Interferon regulatory factor 4 negatively regulates the production of proinflammatory cytokines by macrophages in response to LPS (original) (raw)
Abstract
A member of the IFN regulatory factor (IRF) family of transcription factors, IRF-4 is expressed in lymphocytes and macrophage/dendritic cells. Studies using IRF-4-deficient mice have revealed the critical roles of IRF-4 in lymphocyte responses. However, the role of IRF-4 in innate immune responses is not clearly understood. Here, we demonstrate that IRF-4 negatively regulates the production of proinflammatory cytokines by macrophages in response to Toll-like receptor (TLR) stimulation. Mice lacking IRF-4 are sensitive to LPS-induced shock, and their macrophages produce high levels of proinflammatory cytokines, including TNF-α and IL-6, in response to TLR ligands. The inhibitory role of IRF-4 in response to TLR stimulation was confirmed by the down-regulation of IRF-4 expression in normal macrophages by using the small interfering RNA technique and by the overexpression of IRF-4 in macrophage line RAW264.7. Activation of the important signaling pathways for cytokine production, NF-κB and JNK (c-Jun N-terminal kinase), was enhanced after LPS stimulation in IRF-4-/- macrophages. These results imply that IRF-4 negatively regulates TLR signaling and is inhibitory to the production of proinflammatory cytokines in response to TLR stimulation.
Keywords: c-Jun N-terminal kinase, NF-κB, Toll-like receptor
IFN regulatory factors (IRFs) are a family of transcription factors that bind a specific DNA motif known as the IFN-stimulated response element (ISRE) and play critical roles in a variety of immune processes (1). One of the members, IRF-4, is expressed specifically in lymphocytes and macrophage/dendritic cells (2–6). The critical roles of IRF-4 in the development and function of B and T cells have been established (7–11). IRF-4 is also expressed in macrophages (4, 5). It was reported that the stimulation of macrophages with LPS induces translocation of IRF-4 from the cytosol to the nucleus (4). IRF-4 forms a complex with IRF-8 [also known as IFN consensus sequence-binding protein (ICSBP)] in macrophages, binds to the ISRE, and represses the activity of the α/β-IFN-induced cytokine ISG15 promoter (5). It was also reported that IRF-4 forms a complex with Ets-like protein PU.1 and synergizes with it to enhance transcription of IL-1β by means of the PU.1/IRF composite motif (4, 12). These studies suggested roles of IRF-4 in cytokine production by macrophages. However, studies were performed primarily by using the EMSA and promoter reporter analysis, and the function of IRF-4 in transcription of endogenous genes in macrophages and the role in vivo has not been clearly evaluated.
We previously reported our study on Leishmania major infection using IRF-4-/- mice and showed that CD4+ T cells could give rise to specific Th1 cells and that mice became protected against L. major in the absence of IRF-4 (9). In these experiments, mice were infected in the hind footpads with the intracellular pathogen L. major. Interestingly, the footpad swelling of IRF-4-/- mice at 2–5 weeks of L. major infection was consistently less than in C57BL/6 mice, although T cell responses of IRF-4-/- mice never exceeded those in C57BL/6 mice. These studies suggested to us that the innate immune responses of IRF-4-/- mice might be enhanced, resulting in higher resistance to the intracellular pathogens within macrophages. To this end, we investigated the function of macrophages in mice lacking IRF-4. In this study, we show that IRF-4-/- mice are more sensitive to LPS shock than IRF-4+/+ mice. Macrophages from IRF-4-/- mice produced higher levels of cytokines, including TNF-α and IL-6, in response to LPS and other Toll-like receptor (TLR) ligands. IRF-4-/- macrophages showed enhanced activation of NF-κB and the mitogen-activated protein kinase (MAPK) JNK (c-Jun N-terminal kinase) in response to LPS. These results imply that IRF-4 regulates the innate immune responses by negatively modulating TLR signaling pathways.
Materials and Methods
Mice. IRF-4-/- mice were initially mated to C57BL/6 mice and were maintained by intercrossing in the Laboratory Animal Center for Animal Research at Nagasaki University as described in refs. 9 and 10. C57BL/6 (B6) mice were purchased from SLC (Hamamatsu, Japan) and were used as control IRF-4+/+ mice. IRF-4/TNF-α receptor (TNF-αR) double knockout (DKO) mice were generated by mating IRF-4-/- mice to B6-background TNF-αR knockout mice (13). Mice were used at 6–8 weeks old. For lethality tests, mice were injected i.p. with LPS (20 μg/g of body weight), and sera were collected 0 and 2 h later to determine the cytokine levels. The animal experiments reported herein were conducted according to the Guidelines of the Laboratory Animal Center for Biomedical Research at Nagasaki University.
Cytokine Assay. Macrophages were collected from the peritoneal cavity of mice 4 days after i.p. inoculation of 0.5 ml of 2,4,10,14-tetramethyl-pentadecane (ICN). In some experiments (Fig. 5_A_), macrophages were recovered from the peritoneal cavity of untreated mice. These cells were cultured overnight in complete RPMI medium 1640 (9), and adherent cells that were collected (1–2 × 105 per well) were cultured in 96-well, flat-bottom plates for 24 h. For the inhibition of JNK activity, the inhibitor of JNK, SP600125 (Tocris Cookson, Ellisville, MO) (14), was added 2 h before the addition of LPS (100 ng/ml).
