Double-Stranded RNA Mycovirus from Fusarium graminearum (original) (raw)
Abstract
Double-stranded RNA (dsRNA) viruses in some fungi are associated with hypovirulence and have been used or proposed as biological control agents. We isolated 7.5-kb dsRNAs from 13 of 286 field strains of Fusarium graminearum isolated from maize in Korea. One of these strains, DK21, was examined in more detail. This strain had pronounced morphological changes, including reduction in mycelial growth, increased pigmentation, reduced virulence towards wheat, and decreased (60-fold) production of trichothecene mycotoxins. The presence or absence of the 7.5-kb dsRNA was correlated with the changes in pathogenicity and morphology. The dsRNA could be transferred to virus-free strains by hyphal fusion, and the recipient strain acquired the virus-associated phenotype of the donor strain. The dsRNA was transmitted to approximately 50% of the conidia, and only colonies resulting from conidia carrying the mycovirus had the virus-associated phenotype. Partial nucleotide sequences of the purified dsRNA identify an RNA-dependent RNA polymerase sequence and an ATP-dependent helicase that are closely related to those of Cryphonectria hypovirus and Barley yellow mosaic virus. Collectively, these results suggest that this dsRNA isolated from F. graminearum encodes traits for hypovirulence.
Double-stranded RNA (dsRNA) mycoviruses have been described for a wide variety of fungi and plant-pathogenic fungi (7, 9, 27, 42, 44, 55, 58). These dsRNA mycoviruses are classified into three families based on number of genome segments, capsid structure, and nucleotide sequences, with some dsRNA mycoviruses remaining unclassified (8, 20, 24, 31, 39, 45). The isometric dsRNA mycoviruses are classified into two families, Totiviridae and Partitiviridae, consisting of viruses that are enclosed in nonenveloped isometric particles of 25 to 50 nm in diameter and typically cause latent infections in their host fungi (20). Totiviruses have a nonsegmented genome, while partitiviruses have segmented genomes. Hypoviruses (family Hypoviridae) are the unusual exception in that they can result in considerable morphological and physiological changes, including cytological alterations, changes in colony morphology and growth rate, and persistently attenuate novel virulence-related phenotypes (hypovirulence) (2, 7, 13, 25, 28, 34, 42). Hypoviruses lack conventional virions, and their dsRNAs are enclosed in host-encoded vesicles (17).
Among Fusarium species, dsRNA mycoviruses have been reported to be present in F. poae and F. solani f. sp. robiniae (14, 39, 40). dsRNA elements of the same electrophoretic mobility isolated from vegetatively compatible strains of F. poae were similar or identical in many cases. Similar-sized dsRNA elements of vegetatively incompatible strains of the fungus could contain both homologous and nonhomologous dsRNAs (14). Morphological alterations or signs of degeneration of F. poae, however, were not observed in any of the dsRNA-containing isolates.
We have isolated Fusarium graminearum Schwabe [telemorph: Gibberella zeae (Schwein.) Petch] and occasionally found colonies with unusual morphology, e.g., slow growth, ameboid mycelia, and increased pigmentation, similar to that described earlier for some F. graminearum isolates from China (60). Phenotypic characteristics of F. graminearum have not been reliably attributed to the presence of virus. In this study, we screened isolates of F. graminearum recovered from Korean maize for dsRNA mycoviruses. Our objectives in this study were (i) to determine if dsRNA mycoviruses were present in field isolates of F. graminearum, (ii) to determine if the dsRNAs result in discernible changes in fungal phenotype, and (iii) to conduct sequence analysis of the identified dsRNA. Results of the experiments in this work showed for the first time that the presence of the dsRNA mycovirus results in changes in morphological and pathogenicity phenotypes of F. graminearum. Partial nucleotide sequence analysis also showed that the dsRNA contained sequences that had identities with RNA-dependent RNA polymerases of plus-strand RNA viruses.
MATERIALS AND METHODS
Maize samples.
Samples of maize seeds, approximately 500 g each, were collected from 40 fields in eight corn-producing regions in the Gangwon province of Korea during November 1999.
Isolation of fungi.
One hundred seeds of each sample were soaked in 2% NaOCl for 2 min, rinsed in sterile distilled water, transferred to potato dextrose agar (PDA) (Difco Laboratories, Detroit, Mich.), and incubated for 4 to 7 days at 25°C. Fusarium isolates, with carmine red pigmentation, were transferred to homemade PDA (20% potato extract, 2% dextrose, 1.5% agar), carnation leaf agar (18), or both, incubated under fluorescent lamps (cool white type; 5,000 lx) at 25°C, and identified to the species level by the procedures of Nelson et al. (38). We recovered 809 Fusarium isolates from the maize seed. Isolates were stored as spore suspensions in 15% glycerol at −80°C and subcultured on PDA as needed. We screened 286 F. graminearum isolates for the presence of dsRNA. dsRNA virus-containing strain DK21 has been deposited in the Korean Collection of Type Cultures at the Genetic Resources Center, Korea Research Institute of Bioscience and Biotechnology, Daejon, Korea, under accession number 26916.
Culture conditions, dsRNA extraction, and enzymatic digestions.
