Identification of the hydrophobic strand in the A–B loop of leptin as major binding site III: implications for large-scale preparation of potent recombinant human and ovine leptin antagonists (original) (raw)
Abstract
Interaction of leptin with its receptors resembles that of interleukin-6 and granulocyte colony-stimulating factor, which interact with their receptors through binding sites I–III. Site III plays a pivotal role in receptors' dimerization or tetramerization and subsequent activation. Leptin's site III also mediates the formation of an active multimeric complex through its interaction with the IGD (immunoglobulin-like domain) of LEPRs (leptin receptors). Using a sensitive hydrophobic cluster analysis of leptin's and LEPR's sequences, we identified hydrophobic stretches in leptin's A–B loop (amino acids 39–42) and in the N-terminal end of LEPR's IGD (amino acids 325–328) that are predicted to participate in site III and to interact with each other in a β-sheet-like configuration. To verify this hypothesis, we prepared and purified to homogeneity (as verified by SDS/PAGE, gel filtration and reverse-phase chromatography) several alanine muteins of amino acids 39–42 in human and ovine leptins. CD analyses revealed that those mutations hardly affect the secondary structure. All muteins acted as true antagonists, i.e. they bound LEPR with an affinity similar to the wild-type hormone, had no agonistic activity and specifically inhibited leptin action in several leptin-responsive in vitro bioassays. Alanine mutagenesis of LEPR's IGD (amino acids 325–328) drastically reduced its biological but not binding activity, indicating the importance of this region for interaction with leptin's site III. FRET (fluorescence resonance energy transfer) microscopy experiments have documented that the transient FRET signalling occurring upon exposure to leptin results not from binding of the ligand, but from ligand-induced oligomerization of LEPRs mediated by leptin's site III.
Keywords: A–B loop of leptin, antagonists, fluorescence resonance energy transfer (FRET), hydrophobic strand, immunoglobulin-like domain (IGD), site-directed mutagenesis
Abbreviations: APB, acceptor photobleaching; CFP, cyan fluorescent protein; ch, chicken; CHO, Chinese-hamster ovary; CRH, cytokine receptor homology domain; ERK, extracellular-signal-regulated kinase; fluorescence resonance energy transfer (FRET); G-CSF, granulocyte colony-stimulating factor; h, human; HCA, hydrophobic cluster analysis; HEK-293T cells, human embryonic kidney 293T cells; IBs, inclusion bodies; IL-6, interleukin-6; IGD, immunoglobulin-like domain; LBD, leptin-binding domain; LEPR, leptin receptor; m, mouse; MAPK, mitogen-activated protein kinase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2_H_-tetrazolium bromide; SPR, surface plasmon resonance, STAT3, signal transducer and activator of transcription 3; TFA, trifluoroacetic acid; v, viral; WT, wild-type; YFP, yellow fluorescent protein
INTRODUCTION
Although leptin and LEPR (leptin receptor) were cloned a decade ago [1,2] and leptin's three-dimensional structure was solved [3], the mechanism of leptin-induced LEPR activation was not yet elucidated and several putative models have been proposed [4]. The extracellular domain of mature h (human). LEPR consists of 820 amino acids divided into several subdomains: (i) N-terminal CRH1 (cytokine receptor homology domain 1), amino acids 1–307; (ii) IGD (immunoglobulin-like domain), amino acids 308–406; (iii) CRH2, amino acids 407–616; and (iv) two consecutive F3 domains, amino acids 617–820. Fong et al. [5] localized the LBD (leptin-binding domain) as the CRH2. This domain from human [6] and ch (chicken) LEPR [7] was subcloned in our laboratory and expressed as a recombinant protein, which showed a 1:1 molar interaction with leptin. Only recently, the pioneering work of Tavernier and co-workers [8] revealed that leptin binding to its receptor resembles the interaction between IL-6 (interleukin-6) and its receptor [9–11] and formulated the existence of a putative leptin site III as a major site responsible for the formation of the active 2:2 or 2:4 leptin–LEPR complex. IGD of LEPR was documented to be essential for leptin-induced activation of the LEPR. Its removal attenuated activation, but not binding of the ligand [8]. This model was further substantiated by the extensive mutagenesis of mouse and human leptins, which led to identification of Ser120 and Thr121, located in the N-terminal part of helix D, as contributors to site III [12]. Mutation of these amino acids to alanine converted leptin into a leptin antagonist.
To determine whether, in addition to the N-terminal part of helix D, other parts of the leptin molecule also contribute to the interaction between leptin and LEPR's IGD, we carefully analysed the known structures of IL-6–receptor complexes {v (viral) IL-6–gp130 [9] and IL-6–IL-6Rα–gp130 complex [10]}, in which site III was first identified. This site is formed by the extensive interface between the A–B loop and the N-terminal part of helix D in vIL-6, and the edge of the gp130 IGD β-sheets, involving strands F and G. In the present study, we compared the leptin and LEPR sequences with those of IL-6 and gp130, using the sensitive bidimensional HCA (hydrophobic cluster analysis) [13] and predicted in leptin and LEPR two short β-strands, which are observed to interact with each other in the vIL-6–gp130 complex [10]. They are located in the A–B loop of the cytokine and in the N-terminal part of the receptor's IGD core. To verify this hypothesis and to test its generality, we prepared and purified to homogeneity several ovine and human recombinant leptin alanine muteins of the A–B loop β-strand and documented that they act as potent competitive antagonists. In parallel, the alanine mutagenesis of the hydrophobic strand in LEPR's IGD resulted in a remarkably reduced activity. Preliminary results of the present study were presented at the Nature Biotechnology Winter Symposium [14].
MATERIALS AND METHODS
Materials
Recombinant chLBD [7] and ovine and human leptins were prepared in our laboratory as described previously [15,16]. Restriction enzymes used in the molecular biology experiments were from MBI Fermentas (Vilnius, Lithuania) and New England Biolabs (Beverly, MA, U.S.A.). High purity DNA primers were obtained from Sigma (Rehovot, Israel). RPMI 1640 (medium), IL-3, nalidixic acid and MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2_H_-tetrazolium bromide; Thiazolyl Blue) were purchased from Sigma Chemical (St. Louis, MO, U.S.A.), fetal calf serum was from Bio Lab (Jerusalem, Israel) and a Superdex™75 HR 10/30 column and Q-Sepharose were from Pharmacia LKB Biotechnology AB (Uppsala, Sweden). A research-grade CM5 sensor chip, NHS (_N_-hydroxysuccinimide), EDC [_N_-ethyl-_N_′-(3-dimethylaminopropyl)-carbodi-imide hydrochloride], ethanolamine/HCl and HBS-EP running buffer (10 mM Hepes, 150 mM NaCl, 3.4 mM EDTA and 0.005%, v/v, surfactant P20 at pH 7.4) were purchased from Biacore AB (Uppsala, Sweden). All other chemicals were of analytical grade.
HCA
HCA [13,17] was used for sequence analysis. This two-dimensional method is based on the physicochemical and topological properties underlying the fold of globular domains and allows direct access, from a single sequence, to the gravity centres of regular secondary structures. Indeed, the positions of hydrophobic clusters defined using HCA, which distinguish simple binary patterns, mainly correspond to those of regular secondary structures [18,19]. This information can be used to efficiently compare highly divergent sequences (as secondary structure is often much more conserved than sequence) and identify remote relationships [20,21]. This method was used, among others, over 10 years ago to identify domain duplication in the cytokine family of receptors [22]. Three-dimensional structures were manipulated using a Swiss-PdbViewer [23].
