Type III Secretion of Salmonella enterica Serovar Typhimurium Translocated Effectors and SseFG (original) (raw)
Abstract
The type III secretion system (TTSS) encoded by Salmonella enterica serovar Typhimurium pathogenicity island 2 (SPI2) is employed by Salmonella enterica for interaction with host cells during the intracellular phase of pathogenesis. This TTSS secretes a set of SPI2-encoded proteins in vitro and translocates Salmonella serovar Typhimurium translocated effectors (STE) that are encoded by genes outside of SPI2 into host cells. Using an epitope-tagging approach, we analyzed secretion of proteins by the TTSS of SPI2 and identified SseF and SseG as further secreted substrate proteins. Three members of the STE family, SifA, SifB, and SseJ, were secreted under conditions that also induce secretion of SPI2-encoded substrate proteins.
Type III secretion systems (TTSS) are complex molecular machines found in a large number of gram-negative pathogens that mediate, in a contact-dependent manner, the translocation of effector proteins from the bacterial cytoplasm into a eukaryotic host cell (12). However, under certain in vitro conditions, a TTSS can also secrete proteins into a culture medium.
Invasion of eukaryotic cells and intracellular survival and replication in infected host cells are two hallmarks of Salmonella enterica serovar Typhimurium pathogenesis. In Salmonella serovar Typhimurium, two TTSS are involved in these interactions with eukaryotic cells. Both TTSS of Salmonella serovar Typhimurium are encoded by genes on pathogenicity islands. The TTSS encoded by Salmonella serovar Typhimurium pathogenicity island 1 (SPI1) mediates the invasion by Salmonella serovar Typhimurium of nonphagocytic cells such as epithelial cells of the intestinal mucosa and is involved in enteropathogenesis (reviewed in references 7 and 20). The second TTSS encoded by SPI2 is not involved in invasion but is required for the intracellular phenotypes of Salmonella, such as intracellular survival and replication (for a review, see reference 10).
Until recently, the identities of substrate proteins of the TTSS of SPI2 were unknown. We have identified culture conditions that induce the expression of SPI2 genes in vitro (5). Furthermore, growth conditions were defined that trigger the secretion of SPI2 substrate proteins in vitro (2). Under these conditions, three secreted proteins, SseB, SseC, and SseD, have been detected that are associated with the bacterial cell surface after secretion (2, 13, 15). A role for these proteins in translocation of further effector proteins has been assumed. Further genes in SPI2 are clustered within a group of genes for secreted proteins and their chaperones. It has been proposed that SseE, SseF, and SseG are substrate proteins of the TTSS of SPI2 (11), but so far, there is no experimental evidence for this hypothesis. Recent work by Guy et al. indicated that SseF and SseG are required for an SPI2-related cellular phenotype, i.e., the formation of Salmonella serovar Typhimurium-induced filaments in infected epithelial cells (8).
A set of effector proteins of SPI2 termed Salmonella translocated effectors (STE) has been identified by virtue of the N-terminal conserved domain (14). Studies using fusions to the reporter CyaA indicated that intracellular Salmonella translocates STE into the host cells via the TTSS of SPI2. All STE are encoded by genes outside the SPI2 locus. Several of these loci are associated with prophage genes, indicating that these genes may be part of the variable assortment of virulence factors of Salmonella serovar Typhimurium.
We were interested in analyzing the secretion of STE proteins and other putative substrate proteins of the TTSS of SPI2 under in vitro conditions. In this study, the secretion of SifA, SifB, and SseJ as well as of SseF and SseG by the TTSS of SPI2 is reported.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Salmonella serovar Typhimurium strains used in this study are derivatives of Salmonella serovar Typhimurium ATCC 14028. Mutant strains of Salmonella serovar Typhimurium P8G12 (ssrB::mTn_5_ [18]) and NP ssaV (ssaV::aphT [6]) have been described before. Escherichia coli strains XL-1 Blue (Stratagene) and DH5α (Gibco-BRL) were used for the propagation of plasmids.
The composition of minimal medium has been described before (5). Briefly, N-salts medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 100 mM Bis-Tris/HCl (pH 7.0), 30 μM MgCl2, 38 mM glycerol, and 0.1% Casamino Acids] containing a low (30 μM MgCl2) or high (10 mM MgCl2) concentration of Mg2+ was used. Minimal medium containing a high (PCN) or low (PCN-P) concentration of phosphate was used as described before (5). For PCN-P medium at pH 5.8, 80 mM MOPS (morpholinepropanesulfonic acid) was replaced by 80 mM MES (morpholineethanesulfonic acid). All minimal media were prepared with double-distilled H2O (H2Odd). If required, medium was supplemented with 50 μg of carbenicillin/ml to maintain plasmids.
