Structure of a Mycobacterium tuberculosis NusA–RNA complex (original) (raw)
Abstract
NusA is a key regulator of bacterial transcriptional elongation, pausing, termination and antitermination, yet relatively little is known about the molecular basis of its activity in these fundamental processes. In Mycobacterium tuberculosis, NusA has been shown to bind with high affinity and specificity to BoxB–BoxA–BoxC antitermination sequences within the leader region of the single ribosomal RNA (rRNA) operon. We have determined high-resolution X-ray structures of a complex of NusA with two short oligo-ribonucleotides derived from the BoxC stem–loop motif and have characterised the interaction of NusA with a variety of RNAs derived from the antitermination region. These structures reveal the RNA bound in an extended conformation to a large interacting surface on both KH domains. Combining structural data with observed spectral and calorimetric changes, we now show that NusA binding destabilises secondary structure within rRNA antitermination sequences and propose a model where NusA functions as a chaperone for nascently forming RNA structures.
Keywords: antitermination, KH domain, RNA binding, NusA
Introduction
NusA is an essential bacterial transcription factor involved in several transcriptional regulatory processes including pausing (Landick and Yanofsky, 1987; Chan and Landick, 1993), readthrough (Linn and Greenblatt, 1992) and termination (Farnham et al, 1982; Schmidt and Chamberlin, 1987). Along with this role as a general bacterial elongation factor, NusA is also a key regulator of the transcriptional antitermination observed in Escherichia coli bacteriophage systems as well as bacterial ribosomal RNAs (rRNA) (Berg et al, 1989; Friedman and Court, 1995; Weisberg and Gottesman, 1999).
Antitermination in bacterial ribosomal operons (rrns) is mediated by consensus RNA recognition sequences referred to as BoxA, BoxB and BoxC, analogous to the sequence elements that direct antitermination of λ transcripts. In rrns, these sequences are located just downstream of the ribosomal promoters close to the 5′ end of the pre-rRNA transcript. In Mycobacterium tuberculosis (M. tb), this is 1 nucleotide downstream of the point of initiation from the Pcl1 promoter (Verma et al, 1999). NusA and the other antitermination factors NusB, NusE (ribosomal protein S10) and NusG combine with these consensus sequences and interact with RNA polymerase, rendering it insensitive to termination by rho-dependent terminators that occur throughout the long (5.5 kb) pre-rRNA transcript. It has been proposed that an antitermination mechanism exists in bacterial rrns to overcome the transcriptional polarity associated with transcription–translation uncoupling and is a requirement to maintain the balanced expression of the 16S and 23S structural genes.
BoxB forms a stem–loop, required for λ N-mediated antitermination but dispensable for rrn antitermination (Gourse et al, 1986). BoxA is a highly conserved sequence with consensus UGCUCUUUAACA and has been demonstrated to bind to NusB (Luttgen et al, 2002) and to a NusB–NusE complex (Nodwell and Greenblatt, 1993; Luttgen et al, 2002). The BoxC region is less well characterised but, in the rrn of M. tb, a specific binding site for NusA that includes BoxC has been identified (Arnvig et al, 2004).
The structures of NusA from Thermotoga maritima and M. tb have been determined (Gopal et al, 2001; Worbs et al, 2001; Shin et al, 2003). The protein contains an N-terminal domain (NtD) that mediates the interaction of the protein with RNA polymerase. The NtD is coupled through a short flexible linker to three C-terminal binding domains, a single S1 domain followed by two copies of a K homology domain (KH). A model has been proposed where these two types of recognised RNA binding motif form an extended RNA binding interface (Worbs et al, 2001), but to date no structure of a NusA–RNA complex has been determined.
In this study, we have determined the structure of M. tb NusA in complex with two short oligo-ribonucleotides derived from the BoxC region of the M. tb antitermination sequence. This same sequence contains the leader sequence half of the RNaseIII processing site. We have also investigated the interaction of the antitermination sequences with NusA. The structure reveals that the RNA is bound exclusively to the two KH domains of NusA in an entirely extended conformation. Our solution data indicate that the unbound RNA forms a hairpin loop that is disrupted by the interaction with NusA. The mechanistic implications of this protein-induced RNA melting are discussed.
Results
Characterisation of the NusA binding site
The antitermination sequences from the M. tb rrn encompassing the BoxB, BoxA and BoxC elements are contained within a 63-nucleotide RNA sequence located close to the 5′ end of the ribosomal transcript, shown schematically in Figure 1A together with other RNAs used in this study. Nuclease protection assays using RNase TI, to probe for unpaired guanine residues, and RNase CVI, to probe for regions of base pairing, were carried out on a 43-nucleotide sequence (RNA43) containing just the BoxA and BoxC sequences (Figure 1B). The pattern of digestion reveals that there are strong TI cleavage sites at ribonucleotides 30, 32, 34, 43, 53 and 62 and that CVI cleavage is limited to nucleotides 35–41 and 44–49. Overall, the data from these protection assays are consistent with a structure that contains 13 base pairs arranged into two stem–loops (Figure 1B) similar to that observed in a longer RNA derived from this region (Arnvig et al, 2004).
Figure 1.
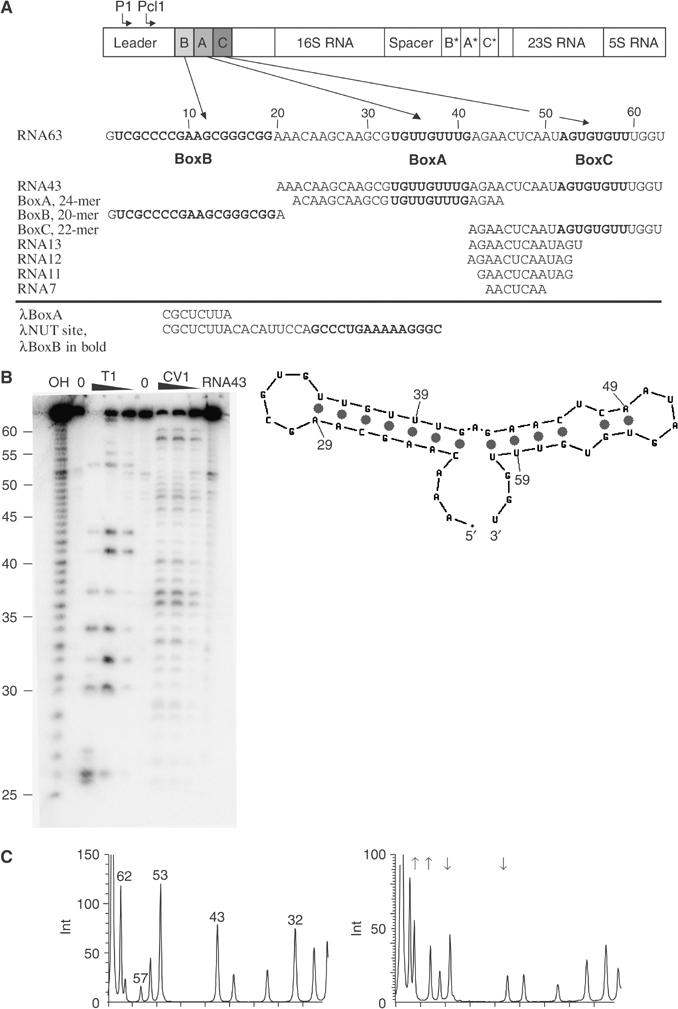
(A) Schematic representation of the M. tb rrn, indicating the position of the P1 and Pcl1 promoters and the location of the BoxA, BoxB and BoxC sequences in the leader and spacer regions. The RNA sequence corresponding to the entire leader rrn antitermination region is shown below together with the name, sequence and location of ribo-oligonucleotides used in this study. BoxB, BoxA and BoxC sequences are highlighted in bold. The sequence elements of the λ nut site are shown for comparison. (B, C) The results of ribonuclease protection assays. (B) The products of T1 and CV1 digestion of RNA43 separated by urea denaturing electrophoresis. The RNA has been digested with increasing concentrations of T1 and CV1 indicated above each track. The numbering is the same as in panel A (left). The secondary structure of RNA43 derived from nuclease protection assays combined with the prediction from the mfold algorithm version 3.1 is shown (right). (C) T1 digests of RNA43 (left) and RNA43–NusAΔNt (right) analysed by integration of the band intensities from a phosphorimaged gel. Arrows indicate the positions of protected and hypersensitive bases that result in a decrease or increase in the integrated band intensity.