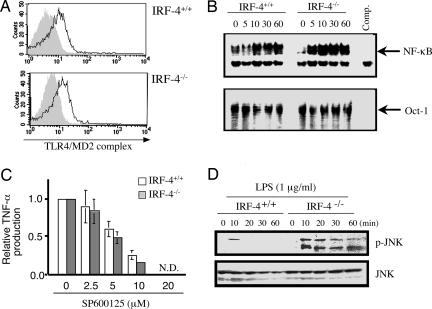
Fig. 5.
Altered LPS signaling in IRF-4-/- macrophages. (A) Peritoneal macrophages from IRF-4+/+ and IRF-4-/- mice were stained with FITC-anti-TLR4/MD2 mAb (solid line) or without Ab (gray). (B) Peritoneal macrophages were stimulated with LPS (1 μg/ml) for 0–60 min, and the nuclear extract was prepared. Mobility-shift assay was performed with NF-κB and Oct-1 probes. (C) Peritoneal macrophages from IRF-4+/+ (open bars) and IRF-4-/- (filled bars) mice were treated with the indicated concentration of SP600125 (JNK-specific inhibitor) for 2 h and then stimulated with LPS (1 μg/ml) for an additional 24 h in the presence of the inhibitor. The relative TNF-α levels in the supernatant were expressed as the ratio to the TNF-α levels in the absence of the inhibitor. (D) Peritoneal macrophages were stimulated with LPS (1 μg/ml) for 0–60 min. Cell lysates (30 μg per lane) were separated by 12.5% SDS/PAGE and blotted. The blot was probed with anti-phospho-JNK Ab, stripped, and reprobed with anti-JNK Ab.
The levels of cytokines in the supernatants were measured by a sandwich ELISA as described in ref. 9. TNF-α was measured by using MP6-XT22 mAb and biotinylated MP6-XT3 mAb. IL-6 was measured by using MP5–20F3 mAb and biotinylated MP5–32C11 mAb. The mAb for ELISA were from BD Biosciences. Recombinant mouse IL-6 and TNF-α (PeproTech, London) were used as standards. The values of TNF-α, IL-12 p70 in sera, and IL-10 in culture supernatant were determined by using ELISA kits (eBioscience, San Diego).
Flow Cytometry. Cells were stained with FITC-anti-F4/80 (Serotec), phycoerythrin-anti-Gr-1 (eBioscience), biotin-anti-CD11b mAb (BD Biosciences) plus avidin-Red670, or FITC-anti-TLR4/MD2 complex mAb (MBL, Nagoya, Japan). Stained cells were analyzed by using a FACScan (BD Biosciences).
Nuclear Extract Preparation and EMSA. Nuclear extracts were prepared as described in ref. 15. Briefly, cells resuspended in buffer A containing protease inhibitor mixture and were lysed in 0.6% Nonidet P-40. After centrifugation, nuclei were resuspended in buffer C, and nuclear extracts were separated by centrifugation and stored in aliquots at -80°C. Protein concentration was determined by using a Bio-Rad protein assay kit.
dsDNA probes that bind to the NF-κB site and to the Oct-1 site (Santa Cruz Biotechnology) were radiolabeled with [γ32]ATP, and EMSA was performed by using 2 μg of the nuclear extract. All EMSA reactions were incubated for 30 min, and DNA–protein complexes were separated by polyacrylamide gels and visualized by autoradiography.
Northern Blotting and RNA Interference (RNAi) Assay. Total RNA was extracted by using Isogen (Nippon Gene, Tokyo). RNA samples (10 μg per lane) were fractionated by 1% agarose-formaldehyde gel electrophoresis, transferred to nylon membranes, and probed with radiolabeled murine suppressor of cytokine signaling (SOCS) 3, IL-1 receptor-associated kinase (IRAK) M, or glyceraldehyde 3-phosphate dehydrogenase (G3PDH) cDNA (16–19). Hybridization was carried out at 42°C overnight, and the membrane was exposed by using a BAS5000 imaging system. For RNAi assay, dsRNA was synthesized by using a Silencer small interfering RNA (siRNA) construction kit (Ambion, Austin, TX) according to the manufacturer's instructions. Sequences of siRNA template oligonucleotides are shown in Table 1, which is published as supporting information on the PNAS web site. Macrophages (2 × 105) were transfected with dsRNA (10 nM) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 4–5 h, cells were washed and then cultured in the presence or absence of LPS.
Real-Time RT-PCR. The mRNA expression was measured by automated real-time RT-PCR using the Applied Biosystems PRISM 7900HT sequence detection system according to the manufacturer's instructions. Briefly, complementary DNA was generated from 500 ng of total RNA by using random hexamers and was amplified by real-time PCR using specific primers (Table 2, which is published as supporting information on the PNAS web site) in the buffer containing the dsDNA-specific fluorescent dye SYBR green. The threshold cycle of each PCR was converted to a DNA equivalent by reading against standard curves generated by amplifying dilutions of plasmid containing the relevant target sequences. The mRNA expression was determined as the ratio of the each DNA to G3PDH.
Transfection of IRF-4 cDNA to the RAW264.7 Macrophage Cell Line. Mouse IRF-4 cDNA was cloned into pcDNA3 (Invitrogen). RAW264.7 cells (1 × 108 per ml) were resuspended in a specified electroporation buffer (buffer V). Cells (0.1 ml) were transfected with the plasmid DNA (2 μg) in a 2.0-mm electroporation cuvette according to protocol T-24 by using a Nucleofector apparatus (Amaxa, Gaithersburg, MD). Five hours later, cells were washed, seeded in 96-well, flat-bottom plates (1 × 105 cells per well), and cultured for 8 h in the presence or absence of LPS.