To determine if dsRNA mycoviruses were present in field isolates of F. graminearum, each isolate was grown in 100 ml of complete medium (CM) broth (16) for 4 to 5 days at 25°C in a orbital shaker (100 rpm). Mycelia were collected by filtration through four layers of cheesecloth, followed by washing with distilled water. Mycelia were dried by blotting with paper towels and stored frozen at −70°C until extracted (54). Extraction of nucleic acids and purification of dsRNA by cellulose chromatography were performed as previously described (54). The quality and relative concentration of dsRNA were checked by electrophoresis on 1% agarose gels at 4°C and visualized following ethidium bromide staining and illumination with 350-nm UV light. RNase A digestion under high- and low-ionic-strength buffer conditions and DNase digestion of extracted nucleic acids were carried out as described by Vilches and Castillo (57).
Mycelial growth and sporulation.
Mycelial growth of dsRNA-containing isolates was measured on CM after incubation for 3 days at 25°C (33). Conidia were grown in carboxymethyl cellulose (CMC) broth (15 g of CMC, 1 g of yeast extract, 0.5 g of MgSO4, 1 g of NH4NO3, and 1 g of KH2PO4 per liter) for 9 days at 25°C in a rotary shaker (100 rpm). One thousand conidia in 100 μl were inoculated into 5 ml of CMC broth in a test tube (1.7 by 17 cm). Sporulation was estimated after incubation for 8 days as described by Ann and Lee (3). Twenty independent experiments were performed to obtain means and standard deviations.
Characteristics of dsRNA obtained from F. graminearum strain DK21.
To further characterize the molecular nature and the role(s) of the dsRNAs in their fungal hosts, one F. graminearum strain, DK21, containing a 7.5-kb dsRNA and with altered fungal morphology, was arbitrarily selected and used for the following assays.
(i) Transmission of dsRNA.
To determine if the dsRNAs were transmitted through the conidia, we collected conidia from F. graminearum DK21 and plated them on 2% water agar. After incubation for 8 h at 25°C, colonies originating from a germinated conidium, as determined microscopically, were transferred to fresh CM, and the presence of the dsRNA was determined.
(ii) Pathogenicity.
To determine if the dsRNAs affect virulence of the host fungus, conidial suspensions of dsRNA-containing and dsRNA-free isolates were inoculated onto wheat plants. Conidia were grown in CMC broth for 9 days at 25°C. Conidia were harvested, filtered through cheesecloth, and diluted with sterile water if necessary. Fifty-milliliter portions of spore suspension (105 or 103 conidia/ml) were prepared and sprayed onto two sheaves each of the OlgruMil and GumgangMil wheat varieties; each sheaf contained at least 50 heads. After being sprayed, the plants were placed in a plastic bag for 3 days to maintain high relative humidity. Approximately 2 weeks after inoculation, plants were harvested and disease severity was rated on a scale of 0 to 5, as follows: 0, no disease; 1, weak head blight with less than one-third of a head infected; 2, less than half of a head infected; 3, more than half of, but not the entire, head infected but few (<10%) completely infected heads observed; 4, more than half of, but not the entire, head infected and more than 10% of heads completely infected; and 5, entire head usually infected. The mean of the disease severity from three independent experiments was used as the virulence value.
(iii) Mycotoxin analyses.
dsRNA-containing and dsRNA-free F. graminearum subcultures of DK21 were grown on sterile rice for 2 weeks at 25°C as previously described (51) to see whether the presence of the dsRNAs affected mycotoxin production by F. graminearum. The mycelial mass and substrate were dispersed onto a screen-bottom tray and allowed to air dry in a ventilated hood (25°C) for 5 days. Ten grams of dried substrate was ground in a blender and extracted as described by Tanaka et al. (51). Briefly, each ground sample (10 g) was extracted with 100 ml of acetonitrile-water (3:1, vol/vol) in a 500-ml Erlenmeyer flask for 30 min on a rotary shaker (150 rpm), and the extract was filtered through Whatman (Maidstone, Kent, England) no. 1 filter paper. A 50-ml portion of the filtrate was defatted with the same volume of _n_-hexane. The aqueous phase was mixed with 100 ml of ethanol and evaporated to dryness. The residue was dissolved in 3 ml of methanol and applied directly to a Florisil column (2 by 20 cm) containing 10 g of Florisil (60/100 mesh; Fisher Scientific Co., Pittsburgh, Pa.). The column was washed with 100 ml of _n_-hexane and eluted with 100 ml of chloroform-methanol (9:1, vol/vol). The eluate was concentrated to dryness under reduced pressure, and the residue was redissolved in 2 ml of methanol. Mycotoxins were analyzed with a Shimadzu (Kyoto, Japan) QP-5000 gas chromatograph-mass spectrometer as previously described (43, 49).
(iv) Preparation of fungal protoplasts, transformation, and anastomosis.
The plasmid pUCHI (53) was used for transformation. Preparation and inoculation of fungal protoplasts were performed as previously described (53). Fungal transformation was performed with the virus-free F. graminearum strain DK. One milliliter of the transformed mixture was mixed with 10 ml of regeneration medium (1 g of yeast extract, 1 g of casein enzymatic hydrolysate, 342 g of sucrose, and 18 g of agar per liter), and poured into a petri dish. After incubation at 25°C overnight, the plate was overlaid with 10 ml of 1% agar containing hygromycin B (150 ppm). The resulting dsRNA mycovirus-free hygromycin B-resistant transformant, T-DK, was used for studies of hyphal anastomosis.