Generation of epitope-tagged SPI2 and STE proteins.
PCR was performed using the Expand high fidelity system (Roche) in order to minimize the error rate of the amplification procedure. Approximately 100 ng of genomic DNA of the Salmonella serovar Typhimurium wild-type strain was used as a template for amplification. DNA manipulations were performed according to standard procedures (17). Genomic DNA, plasmids, PCR products, and DNA fragments were purified using Qiagen kits according to the instructions of the manufacturer.
A 231-bp _Sma_I/_Xba_I fragment encoding the M45 epitope of the adenovirus protein E4-6/7 (16) was kindly provided by W.-D. Hardt (Munich). The fragment was inserted in _Sma_I/_Xba_I-digested plasmid pBluescript SKII to obtain plasmid p2062.
A 450-bp fragment containing the SPI2 promoter Pro_sseA_ was obtained by PCR using primers Pro_sseA_-For-_Hin_dIII and Pro_sseA_-Rev-Eco_RI. The PCR product for Pro_sseA was digested with _Hin_dIII and _Eco_RI, gel purified, and ligated to _Hin_dIII/_Eco_RI-digested p2062 to obtain p2064.
Various regions of SPI2 or STE genes were amplified by PCR using the primers specified in Table 1 to introduce restriction sites at the 5′ and 3′ ends. These fragments were digested with _Eco_RI and _Eco_RV, gel purified, and ligated to the _Eco_RI/_Sma_I-digested plasmid p2064. The characteristics of the resulting constructs are depicted in Fig. 1. These constructs express SPI2 or STE proteins with C-terminal fusions to the M45 epitope tag.
TABLE 1.
Oligonucleotides used in this study
| Designation | Sequencea |
|---|---|
| Pro_sseA_-For-_Hin_dIII | 5′-AGGAAGCTTAAGAAGAGAACAACGGCAAG-3′ |
| Pro_sseA_-Rev-_Eco_RI | 5′-CACGAATTCACGATAGATAATTAACGTGC-3′ |
| SseB-Rev-_Eco_RV | 5′-AGTGATATCTGAGTACGTTTTCTGCGC-3′ |
| SscB-For-_Eco_RI | 5′-CGGAATTCAATAGGTATGATGATGAAAG-3′ |
| SseF-Rev-_Eco_RV | 5′-ACGATATCTCCCCGAGATGTATGATCAG-3′ |
| SseG-Rev-_Eco_RV | 5′-ACGGATATCCGGCGCACGTTGTTCTGGCG-3′ |
| SifA-Pro-For-_Eco_RI | 5′-CCGGAATTCGTGASTATAAGCGATTAATTGC-3′ |
| SifA-For-_Eco_RI | 5′-CCGGAATTCATTTTTACTCCAGTATAAG-3′ |
| SifB-Pro-For-_Eco_RI | 5′-CCGGAATTCTGCCCTACCGCTAAAC-3′ |
| SifB-Rev-_Eco_RV | 5′-ACGGATATCACTCTGGTGATGAGCCTC-3′ |
| SseJ-Pro-For-_Eco_RI | 5′-CCGGAATTCACATAAAACACTAGCAC-3′ |
| SseJ-Rev-_Eco_RV | 5′-ACGGATATCTTCAGTGGAATAATGATGAGC-3′ |
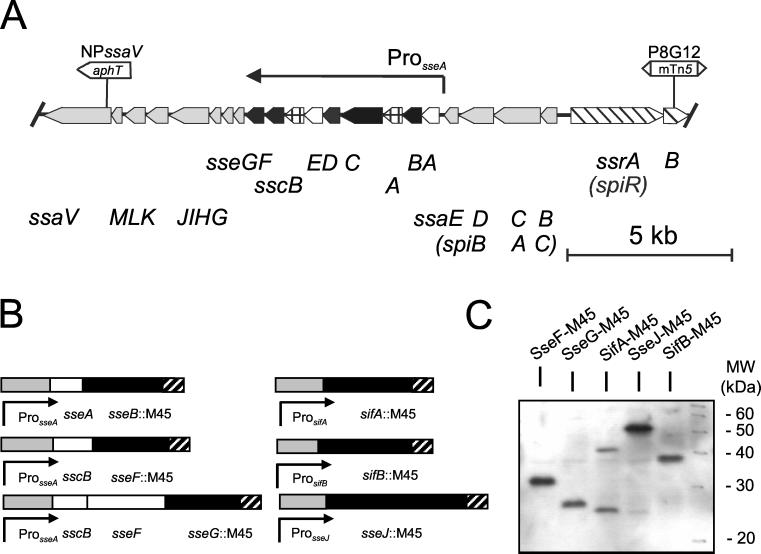
FIG. 1.