Western Blot. Cells were lysed in buffer (0.5% Triton X-100/50 mM Tris/150 mM NaCl/1 mM Na3VO4/5 mM EDTA/1 mM PMSF) containing protease inhibitor mixture “complete,” Pefabloc SC AEBSF, and phosphatase inhibitor mixture I and II. The lysate (30 μg) was size-fractionated on 12.5% SDS/PAGE and transferred to a poly(vinylidene difluoride) membrane. The blot was incubated with anti-phospho-JNK or anti-JNK Ab (Cell Signaling Technology, Beverly, MA). The membrane was incubated with horseradish peroxidase-goat anti-rabbit Ig Ab (MBL) and visualized by using ECL (enhanced chemiluminescence) reagent (Amersham Pharmacia).
Statistics. Significance levels in cytokine production were determined by Mann–Whitney's U test for unpaired observations. Results were considered significant when P < 0.05.
Results
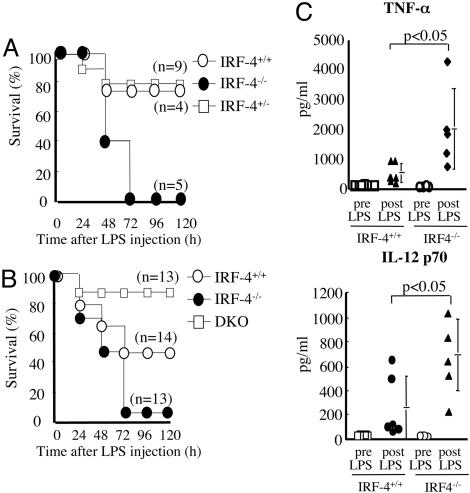
IRF-4-/- Macrophages Produce Excessive Cytokines in Response to TLR Ligands. We injected LPS i.p. into IRF-4+/+, IRF-4+/-, and IRF-4-/- mice and monitored their survival (Fig. 1_A_). All IRF-4-/- mice died within 72 h after LPS challenge, whereas >75% of the IRF-4+/+ and IRF-4+/- mice survived >120 h. Because overproduction of TNF-α is one of the major factors involved in the lethal effect of LPS (20), we injected LPS i.p. into C57BL/6 (IRF-4+/+), IRF-4-/-, or IRF-4/TNF-α receptor DKO mice. The majority of IRF-4-/- mice (12/13) died within 5 days of LPS challenge, whereas 43% of IRF-4+/+ mice (6/14) and >84.6% of the DKO mice (12/13) survived >120 h (Fig. 1_B_), indicating that the lethal effect of LPS is mediated by TNF-α. Serum levels of TNF-α and IL-12 in IRF-4-/- mice 2 h after LPS challange were ≈4 and ≈2.4 times higher, respectively, than in the IRF-4+/+ mice (Fig. 1_C_).
Fig. 1.
Hypersensitivity of IRF-4-/- mice to LPS in vivo.(A) IRF-4-/- mice and their littermates were injected i.p. with LPS. (B) IRF-4-/-, C57BL/6 (IRF-4+/+), and IRF-4-/- TNF-α receptor-/- DKO mice were injected i.p. with LPS. (C) Serum from IRF-4+/+ and IRF-4-/- mice was collected 2 h after LPS challenge. TNF-α and IL-12 p70 levels were measured by ELISA. Each circle represents an individual mouse, and the bars represent the average of each group.
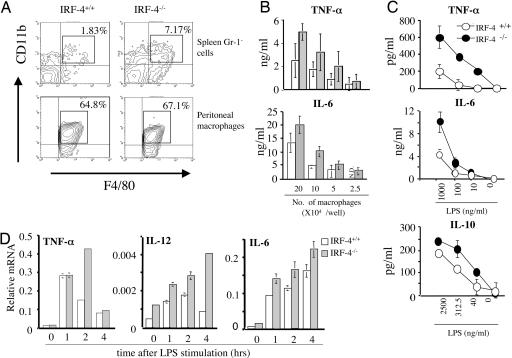
The increased levels of the proinflammatory cytokines in the serum of IRF-4+/+ mice could be due to the increase in the number of producing cells and to the increased production of the cytokines by individual cells. In fact, the proportion of Gr-1-CD11b+F4/80+ cells (macrophage) in the spleen of IRF-4-/- mice (7.17%) was increased when compared with IRF-4+/+ mice (1.83%) (Fig. 2_A_). Because the total numbers of spleen cells in IRF-4+/+ (12.4 ± 1.8 × 107, n = 5) and IRF-4-/- (15.0 ± 3.2 × 107, n = 5) mice were similar, the absolute number of macrophages was increased in the IRF-4-/- spleen. We next investigated the ability of macrophages to produce proinflammatory cytokines in response to LPS in vitro (Fig. 2 B and C). We used peritoneal adherent macrophages for the functional assays. These cells from IRF-4-/- and IRF-4+/+ mice expressed similar levels of CD11b and F4/80 markers. However, these macrophages from IRF-4-/- mice produced greater amounts of TNF-α and IL-6 in response to LPS when compared with IRF-4+/+ macrophages. We also examined the ability of macrophages to produce IL-10, an antiinflammatory cytokine. IRF-4-/- macrophages produced slightly higher levels of IL-10 in response to LPS when compared with IRF-4+/+ macrophages, suggesting that IL-10 does not participate in the higher sensitivity of IRF-4-/- mice to LPS shock. These results demonstrated that enhanced production of proinflammatory cytokines in IRF-4-/- mice is due to both the increased number of macrophages and to the augmented ability of each macrophage to produce proinflammatory cytokines.