To transfer the dsRNA from DK21 to the hygromycin B-resistant transformant, T-DK, mycelial plugs of T-DK were placed around colonies of 13-day-old DK21 on CM and allowed to incubate for at least 4 days after the colonies of the two strains made contact. Mycelial plugs from the edges of the T-DK colonies were transferred to a CM plate containing hygromycin (100 ppm). Hygromycin-resistant colonies were selected as candidates for dsRNA transmission. dsRNA was extracted from each colony and visualized on a 1% agarose gel.
(v) Cloning and sequence analysis.
Five micrograms of gel-purified dsRNA was heat denatured at 99°C for 5 min and primed with random hexamers. First- and second-strand cDNA synthesis reactions and cDNA cloning procedures were performed as described by Gubler and Hoffman (23) and Hillman et al. (26). dsRNA-specific, 25-base oligonucleotide primers (8F, 5′-AAAAGTGTCCTTGACCAAAA-3′; 8R, 5′-AAGCCTTTCAAGCAGTTGTG-3′; 11F, 5′-AGTTTCGCCAGGTCTTTCCA-3′; and 11R, 5′-CTATAGTCCTTTAGATAAGC-3′) were designed based on the sequences of the initial cDNA clones and used for first-strand cDNA synthesis. About 250 ng of the first-strand cDNA was amplified by PCR in a final volume of 100 μl containing 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, a 125 μM concentration of each deoxynucleoside triphosphate, 50 pmol of designed and random primers, and 2.5 U of Taq DNA polymerase (TaKaRa Shuzo Co. Ltd., Shiga, Japan). Amplification was carried out using 1 cycle of 94°C for 1 min, 42°C for 1 min, and 72°C for 5 min followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. After the cycles were completed, the reaction mixtures were held at 72°C for 10 min and then at 4°C. PCR products were purified and ligated into the TOPO cloning vector (Invitrogen, Carlsbad, Calif.) or pT7 Blue-2 T-vector (Novagen, Darmstadt, Germany). Recombinant plasmids containing cDNA inserts were sequenced by the dideoxynucleotide chain termination method by using the ABI Prism Terminator Cycle Sequencing Ready Reaction Kit and ABI Prism 377 Genetic Analyzer (Perkin-Elmer, Foster City, Calif.), located at the National Instrumentation Center for Environmental Management of Seoul National University, according to the manufacturer's instructions. All clones were sequenced in both directions, and at least three different DNA clones representing each amplified DNA fragment were sequenced. Polyprotein-related sequences were initially identified using the BLAST search program of GenBank (61). Multiple-sequence alignment was performed with the CLUSTAL W program (52).
(vi) Northern blotting.
dsRNAs were resolved on a 1% agarose gel in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0), and denatured by soaking the gel in 50 mM NaOH for 30 min at room temperature. Gels were neutralized with 1.5 M Tris-HCl (pH 7.5) containing 0.5 M NaCl for 20 min. The RNAs were transferred to a nylon membrane (Sigma, St. Louis, Mo.) by capillary blotting with 2× SSC (0.3 M NaCl, 30 mM sodium citrate, pH 7.0) for 10 min and cross-linked to the membrane by UV irradiation for 1 min. dsRNA-specific probes were prepared with the NEBlot Phototope kit, and viral bands were detected by chemiluminescence (Phototope-Star Detection kit; New England BioLabs, Boston, Mass.).
Nucleotide sequence accession numbers.
The partial sequences of the dsRNA isolated from DK21 have been deposited in GenBank under accession numbers AF443212 (pDK-2) and AF443213 (pDK-1).
RESULTS
Natural occurrence of 7.5-kb dsRNA in F. graminearum isolated from corn.
We found that 13 of 286 isolates of F. graminearum from maize contained dsRNA. CF-11 cellulose column chromatography and enzyme digestions using RNase A and DNase confirmed the dsRNA nature of the genome. The purified viral genome was resistant to DNase and RNase A at a high salt concentration (0.3 M NaCl) (data not shown).
Morphological differences between a dsRNA-free strain and a dsRNA-containing strain.
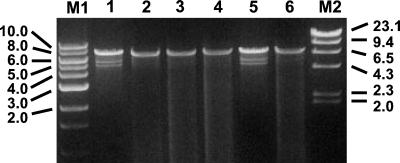
We detected significant differences in mycelial growth rates (Table 1). A 7.5-kb dsRNA was present in all of the F. graminearum isolates with altered phenotypes (Fig. 1). dsRNA-free derivatives of strain DK21 covered the entire plate after 3 days. In contrast, the dsRNA-containing subcultures took more than 9 days to do so. We did not observe a significant difference, however, in the production of conidia. dsRNA-free and dsRNA-containing subcultures produced 135 and 133 conidia per ml of CMC broth, respectively. In addition to the 7.5-kb dsRNA, smaller dsRNAs, of 5.5 to 6 kb, were also found in some isolates (Fig. 1). The presence of these smaller RNAs, however, did not change the altered phenotypes of the F. graminearum isolates.
TABLE 1.