Preparation of leptin mutants
To prepare leptin mutants, the pMon3401 expression plasmids encoding WT (wild-type) ovine or human leptin [15,16] were used as starting material. To make double distant mutations, ovine leptin mutein Y110A (Tyr110→Ala) and human leptin mutein D39A/L40A/F41A/I42A were used. The leptin inserts were modified with the Stratagene QuickChange mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) according to manufacturer's instructions, using two complementary primers (see Supplementary Table S1 at http://www.BiochemJ.org/bj/391/bj3910221add.htm). The procedure included 18 PCR cycles using Pfu polymerase. The mutated construct was then digested with DpnI restriction enzyme, which is specific to methylated and hemi-methylated DNA (target sequence: 5′-Gm6ATC-3′), in order to digest the template and to select for mutations containing synthesized DNA. The plasmids were then transfected into XL1-competent cells. Five colonies of each mutant were screened for mutation, using the specific restriction site designed, and revealed at least 80% efficiency. Two colonies of each mutant were sequenced and confirmed to contain the mutation but no unwanted misincorporation of nucleotides. XL1-competent cells were transformed with the mutated plasmids and grown in 5–10 ml of Luria–Bertani (broth) medium and plasmids were isolated. Mon105-competent cells were then transformed with the mutated plasmids and used for expression.
Site-directed mutagenesis of the IGD
m (mouse) LEPRb cDNA (kindly provided by Dr C. Bjørbaek, Harvard Medical School, Boston, MA, U.S.A.) was subcloned into the mammalian expression vector pEYFP-N1 (where EYFP stands for enhanced yellow fluorescent protein) (ClonTech Ozyme, Saint Quentin en Yvelines, France). Mutagenesis of the different residues in the IGD was performed using the Stratagene QuickChange kit, as already described. Designed primers are shown in Supplementary Table S1 at http://www.BiochemJ.org/bj/391/bj3910221add.htm. Amino acids VFTT (325–328) of the LEPR were changed to encode the aliphatic amino acid alanine. The mutagenesis procedure included 18 PCR cycles using Pfu polymerase. Mutated sequences were confirmed by DNA sequencing.
Expression, refolding and purification of human and ovine leptins and their mutants
The recombinant WT or mutated human leptins with an extra methionine–alanine (methionine is cleaved by the bacteria) at the N-terminus were expressed in a 2.5 litre culture. IBs (inclusion bodies) were then prepared as described previously [15,16] and frozen. Subsequently, IBs obtained from 2.5 litres of bacterial culture were solubilized in 300 ml of 4.5 M urea and 40 mM Tris base containing 10 mM cysteine. In the case of ovine leptin or its mutants, the IBs obtained from 1.0 litre of bacterial culture were solubilized in a similar manner in 200 ml. The pH of the solution was adjusted to 11.3 with NaOH. After 2 h of stirring at 4 °C, three volumes of 0.67 M arginine were added to a final concentration of 0.5 M and stirred for an additional 1.5 h. Then, the solution was dialysed against 10 litres of 10 mM Tris/HCl, pH 9 (human leptin muteins), or 10 mM Tris/HCl, pH 8 (ovine leptin muteins), for 60 h, with five or six external solution exchanges. The protein was then applied at maximal flow rate (400–500 ml/h) on to a Q-Sepharose column (30 ml bead volume) and pre-equilibrated with the respective buffer. The breakthrough fraction, which contained no leptin, was discarded, and the absorbed protein was eluted in a stepwise manner (50, 100, 150 and 400 mM NaCl in 10 mM Tris/HCl, pH 9 or 8). Fractions (50 ml) were collected and protein concentration was determined by absorbance A at 280 nm.
Determination of purity and monomer content
SDS/PAGE was performed as described by Laemmli [24] in a 15% (w/v) polyacrylamide gel under reducing conditions. The gel was stained with Coomassie Brilliant Blue R. Gel filtration chromatography was performed on a Superdex™ 75 HR 10/30 column with 0.2 ml aliquots of the Q-Sepharose-column-eluted fraction using TN buffer (25 mM Tris/HCl and 150 mM NaCl, pH 8). Reverse-phase chromatography was performed using a Vydax column developed with a gradient of 0.1% (v/v) TFA (trifluoroacetic acid) in water (solvent A) and 0.1% TFA in methyl cyanide (solvent B).
Determination of CD spectra
The CD spectra were measured with an AVIV model 62A DS CD spectrometer (Aviv Associates, Lakewood, NJ, U.S.A.) using a 0.020 cm rectangular QS Hellma cuvette as described previously [7].
Various binding assays
Determination of the leptin–chLBD complex stoichiometry by gel filtration chromatography, kinetic measurements of chLBD–leptin interactions using SPR (surface plasmon resonance) methodology [25] and binding assays using radioactive leptin were performed according to procedures described in detail in our recent paper [7]. Binding experiments were also performed using CHO (Chinese-hamster ovary) cells transiently transfected with WT or mutated mLEPR cDNA using ExGen 500 (Euromedex, Souffelweyersheim, France) according to the manufacturer's instructions. Cellular homogenate (200 μl corresponding to 106 cells) was incubated with 500000 c.p.m. of 125I-human leptin in the absence (total binding) or presence (5000 ng/tube) of unlabelled leptin, the latter defined as non-specific binding. Specific binding was calculated as the difference between the total and non-specific binding.
FRET (fluorescence resonance energy transfer) microscopy experiments
Preparation of mLEPRb-CFP (where CFP stands for cyan fluorescent protein) and mLEPRb-YFP plasmids, as well as the FRET microscopy measurements, using APB (acceptor photobleaching), in HEK-293T (human embryonic kidney 293T) cells co-transfected with those constructs, are described in detail in our recent study [26].
BAF/3 proliferation assays
The proliferation rate of leptin-sensitive BAF/3 cells stably transfected with the long form of hLEPR was used to estimate self and antagonistic activity of leptin muteins, using the previously described MTT method [27]. To determine antagonistic activity, 6.25×10−11 M WT homologous leptin was added to each well, which also contained different concentrations of muteins. The average absorbance in wells without leptin (negative control) was used as a blank value and subtracted from other absorbance values to yield the corrected absorbance values. The average absorbance in wells with WT leptin, after subtracting the negative control, was used as a positive control to calculate percentage inhibition. The inhibition curves were drawn using the Prizma (4.0) non-linear regression sigmoidal one-site inhibition program (Prisma, GraphPad Prism™ Version 4.0; GraphPAD Software, San Diego, CA, U.S.A.) and the IC50 values were calculated.
Determination of biological activity by activating luciferase reporter gene
CHO cells were cultured and transfected as described previously [26]. To test the biological activity, transfected cells were grown for 48 h and then subsequently incubated for 24 h in the presence of human leptin in the presence or absence of selected leptin muteins in GC3 medium. The plates were washed with PBS and the enzymatic activity was determined as described previously [29]. The results were expressed as fold induction, after the luciferase activity had been normalized by correcting for β-galactosidase activity, as explained previously [29]. The same method was used to compare the leptin-inducible activity in COS cells transiently transfected with either WT or V325A/F326A/T327A/T328A mutant of mLEPR.