Generation of fusion proteins of SPI2 substrate proteins with the M45 tag. (A) Genetic organization of SPI2 genes and position of mutations relevant for this study. Genes encoding the TTSS, substrate proteins, and chaperones are indicated by shaded, filled, and cross-lined thick arrows, respectively. Genes encoding the regulatory system of SPI2 and proteins of unknown function are shown as slanted-line and open thick arrows, respectively. The positions of Pro_sseA_ and the putative transcriptional unit under the control of Pro_sseA_ are indicated. (B) Schematic representation of fusion constructs used in this study. Gene fusions with the M45 epitope tag are indicated by filled symbols. Black bars indicate genes fused to the M45 tag (hatched bars). Shaded and open bars indicate the promoter and further genes of the construct, respectively. (C) Detection of M45 fusion proteins by monoclonal antibodies against the M45 epitope. Wild-type Salmonella serovar Typhimurium harboring various plasmids for the expression of fusion proteins were grown under conditions inducing SPI2 gene expression (see Fig. 2 for details). Proteins of total cell lysates were separated by SDS-PAGE and transferred onto nitrocellulose membranes. Fusion proteins were detected by using the αM45 antibody.
If specific promoters of STE genes were used instead of Pro_sseA_, PCR was performed with forward primers introducing an _Eco_RI restriction site. PCR fragments containing specific promoters were ligated to the _Eco_RI/_Sma_I-digested plasmid p2062 to obtain gene fusions to the M45 epitope tag.
In order to obtain expression of epitope fusion proteins from a vector with low copy numbers, the inserts of various plasmids were transferred to pWSK29 (21) as listed in Table 2.
TABLE 2.
Plasmids used in this study
| Designation | Characteristics | Source or reference |
|---|---|---|
| pSK+ | High-copy-number vector, Ampr | Stratagene |
| pWSK29 | Low-copy-number vector, Ampr | 21 |
| p2062 | M45 epitope tag in pSK+ | This study |
| p2064 | Pro_sseA_ in p2062 | This study |
| p2095 | Pro_sseA sscB sseF_::M45 in pWSK29 | This study |
| p2096 | Pro_sseA sscB sseFG_::M45 in pWSK29 | This study |
| p2104 | Pro_sifA sifA_::M45 in pWSK29 | This study |
| p2126 | Pro_sseA sseAB_::M45 in pWSK29 | This study |
| p2129 | Pro_sseJ sseJ_::M45 in pWSK29 | This study |
| p2131 | Pro_sifB sifB_::M45 in pWSK29 | This study |
Preparation of surface-associated and secreted proteins.
Bacterial strains were grown in Luria broth (LB) for 8 h at 37°C. Bacteria were washed twice in 0.9% NaCl and used to inoculate a 400-ml culture of PCN-P medium at pH 5.8. Cultures were grown overnight in 2-liter glass flasks without baffles with agitation at 200 rpm. Bacteria were pelleted by centrifugation at 6,000 × g for 20 min and resuspended in 20 ml of phosphate-buffered saline. The supernatant was passed through a 0.2-μm filter and stored on ice for further use. The pellet was resuspended in 5 ml of phosphate-buffered saline in a 50-ml centrifuge tube (Falcon) and mixed vigorously on a Vortex mixer (Vortex Genie 2; Scientific Industries) for 60 s to detach bacteria-associated surface structures. Bacteria were pelleted again, and the supernatant (detached fraction) was passed through a 0.2-μm filter to remove residual bacteria. Protein from the culture supernatant and the detached fraction was recovered by precipitation with trichloroacetic acid (10% [wt/vol] final concentration) on ice for 3 h and centrifugation at 12,000 × g for 45 min. The pellet was washed twice with 15 ml of acetone and recovered by centrifugation at 10,000 × g for 30 min. The final pellet was air dried. To remove salts, the pellet was subjected to methanol-chloroform precipitation (22). For this purpose, the pellet was resuspended in 0.1 ml of H2Odd and 0.4 ml of methanol was added to the sample. The solution was mixed and centrifuged (10 s, 9,000 × g) for recovery of the sample. A total of 0.1 ml of chloroform was added to the sample, mixed, and centrifuged as described above. For phase separation, 0.3 ml of H2Odd was added and the sample was mixed vigorously and centrifuged for 10 min at 9,000 × g. The upper phase was carefully removed and discarded. A further 0.3 ml of methanol was added to the remaining chloroform phase and the interphase with the precipitated protein. The samples were mixed and centrifuged again for 2 min at 9,000 × g. The supernatant was removed and the pellet was vacuum dried and finally resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-HCl [pH 6.8], 4% SDS, 2% β-mercaptoethanol, 12.5% glycerol, and 0.001% bromophenol blue).