Characteristics of dsRNA-containing and dsRNA-free isolates of DK21
| Isolatea | dsRNA | Morphologyb | Colony diam (cm)c |
|---|---|---|---|
| dsRNA-containing DK21 | + | Dark red, irregular | 0.86 ± 0.25d |
| dsRNA-free DK21 | − | Pink, circular | 2.34 ± 0.13 |
| T-DK B2 | + | Dark red, irregular | 1.01 ± 0.33d |
| T-DK C5 | + | Dark red, irregular | 0.98 ± 0.51d |
| T-DK D1 | + | Dark red, irregular | 0.88 ± 0.08d |
| T-DK F1 | − | Pink, circular | 2.38 ± 0.04 |
FIG. 1.
dsRNA molecules purified from F. graminearum isolates and separated on a 1% agarose gel. Lanes M1 and M2, 1-kb ladder (New England BioLabs) and λ DNA digested with _Hin_dIII, respectively; lanes 1 to 6, isolates YWD3, YWD6, DK21 YDP16, YWD5, and JB53, respectively. Numbers on the left and right indicate sizes in kilobases.
Effects of dsRNAs on pathogenicity and mycotoxin production.
Head blight symptoms were observed on all wheat plants inoculated with both the dsRNA-free and dsRNA-containing isolates. The disease severity of the dsRNA-containing single-conidial isolates, however, was significantly less than that of the dsRNA-free isolates. Disease severities of dsRNA-free isolates on inoculated wheat sheaves were 2.3 ± 1.01 and 4.7 ± 0.36, whereas those of dsRNA-containing isolates were 1.7 ± 0.71 and 2.9 ± 0.80, when inoculated with suspensions of 103 and 105 conidia per ml, respectively. No symptoms were associated with the control inoculation. Deoxynivalenol (DON) was detected at significantly lower levels in dsRNA-containing derivatives than in dsRNA-free isolates. The relative levels of DON in the single-conidial isolate of dsRNA-free and dsRNA-containing strains were 76 to 84 ppm and 3 to 7 ppm, respectively.
Transmission of dsRNAs to conidia.
Single-conidial derivatives of strain DK21 could be placed into two groups. The growth rates of isolates in one group were higher than those of isolates in the other group and resembled those of the dsRNA-free derivatives. Mycelial growth and pigmentation also were different (Fig. 2; Table 1). dsRNA profiles of 55 single-conidial isolates were tested. The 28 isolates that lacked dsRNA all had the wild-type dsRNA-free phenotype.
FIG. 2.
Cultural morphology of isolate DK21 and pathogenicity test. A dsRNA-containing single-conidial isolate (A) grew slowly and was deep red. A dsRNA-free isolate (B) grew fast and was pink. Through anastomosis, hygromycin-resistant colonies (C to F) that showed different cultural appearances on CM plates were selected as candidates for dsRNA transmission. The slow-growing colonies (C, D, and E) were hygromycin resistant and contained dsRNA, and the fast-growing one (F) was dsRNA free. For pathogenicity tests, conidial suspensions were inoculated onto wheat plants. Plants infected with dsRNA-free spores showed more severe symptoms than those infected with dsRNA-containing spores. CK, water-sprayed control plants.
Transmission of dsRNA following hyphal anastomosis.
We recovered several colonies containing dsRNA when we attempted to transmit dsRNA from DK21 to T-DK. Colonies T-DK B2, T-DK C5, T-DK C7, T-DK D1 to D3, and T-DK F1 to F6, which were hygromycin resistant, were further studied. T-DK B2, T-DK C5, T-DK C7, and T-DK D1 to D3 had phenotypes similar to those of strains known to carry the dsRNA, while the phenotypes of T-DK F1 to F6 resembled those of other dsRNA-free isolates. Isolates T-DK B2, T-DK C5, T-DK C7, and T-DK D1 to D3 contained dsRNA, but T-DK F1 to F6 did not and are simply subcultures of T-DK (Fig. 2). When the dsRNA was introduced into strain T-DK, it altered the colony morphology. This result suggests that the unusual cultural morphology of DK21 was due to the dsRNA mycovirus infection.
Partial sequence analysis of 7.5-kb dsRNA.
Two cDNA sequences, one of 825 bp and the other of 1,080 bp in length (pDK-1 and pDK-2; GenBank accession no. AF443213 and AF443212, respectively), were obtained from overlapping cDNA clones, i.e., pDK0112-12 (511 bp), pDK1229-1 (157 bp), and pDK1026-8 (283 bp) for the 825-bp fragment and pDK1026-11 (545 bp) and pDK1229-4 (570 bp) for the 1,080-bp fragment. The sequences from the dsRNAs have identity to RNA-dependent RNA polymerase (RdRp) and ATP-dependent helicase sequences of several viruses, including Cryphonectria hypovirus and Barley yellow mosaic virus (Fig. 3). The tentative superposition of the conserved motifs of RdRp is designated as described by Koonin (30), and the conserved helicase motif is shown as suggested by Gorbalenya et al. (21) and Smart et al. (48).
FIG. 3.