SH-SY5Y human neuroblastoma cell bioassay
Cell culture and retinoic acid-induced differentiation of SH-SY5Y human neuroblastoma cells were performed as described in [30]. To determine the effect of leptin or leptin muteins, SH-SY5Y cells were starved in serum-free Dulbecco's modified Eagle's medium for 16 h and pretreated for 15 min in the presence or absence of various concentrations of three selected leptin muteins and then stimulated for 10 min with human leptin. Post-stimulation detection of phospho-STAT3 (signal transducer and activator of transcription 3), total STAT3, phospho-MAPK (where MAPK stands for mitogen-activated protein kinase) or total MAPK (ERK 42,44; where ERK stands for extracellular-signal-regulated kinase) by immunoblotting was also performed as recently published [30].
RESULTS
Sequence-structure analysis of leptin and the IGD fragment of LEPR
In the IL-6–gp130 complexes, heterotetrameric structures are formed through the interaction of the IL-6 site III found in one IL-6–gp130 dimeric assembly with the IGD of the other assembly [9–11] (see the structure in the bottom-left of Figure 1 and also Figure 1 in [9]). We sought to identify such an interaction site in the leptin–LEPR complex, based on comparison with the vIL-6–gp130 (PDB 1i1r, [9]) and IL-6–IL-6Rα–gp130 (PDB 1p9m, [10]) structures. We focused on the interactions occurring in the vIL-6–gp130 structure, in which the structure of site III is completely ordered. In contrast, only a few IL-6 residues of site III are visible on the electron-density map of the IL-6–IL-6Rα–gp130 complex [9]. A similar disordered loop was also observed in the isolated structure of human leptin (PDB 1ax8, [3]). In the vIL-6–gp130 structure, site III is formed by an extensive interface between the tips of the vIL-6 bundle, comprising an A–B loop and the N-terminal end of helix D. This site III interacts with an edge of one of the IGD β-sheets. Of particular interest in A–B loop of vIL-6 (which is usually disordered in non-complexed cytokines, as well as in the non-complexed leptin) are residues 46–51. They indeed form a β-strand-like interaction with four residues of gp130 IGDs (see the structure in the bottom-left of Figure 1 and also Figure 1 in [9]). We thus wondered if a similar interaction occurs in the leptin–LEPR complex.
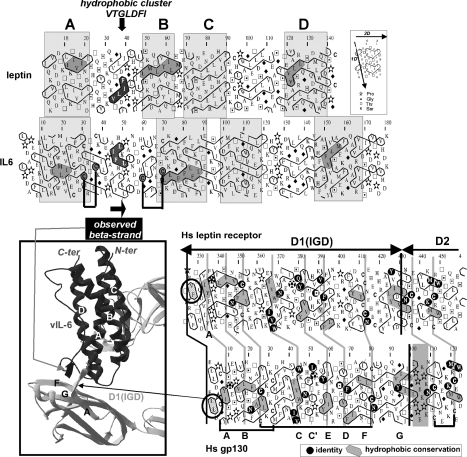
Figure 1. Comparison of the HCA plots of human leptin and vIL-6 (top panel) and of hLEPR and gp130 (bottom right panel).
View of the IL-6 site III structure within the hetero-tetrameric complex formed by gp130 is shown (in grey) and that of the vIL-6 (in black) ([9]; PDB identifier 1I1R), in the three-dimensional inset (bottom left panel). The IL-6's site III interacts with the gp130's IGD. The region including the short β-strand of the A–B loop of vIL-6 interacts with a short extended region preceding the IGD strand βA. The corresponding expressions of these short β-strands on the two-dimensional representations of the sequences (HCA plots) are shown with grey (vIL-6) and black (gp130) arrows. In the HCA plots, the sequence is shown on a duplicated α-helical net, in which the hydrophobic amino acids (V, I, L, F, M, Y and W) are circled. These form two-dimensional hydrophobic clusters, the positions of which have been shown to mainly match those of regular secondary structures [18]. Despite low levels of sequence identity, comparison of the HCA plots indicates the presence of conserved hydrophobicity (shaded in grey) within clusters, indicative of similar secondary structures. Although the experimental structures of leptin are known (PDB identifier 1AX8), a poor density was observed for the first part of the leptin A–B loop (no residues are visible from Ser25 to Gly38), indicating a high flexibility of the loop in the uncomplexed form of the cytokine. The cluster associated with the β-strand of the vIL-6 A–B loop, which is involved in the interaction with gp130, is shown in black. The two disulphide bonds located at the beginning and at the end of this loop are indicated by black lines. The only hydrophobic cluster of the leptin A–B loop, which is highly typical of an extended structure, is also shown in black. Compared with LEPR and gp130 IGDs (bottom panel), a small cluster (encircled), with two consecutive hydrophobic amino acids, precedes the strand βA in both sequences. It corresponds, in the gp130 structure (bottom left view), to the extra β-strand interacting with the vIL-6 A–B loop.
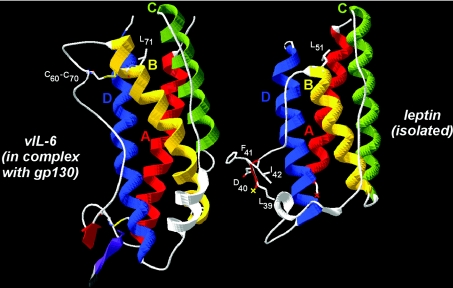
Sequence analysis showed that the A–B loops of the IL-6 family and leptin differ in amino acid composition and length and are thus difficult to align. Furthermore, as no relevant template is available for this loop, modelling of the atomic structure of the leptin A–B loop within the cytokine–receptor complex was impossible. However, HCA, a sensitive method for predicting secondary structure from a single sequence, indicated the presence of a hydrophobic cluster in the middle of leptin's A–B loop, typical of an extended (β-strand) structure and ranging from Val36 to Ile42 (VTGLDFI sequence). This predicted β-strand might play a role similar to that of the vIL-6 β-strand, located in the middle of a larger A–B loop and consisting of the IFHLKL hydrophobic cluster (Figure 1, top panel). The observed structure of the isolated leptin, after superimposition on the vIL-6 structure, further supported the likelihood of this hypothesis (Figure 2). Helix B (yellow) is one turn shorter in leptin than in IL-6, and the part of IL-6's loop located between two bridged cysteine residues (Cys60–Cys70) is absent from leptin. Consequently, the length of the chain separating the end of the β-strand cluster (shaded black in Figure 1, top panel, red in Figure 2) and the connection to the conserved core of helix B (Thr50 in leptin and His59 in vIL-6) are similar in both sequences. This suggests that the 39–42 peptide of leptin may well superimpose with the gp130-interacting β-strand of vIL-6, after this part of the leptin polypeptide chain has been appropriately rotated relative to its position in the non-complexed cytokine. This rotation might occur upon receptor binding through an induced fit mechanism and might be accompanied by a correlated displacement of the C–D loop.
Figure 2. Comparison of the experimental structures of vIL-6 (PDB code 1i1r; chain B) and leptin (PDB code 1ax8), as found complexed with gp130 and isolated respectively.
Superimposition was made on the basis of conserved blocks (helices, boxed on the HCA plots in Figure 1). Helix B is shorter in leptin than in vIL-6 (the position of a conserved leucine is shown in white) and the loop linking Cys60 to Cys70 is absent. The position of the leptin 39–42 peptide is shown, highlighting a position similar to that of the vIL-6 β-strand [shown in red, associated with the N-terminal sequence of gp130 IGD (violet)], provided a conformational change in this region occurs upon receptor binding. Red and yellow crosses indicate the beginning and end of the missing leptin chain within the A–B loop (no residues visible in the electron-density map).