RESULTS
Generation of epitope-tagged derivatives of STE proteins and SPI2-encoded proteins.
Secreted and translocated substrate proteins of the TTSS of SPI2 are encoded by genes both within (see Fig. 1A) and outside of the SPI2 locus. We studied the secretion of both classes of substrate proteins. It has been reported before that only small amounts of proteins were secreted by the TTSS of SPI2 (2). To facilitate the detection of such proteins, a universal approach was chosen for labeling candidate substrate proteins with an epitope tag. The M45 epitope tag, an 18-amino-acid peptide of the adenovirus protein E4-6/7, has previously been used to generate fusions to substrate proteins of the SPI1 TTSS. Secretion into the growth medium, as well as translocation into eukaryotic cells, has been demonstrated using C-terminal fusions of candidate proteins to the M45 epitope and by detection with a monoclonal antibody against the M45 epitope (for examples, see references 4 and 9). We established a vector system that allowed the simple construction of fusions between genes for candidate substrate proteins and the M45 epitope. Furthermore, we generated fusions of various STE and other putative substrate proteins of the TTSS of SPI2. C-terminal fusions were constructed with full-length proteins (Fig. 1B). Functional fusions were obtained for SifA, SifB, SseJ, SseB, SseF, and SseG but not for SseI. Using the antibody against the M45 epitope in Western blot analyses, we detected single bands of the predicted molecular weights for SseF, SseG, SseJ, and SifB (Fig. 1C). However, two bands corresponding to approximately 41 and 25 kDa were obtained with the construct encoding SifA-M45 (Fig. 1C). The presence of the 25-kDa band may indicate an instability of the fusion protein. It has been previously observed that protein fusions of SifA are unstable or susceptible to degradation (3, 14).
Expression of M45 fusion proteins.
Various vector constructs were analyzed for regulated expression of gene fusions. For this purpose, we analyzed levels of fusion proteins synthesized under growth conditions that were previously described as inducing or repressing SPI2 gene expression. Limitation of the amount of phosphate or magnesium in the growth medium was shown to induce the expression of genes under the regulatory control of SsrAB, the two-component system of SPI2 (5). Strains harboring plasmids for the expression of M45-tagged proteins were grown in LB, minimal medium with high (PCN) or limiting (PCN-P) amounts of phosphate, and minimal medium with high or limiting concentrations of magnesium. The amounts of fusion proteins synthesized were analyzed by Western blotting of total cell lysates using an antibody against the M45 epitope.
We first analyzed the expression of sseB::M45 under the control of the SPI2 promoter Pro_sseA_. Fusion proteins expressed by a high-copy-number vector were detected after growth under all medium conditions investigated, as well as in the background of an ssrB mutation, indicating that expression was not coordinately regulated (data not shown). When the same fusion construct was analyzed in the background of the low-copy-number vector pWSK29, the epitope-tagged protein was only observed in minimal medium starved of phosphate or magnesium. In the background of an ssrB strain (P8G12, ssrB::mTn_5_), no epitope-tagged protein was observed (Fig. 2A). A mutation in ssaV, a structural component of the TTSS of SPI2, had no effect on the levels of the fusion protein (Fig. 2A). The effects of medium composition and mutations in ssrB or ssaV on the levels of SseB-M45 were comparable to those of previous observations and indicated that constructs generated based on low-copy-number vector pWSK29 were regulated in a fashion similar to that of the chromosomal alleles. Therefore, for all subsequent studies, gene fusions were constructed in the background of pWSK29. Furthermore, promoter Pro_sseA_ was used for the expression of sseF::M45 and sseG::M45 fusions, and similar effects of growth medium on protein levels were observed (Fig. 2A).