Conserved sequence motifs in the RNA-dependent RNA polymerases (RdRp) (A) and helicases (B) of positive-strand RNA viruses, related dsRNA viruses, and the 7.5-kb dsRNA of strain DK21. The tentative superpositions of the conserved motifs correspond to the motifs described by others (21, 30, 48). The number of amino acid residues between the motifs and the distances from the protein termini are shown (question marks show that numbers are unknown). In the consensus line above the sequences, “&” designates a bulky hydrophobic residue (either aliphatic or aromatic). GenBank accession numbers are as follows: CHV1-EP713 (Cryphonectria hypovirus 1), AAA67458; LRV2-1 (Leishmania RNA virus 1-1), AAB50024; ScV-L-A (Saccharomyces cerevisiae virus L-A), AAA50321; DaV (Diaporthe ambigua RNA virus 1), AAF22958; FusoV (Mycovirus FusoV from Fusarium solani), BAA09520; HvV190S (Helminthosporium victoriae virus 190S), AAB94791; RsV (Rhizoctonia solani virus), AF133290; CHV1-713 (Cryphonectria hypovirus EP713), AAA67458; CHV2-NB58 (Cryphenectria hypovirus 2-NB58), AAA20137; CHV3-GH2 (Cryphonectria hypovirus 3-GH2), AF13604; and BaYMV (Barley yellow mosaic virus), CAA49412.
These two DNA sequences were used as probes for Northern blot analyses to confirm that they originated from the 7.5-kb dsRNA genome. Each probe specifically hybridized to the 7.5-kb dsRNA in dsRNA-containing isolates T-DK B2, T-DK C5, and T-DK D3 (Fig. 4). No signal was observed with either probe in the lanes with virus-free isolates T-DK F1 and T-DK F3. Thus, the sequences that we obtained are from the 7.5-kb dsRNA mycovirus genome. Smaller dsRNAs (5.5 to 6 kb) observed in some F. graminearum isolates were detected in Northern blots using both the pDK-1 and pDK-2 probes (data not shown), indicating that these smaller dsRNAs contain sequences similar or identical to the 7.5-kb dsRNA.
FIG. 4.
Northern blot analysis. Total RNA was extracted from dsRNA-free and -containing derivatives of DK21 and several colonies obtained from anastomosis. The extracts were treated with DNase and blotted to a nylon membrane. Gels were probed with a dsRNA-specific probe (A) and with ethidium bromide (B). The arrowhead indicates the position of the dsRNA. Lanes: 1, λ/_Hin_dIII; 2, prebiotinylated DNA marker; 3, dsRNA-free derivative; 4, dsRNA-containing derivative; 5 and 6, dsRNA-free derivatives obtained from hyphal anastomosis T-DK F2 and T-DK F4, respectively; 7 to 9, dsRNA-containing derivatives obtained from hyphal anastomosis T-DK B2, T-DK C5, and T-DK D1, respectively. Numbers on the left indicate sizes in kilobases.
DISCUSSION
We found that a 7.5-kb dsRNA mycovirus isolated from F. graminearum strain DK21 is associated with altered fungal morphology. Loss of dsRNA through conidial passage resulted in faster mycelial growth and less pigmentation, whereas its acquisition through hyphal anastomosis restored the abnormal dsRNA-containing DK21 phenotypes.
Since the 7.5-kb dsRNA-containing F. graminearum isolates grew slowly compared to dsRNA-free isolates, we expected infected strains to have drastically reduced symptoms on inoculated wheat plants. Although the dsRNA-containing isolates had significantly reduced virulence, some symptom development still occurred. Since the dsRNA of F. graminearum isolates that contain the 7.5-kb dsRNA was transferred to only about half of the conidia, only about half of the spores in the suspension prepared from dsRNA-containing isolates probably have dsRNA. Isolates carrying the dsRNA also produce much less mycotoxin (DON) than do the dsRNA-free isolates. Trichothecene production has a role in virulence; therefore, dsRNA-containing F. graminearum isolates caused much slower disease development on infected wheat plants. Development of disease, however, is a complicated phenomenon, so additional studies are required to confirm these results.
A hypovirulence phenotype associated with dsRNA mycoviruses has been reported for several plant pathogenic fungi, including Ophiostoma novo-ulmi and Cryphonectria parasitica (1, 35, 41, 56). O. novo-ulmi, the causal agent of Dutch elm disease, may contain 12 unencapsidated mitochondrial dsRNAs that result in dramatic changes in the fungal host, including slow growth, formation of colonies with abnormal, irregular margins, and a reduction in the number of viable asexual spores (6, 13, 46, 47). Extensive studies of the biological role of dsRNA have been conducted with C. parasitica, the chestnut blight fungus. dsRNA-containing isolates of C. parasitica have reduced levels of virulence (hypovirulence), suppressed sporulation, altered colony morphology, reduced pigmentation, increased oxalate accumulation, and altered cellulase and laccase activities (2, 12, 25). The dsRNA could be cytoplasmically transmitted from one strain of C. parasitica to another during hyphal fusion (anastomosis). Furthermore, application of a compatible hypovirulent strain to an existing canker resulted in the conversion of the resident virulent strain to the hypovirulence phenotype and consequent healing of the canker. This observation led to the development of several programs that employed the artificial introduction of hypovirulent strains as a practical means of controlling chestnut blight (22). These characteristics and transmission of dsRNA from one strain to another during hyphal anastomosis provided the basis for potential biological control of many fungal diseases (1, 11). However, it should be noted that there are many vegetative compatibility groups reported (5, 37) for F. graminearum, which thus serve as a major constraint for dsRNA transmission. Studies on the population structure of the vegetative compatibility groups and disease development will further extend the possible implication of this dsRNA for biological control.