Our analysis of IGD sequences in LEPR and gp130 readily led to identification of a common short cluster upstream of strand βA, typical of an extended structure (Figure 1, bottom panel). The N-terminal sequence of gp130 (ELLD), forming a short extended structure (see the three-dimensional inset in Figure 1), can indeed be aligned with the LEPR VFTT sequence, which is strongly predicted as a β-strand (the two threonine residues after the VF cluster strongly strengthen the β-strand prediction). As in cytokines' A–B loop, the local conformation of this strand located outside the IGD core might be induced upon ligand binding.
Purification and chemical characterization of leptin mutants
Six human leptin and four ovine leptin muteins of the 39–42 amino acid sequence were prepared. To compare the muteins of the A–B loop with the mutein of the N-terminal part of helix D (S120A/T121A) reported recently by Peelman et al. [12], we also prepared human leptin mutein S120A/T121A and a combined mutein L39A/D40A/F41A/I42A/S120A/T121A. Furthermore, to evaluate the role of the neighbouring residue Tyr119, two additional muteins of ovine leptin: Y119A and L39A/D40A/Y119A were also prepared. The refolded and dialysed fractions of the human leptin muteins (see the Materials and methods section) were purified on a Q-Sepharose anion-exchange column equilibrated with 10 mM Tris/HCl buffer (pH 9), using a non-continuous NaCl gradient. Fractions eluted with 50 mM NaCl, consisting of >95% pure monomer, were pooled, dialysed against NaHCO3 (pH 8) at a 4:1 (w/w) protein/salt ratio and freeze-dried. Ovine leptin mutants were purified in a similar manner, using Tris/HCl buffer (pH 8). The yields of human and ovine leptin mutants varied between 160 and 220 mg from 2.5 litres of bacterial culture and between 80 and 120 mg from 1.0 litre of bacterial culture respectively.
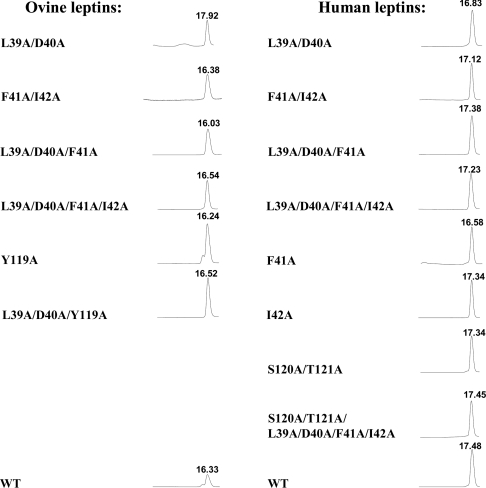
The purity and homogeneity of the purified mutants were documented by three independent methods. Gel filtration at pH 8 under native conditions yielded a single monomeric peak consisting of >95% monomers, corresponding to a molecular mass of approx. 16 kDa (Figure 3). SDS/PAGE under reducing conditions yielded only one band of approx. 16 kDa, and reverse-phase chromatography also yielded a single peak (results not shown). The secondary structures of human and ovine leptins and their mutants calculated from the CD spectra are shown in Supplementary Table S2 (http://www.BiochemJ.org/bj/391/bj3910221add.htm). A high content of α-helix (52–63%), 8–11% β-sheets and 14–18% β-turns were clearly characteristic for all proteins, indicating proper refolding. The only exceptions were ovine F41A/I42A and F41A/I42A/Y119A muteins, in which some disruption of the proper refolding was found. Molar absorption coefficients calculated as described by Pace et al. [31] were used to calculate the specific molar absorption coefficient at 280 nm for a 0.1% solution assuming extra alanine at the N-terminus (see Supplementary Table S3 at http://www.BiochemJ.org/bj/390/bj390ppppadd.htm). The stability of both human and ovine leptins and their mutants in solution was tested at 4 and 37 °C. Both WT leptins and their mutants could be stored at both temperatures as sterile 0.2 mM solutions for at least 30 days at pH 6 or 8 without undergoing any changes in their monomeric content and retaining their activity in the BAF/3 bioassay.
Figure 3. Purity determination of the isolated human and ovine leptin mutants by gel filtration analysis.
Aliquots (200 μl) of 0.5–1 mg/ml of the purified muteins (except 0.2 mg of WT ovine leptin) were applied to a Superdex™ 75 HR 10/30 column pre-equilibrated with TN buffer, which was developed at 0.8 ml/min. All peaks correspond to approx. 16 kDa as calibrated with BSA (66 kDa), chLBD (24 kDa), ovine growth hormone (21.5 kDa) and WT leptins (16 kDa). The differences in the RT values result from a use of different Superdex™ columns with slightly different bead volumes. All muteins consisted of >98% monomers, except for ovine Y119A and ovine L39A/D40 that had small amounts of dimers and oligomers respectively. For other details, see text.
Detection of chLBD–leptin or chLBD–leptin mutein complexes by gel filtration
To characterize the binding stoichiometry between human or ovine leptins or their mutants and chLBD, the respective ligands and chLBD were mixed in different molar ratios and separated by gel filtration using an analytical Superdex™ 75 column to determine the molecular mass of the binding complex under non-denaturing conditions. The experiments were performed using a constant 5 μM of the respective ligand and 2.5, 5 or 10 μM of chLBD. The results of typical chLBD–WT leptin interactions (results not shown) indicate that both species of leptin bind the chLBD in a 1:1 molar ratio, as shown in [7]. This stoichiometry was evidenced by the observation of a single peak, when the components were mixed at 1:1 molar ratio, and the appearance of an additional peak when there was an excess of one of them. The calculation of the complex's molecular mass, based on the peak's retention time, was approx. 41 kDa in all cases, close to the predicted value of 40.5 kDa. An almost identical interaction pattern was also observed with all 14 muteins (results not shown), indicating that mutations did not affect muteins' ability to form 1:1 complexes with chLBD.
Binding experiments
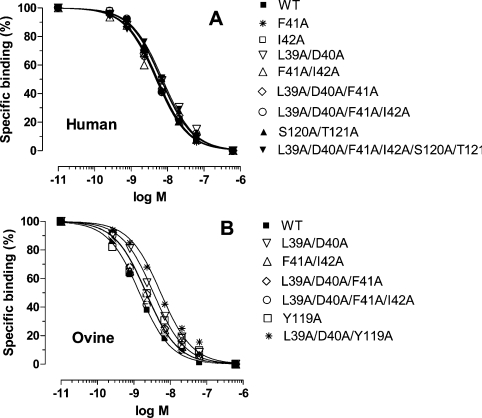
Iodinated human leptin served as a ligand in all competitive experiments and the respective WT human and ovine leptins and their alanine muteins as competitors. Freshly prepared homogenate of BAF/3 cells stably transfected with the long form of hLEPR was used as the receptor source. Homogenate from 1.8×106 cells per tube gave 6–7% specific binding. The inhibition curves (average of two experiments) are presented in Figure 4, and the respective IC50 values are summarized in Supplementary Table S4 (http://www.BiochemJ.org/bj/391/bj3910221add.htm). The differences among the IC50 values were not statistically significant and all were within the 95% confidence limit, indicating that mutation did not change the binding properties.
Figure 4. Radio receptor assay using the homogenate of BAF3 cells stably transfected with the long form of hLEPR.
125I-human leptin (6–8×105 c.p.m./tube) was used as a ligand and human (A) or ovine (B) leptins or their analogues as competitors. The results are averages for two experiments. The specific binding varied from 6.5 to 8.2×104 c.p.m. and was normalized as 100. The non-specific binding was (4.0–4.2)×104 c.p.m. For other details, see text.