Fig. 2.
Increased macrophages and their hyperresponse to LPS in vitro in IRF-4-/- mice. (A)(Upper) CD11b and F4/80 expression of spleen cells is shown after gating on the Gr-1- population. The number in the upper right corner of each profile indicates the percentage of Gr-1-CD11b+F4/80+ cells in total spleen cells. (Lower) Peritoneal adherent macrophages were stained for CD11b and F4/80. The proportion of the cells within the box is indicated. (B) Graded numbers of peritoneal macrophages (2.5–20 × 104) from IRF-4+/+ (open bars) and IRF-4-/- (gray bars) mice were stimulated with LPS (1 μg/ml). The production of cytokines in the supernatant was determined by ELISA. (C) Peritoneal macrophages (1 × 105 per well for TNF-α and IL-6 and 2 × 105 per well for IL-10) from IRF-4+/+ (open circles) and IRF-4-/- (filled circles) mice were stimulated with varying amounts of LPS for 24 h. (D) The expression levels of TNF-α, IL-12, and IL-6 were determined by real-time RT-PCR in IRF-4+/+ or IRF-4-/- peritoneal macrophages 0–4 h after stimulation with LPS (1 μg/ml).
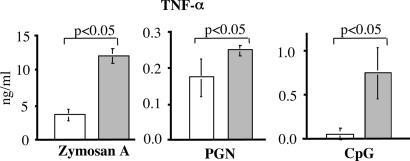
Production of TNF-α is regulated at both transcriptional and posttranscriptional levels (21–23). The expression levels of TNF-α, IL-12, and IL-6 mRNA in IRF-4+/+ and IRF-4-/- macrophages was compared by real-time RT-PCR (Fig. 2_D_). In IRF-4-/- macrophages, the TNF-α mRNA level continued to increase up to 2 h after LPS stimulation and significantly exceeded that in IRF-4+/+ macrophages. The levels of IL-12 and IL-6 mRNA were also higher in IRF-4-/- macrophages than in IRF-4+/+ macrophages. We next determined cytokine production of IRF-4-/- macrophages in response to other TLR ligands (Fig. 3). IRF-4-/- macrophages produced higher levels of TNF-α in response to the TLR2 ligands zymosan A and Staphylococcus aureus peptidoglycan and TLR9-ligand CpG. Therefore, IRF-4-/- macrophages produce increased levels of cytokines in response to stimulation through TLR2, TLR4, and TLR9.
Fig. 3.
Hyperresponse of IRF-4-/- macrophages to other TLR ligands. Peritoneal macrophages (2 × 105 per well) from IRF-4+/+ (open bars) and IRF-4-/- (filled bars) mice were stimulated with zymosan A (1.5 × 106 bioparticles per ml), bacterial peptidoglycan (PGN) (1.5 × 106 bioparticles per ml), or CpG (300 nM) for 24 h. The TNF-α levels in the supernatant were determined by ELISA.
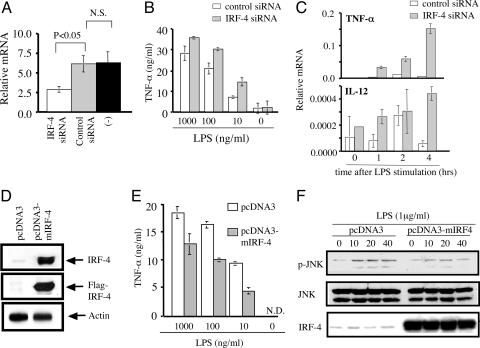
The Expression Level of IRF-4 in Macrophages Affects Their Response to LPS. The functional differences in macrophages from IRF-4+/+ and IRF-4-/- mice might be due to a direct effect of IRF-4 expression. Alternatively, they could indirectly reflect the alteration in the development of macrophages in the absence of IRF-4. To distinguish between these possibilities, we inhibited the expression of IRF-4 in normal mature peritoneal macrophages by the siRNA technique. Transfection of IRF-4-specific, but not control, siRNA resulted in the reduction of the steady-state levels of IRF-4 mRNA (Fig. 4_A_). These macrophages produced higher levels of TNF-α in response to LPS (Fig. 4_B_). Furthermore, the levels of TNF-α and IL-12 mRNA were significantly increased in these macrophages (Fig. 4_C_). We next examined the effect of the overexpression of IRF-4 by using the mouse macrophage cell line RAW264.7, which normally expresses IRF-4 at low levels (Fig. 4_D_). RAW264.7 cells that were transfected with the mouse IRF-4 cDNA construct expressed high levels of IRF-4 protein. The transfection efficiency was ≈44% as determined by the ratio of GFP-positive RAW264.7 after transfection of the GFP cDNA construct (data not shown). As expected, the production of TNF-α by RAW264.7 in response to LPS was reduced in cells overexpressing IRF-4 (Fig. 4_E_). Taken together, these results imply that IRF-4 expressed in macrophages negatively regulates the production of TNF-α in response to LPS.
Fig. 4.