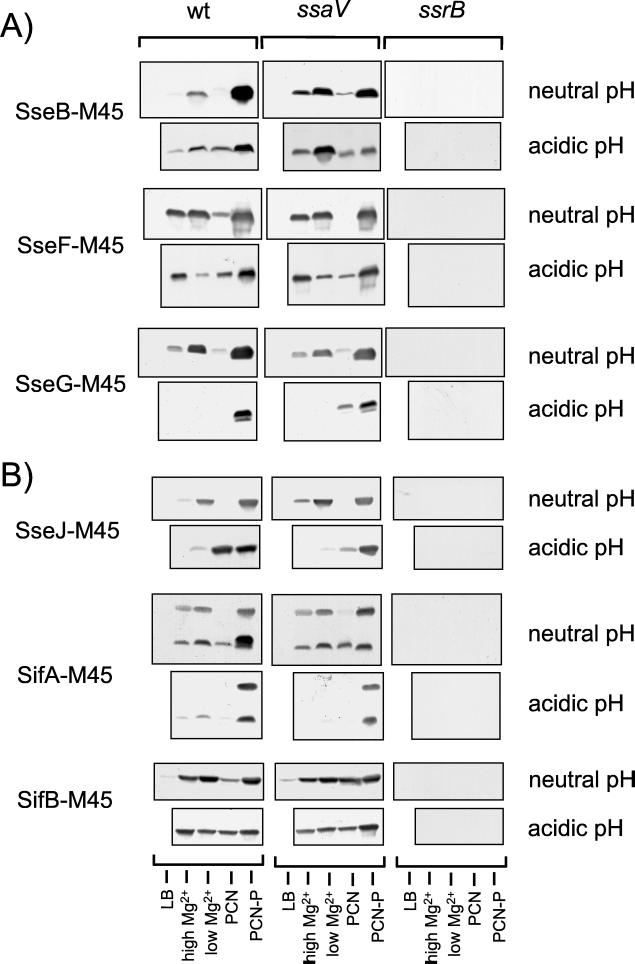
FIG. 2.
Effect of growth conditions on expression of STE-M45 fusions. Salmonella serovar Typhimurium wild-type (wt) strain, a mutant strain defective in secretion by the SPI2 secretion system (ssaV), and a mutant strain (P8G12) defective in the regulatory system SsrAB of SPI2 (ssrB) harboring various plasmids for the expression of fusion proteins with the M45 epitope were grown in LB, N-salts medium containing 10 mM Mg2+ (high Mg2+) or 30 μM Mg2+ (low Mg2+), PCN medium without nutritional limitation (PCN), or PCN medium containing a limiting (0.34 mM) amount of phosphate (PCN-P). Equal amounts of bacteria as adjusted by optical density at 600 nm were concentrated by centrifugation and lysed by boiling for 5 min in SDS-PAGE sample buffer. For Western blotting, protein was separated by SDS-PAGE, transferred onto nitrocellulose membranes, and detected using a monoclonal antibody against the M45 epitope and a goat anti-mouse IgG horseradish peroxidase conjugate as primary and secondary antibodies, respectively. (A) Gene fusions under control of the promoter Pro_sseA_. (B) Gene fusions under control of the individual promoters of STE genes.
We then analyzed the dependence of levels of STE proteins SifA, SifB, and SseJ on growth under various medium conditions and on the background of a mutation in the SPI2-encoded regulator SsrB. As observed for fusions under the control of Pro_sseA_, no fusion proteins were detected in the ssrB background, indicating that functional SsrB was absolutely required for the expression of all of these constructs (Fig. 2B). Effects on the levels of SifA and SseJ of medium conditions imposing phosphate or magnesium starvation were similar to the effect of similar constraints on SPI2-encoded SseB. These observations indicate that the promoters of sifA and sseJ have similar characteristics to those within SPI2. In contrast to SifA and SseJ, SifB was also detected in bacteria grown in rich or minimal medium with nonlimiting amounts of phosphate or magnesium.
After growth in various minimal media in acidic pH conditions, protein levels of M45 fusion proteins were rather heterogeneous. In acidic pH conditions, effects of nutritional limitations on protein levels of SseB-M45 are less pronounced than at neutral pH. In contrast, high levels of SseG-M45 and SifA-M45 were only detected in cultures grown in minimal medium with phosphate limitation (PCN-P [pH 5.8]). The different levels of SseF-M45 and SseG-M45 after growth in media of acidic pH are remarkable, since the corresponding gene fusions are both under the control of Pro_sseA_.
For all fusion proteins, high levels were detected in lysates of cultures grown in PCN-P (pH 5.8). As these growth conditions also induce the secretion of SseBCD (15), subsequent analyses of the secretion of the fusion proteins were performed with cultures grown under these conditions.
M45-tagged SseB is secreted in vitro.