SPR determination of the interaction between chLBD with leptin or its mutants
To further characterize the binding capacities of chLBD with human or ovine leptins and the eight most potent respective muteins, SPR using the respective leptin (or leptin mutein) immobilized on a sensor chip by amine coupling and binding of soluble chLBD was employed. The most acceptable interactions were obtained by comparison with a 1:1 theoretical model using χ2 analysis. The calculated data (mean of two experiments) are presented in Table 1. Up to 3-fold difference in the kinetics constants was observed, but the respective _K_d values varied to a lesser degree and the differences were not statistically significant.
Table 1. Calculation of kinetics and thermodynamic constants for the interaction of immobilized WT leptins and selected muteins with soluble chLBD measured by SPR.
Results are means±S.D. for two experiments.
| ×105_k_on | _k_off | ×10−8_K_d | ||
|---|---|---|---|---|
| Mutant | (mol−1·s−1) | (ms−1) | (M) | χ2* |
| Human leptin | ||||
| WT leptin | 1.49±0.34 | 4.3±0.1 | 2.88±0.53 | 1.71–1.87 |
| L39A/D40A | 5.45±0.55 | 12.5±1.5 | 2.29±0.09 | 3.15–3.83 |
| F41A/I42A | 3.62±0.15 | 11.5±0.4 | 3.18±0.21 | 1.73–4.29 |
| L39A/D40A/F41A | 3.58±0.54 | 10.2±0.8 | 2.85±0.48 | 2.53–4.73 |
| L39A/D40A/F41A/I42A | 1.35±0.47 | 6.8±1.0 | 5.03±3.96 | 0.43–0.54 |
| Ovine leptin | ||||
| WT leptin | 1.62±0.94 | 3.7±0.8 | 2.28±2.15 | 0.58–1.57 |
| L39A/D40A | 2.37±0.03 | 5.7±0.1 | 2.40±0.06 | 2.31–5.18 |
| F41A/I42A | 2.50±0.15 | 3.2±0.2 | 1.28±0.03 | 1.80–3.54 |
| L39A/D40A/F41A | 1.81±0.06 | 2.5±0.2 | 1.38±0.13 | 1.74–4.57 |
| L39A/D40A/F41A/I42A | 1.09±0.18 | 3.2±0.1 | 2.93±0.56 | 1.24–3.18 |
FRET measurements
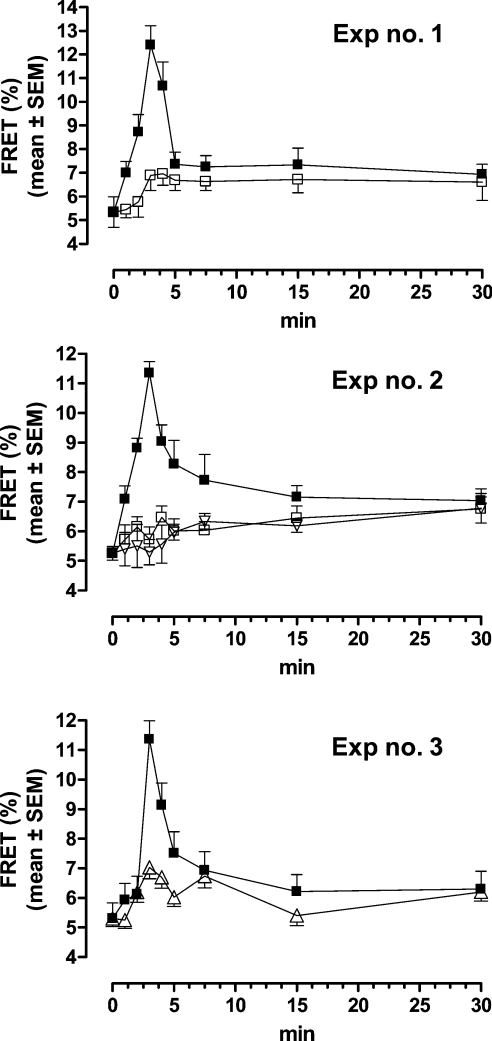
In order to elucidate the mechanism of leptin–LEPR interaction, we compared the effect of mutated and WT leptin on LEPRb configuration at the plasma membrane by following the time-course FRET microscopy measurements. HEK-293T cells were co-transfected with mLEPRb-CFP and mLEPRb-YFP and stimulated for different time points with 400 ng/ml human leptin, human leptin mutein L39A/D40A/F41A, respective ovine leptin mutein or with simultaneously added human leptin (400 ng/ml) and human leptin mutein L39A/D40A/F41A (4000 ng/ml). As shown in Figure 5, in contrast with exposure of the cells to leptin, which resulted in a transient elevation of FRET, confirming the previous results [26], exposure to human or ovine leptin muteins L39A/D40A/F41A (experiments 1 and 2) did not cause any significant FRET elevation. Furthermore, simultaneous addition of human leptin with 10-fold excess of antagonist (experiment 3) almost totally blocked leptin effect.
Figure 5. Time-dependent leptin- or leptin antagonist-induced FRET efficiency curve as measured by APB FRET.
Time-dependent leptin-induced FRET efficiency in cells transfected with LEPRb-CFP and LEPRb-YFP and stimulated by 400 ng/ml human leptin (■), human leptin mutein L39A/D40A/F41A (□) or ovine leptin mutein L39A/D40A/F41A (▽) (experiments 1–3; not all results are shown) or by simultaneously added human leptin (400 ng/ml) and human leptin mutein L39A/D40A/F41A (4000 ng/ml) (△) (experiment 3). The results are presented as means±S.E.M.
Biological activity in vitro in the BAF/3 bioassay
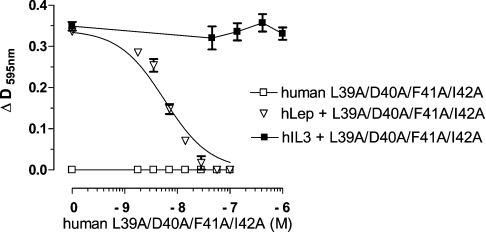
Human and ovine leptin exhibited almost identical activity in the BAF/3 bioassay with their respective EC50 values of 28 and 37 pM, similarly to previously published results [16,27]. Most of the human or ovine leptin mutants were devoid of any agonistic activity, although in a few a weak agonistic activity (<1%) was observed (Table 2). To determine the antagonistic activity, BAF/3 cells were stimulated with 62.5 pM human or ovine leptin in the presence (1–125 nM) or absence of the respective mutants. The experiments were repeated three to seven times and the average IC50 values were calculated (Table 2). All muteins (except ovine Y119A) exhibited antagonistic activity and no major differences among human and ovine leptin mutants were observed. Both human and ovine L39A/D40A/F41A and L39A/D40A/F41A/I42A muteins were more potent than the double muteins L39A/D40A and F41A/I42A. The lower antagonistic activity of ovine L39A/D40A (and L39A/D40A/Y119A) as compared with its human counterpart could be attributed to some distortion in the proper refolding of the former, as deduced from the CD data (Supplementary Table S2 at http://www.BiochemJ.org/bj/391/bj3910221add.htm). Single-point muteins (F41A and I42A) also exhibited antagonistic activity, although their respective IC50 values were higher than the double, triple and quadruple muteins and both exhibited low agonistic activity. Therefore we conclude that the single-point mutations probably lowered the affinity between leptin's site III and the IGD, drastically attenuating the agonistic activity and converting those muteins into both partial antagonists and weak agonists. In contrast, the mutation in Tyr119 markedly reduced (over 95%) the agonistic activity, despite the fact that binding to the whole receptor (see Figure 4) was not changed, indicating the importance of Tyr119 to the proper interaction of site III with IGD. However, as the neighbouring residues Ser120 and Tyr121 participating in site III [12], as well as the more distant residues LDFI (amino acids 39–42), were unchanged, no antagonistic activity was observed. When, in addition to Tyr119, Leu39 and Asp40 were also mutated to alanine, the low agonistic activity was lost and the triple L39A/D40A/Y119A mutein became a moderate antagonist. To compare the antagonistic activity of the double, triple and quadruple muteins of the 39–42 sequence with recently reported human leptin mutein S120A/T121A [12], the latter was also prepared and tested. In the BAF/3 bioassay, it was less potent and even exhibited some low (<1%) agonistic activity, whereas the combined human L39A/D40A/F41A/I42A/S120A/T121A mutein did not differ from the respective quadruple mutein. To verify the specificity of inhibition, the proliferation of BAF/3 cells was stimulated by 55 pM IL-3 in the presence or absence of human and ovine leptin muteins. In all cases (except the Y119A mutein, in which some additional agonistic activity was observed), no inhibition was observed, even at 1.25 μM concentration of all tested muteins. A typical experiment is presented in Figure 6.
