ministry of Health grant FIS 00/3145; and J.A.M. was supported with grant 5 P30 HD28822 from the Mount Sinai Child Health Research Center.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for the human genetic map)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human MYH9 [accession number 3135984], Hs MYHIIA [accession number P35579], Rn MYHA [accession number AAA74950], Rn neur MHC [accession number S21801], Gg MYHIIA [AAA48974], Xl MYHIIA [accession number AAC83556], Hs MYHIIB [accession number AAA99177], Rn MYHB [accession number AAF61445], Bt MYHB [accession number BAA36494], Gg MYHIIB [accession number AAA48988], Xl MYHB [accession number AAA49915], Gg SMMHC [accession number P10587], Oc SM2 [accession number P35748], Mm SM2 [accession number JC5421], Dm NMII [accession number AAB09049], and Ce NMYII [accession number AAA83339])
- Genome Database, The, http://gdbwww.gdb.org/ (for microsatellite accession numbers)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MHA [MIM 155100], FTNS [MIM 153640], SBS [MIM 605249], and EPS and APSM [MIM 153650])
- Tandem Repeats Finder, http://c3.biomath.mssm.edu/trf.html
References
- Alport AC (1927) Hereditary familial congenital hemorrhagic nephritis. Br Med J 1:504–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin CL, Gregory MC, Border WA (1986) Alport syndrome. In: Schrier RW, Gottschalk CW (eds) Strauss and Welt’s diseases of the kidney. Little Brown, Boston, pp 617–641 [Google Scholar]
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA (1995) The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 11:369–375 [DOI] [PubMed] [Google Scholar]
- Bain BJ (1985) Platelet count and platelet size in males and females. Scand J Haematol 35:77–79 [DOI] [PubMed] [Google Scholar]
- Barker DF, Hostikka SL, Zhou J, Chou LT, Oliphant AR, Gerken SC, Gregory MC, Skilnick MH, Atkin CL, Tryggvason K (1990) Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 248:1224–1227 [DOI] [PubMed] [Google Scholar]
- Benson G (1999) Tandem repeat finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beohar N, Kawamoto S (1998) Transcriptional regulation of the human nonmuscle myosin II heavy chain-A gene. J Biol Chem 273:9168–9178 [DOI] [PubMed] [Google Scholar]
- Choy YS, Dabora SL, Hall F, Ramesh V, Niida Y, Franz D, Kasprzyk-Obara J, Reeve MP, Kwiatkowski DJ (1999) Superiority of denaturing high performance liquid chromatography over single-stranded conformation and conformation-sensitive gel electrophoresis for mutation detection in TSC2. Ann Hum Genet 63:383–391 [DOI] [PubMed] [Google Scholar]
- Colville D, Wang YY, Jamieson R, Collins F, Hood J, Savige J (2000) Absence of ocular manifestations in autosomal dominant Alport syndrome associated with haematological abnormalties. Opthalmic Genet 21:217–225 [PubMed] [Google Scholar]
- Dominguez R, Freyzon Y, Trybus KM, Cohen C (1998) Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell 94:559–571 [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Sahud MA, Piel CF, Goodman JR (1972) Hereditary macrothromboctopathia, nephritis and deafness. Am J Med 52:299–310 [DOI] [PubMed] [Google Scholar]
- Greinacher A, Bux J, Kiefel V, White JG, Mueller-Eckhardt C (1992) May-Hegglin anomaly: a rare cause of thrombocytopenia. Eur J Pediatr 151:668–671 [DOI] [PubMed] [Google Scholar]
- Greinacher A, Mueller-Eckhardt C (1990_a_) Hereditary types of thrombocytopenia with giant platelets and inclusion bodies in the leukocytes. Blut 60:53–60 [DOI] [PubMed] [Google Scholar]
- Greinacher A, Nieuwenhuis HK, White JG (1990_b_) Sebastian platelet syndrome: A new variant of hereditary macrothrombocytopenia with leukocyte inclusions. Blut 61:282–288 [DOI] [PubMed] [Google Scholar]
- Harrington WF, Rodgers ME (1984) Myosin. Annu Rev Biochem 53:35–73 [DOI] [PubMed] [Google Scholar]
- Hegglin R (1945) Gleichzeitige konstitutionelle Veranderungen an Neurtophilen und Thrombocyten. Helv Med Acta 12:439–440 [PubMed] [Google Scholar]
- Houdusse A, Sweeney HL (2001) Myosin motors: missing structures and hidden springs. Curr Opin Struct Biol 11:182–194 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47:110–119 [DOI] [PubMed] [Google Scholar]
- Kawamoto S (1994) Evidence for an internal regulatory region in a human nonmuscle myosin heavy chain gene. J Biol Chem 269:15101–15110 [PubMed] [Google Scholar]
- Kelley MJ, Jawien W, Ortel TL, Korczak JF (2000) Mutation of MYH9, encoding non-muscle myosin heavy chain A, in May-Hegglin anomaly. Nat Genet 26:106–108 [DOI] [PubMed] [Google Scholar]
- Kunishima S, Kojima T, Matsushita T, Tanaka T, Tsurusawa M, Furukawa Y, Nakamura Y, Okamura T, Amemiya N, Nakayama T, Kamiya T, Saito H (2001) Mutations in the NMMHC-A gene cause autosomal dominant macrothrombocytopenia with leukocyte inclusions (May-Hegglin anomaly/Sebastian syndrome). Blood 97:1147–1149 [DOI] [PubMed] [Google Scholar]
- Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN (2000) Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am J Hum Genet 67:1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, Brown SD (1997) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet 16:188–190 [DOI] [PubMed] [Google Scholar]
- Lu RC, Wong A (1989) Glutamic acid-88 is close to SH-1 in the tertiary structure of myosin subfragment 1. Biochemistry 28:4826–4829 [DOI] [PubMed] [Google Scholar]
- Martignetti JA, Heath KE, Harris J, Bizzaro N, Savoia A, Balduini CL, Desnick RJ (2000) The gene for May-Hegglin anomaly localizes to a <1-Mb region on chromosome 22q12.3-13.1. Am J Hum Genet 66:1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R (1909) Leukocyteneinschlusse. Deutsch Arch Klin Med 96:1–6 [Google Scholar]
- May-Hegglin/Fechtner Syndrome Consortium (2000) Mutations in MYH9 result in May-Hegglin anomaly, Fechtner and Sebastian syndromes. Nat Genet 26:103–105 [DOI] [PubMed] [Google Scholar]
- Melchionda S, Ahituv N, Bisceglia L, Sobe T, Glaser F, Rabionet R, Arbones ML, Notarangelo A, Di Iorio E, Carella M, Zelante L, Estivill X, Avraham KB, Gasparini P (2001) MYO6, the human homologue of the gene responsible for deafness in snell's waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet 69:635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhawech P, Saleem A (2000) Inherited giant platelet disorders. Classification and literature review. Am J Clin Pathol 113:176–190 [DOI] [PubMed] [Google Scholar]
- Moxey-Mims MM, Young G, Silverman A, Selby DM, White JG, Kher KK (1999) End-stage renal disease in two pediatric patients with Fechtner syndrome. Pediatr Nephrol 13:782–786 [DOI] [PubMed] [Google Scholar]
- Nadeau JH (2001) Modifier genes in mice and humans. Nat Rev Genet 2:165–174 [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Stoeckert CJ (1990) A simple, efficient method for the separate isolation of RNA and DNA from the same cells. Biotechniques 8:154–156 [PubMed] [Google Scholar]
- Nurden P, Nurden A (1996) Giant platelets, megakaryocytes and the expression of glycoprotein Ib-IX complexes. C R Acad Sci III 319:717–726 [PubMed] [Google Scholar]
- O’Neil KT, DeGrado WF (1990) A thermodynamic scale for the helix forming tendencies of the commonly occurring amino acids. Science 250:646–651 [DOI] [PubMed] [Google Scholar]
- Peterson LC, Rao KV, Crosson JT, White JG (1985) Fechtner syndrome— a variant of Alport's syndrome with leukocyte inclusions and macrothrombocytopenia. Blood 65:397–406 [PubMed] [Google Scholar]
- Rocca B, Laghi F, Zini G, Maggiano N, Landolfi R (1993) Fechtner syndrome: report of a third family and literature review. Br J Haematol 85:423–426 [DOI] [PubMed] [Google Scholar]
- Schiffer M, Edmundson AB (1967) Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J 7:121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR (1999) Myosins. Oxford University Press, Oxford [Google Scholar]
- ——— (2000) Myosins: a diverse superfamily. Biochim Biophys Acta 1496:3–22 [DOI] [PubMed] [Google Scholar]
- Simons M, Wang M, McBride W, Kawamoto S, Yamakawa K, Gdula D, Adelstein RS, Weir L (1991) Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res 69:530–539 [DOI] [PubMed] [Google Scholar]
- Tiepold M, Kliche W, Pfannstiel J, Faulstich H (2000) Stepwise modulation of ATPase activity, nucleotide trapping, and sliding motility of myosin S1 by modification of the thiol region with residues of increasing size. Biochemistry 39:1305–1315 [DOI] [PubMed] [Google Scholar]
- Toothaker LE, Gonzalez DA, Tung N, Lemons RS, Le Beau MM, Arnaourt MA, Clayton LK, Tenen DG (1991) Cellular myosin heavy chain in human leukocytes: isolation of 5′ cDNA clones, characterization of the protein, chromosomal localization and upregulation during myeloid differentiation. Blood 78:1826–1833 [PubMed] [Google Scholar]
- Toren A, Amariglio N, Rozenfeld-Granot G, Simon AJ, Brok-Simoni F, Pras E, Rechavi G (1999) Genetic linkage of autosomal-dominant Alport syndrome with leukocyte inclusions and macrothrombocytopenia (Fechtner syndrome) to chromosome 22q11-13. Am J Hum Genet 65:1711–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toren A, Rozenfeld-Granot G, Rocca B, Epstein CJ, Amariglio N, Laghi D, Landolfi R, Brok-Simoni F, Carlsson LE, Rechavi G, Greinacher A (2000) Autosomal dominant giant platelet syndromes: a hint of the same genetic defect as in Fechtner syndrome owing to a similar linkage to chromosome 22q11-13. Blood 96: 3447–3451 [PubMed] [Google Scholar]
- Velasco F, Espinosa, Torres A (2000) Fechtners syndrome. Haematologica 85:988 [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, Friedman TB (1998) Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280:1447–1451 [DOI] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, Kelley PM, Kimberling WJ, Wagenaar M, Levi-Acobias F, Larget-Piet D, Munnich A, Steel KP, Brown SDM, Petit C (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374:60–61 [DOI] [PubMed] [Google Scholar]
- Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, Ayadi H, Petit C (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 16:191–193 [DOI] [PubMed] [Google Scholar]