The pDK-1 and pDK-2 DNA fragments possess conserved RdRp motifs (III and IV), and motifs IV to VI of the helicase genes, respectively (32). Although overall sequence similarity among RNA viruses is poor, the RdRp and helicase genes are relatively highly conserved. Thus, sequence similarities of the RdRp and helicase genes often are used for phylogenetic analyses of RNA viruses (30, 32). Although the deduced amino acid sequence of the 7.5-kb dsRNA RdRp gene contains more sequence upstream of motif III, we could not identify any conserved motifs in this region (data not shown). Motifs IV to VI of RdRp are regarded as core motifs, are conserved across all RdRp classes, and may have a role in binding nucleoside triphosphate substrates (30). As the alignment of motifs I to III is more tentative, it is not surprising that there is no clear conserved motif upstream of motif III in the 7.5-kb dsRNA. RNA viruses that belong to superfamilies I and II contain seven conserved motifs within their helicase genes (30, 32). pDK-2 contained motifs IV to VI of the helicase gene at the N terminus. Although we have not sequenced the regions containing all seven motifs, the degree of sequence similarity left virtually no doubt that the 7.5-kb dsRNA possesses helicase sequences. Experiments to more accurately identify the phylogenic relationships with other RNA viruses and to prove the polymerase activity of this region are under way.
F. graminearum grows rapidly on PDA, with dense arial mycelia and pigmentation ranging from carmine red to tan in the center and white at the margins, and it is an important plant pathogen that causes head and seedling blight of small grains such as wheat and barley, stalk and ear rot of corn, and stem rot of carnation (15, 29, 50). Head blight and ear rot reduce the grain yield, and harvested grain often is contaminated with mycotoxins that have a trichothecene structure, including DON, nivalenol, and zearalenone (36). Direct economic losses that can result from the pathogen, including lower crop yields and poor grain quality, have become common (59), as has reduced animal performance when the product is used for feed (10). There also is concern about the public health implications of exposure to Fusarium mycotoxins, such as feed refusal, vomiting, and skin necrosis (4, 10, 19). If the 7.5-kb dsRNA in strain DK21 is transferable to dsRNA-free isolates through hyphal fusion with a high incidence, and if the virulence level and the mycotoxin production are reduced due to the dsRNA as described above, biological control of diseases caused by F. graminearum could be achieved.
In summary, we have identified a new fungal dsRNA mycovirus that results in changes in the morphological and pathogenicity phenotypes of the fungal host. In addition to the Ophiostoma and Cryphonectria hypoviruses, this is only the third such example of a virus with these biologically and economically important characteristics.
Acknowledgments
This research was supported in part by grant 2000-2-22100-004-3 from the Korea Science & Engineering Foundation and by grants (M101KG010001-01K070104910 and M101KG010001-01K070102810) from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology of the Republic of Korea. Y.-M.C. and J.-J.J. were supported by graduate fellowships from the Ministry of Education through the Brain Korea 21 Project.
REFERENCES
- 1.Anagnostakis, S. L. 1987. Chestnut blight: the classical problem of an introduced pathogen. Mycologia 79**:**23-37. [Google Scholar]
- 2.Anagnostakis, S. L. 1984. The mycelial biology of Endothia parasitica. I. Nuclear and cytoplasmic genes that determine morphology and virulence, p. 353-366. In D. H. Jennings and A. D. M. Rayner (ed.), The ecology and physiology of the fungal mycelium. Cambridge University Press, Cambridge, United Kingdom.
- 3.Ann, I.-P., and Y.-H. Lee. 2001. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant-Microbe Interact. 14**:**496-507. [DOI] [PubMed] [Google Scholar]
- 4.Beardall, J. M., and J. D. Miller. 1994. Diseases in humans with mycotoxins as possible causes, p. 487-539. In J. D. Miller and H. L. Trenbolm (ed.), Mycotoxins in grain-compounds other than aflatoxin. Eagan Press, St. Paul, Minn.
- 5.Bowden, R. L., and J. F. Leslie. 1992. Nitrate-nonutilizing mutants of Gibberella zeae (Fusarium graminearum) and their use in determining vegetative compatibility. Exp. Mycol. 16**:**308-315. [Google Scholar]
- 6.Brasier, C. M. 1983. A cytoplasmically transmitted disease of Ceratocystis ulmi. Nature 305**:**220-223. [Google Scholar]
- 7.Buck, K. W. 1986. Fungal viruses. CRC Press, Boca Raton, Fla.
- 8.Castillo, A., and V. Cifuentes. 1994. Presence of double-stranded RNA and virus-like particles in Phaffia rhodozyma. Curr. Genet. 26**:**364-368. [DOI] [PubMed] [Google Scholar]
- 9.Castro, M., K. Kramer, L. Valdivia, S. Ortiz, J. Benavente, and A. Castillo. 1999. A new double-stranded RNA mycovirus from Botrytis cinerea. FEMS Microbiol. Lett. 175**:**95-99. [DOI] [PubMed] [Google Scholar]
- 10.Charmley, L. L., A. Rosenberg, and H. L. Trenholm. 1994. Factors responsible for economic losses due to Fusarium mycotoxin contamination of grains, foods and feedstuffs, p. 471-486. In J. D. Miller and H. L. Trenbolm (ed.), Mycotoxins in grain: compounds other than aflatoxin. Eagan Press, St. Paul, Minn.