Ribonuclease protection experiments were also performed in the presence of NusAΔNt, a derivative of the M. tb NusA protein that has the first 104 residues deleted. Deletion of these N-terminal residues removes the flexibly linked RNA polymerase interaction domain but does not alter the RNA binding activity of NusA in any of our assays. A comparison of the cleavage pattern that results from T1 digestion of RNA43 and the NusAΔNt–RNA43 nucleoprotein complex is shown in Figure 1C. The addition of NusAΔNt causes several changes in the T1 digestion pattern of RNA43. Cleavages at nucleotides 43 and 53 in the 5′ arm of the BoxC stem–loop are significantly decreased when the protein is bound, indicating that these nucleotides are protected from digestion when in complex with NusA. However, the intensity of bands corresponding to cleavages at nucleotides 57 and 61 in the 3′ arm of the BoxC stem–loop is increased in the nucleoprotein complex compared to free RNA, indicating that these guanines are unpaired when the protein is bound.
Evidence for secondary structure and base pairing within the antitermination sequences also comes from thermal denaturation experiments monitored by CD spectroscopy. Figure 2A–C shows the near-UV CD spectra recorded at 5 and 90°C of RNA43 together with two other ribo-oligonucleotides, BoxC-loop and RNA11. These ribo-oligonucleotides correspond to the whole of the proposed BoxC stem–loop and the 5′ arm of the stem–loop only. The spectra recorded at 5°C are characterised by a large positive maximum centred at around 269 nm. The values of Δɛ per nucleotide at the peak maxima range from 6 to 10, indicating that there is a degree of structuring in all of the RNAs. The spectra recorded at 90°C have a much lower intensity and, overall, heating induces around 60–70% reduction in the CD intensity, indicating disruption of base stacking and any other secondary structures. Figure 2D shows the melting profiles of RNA43, BoxC-loop and RNA11 monitored by CD at 269 nm (CD269). The melting profile of RNA43 is biphasic, containing a transition midpoint (_T_m) at 44°C and a Van't Hoff Δ_H_unfolding of 26 kcal mol−1. The cooperative nature of the transition indicates the presence of base pairing within the RNA. The melting of BoxC-loop produces a similar curve, this time with a _T_m at 42°C and Δ_H_unfolding=35 kcal mol−1, indicating a similar disruption of base pairing within this RNA as in RNA43. Additionally, the coincidence of the transition curves and similarity of the _T_m values means that the melting of the two stem–loops of RNA43 is likely to involve independent folding events that have a similar _T_m. In the case of RNA11, there is a large decreasing temperature dependence of the CD with increasing temperature but no single transition is apparent. Van't Hoff analysis of the RNA11 transition gives a Δ_H_unfolding of only 10 kcal mol−1. A non-cooperative melting curve of this type is typical for single-stranded nucleic acids that contain considerable stacking interactions but without base pairing (Isaksson et al, 2004). The results of these melting experiments demonstrate that the RNA43 and BoxC-loop RNAs contain significant base pairing whereas RNA11, although containing stacked bases, does not contain base pairs. Combined with the nuclease protection experiments, these data suggest that NusA disrupts the BoxC stem–loop protecting nucleotides on the 5′ arm of the BoxC hairpin from nuclease digestion and causing hypersensitivity in nucleotides on the 3′ arm.
Figure 2.
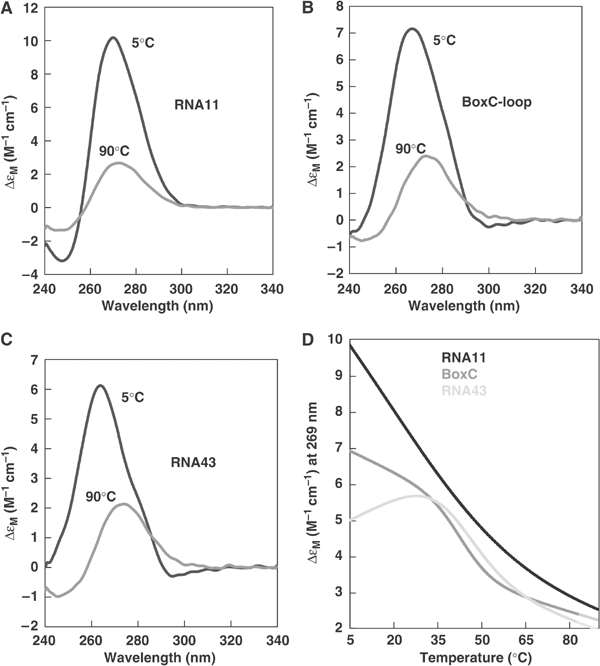
Analysis of RNA secondary structure by thermal denaturation. (A–C) Near-UV CD spectra recorded at 5°C upper curve and 90°C lower curve of (A) RNA11, (B) BoxC-loop and (C) RNA43. The spectra are expressed as Δɛ per nucleotide. (D) Thermal denaturation profile (CD269) of RNA43 (—), BoxC-loop (—) and RNA11 (—).
The NusA–BoxC interaction
In order to investigate the possibility of stem–loop disruption by NusA, we examined the effect of NusA on the UV absorbance and near-UV CD spectra of BoxC-loop and RNA11. In addition, the thermodynamics of the interaction were characterised using isothermal titration calorimetry (ITC).
The UV absorbance spectra of BoxC-loop and RNA11, before and after the addition of NusAΔNt, are presented in Figure 3A. The contribution of the protein to the absorbance spectrum is small and after subtraction, it is apparent that addition of NusAΔNt induces hyperchromicity of the BoxC-loop RNA giving rise to a significant increase in the intensity of the absorbance spectrum, whereas addition of NusAΔNt to RNA11 results in only a slight increase in the spectral intensity. As hyperchromicity in nucleic acids is largely associated with the base unstacking observed during thermal denaturation, it is a reasonable assumption that the enhancement in the extinction of BoxC-loop upon interaction with NusAΔNt is also likely to originate from base unstacking, consistent with a loss of base pairing within the RNA in the bound conformation. The small changes observed in the RNA11 spectrum are also likely to result from changes in base stacking in the single-stranded nucleotide conformation upon interaction with NusA.
Figure 3.