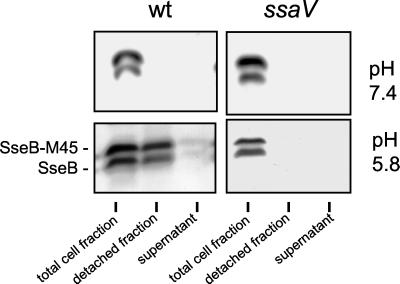
To analyze whether M45-tagged proteins are secreted in vitro by the TTSS of SPI2, we first performed secretion experiments with SseB, a known substrate protein (3). Salmonella serovar Typhimurium wild-type and ssaV mutant strains, both containing plasmid-borne sseB::M45, were grown in PCN-P medium in neutral and acid pH conditions. The distribution of putative substrate proteins was analyzed in the bacterial pellet, in the culture supernatant, and in the protein fraction that was detached from the bacterial cell surface by mechanical forces. Previous analyses of SseB, SseC, and SseD showed that, after secretion, these proteins mainly accumulate in the cell surface-associated fraction and are present only in small amounts in the culture supernatant (2, 13). The native SseB protein and the SseB-M45 protein were detected in the detached fraction of the wild-type strain grown in acidic pH conditions, indicating that both proteins are secreted and located on the bacterial cell surface. No secretion of SseB or SseB-M45, at neutral pH or by the ssaV strain, was observed (Fig. 3).
FIG. 3.
Secretion of SseB and the SseB-M45 fusion protein by the TTSS of SPI2. Secretion of native and M45-tagged SseB was analyzed for Salmonella serovar Typhimurium wild-type (wt) strain and a mutant strain defective in the TTSS of SPI2 (ssaV). Bacteria were grown in 400 ml of PCN-P at pH 7.4 or pH 5.8 to induce expression of the TTSS and secretion of substrate proteins. Protein present on the bacterial cell surface and in the culture supernatant was recovered as described in Materials and Methods. For analysis of the bacterial pellet, equal amounts of bacterial cells as adjusted by optical density at 600 nm were analyzed. Equal amounts of bacterial cells and culture supernatant were processed to analyze protein in the detached and supernatant fractions, respectively. Under the assay conditions, all cultures grew to a comparable cell density. Western blots were incubated with a polyclonal antiserum raised against recombinant SseB (2) and goat anti-rabbit IgG horseradish peroxidase conjugate as primary and secondary antibodies, respectively.
SseF and SseG are secreted target proteins of the TTSS of SPI2.
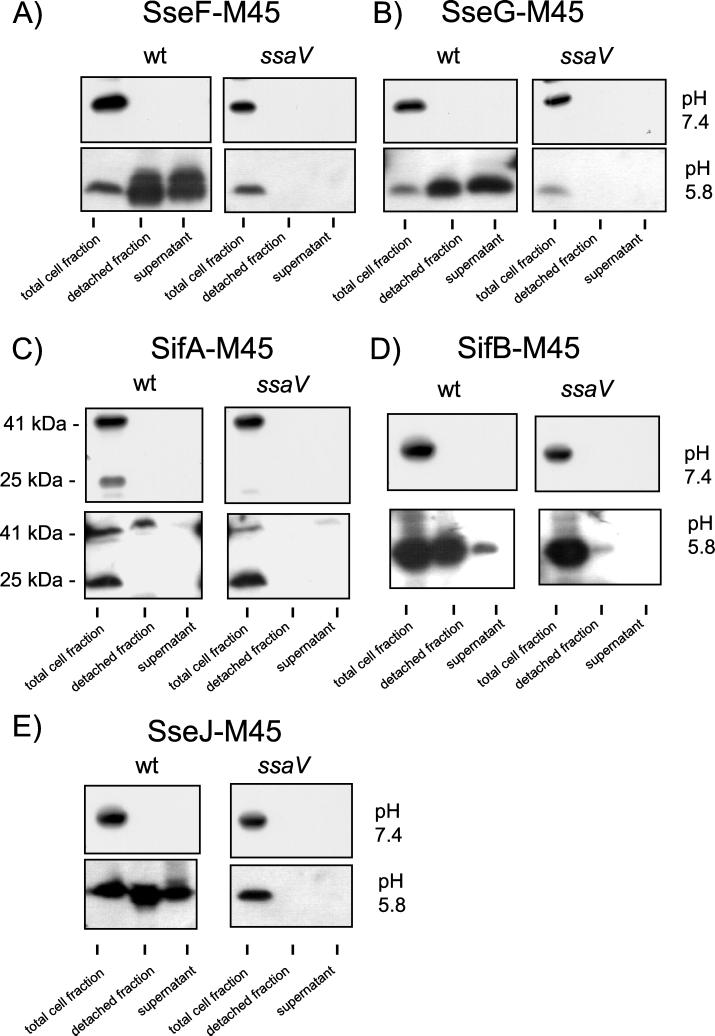
The SPI2-encoded proteins SseF and SseG have been suggested to be substrate proteins of the TTSS of SPI2 (11). However, the secretion or translocation of these proteins has not been demonstrated so far. Here, we analyzed secretion of SseF and SseG in vitro. After growth of Salmonella serovar Typhimurium wild-type harboring plasmids for the expression of sseF::M45 or sseG::M45 in PCN-P at pH 5.8, both SseF-M45 and SseG-M45 were detected in the protein fraction detached from the cell surface as well as in the culture supernatant (Fig. 4A and B). When these strains were grown under SPI2-inducing conditions at neutral pH or in the background of a mutation in ssaV, SseF-M45 or SseG-M45 were detected neither in the detached fraction nor in the culture supernatant (Fig. 4A and B). Taken together, these observations indicate that SseF and SseG are secreted substrate proteins of the TTSS of SPI2.