- 11.Chen, B., G. H. Choi, and D. L. Nuss. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264**:**1762-1764. [DOI] [PubMed] [Google Scholar]
- 12.Choi, G. H., and D. L. Nuss. 1992. A viral gene confers hypovirulence-associated traits to the chestnut blight fungus. EMBO J. 11**:**473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole, T. E., B. Muller, Y. Hong, C. M. Brasier, and K. W. Buck. 1998. Complexity of virus-like double-stranded RNA elements in a diseased isolate of the Dutch elm disease fungus, Ophiostoma novo-ulmi. J. Phytopathol. 146**:**593-598. [Google Scholar]
- 14.Compel, P., I. Papp, M. Bibo, C. Fekete, and L. Hornok. 1999. Genetic interrelationships and genome organization of double-stranded RNA elements of Fusarium poae. Virus Genes 18**:**49-56. [DOI] [PubMed] [Google Scholar]
- 15.Cook, R. J. 1981. Fusarium diseases of wheat and other small grains in North America, p. 39-52. In P. E. Nelson, T. A. Toussoun, and R. J. Cook (ed.), Fusarium diseases, biology, and taxonomy. The Pennsylvania State University Press, University Park.
- 16.Correll, J. C., C. J. R. Klittich, and J. F. Leslie. 1987. Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 77**:**1640-1646. [Google Scholar]
- 17.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35**:**1-29. [DOI] [PubMed] [Google Scholar]
- 18.Fisher, N. L., L. W. Burgess, T. A. Toussoun, and P. E. Nelson. 1982. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72**:**151-153. [Google Scholar]
- 19.Foster, B. C., H. L. Trenholm, D. W. Friend, B. K. Thompson, and K. E. Hartin. 1986. Evaluation of different sources of deoxynivalenol (vomitoxin) fed to swine. Can. J. Anim. Sci. 66**:**1149-1154. [Google Scholar]
- 20.Ghabrial, S. A. 1998. Origin, adaptation and evolutionary pathways of fungal viruses. Virus Genes 16**:**119-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17**:**4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffin, G. J. 1986. Chestnut blight and its control. Hortic. Rev. 8**:**291-335. [Google Scholar]
- 23.Gubler, U., and B. J. Hoffman. 1983. A simple and very efficient method for generating cDNA libraries. Gene 25**:**163-269. [DOI] [PubMed] [Google Scholar]
- 24.Hillman, B. I., B. T. Halpern, and M. P. Brown. 1994. A viral dsRNA element of the chestnut blight fungus with a distinct genetic organization. Virology 201**:**241-250. [DOI] [PubMed] [Google Scholar]
- 25.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80**:**950-956. [Google Scholar]
- 26.Hillman, B. I., Y. Tian, P. J. Bedker, and M. P. Brown. 1992. A North American hypovirulent isolate of the chestnut blight fungus with European isolate-related dsRNA. J. Gen. Virol. 73**:**681-686. [DOI] [PubMed] [Google Scholar]
- 27.Huang, S., and S. A. Ghabrial. 1996. Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc. Natl. Acad. Sci. USA 93**:**12541-12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jian, J., D. K. Lakshman, and S. M. Tavantzis. 1997. Association of distinct double-stranded RNAs with enhanced or diminished virulence in Rhizoctonia solani infecting potato. Mol. Plant-Microbe Interact. 10**:**1002-1009. [Google Scholar]
- 29.Kommedahl, T., and C. E. Windels. 1981. Root-, stalk-, and ear-infecting Fusarium species on corn in the USA, p. 94-103. In P. E. Nelson, T. A. Toussoun, and R. J. Cook (ed.), Fusarium diseases, biology, and taxonomy. The Pennsylvania State University Press, University Park.
- 30.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72**:**2197-2206. [DOI] [PubMed] [Google Scholar]
- 31.Koonin, E. V., G. H. Choi, D. L. Nuss, R. Shapira, and J. C. Carrington. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. USA 88**:**10647-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28**:**375-430. [DOI] [PubMed] [Google Scholar]
- 33.Kousik, C. S., J. P. Snow, and R. A. Valverde. 1994. Comparison of double-stranded RNA components and virulence among isolates of Rhizoctonia solani AG-11A and AG-11B. Phytopathology 84**:**44-49. [Google Scholar]
- 34.Lemke, P. A. 1979. Viruses and plasmids in fungi. Marcel Dekker, New York, N.Y.
- 35.MacDonald, W. L., and D. W. Fulbright. 1991. Biological control of chestnut blight: use and limitation of transmissible hypovirulence. Plant Dis. 75**:**656-661. [Google Scholar]
- 36.Marasas, W. F. O., P. E. Nelson, and T. A. Toussoun. 1984. Toxigenic Fusarium species: identity and mycotoxicology. The Pennsylvania State University Press, University Park.
- 37.Moon, J.-H., Y.-H. Lee, and Y.-W. Lee. 1999. Vegetative compatibility groups in Fusarium graminearum isolates from corn and barley in Korea. Plant Pathol. J. 15**:**53-56. [Google Scholar]
- 38.Nelson, P. E., T. A. Toussoun, and W. F. O. Marasas. 1983. Fusarium species: an illustrated manual for identification. The Pennsylvania State University Press, University Park.