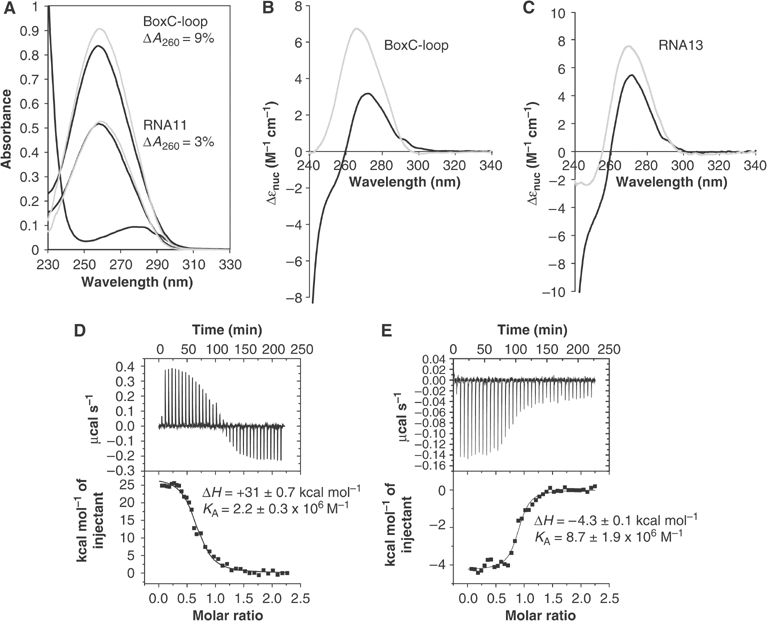
(A–C) NusAΔNt-induced UV absorbance and CD spectral changes. (A) The UV absorbance spectra of BoxC-loop (black) and BoxC-loop upon addition of NusAΔNt (grey) (upper set of curves); RNA11 (black) and RNA11 upon addition of NusAΔNt (grey) (lower set of curves); and NusAΔNt (black) (lowest curve). The spectra were recorded at an RNA concentration of 4 μM with a two-fold excess of protein added, where appropriate. The contribution of NusAΔNt to the bound spectra has been subtracted. (B) The near-UV CD spectra of 3 μM BoxC-loop (grey) and BoxC-loop plus 13 μM NusAΔNt (black). (C) The near-UV CD spectra of 4 μM RNA13 (grey) and RNA13 plus 9 μM NusAΔNt (black). The spectra are expressed in Δɛ per nucleotide. At this concentration, the contribution from NusAΔNt to the overall CD spectrum above 260 nm is minimal. (D, E) Titration of NusAΔNt with rrn antitermination sequences measured by ITC: (D) BoxC-loop and (E) RNA13. In panels D and E, the top section shows the thermogram and the bottom section shows the line of best fit to the data.
Changes in the near-UV CD spectra of BoxC-loop and RNA11 are also observed upon addition of NusAΔNt (Figure 3B and C). In both cases, binding results in a decrease in the CD intensity. The magnitude of these CD changes is greater than the equivalent UV absorbance changes and so these differences were exploited in order to construct binding isotherms for each interaction (Supplementary data). The data from these titration experiments fit well to a simple one-site heterologous equilibrium, allowing apparent association equilibrium constants in the order of 106–107 M−1 to be derived for the interaction. Notably, while the association constant for the RNA–protein interaction is similar in both cases, the percentage decrease in the spectral intensity is much greater for BoxC-loop (52%) than for RNA11 (29%). As the origin of the CD spectrum is related to the degree of base pairing and stacking within the RNA, the difference in the magnitude of these spectral changes is wholly consistent with the idea that much larger conformational changes occur in BoxC-loop than in RNA11 upon binding to NusA. Furthermore, the large decrease in CD is reminiscent of the changes observed when the RNAs undergo thermal denaturation (cf. Figure 2 with Figure 3B and C), again consistent with a loss of base stacking and RNA secondary structure.
ITC was used to examine the thermodynamics of the interaction between NusAΔNt and several RNAs derived from rrn antitermination sequences. Typical thermograms are shown in Figure 3D and E and Supplementary data. Firstly, only RNAs containing BoxC-loop-derived sequences show significant heat changes. There is no discernable heat change evident in the BoxA or BoxB titration (Supplementary data). The titration of NusAΔNt with BoxC-loop (Figure 3D) is characterised by a significant endothermic heat change, +31 kcal mol−1, together with an accompanying association constant of 2.2 × 106 M−1 for the interaction. The equivalent titration with RNA13 and NusAΔNt is shown in Figure 3E. In this case, the interaction is characterised by an exothermic heat change, −4.3 kcal mol−1, and an association constant of 8.7 × 106 M−1. In general, the association constants derived from the ITC data, measured for the BoxC-loop and RNA13 titrations, are of comparable magnitude, in the range of 106–107 M−1, and similar to the values determined from titrations monitored by CD. However, major differences are apparent between the thermodynamic signature of the NusAΔNt–BoxC-loop interaction and that of the NusAΔNt–RNA13 interaction. The NusAΔNt–BoxC-loop interaction is a strongly endothermic process, characterised by a large positive enthalpic term whereas binding of NusAΔNt to RNA13 is associated with a smaller but negative heat change. Taken together with the observed spectral changes, the likelihood is that these thermodynamic differences are a result of differing degrees of conformational rearrangement in the RNA upon binding to NusA. The strongly endothermic nature of the NusAΔNt–BoxC-loop interaction, hyperchromicity in the UV absorbance spectrum and large reductions in near-UV CD intensity provide strong evidence that NusA induces large-scale disruption of base stacking and pairing in the BoxC-loop, resulting in total destabilisation of the stem–loop structure in the RNA–protein complex. On the basis of these biophysical and biochemical data, complexes of NusA with short 9–13 ribo-oligonucleotides derived from the 5′ arm of BoxC-loop were used in subsequent crystallisation experiments.
Structure of the NusA–RNA complex
Using molecular replacement, we have determined the structure of NusAΔNt in complex with the RNA11 and RNA12 ribo-oligonucleotides. Details of data collection, structure solution and refinement are presented in Table I. The structures of the complexes differ only in the addition of a single ribonucleotide at the 5′ end of RNA12, Ade42. As a result, the base of Gua43 in NusAΔNt–RNA12 is flipped by 180° compared to NusAΔNt–RNA11. However, the axis of the rotation roughly intersects the atoms N9, N5 and O6 of the base and so the O6 of Gua43 contacts the protein in a similar manner in both complexes. The conformation of the other nucleotides is the same in both structures and it is for this reason that we refer to NusAΔNt–RNA12 when discussing Ade42 and Gua43; otherwise, the structure is discussed in terms of the higher resolution (1.55 Å) NusA–RNA11 complex.
Table 1.