Table 2. Antagonistic activity of human (h) and ovine (o) leptin muteins in BAF/3 cells stably transfected with the long form of hLEPR.
Cells were stimulated with 62.5 pM leptin. IC50 values are means±S.E.M. (or means±S.D. where indicated by an asterisk, *) for the number of experiments indicated in parentheses.
| IC50 (nM) | |||
|---|---|---|---|
| Mutein | h-leptin | o-leptin | Agonistic activity |
| L39A/D40A | 17.7±3.76 (4) | 41.8±14.1 (7) | None |
| F41A | 531±447* (2) | Not prepared | Very weak |
| I42A | 53.4±16.5 (4) | Not prepared | Very weak |
| F41A/I42A | 15.6±3.87 (4) | 31.0±19.9 (3) | None |
| L39A/D40A/F41A | 11.9±1.94 (3) | 18.0±6.2 (3) | None |
| L39A/D40A/F41A/I42A | 9.3±1.60 (5) | 9.5±4.6 (3) | None |
| Y119A | Not prepared | No inhibition | Weak (5%) |
| L39A/D40A/Y119A | Not prepared | 98.0±14.6 (3) | Very weak |
| S120A/T121A† | 165±81 (5) | Not prepared | Very weak |
| L39A/D40A/F41A/I42A/S120A/T121A | 14.3±1.34* (2) | Not prepared | None |
Figure 6. Effect of human leptin L39A/D40A/F41A/I42A mutein on leptin-induced or IL-3-induced proliferation of BAF/3 cells stably transfected with the long form of hLEPR.
For details, see text.
Biological activity in vitro in additional bioassays
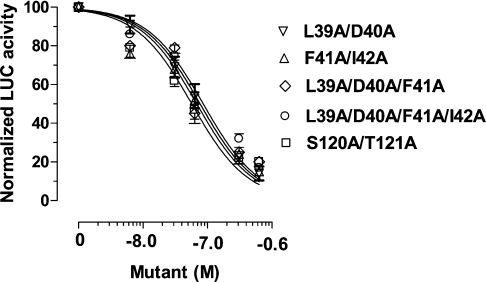
Five selected human leptin muteins (L39A/D40A, F41A/I42A, L39A/D40A/F41A, L39A/D40A/F41A/I42A and S120A/121A) were also tested for their ability to inhibit leptin-inducible transactivation of a luciferase reporter gene in CHO cells transiently transfected with mLEPR. All muteins progressively attenuated the leptin-inducible (6.25 nM) activation of luciferase (Figure 7). The respective IC50 values (in nM) were 65, 62, 76, 85 and 57, namely a 9–13-fold molar antagonist excess. Those differences were not significant and were confined within 95% confidence limit.
Figure 7. Inhibition of leptin-inducible transactivation of luciferase (LUC) reporting gene in CHO cells transiently transfected with mLEPRb and stimulated with 6.25 nM human leptin by five human leptin muteins.
For other details, see text.
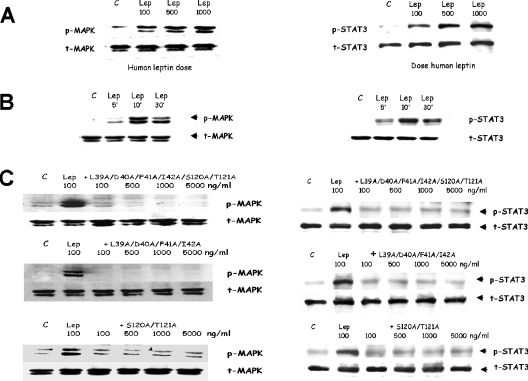
To check whether antagonists inhibit leptin action in cells possessing natural LEPRs, the effect of human leptin muteins S120A/T121A, L39A/D40A/F41A/I42A/S120A/T121A and L39A/D40A/F41A/I42A on leptin-inducible MAPK and STAT3 phosphorylation was tested in SH-SY5Y human neuroblastoma cells. These cells phosphorylate MAPK and STAT3 in a dose- (Figure 8A) and time- (Figure 8B) dependent manner. Pre-treatment with the three human leptin muteins for 15 min blocked the subsequent leptin-inducible phosphorylation of both targets, even at a 1:1 molar agonist/antagonist ratio (Figure 8C).
Figure 8. Inhibition of leptin-inducible phosphorylation of MAPK and STAT3 in SH-SY5Y human neuroblastoma cells by human leptin L39A/D40A/F41A/I42A, S120A/T121A and L39A/D40A/F41A/I42A/S120A/T121A muteins.
(A) Dose-dependent (10 min) and (B) time-dependent (100 ng leptin) leptin-induced phosphorylation of MAPK (ERK 1/2) and STAT3. (C) Dose-dependent inhibition: cells were preincubated with 100–5000 ng/ml of the respective muteins for 15 min and then with 100 ng/ml of leptin for an additional 10 min. For other details, see text. C, control; p-MAPK, phosphorylated MAPK; t-MAPK, total MAPK.
Mutagenesis of the IGD of mLEPR
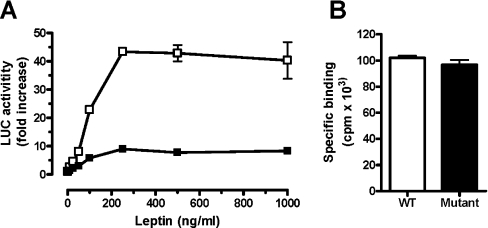
The activity of LEPR mutant V325A/F326A/T327A/T328A was compared with the WT receptor in transiently transfected CHO cells using a luciferase reporter gene assay (Figure 9). The mutation drastically reduced the activity of the receptor. This reduction in activity could not be attributed to the different expression of the receptor as indicated by binding of the 125I-ovine leptin. These results are in agreement with our previously described molecular model and confirm that the VFTT sequence, which is located at the N-terminal end of the IGD, is necessary for the functionality of the receptor and may constitute the domain in which a conformational change occurs after binding of leptin through its site III.
Figure 9. Inhibition of leptin-inducible transactivation of luciferase (LUC) reporting gene.