The levels of IRF-4 expression in macrophages affect the production of TNF-α. (A) Peritoneal macrophages were transfected with IRF-4-specific siRNA, control siRNA, or Lipofectamine 2000 only. Eight hours later, the expression of IRF-4 mRNA was determined by real-time RT-PCR. (B) Peritoneal macrophages were transfected with control siRNA (open bars) or IRF-4 specific siRNA (filled bars), cultured for 5 h, washed, and cultured in the presence of LPS for an additional 24 h. (C) Peritoneal macrophages transfected with control or IRF-4-specific siRNA were cultured with LPS (1 μg/ml) for the indicated period. The expression levels of TNF-α and IL-12 mRNA were determined by real-time RT-PCR analysis. (D) RAW264.7 cells were transfected with pcDNA3 or pcDNA3-mIRF-4. After5hof culture, cell lysate was separated by 12.5% SDS/PAGE. The blot was probed with anti-IRF-4 Ab, stripped, and reprobed with anti-Flag mAb. The same sample was separated by SDS/PAGE, blotted, and probed with anti-actin Ab. (E) RAW264.7 cells were transfected with pcDNA3 or pcDNA3-mIRF-4, cultured for 5 h, and stimulated with LPS for 8 h. (F) RAW-264.7 cells transfected with empty vector (pcDNA3) or with IRF-4 (pcDNA3-mIRF4) were stimulated with LPS. Cell lysate was separated by 12.5% SDS/PAGE. The blot was probed with anti-phospho-JNK Ab, stripped, and reprobed with anti-JNK Ab. The same blot was stripped and reprobed with anti-IRF-4 Ab.
Activation of NF-κB and JNK in Response to LPS Is Augmented in IRF-4-/- Macrophages. To investigate the molecular basis of the enhanced cytokine production by IRF-4-/- macrophages, we first compared the expression of TLR4 by flow cytometry (Fig. 5_A_). IRF-4-/- macrophages expressed TLR4 at levels similar to IRF-4+/+ macrophages. We next investigated the downstream signaling of TLR4. TLR stimulation is linked by adaptor protein MyD88 to the activation of IRAK and TNF receptor-associated factor 6, resulting in the activation of NF-κB and MAPKs (24). We used EMSA to examine the DNA-binding activity of NF-κB in macrophages after LPS stimulation (Fig. 5_B_). NF-κB-binding activity in IRF-4-/- macrophages showed stronger signals than those in IRF-4+/+ macrophages. We did not observe significant differences in control Oct-1. We also examined the activation of MAPK pathways. We confirmed the importance of JNK activation in the LPS response by using the specific inhibitor SP600125, which completely inhibited the production of TNF-α (Fig. 5_C_) (14, 22). Therefore, we examined the activation of JNK after LPS stimulation by using phospho-JNK-specific Ab. In IRF-4+/+ macrophages, JNK1 was transiently phosphorylated after LPS treatment and was quickly inactivated. In contrast, JNK1 were phosphorylated at high levels and remained active >60 min in IRF-4-/- macrophages (Fig. 5_D_). To confirm that this augmentation of JNK activation in IRF-4-/- macrophages reflected the direct effect of IRF-4, we examined the phosphorylation of JNK in a macrophage cell line that overexpresses IRF-4 (Fig. 4_F_). The levels of JNK phosphorylation after LPS stimulation were significantly reduced in RAW264.7 cells that overexpress IRF-4. Taken together, the results showed that IRF-4 is involved in the regulation of the LPS signaling pathway at least in part by affecting the activation of both the JNK and NF-κB pathways.
IRF-4 Is Not Involved in the MyD88-Independent Signaling Pathway and Endotoxin Tolerance. The TLR4 signal bifurcates to MyD88-dependent and MyD88-independent pathways; the former pathway is responsible for the production of proinflammatory cytokines, and the latter is involved in IRF-3 activation resulting in the induction of IFN-inducible genes, such as IP-10 and IRG-1 (24). To determine whether the expression of IRF-4 influences the MyD88-independent pathway of TLR-4 signaling, LPS-induced expression of the IFN-inducible genes (IRG-1 and IP10) was compared in IRF-4+/+ and IRF-4-/- macrophages by real-time PCR (Fig. 7, which is published as supporting information on the PNAS web site). No significant difference was observed in the levels of IRG-1 and IP-10 mRNA.
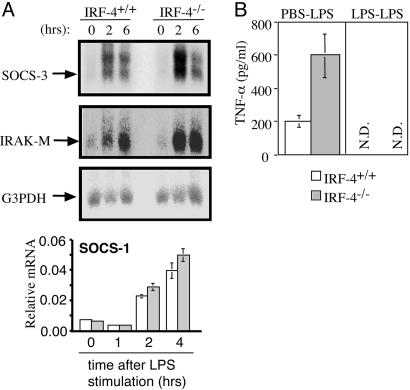
We also examined whether IRF-4 is involved in LPS tolerance, an important negative feedback mechanism to protect the host from endotoxin shock (25). After stimulation of macrophages with LPS, negative regulators of TLR4 signaling such as SOCS-1, SOCS-3, and IRAK-M are induced and protect the host from the lethal effect of excessive innate responses (16–19). Peritoneal macrophages were stimulated with LPS; the expression of SOCS-3 and IRAK-M was investigated by Northern blotting, and the expression of SOCS-1 was investigated by real-time RT-PCR. As shown in Fig. 6_A_, the expression levels of these mRNAs were not significantly reduced in IRF-4-/- macrophages. Furthermore, LPS tolerance was induced in IRF-4-/- mice (Fig. 6_B_). Therefore, the mechanisms of LPS tolerance are not impaired in IRF-4-/- macrophages.