Details of structure determination
| | NusAΔNt–RNA11 | NusAΔNt–RNA12 | | | ------------------------------------------------------------ | ------------------------------ | -------------------------------------- | | A. Data collection | | | | Wavelength (Å) | 0.9795 | 1.5418 | | Resolution (Å) | 30–1.55 | 15–2.25 | | Space group | P4122 | P212121 | | Cell parameters | _a_=_b_=71.4 Å, _c_=115.2 Å | _a_=64.6 Å, _b_=89.5 Å, _c_=100.9 Å | | | α=β=γ=90° | α=β=γ=90° | | | Number of reflections | 205 124 | 99 553 | | Number of unique reflections | 43 238 | 28 167 | | Completeness (%) | 98.2 (96.7)a | 99.4 (99.0)a | | I/σ(I) | 30.3 (3.1)a | 17.2 (2.9)a | | _R_sym (%) | 5.3 (50.5)a | 7.8 (46.3)a | | | | | | B. Contents of model in asymmetric unit | | | | Protein/RNA (molecules) | 1/1 | 2/2 | | Protein (residues/atoms) | 225/1690 | 446/3364 | | RNA (residues/atoms) | 11/232 | 24/508 | | Water | 264 | 208 | | | | | | C. Refinement | | | | _R_work (%) | 18.9 | 20.7 | | _R_free (%) | 22.2 | 27.4 | | | | | | D. Quality of the model | | | | R.m.s.d. bond lengths (Å) | 0.014 | 0.016 | | R.m.s.d. bond angles (deg) | 1.621 | 1.786 | | R.m.s.d. torsions period 1 (deg) | 4.889 | 4.303 | | R.m.s.d. torsions period 3 (deg) | 14.871 | 18.885 | | R.m.s.d. chiral volume (Å3) | 0.103 | 0.107 | | | | | | E. Average B-factors | | | | Average protein main-chain _B_-factor (Å2) | 21.3 | 32.1 | | Average protein side-chain _B_-factor (Å2) | 25.2 | 35.5 | | Average RNA _B_-factor (Å2) | 21.1 | 36.4 | | Average water _B_-factor (Å2) | 31.6 | 40.3 | | aNumbers in parentheses refer to the highest resolution bin. | | |
A ribbon representation together with the molecular surface of the NusAΔNt–RNA11 complex is shown in Figure 4A. At this resolution, the entire 11-mer RNA can be modelled and sugar puckers unambiguously assigned for each nucleotide (Figure 4B). Briefly, the NusAΔNt structure comprises three domains, S1, KH1 and KH2. The domains are arranged in an elongated structure that is kinked by about 100° around the KH1 domain. This arrangement together with the numbering of secondary structure elements is shown in Figure 4C. A comparison of the conformation of NusA in the protein–RNA complex with that of the free protein reveals no significant changes in the structure upon binding RNA. The r.m.s. deviation of the alpha carbon positions between the free and bound forms is 1.0 Å with the largest changes occurring sporadically throughout the S1 and KH2 domains. Similar, small localised changes have been observed in a number of other KH-domain–RNA/DNA complexes solved to date (Lewis et al, 2000; Braddock et al, 2002a). The S1 domain consists of a five-stranded antiparallel β-sheet containing a small α-helix between βS1 and βS2 and a short stretch of 310 helix between βS3 and βS4. The two remaining KH domains are made up of a three-stranded β-sheet that is flanked on one side by an α-helix. The β-sheet contains a helix–turn–helix (HTH) insertion that encompasses the conserved GXXG motif giving it the α(α)ββααβ topology common to all type II KH domains (Grishin, 2001). In comparison, the topological arrangement of secondary structures in type I KH domains is βααββα (Grishin, 2001). The structure presented here is the first one determined of a type II KH domain in complex with RNA. As a consequence of the differing topologies of type I and type II KH domains, there are some small differences between this structure and that of the structures of type I KH-domain–RNA complexes (Lewis et al, 2000; Liu et al, 2001). These differences result in a slightly twisted orientation between the HTH and the three-stranded β-sheet in this structure with respect to the type I structures. However, despite the topological differences between types I and II KH domains, the overall fold remains the same and importantly, the tertiary arrangement of the βααβ structure and position of the GXXG motif remain conserved.
Figure 4.
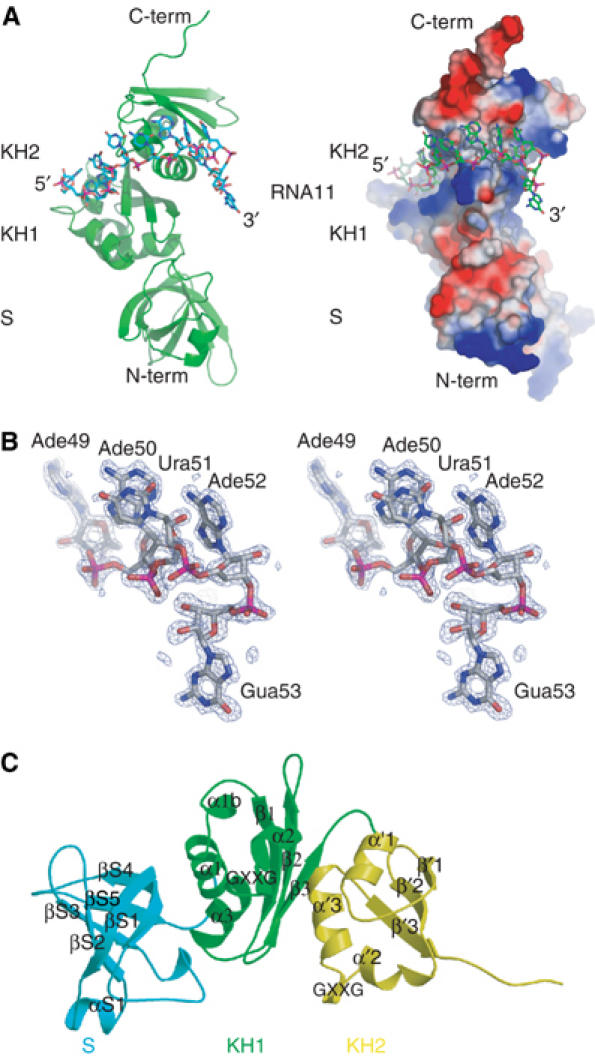
The structure of the NusAΔNt–RNA11 complex. (A) Cartoon representations of the complex. The left- and right-hand panels show the complex in the same orientation. In the left-hand panel, the protein is shown as a green ribbon and the 11-mer RNA is shown in a stick representation associated with the KH1 and KH2 domains. The right-hand panel shows a representation of the molecular surface of NusAΔNt, where the calculated electrostatic potential has been mapped onto the surface of the protein. Regions of electropositive potential are shown in blue and regions of electronegative potential are coloured red. The RNA is shown in a stick representation. (B) Stereo view of the _F_o−_F_c omit map around nucleotides Ade49 to Gua53 at 3σ contouring. (C) The arrangement of domains in NusAΔNt. The individual domains are coloured blue (S1), green (KH1) and yellow (KH2) and are shown in a ribbon representation. β-Strands and α-helices are labelled per domain and in sequential order.
A feature of the NusA structure relevant to its RNA binding activity is that the KH domains are connected by only a six-residue linker. This short linker, in combination with the 100° twist in the protein, brings the two KH domains into close proximity. As a result, the GXXG motifs are separated by only 30 Å and the two KH domains form an extended continuous RNA–protein interface or ‘super KH domain'. In the complex, there is no observable interaction of the RNA with the S1 domain and the completely single-stranded RNA is wound around the surface of the KH1–KH2 domains, steered by interactions with patches of electropositive potential (Figure 4A). Specifically, the 5′ end of the RNA (Ade42-Ade45) is bound in the groove between the HTH and β3 of KH1, Cyt46 binds to the loop between α′1 and β′1 of KH2 while Ura47 and Cyt48 make contacts to the loop between β′2 and α′2 of KH2. The nucleotides at the 3′ end of the RNA (Ade49-Ade52) are bound by the groove between the HTH and β′3 of KH2.