- 39.Nogawa, M., S. T. Kageyama, and M. Okazaki. 1993. A double-stranded RNA mycovirus from the plant pathogenic fungus Fusarium solani f. sp. robiniae. FEMS Microbiol. Lett. 110**:**153-158. [DOI] [PubMed] [Google Scholar]
- 40.Nogawa, M., T. Kageyama, A. Nakatani, G. Taguchi, M. Shimosaka, and M. Okazaki. 1996. Cloning and characterization of mycovirus double-stranded RNA from the plant pathogenic fungus Fusarium solani f. sp. robiniae. Biosci. Biotechnol. Biochem. 60**:**784-788. [DOI] [PubMed] [Google Scholar]
- 41.Nuss, D. L. 1992. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol. Rev. 56**:**561-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nuss, D. L., and Y. Koltin. 1990. Significance of dsRNA genetic elements in plant pathogenic fungi. Annu. Rev. Phytopathol. 28**:**37-58. [DOI] [PubMed] [Google Scholar]
- 43.Park, J. S., K. R. Lee, J. C. Kim, S. H. Lim, J. A. Seo, and Y. W. Lee. 1999. A hemorrhagic factor (apicidin) produced by toxic Fusarium isolates from soybean seeds. Appl. Environ. Microbiol. 65**:**126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peery, T., T. Shabat-Brand, R. Steinlauf, Y. Koltin, and J. Bruenn. 1987. Virus-encoded toxin of Ustilago maydis: two polypeptides are essential for activity. Mol. Cell. Biol. 7**:**470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preisig, O., B. D. Wingfield, and M. J. Wingfield. 1998. Coinfection of a fungal pathogen by two distinct double-stranded RNA viruses. Virology 252**:**399-406. [DOI] [PubMed] [Google Scholar]
- 46.Rogers, H. J., K. W. Buck, and C. M. Brasier. 1987. A mitochondrial target for double-stranded RNA in diseased isolates of the fungus that causes Dutch elm disease. Nature 329**:**558-560. [Google Scholar]
- 47.Rogers, H. J., K. W. Buck, and C. M. Brasier. 1986. Transmission of double-stranded RNA and a disease factor in Ophiostoma ulmi. Plant Pathol. 35**:**277-287. [Google Scholar]
- 48.Smart, C. D., W. Yuan, R. Foglia, D. L. Nuss, D. W. Fulbright, and B. I. Hillman. 1999. Cryphonectria hypovirus 3, a virus species in the family Hypoviridae with a single open reading frame. Virology 265**:**66-73. [DOI] [PubMed] [Google Scholar]
- 49.Sohn, H.-B., J.-A. Seo, and Y.-W. Lee. 1999. Co-occurrence of Fusarium mycotoxins in moldy and healthy corn from Korea. Food Addit. Contam. 16**:**153-158. [DOI] [PubMed] [Google Scholar]
- 50.Sutton, J. C. 1982. Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum. Can. J. Plant Pathol. 4**:**195-209. [Google Scholar]
- 51.Tanaka, T., A. Hasegawa, Y. Matsuki, K. Ishii, and Y. Ueno. 1985. Improved methodology for the simultaneous detection of the Fusarium mycotoxins deoxynivalenol and nivalenol in cereals. Food Addit. Contam. 2**:**125-137. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:**4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turgeon, B. G., R. C. Garber, and O. C. Yoder. 1987. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7**:**3297-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valverde, R. A. 1990. Analysis of double-stranded RNA for plant virus diagnosis. Plant Dis. 74**:**255-258. [Google Scholar]
- 55.Van Alfen, N. K. 1986. Hypovirulence of Endothia (Cryphonectria) parasitica and Rhizoctonia solani, p. 143-162. In K. W. Buck (ed.), Fungal virology. CRC Press, Boca Raton, Fla.
- 56.Van Alfen, N. K., R. A. Jaynes, S. L. Anagnostakis, and P. R. Day. 1975. Chestnut blight: biological control by transmissible hypovirulence in Endothia parasitica. Science 189**:**890-891. [DOI] [PubMed] [Google Scholar]
- 57.Vilches, S., and A. Castillo. 1997. A double-stranded RNA mycovirus in Botrytis cinerea. FEMS Microbiol. Lett. 155**:**125-130. [DOI] [PubMed] [Google Scholar]
- 58.Wickner, R. B. 1992. Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Microbiol. 46**:**347-375. [DOI] [PubMed] [Google Scholar]
- 59.Windels, C. E. 2000. Economic and social impacts of Fusarium headblight changing farms and rural communities in the Northern Great Plains. Phytopathology 90**:**17-21. [DOI] [PubMed] [Google Scholar]
- 60.Xu, Y.-G., J.-Y. Xu, and Z.-D. Fang. 1992. Studies of sectoring in Fusarium graminearum Schw. causing wheat scab. Acta Phytopathol. Sinica 22**:**11-14. [Google Scholar]
- 61.Zhang, J., and T. L. Madden. 1997. PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 7**:**649-656. [DOI] [PMC free article] [PubMed] [Google Scholar]