Fig. 6.
LPS tolerance can be induced in IRF-4-/- macrophages. (A) Peritoneal macrophages were treated with LPS (1 μg/ml). The expression levels of SOCS-3, IRAK-M, and G3PDH were analyzed by Northern blotting. The relative levels of SOCS-1 mRNA were determined by real-time RT-PCR (open bars, IRF-4+/+; gray bars, IRF-4-/-). (B) Peritoneal macrophages were treated with LPS (100 ng/ml) or left untreated. After 24 h, cells were washed and restimulated with LPS (100 ng/ml) for an additional 24 h. N.D., not detected.
Discussion
The role of IRF family transcription factors in macrophage/dendritic cell function has not been clearly understood. In this report, we presented evidence that IRF-4 expressed in macrophages negatively regulates the production of proinflammatory cytokines in response to TLR ligands. Mice lacking IRF-4 showed higher serum levels of TNF-α and IL-12 after LPS inoculation and were more sensitive to the lethal effect of LPS when compared with wild type. Peritoneal macrophages in these mice produced higher levels of proinflammatory cytokines in response to TLR ligands, which we suggest is the direct result of the lack of IRF-4 expression in the macrophages for two reasons. First, normal peritoneal macrophages showed increased production of TNF-α when IRF-4 expression was down-regulated by siRNA. Second, the macrophage cell line RAW264.7, which overexpresses IRF-4, produced reduced levels of TNF-α in response to LPS. These results imply that IRF-4 has inhibitory roles for the production of TNF-α in response to LPS.
Activation of TLRs leads to the recruitment of the adaptor protein MyD88 linking TLRs to IRAK and TNF receptor-associated factor (TRAF) 6, which then activates two signaling pathways, NF-κB and MAPKs (24). It has been reported that TLR stimulation also activates other members of the IRF family: IRF-3, IRF-5, and IRF-7. Activation of IRF-3 and IRF-7 is critical for the production of type I IFNs in response to TLRs (26–28). IRF-5 is more generally involved in the TLR–MyD88 signaling pathway for induction of proinflammatory cytokines (29). Although IRF-7 and IRF-5 interact with MyD88, TRAF6, and IRAK-4 for activation, NF-κB or MAPK pathways were not impaired in mice lacking IRF-7 or IRF-5, indicating that the downstream portions of the IRF-7 and IRF-5 signaling pathways are independent of the NF-κB and MAPK pathways. However, our study suggested that IRF-4 is involved in both the NF-κB and MAPK pathways. In IRF-4-/- macrophages, activation of both NF-κB and JNK was sustained, culminating in the excessive production of the proinflammatory cytokines. We speculate that IRF-4 or its regulated gene products might be involved in activation and/or inactivation of NF-κB and JNK. Further study is required to identify the direct involvement of IRF-4 in these signaling events.
IRF-8, another member of the IRF family of transcription factors, has a structure closely related to that of IRF-4. We have recently shown that IRF-4 and IRF-8 are expressed in the majority of CD4+CD8α- and CD8α+ dendritic cells, respectively, in a mutually exclusive manner (6). IRF-4-/- mice have defects in CD4+CD8α- dendritic cells, whereas IRF-8-/- mice have defects in CD8α+ and plasmacytoid dendritic cells, suggesting that IRF-4 and IRF-8 play critical roles in the development of a distinct dendritic cell subset (6, 30, 31). Macrophages express both IRF-4 and IRF-8, but these IRFs have distinct functions in the production of cytokines. IRF-8-/- macrophages display selective impairment of IL-12 p40 expression but not other cytokines, such as IL-1, IL-6, and TNF-α (32, 33). The promoter of the IL-12 p40 gene contains an Ets site as well as a composite IRF-8-nuclear factor of activated T cells DNA-binding element, both of which play an important role for the expression of IL-12 p40 (34, 35). In contrast, IRF-4-/- macrophages produce excessive cytokines, including IL-6, TNF-α, and IL-10, in response to LPS, suggesting that the regulation of cytokine production by IRF-4 is distinct from that by IRF-8. IRF-4 may directly interact with each of these promoters and repress their transcription. Furthermore, our study suggested that IRF-4 is involved in the modulation of TLR signaling, which includes the NF-κB and JNK pathways. These two mechanisms are not necessarily mutually exclusive in regulating the production of proinflammatory cytokines. Taken together, IRF-4 and IRF-8 are both expressed in macrophage/dendritic cells and appear to have distinct functions in regulating the production of proinflammatory cytokines. The balance of the activities of these two IRF family transcription factors is critical for the fine-tuning of the innate immune response.