FIG. 4.
Secretion of M45-fusion proteins by the TTSS of SPI2. Secretion of M45-tagged substrate proteins was analyzed for Salmonella serovar Typhimurium wild-type (wt) strain and a mutant strain defective in the TTSS of SPI2 (ssaV) as described in the legend to Fig. 3. M45 fusion proteins SseF-M45 (A), SseG-M45 (B), SifA-M45 (C), SifB-M45 (D), or SseJ-M45 (E) were detected in the various fractions by using the antibody against the M45 epitope.
STE proteins are secreted in vitro by the TTSS of SPI2.
We extended the analysis of secretion to members of the newly defined family of STE proteins. Translocation of N-terminal domains of STE proteins fused to CyaA into host cells has been observed, but secretion of full-length STE proteins has not been analyzed (14). By applying in vitro conditions inducing the secretion of SPI2-encoded substrate proteins, we observed that SseJ-M45, SifA-M45, and SifB-M45 were present in a protein fraction on the bacterial cell surface. SseJ-M45 and SifB-M45 were also detected in the culture supernatant. No secretion of the fusion proteins was observed in the ssaV strain or after growth under inducing neutral-pH conditions (Fig. 4C, D, and E).
All of the STE fusion proteins were present in a surface-attached fraction. After secretion, these proteins may aggregate into macromolecular complexes, as was observed previously for the SPI2-encoded substrate proteins SseB, SseC, and SseD (2, 13). Aggregation of proteins after secretion may be due to the composition of the secretion medium, namely, acidic pH conditions and lack of complex nutritional factors such as peptides and polysaccharides.
Only a protein of 41 kDa, representing the full-length SifA-M45 fusion protein, was detected in the detached fraction after growth at pH 5.8. This observation may indicate that an N-terminal truncated form of SifA-M45 is present in the bacterial cytoplasm that is not secreted by the TTSS of SPI2.
DISCUSSION
Recent studies showed that a set of secreted substrate proteins of the TTSS of SPI2 are encoded by genes within the SPI2 locus (2, 13, 15). In addition, a family of STE exists for SPI2, encoded by genes outside the SPI2 locus. The translocation of STE proteins into host cells by the TTSS of SPI2 has been demonstrated indirectly using the CyaA reporter technique (14) and has been postulated based on indirect evidence (3). In this study, we have shown that several STE proteins can be secreted in vitro under conditions that trigger the secretion of SseBCD. The substrate proteins SseBCD are encoded by genes within SPI2 and function as a translocon for the translocation of effector proteins into host cells (15). The data presented here confirm that SifA, SifB, and SseJ are substrate proteins of the SPI2 TTSS.
Our data demonstrate that the SPI2-encoded proteins SseF and SseG are additional secreted substrate proteins of the TTSS of SPI2. At present, the role of these proteins is unclear. Mutants in sseF or sseG are not attenuated in the animal model of salmonellosis and show only slightly reduced replication inside cultured macrophages (11). However, SseF and SseG are required for the formation of _Salmonella_-induced filaments (Sif) in infected epithelial cells (8). These observations suggest that SseF and SseG have functions that are required only in a subset of host cell types. Furthermore, as the functions of SseF and SseG are not absolutely required for the SPI2 phenotype, these proteins may not contribute to the translocon formation as shown for SseBCD (15). These observations support the hypothesis that SseF and SseG are more likely to be members of the family of translocated effector proteins with partially redundant or overlapping functions in the host cell.