Sequence-specific recognition of the RNA
The NusAΔNt–RNA interface incorporates many types of interaction. These include hydrogen bonds to both amino-acid side chains and the protein backbone, electrostatic and polar interactions and to a lesser extent hydrophobic interactions between bases and non-aromatic amino-acid side chains. The details of the protein–RNA interactions are shown schematically in Figure 5. At the 5′ end of the RNA, the exocyclic N6 amino group of Ade42 is hydrogen bonded to the Oɛ2 of Glu199 and the base is involved in hydrophobic interactions with Ala234 and Gly233 in helix α2. The phosphate of Gua43 contacts the backbone nitrogen of Ile257 and the O6 of the base mediates a hydrogen bond with the Nδ2 of Asn230. The bases of the two following ribonucleotides, Ade44 and Ade45, are stacked and are sandwiched by Pro274 and Ile236, which form a hydrophobic clamp around the aromatic rings. Ade44 is further fixed in position by hydrogen bonding between the N1 and N6 of its base and the backbone amide and carbonyl of Ile257 (Figure 6A). This hydrogen bonding arrangement mimics an A:U base pair but one in which the backbone amide and carbonyl substitute for the N3 and O4 of a uracil base. The conformation of the adjacent ribonucleotide, Ade45, is stabilised by hydrogen bonding between the base N1 and the Oγ of Ser273 and by hydrogen bonds between the 2′ OH of the sugar and the Oδ1 of Asp256 and NH2 of Arg217 (Figure 6B). Ade45 contacts residues from KH1 and KH2, Arg217 and Asp256 are located on β2 and β3 of KH1 while Ser273 and Pro274 are on helix α′1 and the loop between α′1 and β′1 in KH2, illustrating how the two KH domains associate to provide a single continuous binding surface. The Ade44-Ade45 stacked base pairs provide a large degree of specificity to the NusA–RNA interaction. The combination of backbone–base hydrogen bonding and stacking interactions around Ade44 creates a binding pocket specific for adenine. In addition, the 2′ hydroxyl-mediated interaction of Ade45 sugar provides the means to discriminate directly between RNA and DNA at this position. Ribonucleotides Cyt46, Ura47 and Cyt48 effectively form a linker between the two KH recognition modules and make fewer contacts with the protein. Nevertheless, several interactions are made that contribute to the overall affinity of the complex (Figure 5).
Figure 5.
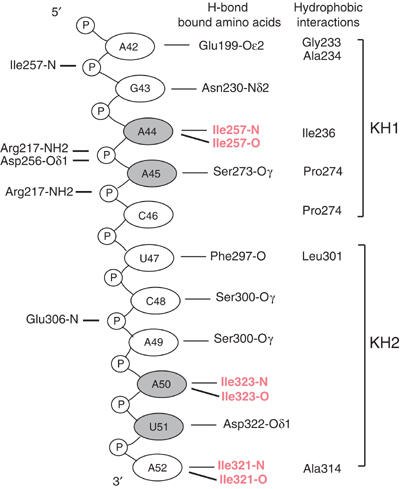
A schematic representation of the RNA–protein contacts in the NusAΔNt–RNA11 complex. Bases represented in grey circles are stacked and hydrogen bonding interactions coloured red are mediated through backbone–base contacts.
Figure 6.
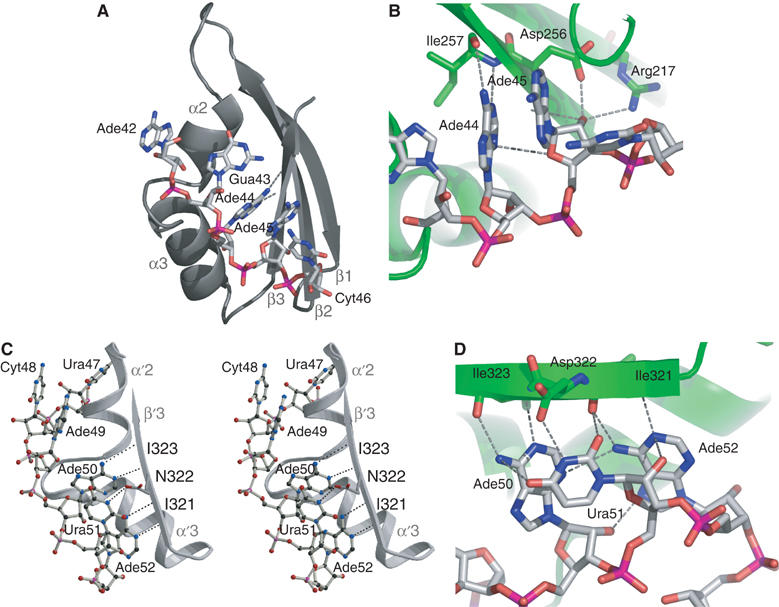
Details of the interaction of the KH domains with RNA11. (A) Interaction of KH1 with nucleotides Ade42 to Ura46 in the α/β groove of helices α2, α3 and β1–β3 of the KH1 domain. (B) A view highlighting the protein–nucleic acid interactions around Ade44 and Ade45. The protein is shown as a green ribbon. The hydrogen bonds of Ade44 to the backbone are shown together with the interactions of the 2′-hydroxy group of Ade45. (C) Stereo view of the interactions of Ade50, Ura51 and Ade52 with the protein. The path of the RNA along the groove of α′2/α′3 and β′3 in KH2 is shown. (D) Highlights of the network of polar interactions around Ade50 to Ade52. The protein and RNA are represented as in panel B.
At the 3′ end of the RNA, the tri-ribonucleotide sequence, Ade50, Ura51 and Ade52, makes up a second sequence-specific motif. Ade50 and Ade52 are hydrogen bonded to the protein backbone through the same A:U-like, base–protein backbone interaction as Ade44 and the N3 of Ura51 is hydrogen bonded to the Oδ1 of Asp322 (Figure 6C). This trinucleotide arrangement is stabilised by a network of polar interactions between the three ribonucleotides (Figure 6D). The network includes contributions from the 2′ hydroxyls of Ade50 and Ura51 that stabilise and indirectly provide specificity to the interaction. The protein residues that interact with the tri-ribonucleotide sequence Ile321, Asp322 and Ile323 are all located in β′3 of KH2. The structural arrangement of this tripeptide exposes the backbone of Ile321 and Ile323 and facilitates hydrogen bonding to the base-pair edges of Ade50 and Ade52. The bases of Ade50 and Ura51 also make a stacking interaction similar to that observed between Ade44 and Ade45. The uracil base of Ura51 is confined to a pocket flanked by the two adenines where it interacts with the side chain of Asp322. The restrictive space within this pocket implies that it would most likely only accommodate a pyrimidine and not a larger purine base.
The polar network and base stacking interactions that hold together the tri-ribonucleotide motif combined with the sequence-specific adenine polypeptide backbone interaction, size restriction in the uracil pocket and hydrogen bonding to Asp322 make an environment that is specific for the RNA sequence A-Y-A (Y=pyrimidine base). The nuclease protection experiments performed on RNA43 locate the Ade50-Ura51-Ade52 sequence in the unpaired region at the top of the BoxC stem–loop, whereas most of the other bases involved in binding to NusA are base paired in the free RNA. The extent of this polar network combined with the fact that the tri-ribonucleotide motif is presented to NusA in an unpaired conformation is suggestive of this motif being of significant functional importance. Moreover, the importance of this tri-ribonucleotide motif is reiterated by the results of experiments that show that binding is severely reduced or abolished in ribo-oligonucleotides that do not contain an intact Ade50-Ura51-Ade52 sequence (Supplementary data).
Comparison with other KH-domain–RNA/DNA complexes
The common feature of all KH-domain–RNA/DNA complexes solved to date is a hydrophobic α/β cleft that interacts with single-stranded RNA or DNA in a sequence-specific manner. In all cases, hydrogen bonds between bases and both amino-acid side chains and the protein backbone stabilise the interaction, as do hydrophobic and electrostatic interactions. To date, no examples of stacking interactions between bases and aromatic amino-acid side chains have been observed in KH–RNA structures. This is also true for the NusAΔNt–RNA complex where hydrogen bonding appears to dominate this interaction. In contrast, in other RNA binding domains such as RRMs (Burd and Dreyfuss, 1994) or the human Puf protein, Pumilio1 (Wang et al, 2002), ring stacking interactions appear to be critically important for specificity and stability.