Although the innate immune response is critical to control the growth of pathogenic microorganisms, excessive inflammatory cytokine production is harmful to the host and can even be fatal, resulting in septic shock. Multiple mechanisms are present to prevent the harmful effect of endotoxin (24). First, the expression levels of TLR/MD2 complexes on the macrophage surface are down-regulated after LPS stimulation (36). Second, the inhibitory molecules of LPS signaling, such as SOCS-1, SOCS-3, and IRAK-M, are induced after LPS stimulation and prevent excessive production of proinflammatory cytokines (16, 18, 37). These mechanisms are called LPS tolerance, because they inhibit the response to the second challenge of the endotoxin. We have presented a third mechanism that prevents LPS-induced shock by inhibiting the TLR signaling pathway. This mechanism is mediated by IRF-4, which is constitutively expressed in macrophages and is independent of SOCS-1-, SOCS-3-, and IRAK-M-mediated modulation of cytokine production. Similar negative regulation of TLR signaling was reported in studies using mice deficient in phosphoinositide 3-kinase (PI3K) and the single Ig IL-1 receptor-related molecule (SIGIRR) (38, 39). The relationship between these inhibitory molecules of TLR signaling is unclear. Further research is warranted to examine the mechanisms of IRF-4-mediated inhibition of TLR signaling and the possible interaction of IRF-4 with other regulatory molecules. Finally, transcription factors of the IRF family, including IRF-3, IRF-4, IRF-5, IRF-7, and IRF-8, play critical positive and negative roles in the development and function of macrophage/dendritic cells, and understanding of the precise mechanisms is important for control of the innate immune response.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. K. Akagawa (National Institute of Infectious Diseases, Tokyo) for providing the RAW264.7 cell line, Mr. M. Ueda and Ms. T. Ikeda for excellent technical assistance, and Dr. G. Massey for editorial assistance with the manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan and by the 21st Century Center of Excellence Program at Nagasaki University.
Author contributions: K.H. and K. Yui designed research; K.H. and A.O. performed research; T.K., K. Yamamoto, T.T., and T.M. contributed new reagents/analytic tools; K.H., H.U., A.K., S.S., T.M., and K. Yui analyzed data; and K.H. and K. Yui wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IRF, IFN regulatory factor; TLR, Toll-like receptor; JNK, c-Jun N-terminal kinase; siRNA, small interfering RNA; MAPK, mitogen-activated protein kinase; DKO, double knockout; SOCS, suppressor of cytokine signaling; IRAK, IL-1 receptor-associated kinase; G3PDH, glyceraldehyde 3-phosphate dehydrogenase.
References
- 1.Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. (2001) Annu. Rev. Immunol. 19**,** 623-655. [DOI] [PubMed] [Google Scholar]
- 2.Eisenbeis, C. F., Singh, H. & Storb, U. (1995) Genes Dev. 9**,** 1377-1387. [DOI] [PubMed] [Google Scholar]
- 3.Matsuyama, T., Grossman, A., Mittrucker, H. W., Siderovski, D. P., Kiefer, F., Kawakami, T., Richardson, C. D., Taniguchi, T., Yoshinaga, S. K. & Mak, T. W. (1995) Nucleic Acids Res. 23**,** 2127-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marecki, S., Atchison, M. L. & Fenton, M. J. (1999) J. Immunol. 163**,** 2713-2722. [PubMed] [Google Scholar]
- 5.Rosenbauer, F., Waring, J. F., Foerster, J., Wietstruk, M., Philipp, D. & Horak, I. (1999) Blood 94**,** 4274-4281. [PubMed] [Google Scholar]
- 6.Suzuki, S., Honma, K., Matsuyama, T., Suzuki, K., Toriyama, K., Akitoyo, I., Yamamoto, K., Suematsu, T., Nakamura, M., Yui, K. & Kumatori, A. (2004) Proc. Natl. Acad. Sci. USA 101**,** 8981-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rengarajan, J., Mowen, K. A., McBride, K. D., Smith, E. D., Singh, H. & Glimcher, L. H. (2002) J. Exp. Med. 195**,** 1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohoff, M., Mittrucker, H. W., Prechtl, S., Bischof, S., Sommer, F., Kock, S., Ferrick, D. A., Duncan, G. S., Gessner, A. & Mak, T. W. (2002) Proc. Natl. Acad. Sci. USA. 99**,** 11808-11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tominaga, N., Ohkusu-Tsukada, K., Udono, H., Abe, R., Matsuyama, T. & Yui, K. (2003) Int. Immunol. 15**,** 1-10. [DOI] [PubMed] [Google Scholar]
- 10.Mittrucker, H. W., Matsuyama, T., Grossman, A., Kundig, T. M., Potter, J., Shahinian, A., Wakeham, A., Patterson, B., Ohashi, P. S. & Mak, T. W. (1997) Science 275**,** 540-543. [DOI] [PubMed] [Google Scholar]
- 11.Yamagata, T., Nishida, J., Tanaka, S., Sakai, R., Mitani, K., Yoshida, M., Taniguchi, T., Yazaki, Y. & Hirai, H. (1996) Mol. Cell. Biol. 16**,** 1283-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marecki, S., Riendeau, C. J., Liang, M. D. & Fenton, M. J. (2001) J. Immunol. 166**,** 6829-6838. [DOI] [PubMed] [Google Scholar]