Our data indicate the existence of a secretion signal for both STE and SPI2-encoded substrate proteins in addition to a translocation signal specific for STE. While the translocation signal for substrate proteins of the TTSS of SPI2 is defined by a conserved amino acid motif, a conserved domain for secretion is not present in these proteins. Such a secretion signal may be located on the mRNA, as reported for substrate proteins of the plasmid-encoded TTSS of Yersinia spp. (1). This model might imply that SPI2-encoded proteins such as SseF and SseG are not translocated into host cells, as SPI2-encoded substrate proteins lack a conserved amino acid domain. We also applied the tagged approach to SpiC (SsaB), a SPI2-encoded protein previously described as a translocated effector (19). Under the assay conditions described here, we could not detect secretion of a SpiC-M45 fusion protein in vitro (unpublished observations).
Rapid secretion of SseB within minutes after shifting Salmonella serovar Typhimurium cultures from SPI2-inducing neutral medium to acidic medium has been observed. Further analysis of the secretion of SseBCD indicated that these proteins are detectable in a detached fraction obtained by mechanical shearing not earlier than 2 h after shifting cultures to acidic pH (15). We have not been able to detect M45-tagged proteins in the detached fraction obtained by the method described here within minutes after shifting cultures to acidic medium (data not shown). We assume that larger amounts of SseBCD and additional substrate proteins of the SPI2 system have to accumulate on the bacterial surface before these proteins can be detached by the mechanical shearing method applied in this study.
In conclusion, we have identified SseF and SseG as further substrate proteins of the TTSS of SPI2 and demonstrated that SPI2-encoded as well as STE proteins are secreted by the TTSS of SPI2 under in vitro conditions. The experimental setup used in this study only detects secretion of substrate proteins, not translocation into host cells. Further work is needed to reveal whether SseF and SseG are also translocated into the host cell by intraphagosomal Salmonella serovar Typhimurium or whether SseF and SseG are components of the translocon that have accessory functions for the translocation of effector proteins.
Acknowledgments
Imke Hansen-Wester and Bärbel Stecher contributed equally to this work.
We are grateful to Jürgen Heesemann for generous support of this work at the Max von Pettenkofer-Insitute in Munich and to Cosima Pelludat and Brad Taylor for critical review of the manuscript. We thank Wolf-Dietrich Hardt for providing the M45 epitope and stimulating discussions and P. Hearing (SUNY, Stony Brook) for providing us with a hybridoma line for the production of monoclonal antibodies against the M45 epitope.
The project was supported by the Deutsche Forschungsgemeinschaft grants HE1964/2-3 and HE1964/4-2.
Footnotes
†
Michael Hensel dedicates this article to Karlheinz Altendorf (Osnabrück) on the occasion of his 60th birthday.
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278**:**1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33**:**806-816. [DOI] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of sifA. EMBO J. 19**:**3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64**:**3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31**:**1759-1773. [DOI] [PubMed] [Google Scholar]
- 6.Deiwick, J., T. Nikolaus, J. E. Shea, C. Gleeson, D. W. Holden, and M. Hensel. 1998. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J. Bacteriol. 180**:**4775-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galan, J. E. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol. 2**:**46-50. [DOI] [PubMed] [Google Scholar]
- 8.Guy, R. L., L. A. Gonias, and M. A. Stein. 2000. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol. 37**:**1417-1435. [DOI] [PubMed] [Google Scholar]
- 9.Hardt, W. D., and J. E. Galan. 1997. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA 94**:**9887-9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36**:**1015-1023. [DOI] [PubMed] [Google Scholar]
- 11.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30**:**163-174. [DOI] [PubMed] [Google Scholar]
- 12.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62**:**379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein, J. R., and B. D. Jones. 2001. Salmonella pathogenicity island 2-encoded proteins SseC and SseD are essential for virulence and are substrates of the type III secretion system. Infect. Immun. 69**:**737-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97**:**7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schröder, and I. Miller. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183**:**6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obert, S., R. J. O'Connor, S. Schmid, and P. Hearing. 1994. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14**:**1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93**:**2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18**:**3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of _Salmonella_-induced enteritidis. Mol. Microbiol. 36**:**997-1005. [DOI] [PubMed] [Google Scholar]
- 21.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100**:**195-199. [PubMed] [Google Scholar]
- 22.Wessel, D., and U. I. Flügge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138**:**141-143. [DOI] [PubMed] [Google Scholar]