All the structures of KH domains bound to single-stranded RNA show conservation of the base-pair-like adenine and protein backbone interaction (Ade44-Ile257, Ade50-Ile323 and Ade52-Ile321 in the NusAΔNt–RNA structure). In the other examples of KH–RNA complexes (Lewis et al, 2000; Liu et al, 2001), adenine–backbone interactions appear to be major determinants of specificity and have been proposed to have a functional significance in the case of splicing factor 1 (SF1) (Liu et al, 2001). A structural overlap of KH1 and ribonucleotides Ade42 to Cyt46 with KH2 and ribonucleotides Ura48 to Gua53 reveals that the adenine bases of Ade44 and Ade50 are superimposable (Figure 7). In both ribonucleotide motifs, the adenine bases make equivalent hydrogen bonds to the protein backbone. The importance of the adenine–backbone interaction is underlined by comparison with other KH structures. A structure-based sequence alignment (Figure 7A) reveals that Ile257 and Ile323 are highly conserved. Moreover, structural superposition of the KH2-bound RNA in the NusAΔNt–RNA complex with the RNA in the Nova KH3–RNA structure (Figure 7C) reveals that the degree of overlap is strongest at the equivalent adenines involved in the backbone interaction (Ade42-Gua43-Ade44-Ade45 in NusA KH1, Cyt48-Ade49-Ade50-Ura51 in NusA KH2, Ura12-Cyt13-Ade14-Cyt15 in Nova KH3 and Ura6-Ade7-Ade8-Cyt9 in SF1). A similar comparison of the NusAΔNt–RNA structure with KH-domain–DNA structures of far-upstream element (_F_USE) _b_inding _p_rotein (FBP) and hnRNP in complex with single-stranded DNA (ssDNA) (Braddock et al, 2002a, 2002b) reveals much larger differences between the protein–nucleic acid interfaces. Although the nucleic acid binding sites in the structures of FBP and hnRNP bound to ssDNA have the same overall orientation as the RNA binding site in NusA, the ssDNA in these complexes displays a right-handed helical geometry with all bases parallel to each other. In contrast, the ribonucleotide conformation in the NusA–RNA structure and the other KH–RNA structures displays a larger variety of torsion angles between base, sugar and phosphate. In fact, in the NusA–RNA complex, only the conserved bases Ade44 and Ade50 have torsion angles corresponding to that of an A-form helix. The strong conservation of protein and nucleic acid conformation at the RNA–protein interface shows that this mode of recognition is likely to be a species-wide feature, common to many KH–RNA complexes. Moreover, the presence of 2′ specific interactions and the fact that conservation of nucleic acid conformation is not extended to KH–DNA complexes is suggestive of this type of interaction being important for KH domains to discriminate between RNA and DNA.
Figure 7.
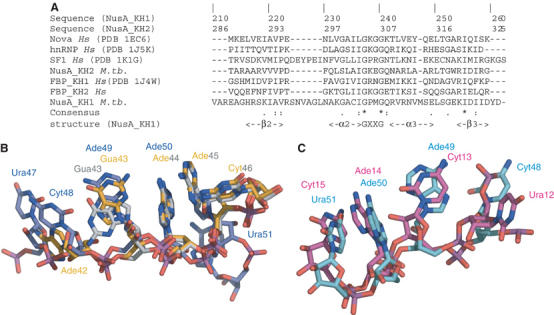
(A) Multiple sequence alignment of the βααβ motif of KH domains. Secondary structure elements were assigned based on the X-ray structure. Residues that are 100% conserved include the glycines of the GXXG motif and isoleucine in β-strand 3 where the backbone is in contact with Ade44 or Ade50. (B) Structural superposition of KH1 in the NusAΔNt–RNA11 complex (grey) with KH1 in the NusAΔNt–RNA12 complex (yellow) and with KH2 (blue). (C) Structural superposition of RNA in KH2 (blue) with RNA bound to Nova (red).
Discussion
Structure of the NusA–RNA complex
In the NusA–RNA complex, the BoxC-loop-derived RNA is bound in an extended conformation contacting both KH domains while not interacting with the S1 domain. The RNA–protein interaction mediated by this double KH domain differs substantially from the interaction observed in the only other structure of a double KH domain bound to a nucleic acid target, the double KH domain of FBP bound to ssDNA from the FUSE (Braddock et al, 2002b). In the FBP–FUSE complex, the KH domains are connected by a flexible 30-residue linker and, in the free protein, the individual domains tumble independently of each other. In the complex, a 5-nucleotide non-interacting nucleic acid spacer separates the two bound DNA recognition sequences and hence, while tethered to each other through the protein–nucleic acid interaction, the two KH domains act independently of one another. In contrast, in NusA, the two KH domains are connected by a much shorter six-residue linker and the two KH domains associate with each other to produce a single continuous binding surface that interacts with an uninterrupted 10-ribonucleotide recognition sequence. In both cases, the coupling of multiple RNA binding domains in either an associative or an independent mode will result in an increase in both the specificity and affinity of the RNA–protein interaction. However, the major difference between these modes of interaction illustrates the versatility and modularity of KH domains. They may act in an associative manner to produce a single long uninterrupted protein–RNA interface, as is the case in NusA, or they can act in an independent fashion and have the effect of coupling shorter, separated RNA recognition sequences together. The idea of RNA binding modules acting either in an independent or an associative manner is not limited to the KH family and has also been observed in proteins containing multiple RRMs (Ding et al, 1999; Handa et al, 1999).
Another important question is what mediates the sequence specificity of the RNA–protein interaction and how much, if any, RNA/DNA discrimination is made by the NusA protein. Much of the NusA–RNA interaction is mediated through hydrogen bonding interactions similar to those observed in other KH–RNA structures (Lewis et al, 2000; Liu et al, 2001). Base–aromatic ring stacking interactions, important in RRM–RNA interactions, are not present in the NusAΔNt–RNA structure and this mode of interaction appears not to be utilised by KH domains at least in the structures solved to date. A major component of the sequence specificity of the NusA–RNA complex appears to be mediated by adenine–backbone interactions. The complex contains three of these adenine-specific interactions at Ade44, Ade50 and Ade52 and a further pyrimidine-specific interaction at Ura51. This adenine–backbone interaction appears to be an important common feature of KH–RNA complexes and is utilised as a mode of recognition by the KH3 domain of Nova-2 (Lewis et al, 2000) and by the KH domain of SF1 (Liu et al, 2001). In addition to base-specific interactions, the interface contains three RNA-specific 2′ OH-mediated interactions. In the first, the 2′ OH of Ade45 provides specificity directly by hydrogen bonding to the Oδ1 of Asp256 and NH2 of Arg217. The remaining two 2′ OH interactions are internucleotide and provide RNA specificity indirectly by contributing to the polar network responsible for stabilising the bound conformation of the trinucleotide sequence Ade50-Ura51-Ade52. Similar internucleotide 2′ OH-mediated interactions are also involved in stabilisation of the RNA conformation in the Sex-lethal–traRNA complex (Handa et al, 1999). More generally, RNA/DNA discrimination may be provided by the ability of bound RNA to adopt a greater range of torsion angles. This is illustrated by the fact that, in the structures of KH–ssDNA complexes (Braddock et al, 2002a, 2002b), the ssDNA has a much more regular arrangement compared with the wide variety of torsional space sampled by the ribonucleotides in the NusAΔNt–RNA complex.
NusA–RNA binding
NusA interacts tightly with several ribo-oligonucleotides derived from the rrn antitermination sequences. However, the interaction only occurs with ribo-oligonucleotides that contain the BoxC-loop region and we can detect no interaction of NusA with either BoxB- or BoxA-derived sequences measured by ITC (Supplementary data) or by gel retardation assays (data not shown). The observation that the NusA binding site is located in the BoxC-loop region is in accord with one previous study (Arnvig et al, 2004) but not with other studies of E. coli NusA, which suggested that in combination with λ N, the BoxA motif is the binding site for NusA (Mogridge et al, 1995). It is likely that this altered binding specificity is the result of mechanistic differences between rrn antitermination and that seen in λ.