Representative experiment (one out of three) in CHO cells transiently transfected with mLEPR (□, WT; ■, mutated at the IGD) and stimulated with 6.25 nM human leptin (A). Binding of 125I-human leptin to CHO cells transfected with WT (empty bars) or mutated (full bars) mLEPR IGD (B). The results are presented as means±S.E.M. (in A) or means±S.D. (in B). In some cases, the variations were too small to be seen. For other details, see text.
DISCUSSION
Class I cytokines interact with their receptor through conserved binding sites (sites I and II). G-CSF (granulocyte colony-stimulating factor) and cytokines of the gp130 family, including IL-6, have an additional binding site (site III), which was recently shown to interact with the receptor IGD in crystal structures of IL-6–gp130 complexes [9,10] in a 2:2 or 2:4 arrangement. A similar type of complex formation is expected for the leptin–LEPR interaction due to the sequence similarities and overall architecture that the two interacting partners share with the hormone and receptors of the G-CSF and gp130 family of cytokines [8,12]. In the present study, using a HCA analysis, we identified a short sequence in the A–B loop of leptin, which could play a role similar to that of a short extended structure, located in the middle of the IL-6 A–B loop, which is involved in the formation of an intermolecular β-sheet with the receptor's IGD. The potential interacting partner was also identified in the LEPR's IGD as amino acids 325–328 (VFTT) and confirmed by bioassay. It is worth noting that these regions are probably mobile and that the β-strand conformation is probably obtained upon interaction of the partners. Indeed, on one hand, the A–B loop is often disordered in non-complexed cytokines, and on the other, the short β-strand at the N-terminus of the receptor IGD is located outside the core of the domain.
Therefore we considered this conserved β-strand of the leptin A–B loop, which exists in leptin in all species, as a target and mutated the LDFI (amino acids 39–42) fragment. Since when preparing the R128Q leptin mutein, we observed that the effect of mutation may be species-dependent [16], the presently described mutations were performed simultaneously in human and ovine leptins. All muteins of this region (seven human and five ovine) were purified to homogeneity in high yield and their secondary-structure analysis revealed proper refolding, except of ovine L39A/D40A and L39A/D40A/Y119A muteins, which exhibited a slightly lower α-helix content. All double-, triple- and quadruple-mutated muteins acted as true antagonists, i.e. they interacted with LEPR with an affinity similar to the WT hormone, as evidenced by SPR (for eight of them), by binding experiments, and formed a 1:1 complex with chLBD similarly to WT leptins. Those muteins were devoid (or almost devoid in the case of S120A/T121A mutein) of biological activity in a BAF/3 leptin-responsive bioassay and specifically inhibited leptin action. Selected human muteins also inhibited leptin-inducible activity in other in vitro bioassays based not only on LEPR-transfected CHO cells but also on cells (SH-SY5Y) that naturally express LEPRs. This indicated that the inhibitory potential of those antagonists is general and not model-dependent.
To compare the activity of our mutants with that of the recently reported S120A/T121A human leptin mutant, which exhibits antagonistic activity [12], we prepared and purified to homogeneity two additional human leptin mutants: S120A/T121A and L39A/D40A/F41A/I42A/S120A/T121A. As evidenced by SPR analysis, both muteins bound to chLBD with affinities similar to the WT human leptin. However, in the BAF/3 bioassay, the S120A/T121A mutant exhibited low (less than 1% as compared with WT human leptin) agonistic activity, and the IC50 for inhibition was more than 150-fold higher than either L39A/D40A/F41A or L39A/D40A/F41A/I42A. In contrast, the IC50 value for the L39A/D40A/F41A/I42A/S120A/T121A was comparable with that for the L39A/D40A/F41A and L39A/D40A/F41A/I42A muteins. However, such a difference was not observed in STAT3-dependent transcriptional activity in the luciferase bioassay, in which all human double, triple and quadruple muteins were equally as potent as S120A/T121A. This indicated that the inhibitory potential may differ with the various in vitro bioassays. Furthermore, whereas a 160–200-fold molar excess of antagonist was required to achieve 50% inhibition in the BAF/3 bioassay, in the luciferase bioassay, the same level of inhibition was achieved with a 9–13-fold antagonist excess. The BAF/3 assay is a very sensitive bioassay, probably because occupation of only a small fraction of the receptors is sufficient to induce a maximal effect in terms of cell proliferation. The case of the Y119A mutant is also interesting. This mutation did not change the binding, but decreased drastically the agonistic activity. This happens probably because this mutation located close to the site III (see the results with S120A/T121A) weakens the interaction with IGD.
Recent study using BRET (bioluminescence resonance energy transfer) in living cells [32] suggested that LEPRs exist as constitutive preformed dimers and are activated as a result of conformational change, induced by leptin stimulation. Using FRET microscopy, we reached a similar conclusion. However further analysis of FRET results indirectly suggested de novo association of monomeric or pre-dimerized receptors in addition to conformational change [26]. The present results, using two muteins that bind to LEPR but are unable to activate it, documented that binding alone, to either single or pre-dimerized receptors, does not lead to increase in FRET efficiency. These results indicate that leptin stimulation leads to de novo association between the receptors, possibly to hexamerization of two inactive trimers (each composed of two LEPRs and one leptin) as suggested by Zabeau et al. [8] or alternatively by tetramerization of two dimers each composed of a 1:1 LEPR–leptin complex. Whichever assumption is correct, FRET results correlate with activation occurring by interaction of leptin's site III with the respective sequence of IGD and not with leptin binding through high-affinity site I.
In conclusion, the muteins of the 39–42 amino acid fragment prepared in the present study act as competitive antagonists to leptin. Those mutations seem to be relevant to all leptins, as evidenced by the recently prepared L39A/D40A/F39A muteins of rat and mouse leptin, which also act as potent antagonists (G. Ben-Yehuda, L. Niv-Spector and A. Gertler, unpublished work). All muteins can be easily prepared in gram amounts and thus can serve as a novel tool for studying leptin function, not only in vitro but also in vivo. Leptin antagonists also offer a novel tool to elucidate the role of leptin in mammalian physiology and pathology, mainly by blocking the action of the endogenous leptin and potentially serving as therapeutic agents, as suggested in a recent review [33].
Online data
Supplementary data
Acknowledgments
We thank Dr C. I. Rosenblum (Merck Company, Rahway, NJ, U.S.A.) for the STAT3-responsive pAH32 luciferase-encoding plasmid, and Dr C. Bjørbaek (Harvard Medical School, Boston, MA, U.S.A.) for the cDNA encoding mLEPR. This work was supported by the Israeli Science Foundation (grant no. 594/02, to A.G.) and by the Binational U.S.A.–Israel Science Foundation (grant no. 2000115 to A.G. and B.H.).