- 13.Pfeffer, K., Matsuyama, T., Kundig, T. M., Wakeham, A., Kishihara, K., Shahinian, A., Wiegmann, K., Ohashi, P. S., Kronke, M. & Mak, T. W. (1993) Cell 73**,** 457-467. [DOI] [PubMed] [Google Scholar]
- 14.Bennett, B. L., Sasaki, D. T., Murray, B. W., O'Leary, E. C., Sakata, S. T., Xu, W., Leisten, J. C., Motiwala, A., Pierce, S., Satoh, Y., et al. (2001) Proc. Natl. Acad. Sci. USA 98**,** 13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumatori, A., Yang, D., Suzuki, S. & Nakamura, M. (2002) J. Biol. Chem. 277**,** 9103-9111. [DOI] [PubMed] [Google Scholar]
- 16.Kinjyo, I., Hanada, T., Inagaki-Ohara, K., Mori, H., Aki, D., Ohishi, M., Yoshida, H., Kubo, M. & Yoshimura, A. (2002) Immunity 17**,** 583-591. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa, R., Naka, T., Tsutsui, H., Fujimoto, M., Kimura, A., Abe, T., Seki, E., Sato, S., Takeuchi, O., Takeda, K., et al. (2002) Immunity 17**,** 677-687. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, K., Hernandez, L. D., Galan, J. E., Janeway, C. A., Jr., Medzhitov, R. & Flavell, R. A. (2002) Cell 110**,** 191-202. [DOI] [PubMed] [Google Scholar]
- 19.Yasukawa, H., Ohishi, M., Mori, H., Murakami, M., Chinen, T., Aki, D., Hanada, T., Takeda, K., Akira, S., Hoshijima, M., et al. (2003) Nat. Immunol. 4**,** 551-556. [DOI] [PubMed] [Google Scholar]
- 20.Beutler, B., Mahoney, J., Le Trang, N., Pekala, P. & Cerami, A. (1985) J. Exp. Med. 161**,** 984-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shakhov, A. N., Collart, M. A., Vassalli, P., Nedospasov, S. A. & Jongeneel, C. V. (1990) J. Exp. Med. 171**,** 35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swantek, J. L., Cobb, M. H. & Geppert, T. D. (1997) Mol. Cell. Biol. 17**,** 6274-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotlyarov, A., Neininger, A., Schubert, C., Eckert, R., Birchmeier, C., Volk, H. D. & Gaestel, M. (1999) Nat. Cell Biol. 1**,** 94-97. [DOI] [PubMed] [Google Scholar]
- 24.Akira, S., Takeda, K. & Kaisho, T. (2001) Nat. Immunol. 2**,** 675-680. [DOI] [PubMed] [Google Scholar]
- 25.Beeson, P. B. (1947) J. Exp. Med. 86**,** 29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda, K., Yanai, H., Mizutani, T., Negishi, H., Shimada, N., Suzuki, N., Ohba, Y., Takaoka, A., Yeh, W. C. & Taniguchi, T. (2004) Proc. Natl. Acad. Sci. USA 101**,** 15416-15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawai, T., Sato, S., Ishii, K. J., Coban, C., Hemmi, H., Yamamoto, M., Terai, K., Matsuda, M., Inoue, J., Uematsu, S., et al. (2004) Nat. Immunol. 5**,** 1061-1068. [DOI] [PubMed] [Google Scholar]
- 28.Honda, K., Yanai, H., Negishi, H., Asagiri, M., Sato, M., Mizutani, T., Shimada, N., Ohba, Y., Takaoka, A., Yoshida, N. & Taniguchi, T. (2005) Nature 434**,** 772-777. [DOI] [PubMed] [Google Scholar]
- 29.Takaoka, A., Yanai, H., Kondo, S., Duncan, G., Negishi, H., Mizutani, T., Kano, S., Honda, K., Ohba, Y., Mak, T. W. & Taniguchi, T. (2005) Nature 434**,** 243-249. [DOI] [PubMed] [Google Scholar]
- 30.Schiavoni, G., Mattei, F., Sestili, P., Borghi, P., Venditti, M., Morse, H. C., III, Belardelli, F. & Gabriele, L. (2002) J. Exp. Med. 196**,** 1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsujimura, H., Nagamura-Inoue, T., Tamura, T. & Ozato, K. (2002) J. Immunol. 169**,** 1261-1269. [DOI] [PubMed] [Google Scholar]
- 32.Giese, N. A., Gabriele, L., Doherty, T. M., Klinman, D. M., Tadesse-Heath, L., Contursi, C., Epstein, S. L. & Morse, H. C., III (1997) J. Exp. Med. 186**,** 1535-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scharton-Kersten, T., Contursi, C., Masumi, A., Sher, A. & Ozato, K. (1997) J. Exp. Med. 186**,** 1523-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, I. M., Contursi, C., Masumi, A., Ma, X., Trinchieri, G. & Ozato, K. (2000) J. Immunol. 165**,** 271-279. [DOI] [PubMed] [Google Scholar]
- 35.Zhu, C., Rao, K., Xiong, H., Gagnidze, K., Li, F., Horvath, C. & Plevy, S. (2003) J. Biol. Chem. 278**,** 39372-39382. [DOI] [PubMed] [Google Scholar]
- 36.Ziegler-Heitbrock, H. W. (1995) J. Inflamm. 45**,** 13-26. [PubMed] [Google Scholar]
- 37.Ding, Y., Chen, D., Tarcsafalvi, A., Su, R., Qin, L. & Bromberg, J. S. (2003) J. Immunol. 170**,** 1383-1391. [DOI] [PubMed] [Google Scholar]
- 38.Fukao, T., Tanabe, M., Terauchi, Y., Ota, T., Matsuda, S., Asano, T., Kadowaki, T., Takeuchi, T. & Koyasu, S. (2002) Nat. Immunol. 3**,** 875-881. [DOI] [PubMed] [Google Scholar]
- 39.Wald, D., Qin, J., Zhao, Z., Qian, Y., Naramura, M., Tian, L., Towne, J., Sims, J. E., Stark, G. R. & Li, X. (2003) Nat. Immunol. 4**,** 920-927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information