The interaction of NusA with the BoxC stem–loop is characterised by its strong endothermic nature, Δ_H_=+31 kcal mol−1. In view of this unfavourable enthalpy and given the sub-micromolar equilibrium dissociation constant, there is a significant favourable entropic term associated with the formation of the NusA–BoxC-loop complex. Binding is also accompanied by large changes in both the UV absorbance spectrum and near-UV CD spectrum of the BoxC-loop RNA. Taken together, these observations are strong indicators that NusA destabilises the secondary structure of the BoxC-loop either by binding preferentially to an unfolded form of BoxC-loop and perturbing the conformational equilibrium or by binding to the folded form and inducing an isomerisation event. Whatever the case, both of these possibilities result in melting of the BoxC stem–loop. Similar hyperchromicity and spectral changes are associated with induced RNA melting by the HIV P7 nucleocapsid upon interaction with the cTAR stem–loop (Beltz et al, 2003), and spectral changes are also associated with the RNA melting activity of CspE, a bacterial cold-shock protein (Phadtare et al, 2004). Interestingly, CspE and other bacterial cold-shock proteins have significant antitermination activity (Bae et al, 2000) and this activity is likely to be directly related to their ability to destabilise RNA secondary structures during the cold-shock response (Phadtare et al, 2002). NusA is also upregulated during the cold-shock response (Bae et al, 2000), indicating that greater levels of the protein are required under conditions where RNA secondary structures are more stable.
NusA-induced secondary structure destabilisation
The observations presented here raise the question of what the significance of this might be in rrn antitermination, rRNA processing and NusA's other functions in transcriptional elongation and pausing. It has previously been demonstrated that NusA is associated with the flap domain of RNAP (Toulokhonov et al, 2001), where it can interact with the 5′ arm of hairpins that form intrinsic terminators (Gusarov and Nudler, 2001; Toulokhonov et al, 2001). In this context, it is suggested that NusA competes for the 5′ arm of the hairpin with a weak upstream RNA binding site on RNAP. This competitive effect then promotes formation of a stem–loop with the nascently forming 3′ arm of the terminator. It is clear from these observations and our data that NusA can interact with nascently forming RNA structures in what could be regarded as an RNA chaperone function. In some cases, this may destabilise an RNA secondary structure and in others promote the formation of stem–loops by a competitive mechanism involving other RNA binding proteins/sites (Gusarov and Nudler, 2001). In the light of this hairpin destabilisation activity, one postulate is that the function of NusA in rrn antitermination is to prevent the formation of weak stem–loops during rRNA transcription. Another attractive possibility is the idea that the KH domains in NusA are involved in a mechanism similar to that proposed for SF1. Here, the interaction of the KH domain of SF1 with RNA is important as an intermediary in the pre-mRNA splicing reaction (Liu et al, 2001). In this case, the branch point site RNA (BPS RNA) is bound on the surface of the KH domain of SFI in an extended conformation, with the base of the catalytic branch point adenylate oriented to make the same two hydrogen bonds to the protein backbone as Ade44, Ade50 and Ade52 do in the NusA–RNA complex. It is suggested that this initial KH–RNA interaction is important to prearrange the conformation of the single-stranded BPS RNA in order to facilitate the formation of the BPS/U2 snRNA duplex and to position the branch point adenylate in the required bulged conformation within this duplex. Given the similarity in the mode of RNA recognition by NusA and SFI, it is tempting to speculate that this specific NusA–RNA complex may represent an intermediary directly involved in rRNA processing. Some weight is lent to this suggestion by the fact that the RNaseIII processing site in the rRNA leader sequence (Verma et al, 1999) is actually part of the NusA recognition site (G43AACUC48). The mechanism of the excision of the 16S rRNA from the rRNA precursor involves the formation of a stretch of double-stranded RNA between this NusA-bound sequence and a complementary sequence in the rRNA spacer region in order for it to be cleaved by RnaseIII and release the 16S rRNA. Just as SF1 is required to present the BPS RNA to the U2 snRNA, NusA may be required to present the leader part of the RNaseIII site to the complementary sequence present in the spacer. If this were the case, the presence of NusA might be required to enhance RNaseIII processing of rRNA transcripts, an idea that remains to be tested.
Materials and methods
Protein expression and purification
The DNA sequence coding for NusAΔNt (residues 105–347) was isolated by PCR amplification from the M. tb genome. The DNA fragment was inserted into the _Nde_I and _Xho_I sites of pET22b (Novagen) in order to produce a C-terminal hexa-histidine fusion. The nucleotide sequence of the expression clone was verified by automated DNA sequencing. NusAΔNt was expressed in the E. coli strain BL21 (DE3) and purified from clarified crude cell extracts using ion exchange, nickel affinity and gel filtration chromatography. The purity and monodispersity of preparations were monitored by ESI-MS, SDS–PAGE and photon correlation spectroscopy. Protein concentration was determined from the absorbance at 280 nm using a molar extinction coefficient derived by summing the contributions from tyrosine and tryptophan residues (9600 M−1 cm−1).
Preparation of RNA and nuclease protection assays
Short sequences of RNA derived from the antitermination region of the M. tb rrn operon (7-mer to 13-mer RNA, BoxA, BoxB and BoxC-loop; Figure 1) were purchased from Curevac, Germany (HPLC purified) or from Eurogentec Ltd, Belgium (gel purified). RNA43 was synthesised by in vitro transcription. The preparation of this RNA and nuclease protection assays were carried out as described earlier (Arnvig et al, 2004). A detailed description of RNA preparation and nuclease protection experiments is provided in Supplementary data.
Crystallisation, data collection, structure determination and refinement
Prior to crystallisation, RNA and protein were dialysed against 20 mM Tris pH 8.0, 150 mM NaCl and 1 mM EDTA. NusAΔNt–RNA11 crystals were obtained by sitting drop vapour diffusion against 0.15 M Li2SO4, 18% PEG 4000 and 0.1 M Tris–HCl, pH 8.5, at 18°C with a protein concentration of 240 μM and a protein to RNA ratio of 1:2 in the drop. For cryo-protection, 2 μl of reservoir solution containing 25% glycerol was added to the drop before the crystal was transferred to the same solution. The crystals grow in the tetragonal space group P4122 with one protein–RNA complex per AU.
Data were collected at beamline 14.2, Daresbury Laboratories at 100 K and processed using the HKL program package (Otwinowski and Minor, 1997). The structure was solved by molecular replacement using the CCP4 program AMORE (Navaza, 2001) with the three C-terminal domains of the M. tb NusA structure as a search model. An automatic water search/refinement was performed with ARP/REFMAC (Murshudov et al, 1997), showing clearly the position of the RNA. The model of the RNA was then manually built in O (Jones et al, 1991) followed by cycles of refinement in REFMAC and rebuilding in O. The data were refined to 1.55 Å resolution using a model containing amino acids 108–333 and all 11 RNA nucleotides. The stereochemical quality of the protein model was assessed with PROCHECK (Laskowski et al, 1993), and RNA torsions and sugar puckers were analysed with AMIGOS (Duarte and Pyle, 1998). Only two residues are outside the preferred φ/ψ regions and fall into flexible loop regions. All sugar puckers are in ranges available to RNA.