References
- 1.Zhang Y., Proenca R., Maffei M., Barone M., Leopold L., Friedman J. M. Positional cloning of the mouse obese gene and its human homologue. Nature (London) 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 2.Tartaglia L. A., Dembski M., Weng X., Deng N., Culpepper J., Devos R., Richards G. J., Campfield L. A., Clark F. T., Deeds J., et al. Identification and expression cloning of a leptin receptor, OB-R. Cell (Cambridge, Mass.) 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F., Basinski M. B., Beals J. M., Briggs S. L., Churgay L. M., Clawson D. K., DiMarchi R. D., Furman T. C., Hale J. E., Hsiung H. M., et al. Crystal structure of the obese protein leptin-E100. Nature (London) 1997;387:206–209. doi: 10.1038/387206a0. [DOI] [PubMed] [Google Scholar]
- 4.Zabeau L., Defeau D., Van der Heyden J., Iserentant H., Vandekerckhove J., Tavernier J. The ins and outs of leptin receptor activation. FEBS Lett. 2003;18:150–161. doi: 10.1210/me.2003-0078. [DOI] [PubMed] [Google Scholar]
- 5.Fong T. M., Huang R. R., Tota M. R., Mao C., Smith T., Varnerin J., Karpitskiy V. V., Krause J. E., Van der Ploeg L. H. Localization of leptin binding domain in the leptin receptor. Mol. Pharmacol. 1998;53:234–240. doi: 10.1124/mol.53.2.234. [DOI] [PubMed] [Google Scholar]
- 6.Sandowski Y., Raver N., Gussakovsky E. E., Shochat S., Dym O., Livnah O., Rubinstein M., Krishna R., Gertler A. Subcloning, expression, purification, and characterization of recombinant human leptin-binding domain. J. Biol. Chem. 2002;277:46304–46309. doi: 10.1074/jbc.M207556200. [DOI] [PubMed] [Google Scholar]
- 7.Niv-Spector L., Raver N., Friedman-Einat M., Grosclaude J., Gussakowsky E. E., Livnah O., Gertler A. Mapping leptin-interacting sites in recombinant leptin-binding domain (LBD) subcloned from chicken leptin receptor. Biochem. J. 2005;390:475–484. doi: 10.1042/BJ20050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zabeau L., Defeau D., Van der Heyden J., Iserentant H., Vandekerckhove J., Tavernier J. Functional analysis of leptin receptor activation using a Janus kinase/signal transducer and activator of transcription complementation assay. Mol. Endocrinol. 2004;18:150–161. doi: 10.1210/me.2003-0078. [DOI] [PubMed] [Google Scholar]
- 9.Chow D., He X., Snow A. L., Rose-John S., Garcia K. C. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–2155. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]
- 10.Boulanger M. J., Chow D. C., Brevnova E. E., Garcia K. C. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 11.Muller-Newen G. The cytokine receptor gp130: faithfully promiscuous. Science STKE 2003. 2003;201:PE40. doi: 10.1126/stke.2003.201.pe40. [DOI] [PubMed] [Google Scholar]
- 12.Peelman F., Van Beneden K., Zabeau L., Iserentant H., Ulrichts P., Defeau D., Verhee A., Catteeuw D., Elewaut D., Tavernier J. Mapping of the leptin binding sites and design of a leptin antagonist. J. Biol. Chem. 2004;279:41038–41046. doi: 10.1074/jbc.M404962200. [DOI] [PubMed] [Google Scholar]
- 13.Callebaut I., Labesse G., Durand P., Poupon A., Canard L., Chomilier J., Henrissat B., Mornon J. P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol. Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niv-Spector L., Gonen-Berger D., Callebaut I., Djiane J., Gertler A. Miami, FL: Nature Publishing Company; 2005. Large-scale preparation of novel leptin antagonists by site-directed mutagenesis of leptin site III: implications on understanding an activation of leptin:leptin receptor complex. The Proceedings of the 2005 Miami Winter Symposium Signal Transduction in Cancer, vol. 16, p. T-11, 5–9 February 2005. [Google Scholar]
- 15.Gertler A., Simmons J., Keisler D. H. Large-scale preparation of biologically active recombinant ovine obese protein (leptin) FEBS Lett. 1998;422:137–140. doi: 10.1016/s0014-5793(97)01613-x. [DOI] [PubMed] [Google Scholar]
- 16.Raver N., Vardy E., Livnah O., Devos R., Gertler A. Comparison of R128Q mutations in human, ovine, and chicken leptins. Gen. Comp. Endocrinol. 2000;126:52–58. doi: 10.1006/gcen.2001.7766. [DOI] [PubMed] [Google Scholar]
- 17.Gaboriaud C., Bissery V., Benchetrit T., Mornon J.-P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 18.Woodcock S., Mornon J. P., Henrissat B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992;5:629–635. doi: 10.1093/protein/5.7.629. [DOI] [PubMed] [Google Scholar]
- 19.Hennetin J., LeTuan K., Canard L., Colloc'h N., Mornon J.-P., Callebaut I. Non-intertwined binary patterns of hydrophobic/nonhydrophobic amino acids are considerably better markers of regular secondary structures than nonconstrained patterns. Proteins. 2003;51:236–244. doi: 10.1002/prot.10355. [DOI] [PubMed] [Google Scholar]
- 20.Callebaut I., Moshous D., Mornon J. P., de Villartay J. P. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callebaut I., Curcio-Morelli C., Mornon J. P., Gereben B., Buettner C., Huang S., Castro B., Fonseca T. L., Harney J. W., Larsen P. R., et al. The iodothyronine selenodeiodinases are thioredoxin-fold family proteins containing a glycoside hydrolase clan GH-A-like structure. J. Biol. Chem. 2003;278:36887–36896. doi: 10.1074/jbc.M305725200. [DOI] [PubMed] [Google Scholar]
- 22.Thoreau E., Petridou B., Kelly P. A., Djiane J., Mornon J. P. Structural symmetry of the extracellular domain of the cytokine/growth hormone/prolactin receptor family and interferon receptors revealed by hydrophobic cluster analysis. FEBS Lett. 1991;282:26–31. doi: 10.1016/0014-5793(91)80437-8. [DOI] [PubMed] [Google Scholar]
- 23.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Johnsson B., Lofas S., Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 26.Biener E., Charlier M., Ramanujan V. K., Daniel N., Eisenberg A., Bjørbaek C., Herman B., Gertler A., Djiane J. Quantitative FRET imaging of leptin receptor oligomerization kinetics in single cells. Biol. Cell. 2005 doi: 10.1042/BC20040511. 10.1042/BC20040511. [DOI] [PubMed] [Google Scholar]
- 27.Raver N., Gussakovsky E. E., Keisler D. H., Krishna R., Mistry J., Gertler A. Preparation of recombinant bovine, porcine, and porcine W4R/R5K leptins and comparison of their activity and immunoreactivity with ovine, chicken, and human leptins. Protein Expr. Purif. 2000;19:30–40. doi: 10.1006/prep.2000.1202. [DOI] [PubMed] [Google Scholar]
- Reference deleted.
- 29.Sotiropoulos A., Moutoussamy S., Renaudie F., Clauss M., Kayser C., Gouilleux F., Kelly P. A., Finidori J. Differential activation of Stat3 and Stat5 by distinct regions of the growth hormone receptor. Mol. Endocrinol. 1996;10:998–1009. doi: 10.1210/mend.10.8.8843416. [DOI] [PubMed] [Google Scholar]
- 30.Benomar Y., Roy A. F., Aubourg A., Djiane J., Taouis M. Cross down-regulation of leptin and insulin receptor expression and signaling in human neuronal cell line. Biochem. J. 2005;388:929–939. doi: 10.1042/BJ20041621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couturier C., Jockers R. Activation of the leptin receptor by a ligand-induced conformational change of constitutive receptor dimers. J. Biol. Chem. 2003;278:26604–26611. doi: 10.1074/jbc.M302002200. [DOI] [PubMed] [Google Scholar]
- 33.Peelman F., Waelput W., Iserentant H., Lavens D., Eyckerman S., Zabeau L., Tavernier J. Leptin: linking adipocyte metabolism with cardiovascular and autoimmune diseases. Prog. Lipid Res. 2004;43:283–301. doi: 10.1016/j.plipres.2004.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data