Crystals of NusAΔNt–RNA12 were obtained in sitting drops equilibrated against 10 mM KH2PO4 and 19% PEG 8000 at 18°C with a protein concentration of 240 μM and a protein to RNA ratio of 1:2. Cryo-protection was achieved in the same way as for NusA–RNA11. The crystals belong to the space group P212121 and contain two protein–RNA complexes per AU. Data were collected at 100 K on an RAXIS image plate detector with a copper-rotating anode as the X-ray source. The structure was solved using the NusA–RNA11 structure as a search model. Refinement and model building were carried out as described for the NusAΔNt–RNA11 complex. The structure was refined to convergence at 2.25 Å resolution using a model containing amino acids 105 (107)–329 and all 12 RNA nucleotides. Again, the protein model is of excellent quality and the sugar puckers are all in the preferred regions for RNA.
UV absorbance and CD spectroscopy
UV absorbance data were recorded on a Cary 400 UV/Vis spectrophotometer. CD data were recorded using a Jasco 715 spectropolarimeter equipped with a Peltier temperature controller. UV hyperchromicity measurements, CD binding and melting experiments were all conducted in 150 mM NaCl, 20 mM Tris–HCl pH 7.8 and 1 mM EDTA. Thermal denaturation of RNAs was carried out by heating samples at a constant rate of 2°C per minute from 5 to 90°C. The melting profile of the RNA was monitored by recording the CD at 269 nm while heating. _T_m's for thermal transitions were determined from derivative plots or by Van't Hoff analysis of the data. Binding of NusAΔNt to ribo-oligonucleotides was also monitored by CD spectroscopy. Typically, titrations were carried out at 18°C with a fixed ribo-oligonucleotide concentration of ∼3 μM and a varying NusAΔNt concentration up to a stoichiometric ratio of 3:1. Binding was monitored by recording CD spectra from 240 to 320 nm after each protein addition. The decrease in the RNA CD at 269 nm caused by addition of NusA was used to construct binding isotherms and these were fitted by nonlinear regression using a single-site model.
Isothermal titration calorimetry
ITC was performed using a VP-ITC microcalorimeter (MicroCal Inc.). Data were analysed using the ‘Origin'-based software provided by the manufacturers. Briefly, NusAΔNt and RNAs were dialysed into 150 mM NaCl, 20 mM Tris–HCl pH 7.8 and 1 mM EDTA. Titrations were carried out at 18°C and in a typical experiment 4–20 μM RNA was loaded into the sample cell and titrated against 40–200 μM NusAΔNt in the injection syringe.
Coordinates
The atomic coordinates of NusAΔNt–RNA12 and NusAΔNt–RNA11 have been deposited in the Protein Data Bank under ID codes 2ATW and 2ASB, respectively.
Supplementary Material
Supplementary Methods
Acknowledgments
We thank Dr Steve Smerdon and Dr Andrew Lane for critical reading of the manuscript.
References
- Arnvig KB, Pennell S, Gopal B, Colston MJ (2004) A high-affinity interaction between NusA and the rrn nut site in Mycobacterium tuberculosis. Proc Natl Acad Sci USA 101: 8325–8330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W, Xia B, Inouye M, Severinov K (2000) Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci USA 97: 7784–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz H, Azoulay J, Bernacchi S, Clamme JP, Ficheux D, Roques B, Darlix JL, Mely Y (2003) Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol 328: 95–108 [DOI] [PubMed] [Google Scholar]
- Berg KL, Squires C, Squires CL (1989) Ribosomal RNA operon anti-termination. Function of leader and spacer region box B-box A sequences and their conservation in diverse micro-organisms. J Mol Biol 209: 345–358 [DOI] [PubMed] [Google Scholar]
- Braddock DT, Baber JL, Levens D, Clore GM (2002a) Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J 21: 3476–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock DT, Louis JM, Baber JL, Levens D, Clore GM (2002b) Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature 415: 1051–1056 [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Chan CL, Landick R (1993) Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol 233: 25–42 [DOI] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev 13: 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte CM, Pyle AM (1998) Stepping through an RNA structure: a novel approach to conformational analysis. J Mol Biol 284: 1465–1478 [DOI] [PubMed] [Google Scholar]
- Farnham PJ, Greenblatt J, Platt T (1982) Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell 29: 945–951 [DOI] [PubMed] [Google Scholar]
- Friedman DI, Court DL (1995) Transcription antitermination: the lambda paradigm updated. Mol Microbiol 18: 191–200 [DOI] [PubMed] [Google Scholar]
- Gopal B, Haire LF, Gamblin SJ, Dodson EJ, Lane AN, Papavinasasundaram KG, Colston MJ, Dodson G (2001) Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 Å resolution. J Mol Biol 314: 1087–1095 [DOI] [PubMed] [Google Scholar]
- Gourse RL, de Boer HA, Nomura M (1986) DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell 44: 197–205 [DOI] [PubMed] [Google Scholar]
- Grishin NV (2001) KH domain: one motif, two folds. Nucleic Acids Res 29: 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Nudler E (2001) Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell 107: 437–449 [DOI] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398: 579–585 [DOI] [PubMed] [Google Scholar]
- Isaksson J, Acharya S, Barman J, Cheruku P, Chattopadhyaya J (2004) Single-stranded adenine-rich DNA and RNA retain structural characteristics of their respective double-stranded conformations and show directional differences in stacking pattern. Biochemistry 43: 15996–16010 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Landick R, Yanofsky C (1987) Isolation and structural analysis of the Escherichia coli trp leader paused transcription complex. J Mol Biol 196: 363–377 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323–332 [DOI] [PubMed] [Google Scholar]
- Linn T, Greenblatt J (1992) The NusA and NusG proteins of Escherichia coli increase the in vitro readthrough frequency of a transcriptional attenuator preceding the gene for the beta subunit of RNA polymerase. J Biol Chem 267: 1449–1454 [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M (2001) Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Luttgen H, Robelek R, Muhlberger R, Diercks T, Schuster SC, Kohler P, Kessler H, Bacher A, Richter G (2002) Transcriptional regulation by antitermination. Interaction of RNA with NusB protein and NusB/NusE protein complex of Escherichia coli. J Mol Biol 316: 875–885 [DOI] [PubMed] [Google Scholar]
- Mogridge J, Mah TF, Greenblatt J (1995) A protein–RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev 9: 2831–2845 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53 (Patt 3): 240–255 [DOI] [PubMed] [Google Scholar]
- Navaza J (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr D 57: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Nodwell JR, Greenblatt J (1993) Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell 72: 261–268 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Phadtare S, Inouye M, Severinov K (2004) The mechanism of nucleic acid melting by a CspA family protein. J Mol Biol 337: 147–155 [DOI] [PubMed] [Google Scholar]
- Phadtare S, Tyagi S, Inouye M, Severinov K (2002) Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J Biol Chem 277: 46706–46711 [DOI] [PubMed] [Google Scholar]
- Schmidt MC, Chamberlin MJ (1987) nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol 195: 809–818 [DOI] [PubMed] [Google Scholar]
- Shin DH, Nguyen HH, Jancarik J, Yokota H, Kim R, Kim SH (2003) Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain. Biochemistry 42: 13429–13437 [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R (2001) Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science 292: 730–733 [DOI] [PubMed] [Google Scholar]
- Verma A, Sampla AK, Tyagi JS (1999) Mycobacterium tuberculosis rrn promoters: differential usage and growth rate-dependent control. J Bacteriol 181: 4326–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM (2002) Modular recognition of RNA by a human pumilio-homology domain. Cell 110: 501–512 [DOI] [PubMed] [Google Scholar]
- Weisberg RA, Gottesman ME (1999) Processive antitermination. J Bacteriol 181: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC (2001) An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell 7: 1177–1189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods