Stable Ribosome Binding to the Endoplasmic Reticulum Enables Compartment-specific Regulation of mRNA Translation (original) (raw)
Abstract
In eukaryotic cells, protein synthesis is compartmentalized; mRNAs encoding secretory/membrane proteins are translated on endoplasmic reticulum (ER)-bound ribosomes, whereas mRNAs encoding cytosolic proteins are translated on free ribosomes. mRNA partitioning between the two compartments occurs via positive selection: free ribosomes engaged in the translation of signal sequence-encoding mRNAs are trafficked from the cytosol to the ER. After translation termination, ER-bound ribosomes are thought to dissociate, thereby completing a cycle of mRNA partitioning. At present, the physiological basis for termination-coupled ribosome release is unknown. To gain insight into this process, we examined ribosome and mRNA partitioning during the unfolded protein response, key elements of which include suppression of the initiation stage of protein synthesis and polyribosome breakdown. We report that unfolded protein response (UPR)-elicited polyribosome breakdown resulted in the continued association, rather than release, of ER-bound ribosomes. Under these conditions, mRNA translation in the cytosol was suppressed, whereas mRNA translation on the ER was sustained. Furthermore, mRNAs encoding key soluble stress proteins (XBP-1 and ATF-4) were translated primarily on ER-bound ribosomes. These studies demonstrate that ribosome release from the ER is termination independent and identify new and unexpected roles for the ER compartment in the translational response to induction of the unfolded protein response.
INTRODUCTION
Eukaryotic cells display a ubiquitous and phylogenetically conserved process of mRNA partitioning whereby mRNAs encoding secretory or integral membrane proteins are localized early in translation to the endoplasmic reticulum (ER) (Blobel and Dobberstein, 1975a,b; Palade, 1975; Blobel et al., 1979; Lingappa and Blobel, 1980). This process serves to compartmentalize protein synthesis; those proteins that require entry into the secretory pathway are synthesized on ER-bound ribosomes (Blobel and Dobberstein, 1975a,b; Palade, 1975; Blobel et al., 1979; Lingappa and Blobel, 1980). The trafficking of ribosomes from the cytosol to the ER membrane is functionally linked to translation; ribosomes engaged in the translation of mRNAs encoding a signal sequence or a transmembrane domain are directed to the ER via the signal recognition particle (SRP) pathway (Blobel and Dobberstein, 1975a,b; Walter et al., 1981; Gilmore and Blobel, 1983; Walter and Johnson, 1994). In this pathway, the termination of protein synthesis is thought to yield ribosome dissociation from the ER membrane to a common cytosolic ribosome pool, thereby completing a cycle of translation-dependent ribosome binding and release (Blobel and Dobberstein, 1975a; Mechler and Vassalli, 1975; Lingappa and Blobel, 1980).
The role of the SRP pathway in the partitioning of ribosome-mRNA complexes between the cytosol and ER compartments is very well established (Blobel et al., 1979; Lingappa and Blobel, 1980; Walter and Johnson, 1994; Alberts et al., 2002). Recent studies in mammalian tissue culture cells, using small interfering RNA (siRNA)-mediated knockdown of individual components of the SRP complex indicate, however, that tissue culture cell lines can be tolerant of marked reductions in SRP levels (Aza-Blanc et al., 2003). In these experiments, cell surface expression of death receptor (DR)-5 and Fas receptor was insensitive to siRNA-mediated reduction in SRP levels, whereas cell surface expression of DR-4 was inhibited, suggesting that secretory and membrane protein precursors can vary dramatically in their requirement for SRP function (Aza-Blanc et al., 2003). Also consistent with these data are the suggestions that mRNAs may undergo direct localization to the ER and by this means differ in their requirement for SRP function (Choi et al., 2000; Diehn et al., 2000; Nicchitta, 2002; Washida et al., 2004). In support of this view, it has been reported that ribosomes remain in stable association with the ER translocation machinery after pharmacological induction of protein synthesis termination, and while in the ER-bound state they are capable of de novo translation of mRNAs encoding secretory and cytosolic proteins (Potter and Nicchitta, 2000, 2002; Seiser and Nicchitta, 2000). This latter observation is consistent with studies conducted in yeast, fly, and mammalian cells, each demonstrating a significant steady-state representation of mRNAs encoding cytosolic proteins on ER-bound polyribosomes (Mechler and Rabbitts, 1981; Mueckler and Pitot, 1981; Kopczynski et al., 1998; Diehn et al., 2000; Lerner et al., 2003). Although the ultimate biological significance of these findings is not yet clear, it seems that the process of mRNA partitioning to the ER is complex and may require multiple, perhaps independent, pathways to achieve the compartmentalization of protein synthesis characteristic of eukaryotic cells.
In the current study, we examined mRNA partitioning and translation in the ER and cytosol compartments after induction of the unfolded protein response (UPR). In mammalian cells, UPR induction elicits activation of the ER-resident eukaryotic initiation factor 2α (eIF2α)-directed kinase PERK, and the subsequent inactivation of the initiation stage of protein synthesis, as well as a stress response transcriptional program requiring the de novo synthesis of several transcription factors, most prominently, ATF4 and XBP-1 (Harding et al., 1999, 2000b; Okada et al., 2002; Rutkowski and Kaufman, 2004). PERK-mediated eIF2α phosphorylation prevents eIF2B-dependent GDP/GTP exchange, thereby compromising the formation of translation initiation complexes (Harding et al., 2000b; Hinnebusch, 2000). As a consequence, a rapid and sustained breakdown of polyribosomes ensues (Brostrom and Brostrom, 1998; Scheuner et al., 2001). UPR induction thus represents a useful and physiologically relevant means for examining the cellular basis for ribosome and mRNA trafficking/exchange on the ER membrane.
The UPR uses mRNA stabilization and transcriptional regulation to impart substantial changes in the cellular content of numerous mRNA species (Travers et al., 2000; Okada et al., 2002; Kawai et al., 2004). Together, the coordinated transcriptional and translational UPR program results in the dramatic remodeling of polyribosome complexes, because ribosome recruitment is enhanced for UPR target mRNAs and diminished for numerous housekeeping transcripts (Harding et al., 2000a; Kawai et al., 2004). It is not known whether this process of polyribosome remodeling proceeds in an unbiased way in the cytosolic and ER compartments of the cell. Indeed, given that enhanced transcription/translation of mRNAs encoding ATF4, XBP-1, and the ER-resident heat shock protein (Hsp) 70 chaperone BiP is a common hallmark of the UPR, the general expectation is that the protein synthesis machinery of the cytosol and ER compartments is under common regulatory control and thus that the translation of mRNAs encoding both stress response cytosolic proteins (i.e., ATF4 and XBP-1) and signal-sequence bearing proteins (i.e., BiP) would be maintained in the cytosol and ER compartments, respectively.
In this report, we have examined ribosome and mRNA distributions between the ER and cytosol compartments after UPR induction and report three primary findings: 1) after termination, the predominant fate of ER-bound ribosomes is continued association with the ER membrane; 2) the protein synthesis machinery of the cytosol and ER compartments of the cell is under distinct regulatory control, with mRNA translation being sustained in the ER compartment and suppressed in the cytosol compartment under conditions of elevated phopho-eIF2a levels; and 3) ER-bound ribosomes can participate in the translation of mRNAs encoding cytosolic and secretory/membrane proteins alike. These data identify novel roles for the ER compartment in the global regulation of cellular protein synthesis.
MATERIALS AND METHODS
Reagents
[35S]Methionine/cysteine and α-[32P]dCTP were from MP Biomedicals (Irvine, CA). [3H]5,6-uridine was from MP Biomedicals. Digitonin and dodecylmaltoside were from Calbiochem (San Diego, CA). Polyuridylic acid (poly-U), polyinosinic acid, polyadenylic acid, and cycloheximide were from Sigma-Aldrich (St. Louis, MO). Cyanogen bromide-activated Sepharose 4B resin and Hybond membranes were from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). Cell culture reagents were from Invitrogen (Carlsbad, CA). RNase OUT, SuperScript RNase H- reverse transcription reaction kit, and TRIzol reagent were from Invitrogen. Random-primed DNA labeling kit was from Roche Diagnostics (Indianapolis, IN). Nuclease-treated rabbit reticulocyte lysate was obtained from Promega (Madison, WI). eIF2α, phospho-eIF2α, XBP-1, and ATF4 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). β-Tubulin antibody (E7) was from the Iowa Developmental Studies Hybridoma Bank (Iowa City, IA).
Cell Culturing and Labeling
Murine plasmacytoma J558L cells were cultured in DMEM supplemented with 10% equine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, at 37°C with 5% CO2. Before experiments, cells were adjusted to 0.5 × 106 cells/ml in 2:1 DMEM supplemented with 10% fetal bovine serum:cultured media (DMEM supplemented with 10% equine serum) and cultured overnight. Murine NIH 3T3 and human embryonic kidney (HEK)293 cells were cultured in DMEM supplemented with 10% calf serum or fetal calf serum, respectively, 100 U/ml penicillin, 100 μg/ml streptomycin at 37°C with 5% CO2.
Subcellular Fractionation
Cells were treated with either 2 or 10 mM dithiothreitol (DTT) (NIH 3T3 and J558, respectively) or 500 nM thapsigargin (NIH 3T3) for the indicated times, where applicable. All cells were treated with 0.2 mM cycloheximide for 5 min at 37°C before harvest. Cytosol and total membrane-derived ER fractions were recovered by sequential detergent extraction using 0.008% digitonin for plasma membrane permeabilization/cytosol release and 2% digitonin or dodecylmaltoside for ER solubilization, using 1.5 × 107 cells (107 cells/ml) as reported previously (Potter and Nicchitta, 2002; Lerner et al., 2003). For the adherent cell line NIH 3T3, the detergent fractionation procedures were performed on cell monolayers.
Rough microsome fractions were obtained as follows: 3-5 × 107 J558 cells were homogenized in a 2-ml Dounce homogenizer (B pestle) at a concentration of 108 cells/ml in a buffer consisting of 10 mM KOAc, 10 mM K-HEPES, pH 7.2, 1.5 mM Mg(OAc)2, 0.2 mM cycloheximide, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 40 U/ml RNase OUT. The cell homogenate was diluted fivefold in 2.5 M sucrose/HKM [25 mM K-HEPES, pH 7.2, 150 mM KOAc, 5 mM Mg(OAc)2, 0.2 mM cycloheximide, 1 mM DTT, 1 mM PMSF, and 10 U/ml RNase OUT]. Then, 1.5 ml of homogenate was loaded above a 0.25-ml cushion of 2.5 M sucrose/HKM and the following gradient layers were added: 1.5 ml of 1.9 M sucrose/HKM, 0.75 ml of 1.3 M sucrose/HKM, and 0.25 ml of HKM. Gradients were centrifuged in a Beckman SW55 rotor at 55,000 rpm for 2.5 h at 4°C. The rough microsome layer, which banded at the 1.3 M/1.9 M sucrose interface, was diluted fourfold in HKM and recovered by centrifugation in a Beckman TLA100.3 rotor at 40,000 rpm for 20 min at 4°C. The rough microsome (RM) pellet was solubilized using 2% dodecylmaltoside lysis buffer and the clarified lysate used for velocity sedimentation experiments (Potter and Nicchitta, 2002).
Velocity Sedimentation
Cell equivalent fractions were loaded onto 10 ml of 15-50% linear sucrose gradients and centrifuged at 151,000 × g as reported previously (Potter and Nicchitta, 2002). Fractions were collected manually, and UV (260-nm) absorbance was monitored. Immunoblots for ER resident membrane proteins were performed as reported previously (Potter and Nicchitta, 2002).
Immunofluorescence Microscopy
HEK293 cells were grown overnight on coverslips (1.5 mm). After different stages of sequential detergent extraction, cells were fixed in 4% paraformaldehyde and permeabilized in 0.1% saponin. Coverslips were incubated with primary antibodies (anti-TRAPα (rabbit polyclonal), 1:50; anti-β-tubulin (mouse monoclonal; E7 [Iowa Developmental Studies Hybridoma Bank]; 1:50) for 30 min and extensively washed with 0.5% bovine serum albumin (BSA)-containing phosphate-buffered saline (PBS). Coverslips were then incubated with secondary antibodies (goat anti-mouse IgG-conjugated AlexaFluor-555 [Invitrogen], 1:100; goat anti-rabbit IgG-conjugated AlexaFluor-647 [Invitrogen], 1:100) and 4,6-diamidino-2-phenylindole (DAPI) (0.5 μg/ml) for 30 min, washed with 0.5% BSA-containing PBS, and fixed with FluorSave reagent (Calbiochem). Micrographs were generated on a Zeiss Axiophot microscope, and images were assembled in Adobe Photoshop 7.0.
Protein Synthesis Assays
Cells (1 × 106) were either treated with 2 mM DTT, 10 mM DTT, 500 nM thapsigargin, or untreated for the indicated times and subsequently radiolabeled by the addition of 50-100 μCi of [35S]methionine to cell culture media (minus methionine) suspensions. Cells were lysed in 1 ml of 1% Triton X-100, 0.05% SDS, 25 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM DTT, and 1 mM PMSF for 30 min on ice and clarified by centrifugation. For total protein synthesis rates, trichloroacetic acid (TCA)-precipitated lysates were processed for SDS-PAGE, and radiolabel incorporation was determined by PhosphorImager analysis. Immunoprecipitation of radiolabeled proteins was performed using the indicated antibodies and protein A- or G-Sepharose resin by standard protocols. Immunoblots of XBP-1, ATF4, and β-actin were performed as described previously (Gass et al., 2002).
Ribosome Quantitation
Cell aliquots (5 × 106) of J558L cells were cultured overnight in 2.5 μCi/ml [3H]5,6-uridine. Cells were harvested, and cytosol and membrane fractions were obtained as described previously (Potter and Nicchitta, 2002; Lerner et al., 2003). RNA was extracted in triplicate using TRIzol reagent (Invitrogen) and resolved on 3% formaldehyde, 1% agarose gels. Bands corresponding to the 28S and 18S rRNA were excised, the gel matrix was melted at 95°C for 3 min, and radioactivity was determined by liquid scintillation spectroscopy by using a liquid scintillation analyzer Tri-Carb 2100TR (PerkinElmer Life and Analytical Sciences, Boston, MA). For quantification of ribosmal RNAs (rRNAs) derived from NIH 3T3 cells, ethidium bromide fluorescence was determined at 610 nm by using a Typhoon 9400 (GE Healthcare).
Poly(U)-Sepharose Binding
Polyuridylic acid was coupled to cyanogen bromide-activated Sepharose 4B resin according to the manufacturer's specifications. Before use, poly(U)-Sepharose was washed twice in 1 bed volume of 2 M KCl, 25 mM K-HEPES, pH 7.2; twice in 1 bed volume of 10 mM KCl, 25 mM K-HEPES, pH 7.2, 13 mM Mg(OAc)2, 0.2 mM cycloheximide, 1 mM DTT, and 10 U/ml RNase OUT; washed once; and resuspended in 1 bed volume of binding buffer [300 mM KCl, 25 mM K-HEPES, pH 7.2, 13 mM Mg(OAc)2, 1% polyvinyl alcohol, 0.2 mg/ml tRNA, 0.2 mM cycloheximide, 1 mM DTT, of 40 U/ml RNase OUT].
Cells (2 × 107) treated with 0.2 mM cycloheximide for 5 min at 37°C were harvested and subsequently lysed in 2% digitonin lysis buffer (Potter and Nicchitta, 2002). Ribosomes were recovered from clarified lysates by centrifugation (Potter and Nicchitta, 2002). Ribosomal pellets were resuspended in 0.5 ml of binding buffer and added to 200 μl of poly(U)-Sepharose beads (50% slurry) and rotated at 37°C for 30 min. The binding mixture was cooled and transferred to 0.8 × 4-cm polypropylene columns (Bio-Rad, Hercules, CA). The resin was washed with 5 bed volumes of wash buffer [300 mM KCl, 25 mM K-HEPES, pH 7.2, 13 mM Mg(OAc)2, 1 mM DTT, 0.2 mM cycloheximide, and 10 U/ml RNase OUT], and ribosomes were eluted with 5 bed volumes of EDTA elution buffer (300 mM KCl, 25 mM K-HEPES, pH 7.2, 5 mM EDTA, 0.2 mM cycloheximide, 10 mM DTT, and 40 U/ml RNase OUT). Fractions were collected (150-200 μl) and measured by UV-spectrometry (260 nm).
For quantitative assays, cells were cultured overnight with 2.5 μCi/ml [3H]5,6-uridine, to allow biosynthetic radiolabeling of rRNA. Ribosomal loadings were normalized for total radioisotope incorporation. The major fractions corresponding to EDTA-eluted ribosomes were combined, adjusted to 1 ml with diethyl pyrocarbonate-treated H2O, and precipitated by the addition of 5 μg of carrier tRNA and TCA to a final concentration of 10%. Samples were collected in triplicate (300 μl) by vacuum filtration onto GC/F glass fiber filters presoaked in 10% TCA. Filters were washed once with 3 ml of 10% cold TCA, twice with 3 ml of cold 95% ethanol and dried. Filter-associated radioactivity was measured by liquid scintillation spectroscopy.
Competition studies were performed as stated above except poly(U)-Sepharose was preincubated with polyinosinic acid, polyadenylic acid, or buffer alone for 30 min at 37°C before addition of ribosomes.
Northern Blot Analysis
Northern blots and quantitative dot blots were conducted by standard protocols (Sambrook et al., 1989). RNA was isolated from gradient fractions by phenol-chloroform extraction, resolved on 3% formaldehyde 1% agarose gels, and transferred in 5× SSC, 10 mM NaOH by downward capillary flow onto Hybond for 2 h. Probes were generated from plasmid-containing cDNAs and included a KpnI fragment of canine GRP94 (975 bp), a full-length clone of murine ATF4 (1.4 kb), NcoI/ApoI fragment of murine XBP-1 (620 bp), a BamHI/KpnI fragment of human GAPDH cDNA (498 base pairs), and PstI/EcoRI fragment of BiP (1.5 kb). Probes were internally labeled with [α-32P]dCTP using a random hexanucleotide primer kit. PhosphorImager plates were scanned using a Typhoon 9400 (GE Healthcare), and data analyses were performed using the ImageQuantTL software (GE Healthcare).
Reverse Transcription Assay
RNA isolated from gradient fractions was suspended in 10 μl of nuclease-free H2O. Then, 50 ng of oligo(dT)20 primer was added, and samples were heated at 65°C for 10 min. Samples were cooled on ice for 2 min, and 8 μl of reverse transcription reaction mix (4 μl of 5× first strand buffer; 2 μl of 0.1 mM DTT; 1 μl of dNTPs (10 mM dATP, dTTP, and dGTP; and 8.33 mM dCTP), 20 U of RNase OUT, 5 μCi of [α-32P]dCTP) was added. Samples were incubated at 42°C for 5 min, and 200 U of SuperScript RNaseH- reverse transcriptase was added. Reactions were conducted at 42°C for 50 min and stopped by incubation at 65°C for 15 min. Yeast carrier tRNA (10 μg) and 1 ml of 10% cold TCA were added, and the samples were precipitated on ice for 10 min. Samples were collected in triplicate (300 μl) by vacuum filtration onto GC/F glass fiber filters presoaked in 10% TCA. Filters were washed once with 3 ml of 10% cold TCA, twice with 3 ml of cold 95% ethanol, and dried. Filter-associated radioactivity was measured by liquid scintillation spectroscopy.
RESULTS
Model Systems for Analysis of RNA Distribution
Previous investigations into the mechanism of termination-elicited ribosome release have relied on pharmacological (pactamycin) inhibition of the initiation stage of protein synthesis (Seiser and Nicchitta, 2000; Potter and Nicchitta, 2002). Given concerns regarding off-target effects of this compound, we sought physiological scenarios yielding the physiological inhibition of initiation and thus examined ribosome/mRNA partitioning during the UPR. Because the UPR provides a prominent example of a regulatory pathway that modulates mRNA entry into polyribosomes, we postulated that ER-bound ribosomes could serve a unique role in the synthesis of stress response proteins (Brostrom and Brostrom, 1998). Two cell types were used in these studies, the murine plasmacytoma J558L and the murine fibroblast NIH 3T3.
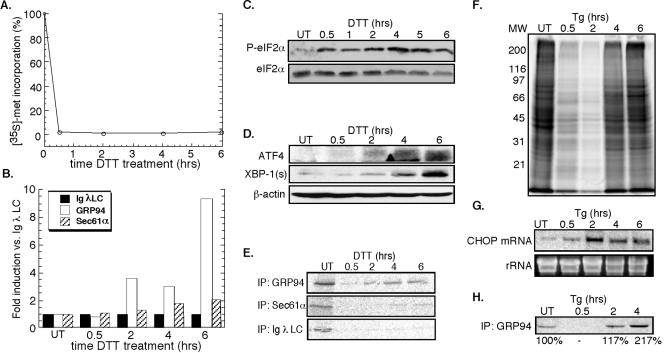
The UPR was elicited by the membrane-permeable reducing agent DTT, demonstrated previously to elicit a rapid and reversible disruption of disulfide bond formation in the ER, or thapsigargin, which inhibits the ER calcium ATPase (Lytton et al., 1991; Braakman et al., 1992a,b; Lodish and Kong, 1993). As shown in Figure 1A, addition of DTT to J558L cultures resulted in a rapid (≤0.5 h) and extended (≥6 h) suppression of cellular protein synthesis, concomitant with an increase in phospho-eIF2α levels (Figure 1C) (Brostrom and Brostrom, 1998; Harding et al., 2000b). Under these conditions and although general protein synthesis is suppressed, mRNAs encoding stress response proteins display relatively enhanced translation. This phenomenon is evident in the data depicted in Figure 1, B and E, where synthesis of GRP94, the ER Hsp90 chaperone, is enhanced relative to the translocon component Sec61α or the secretory protein immunoglobulin (Ig) λ light chain. Also, as a consequence of phospho-eIF2α accumulation, ATF4 mRNA translation was enhanced, with significant protein accumulation occurring by 4 h (Figure 1D) (Harding et al., 2000a). A similar accumulation of XBP-1 protein was evident after DTT treatment (Figure 1D), as reported previously (Shen et al., 2001; Yoshida et al., 2001; Calfon et al., 2002).
Figure 1.
Induction of the UPR in J558L and NIH 3T3 cells. J558L cells were treated with 10 mM DTT (A-D) and NIH 3T3 cells were treated with 500 nM thapsigargin (E-H) for the indicated times. (A) Protein synthesis was measured by the metabolic incorporation of [35S]methionine/cysteine into TCA-insoluble material. (B and E) By similar methods, immunoprecipitation protocols were used to assess J558L Ig light chain, GRP94, and Sec61α synthesis rates. (C) Immunoblot analysis of phospho-eIF2α versus eIF2α levels in DTT-treated J558L cells. (D) ATF4 and XBP-1 expression versus β-actin loading control in DTT-treated J558L cells. (F) SDS-PAGE profile of newly synthesized [35S]methionine/cysteine-labeled proteins in thapsigargin-treated NIH 3T3 cells. (G) Northern blot depicting thapsigargin-induced CHOP mRNA expression in NIH 3T3 cells, with ethidium bromide stain of rRNA as a loading control. (H) Immunoprecipitation of [35S]methionine/cysteine-labeled GRP94 after thapsigargin treatment of NIH 3T3 cells.
In companion studies, treatment of NIH 3T3 cells with DTT yielded a similar and pronounced inhibition of protein synthesis, with maximal inhibition being observed at substantially lower (2 mM) concentrations of DTT (our unpublished data). Presumably, such differences reflect the far higher secretory capacity of the plasmacytoma cell line, relative to the 3T3 fibroblast line. As seen after treatment with DTT, exposure of NIH 3T3 fibroblasts to thapsigargin yielded a suppression of protein synthesis with an accompanying increase in phospho-eIF2α levels (Figure 1F; our unpublished data). In this cell system, transcriptional induction of a downstream target of the ATF4 pathway, CHOP, was also clearly evident by 2 h (Figure 1G). As a further test of UPR induction, we measured synthesis of GRP94, one of the most prominent fibroblast proteins synthesized during ER stress (Kozutsumi et al., 1988). As expected, thapsigargin treatment resulted in a robust increase in the synthesis of GRP94, increasing by more than twofold by 4 h (Figure 1H). It is worth noting the difference in stress response recovery between DTT-treated J558L cells and thapsigargin-treated 3T3 cells (Figure 1, A and F). In DTT-treated J558L cells, protein synthesis was strongly suppressed for ≥6 h, whereas in thapsigargin-treated NIH 3T3 cells, protein synthesis levels have recovered by 6 h. These differences may reflect the different modes of UPR induction used in the two cell lines.
Subcellular Fractionation: Derivation of Cytosol and ER Compartments
In these studies, the coupling of protein synthesis to ribosome exchange on the ER membrane was examined using an established sequential detergent extraction technique (Seiser and Nicchitta, 2000; Potter and Nicchitta, 2002; Lerner et al., 2003). In this protocol, the cytosol fraction is obtained upon treatment of cell suspensions with concentrations of digitonin sufficient to permeabilize the plasma membrane and to release cytosolic ribosomes, without release of the ER-bound ribosome fraction (Adam et al., 1990; Seiser and Nicchitta, 2000; Lerner et al., 2003). Subsequently, ER-bound ribosomes were recovered by detergent solubilization of the total membrane fraction at substantially higher detergent concentrations (Lerner et al., 2003). The efficacy of this methodology was visualized by in situ immunofluorescence microscopy (Supplemental Figure S1). In Supplemental Figure S1B, cells were treated with a physiological salts buffer containing 80 μg/ml digitonin. Under these conditions, complete release of cytosolic tubulin (red) was observed, with no loss of ER structure, as assessed by staining for the resident ER membrane protein TRAPα, (green) or nuclear integrity (DAPI (blue) (compare to Supplemental Figure S1A). Subsequently, ER membranes were solubilized by addition of a high salt buffer containing 2% dodecylmaltoside (or 2% digitonin) using conditions demonstrated previously to maintain ER ribosome-translocon interactions (Supplemental Figure S1C) (Potter and Nicchitta, 2002).
This fractionation method, also described in detail in a previous publication (Lerner et al., 2003), was further examined by cytosol and ER marker protein distributions in J558L cells and NIH 3T3 cells using immunoblot and radiolabeled immunoprecipitations, respectively. As shown in Supplemental Figure S1, D and E, the cytosolic protein tubulin was effectively solubilized and released by digitonin-mediated cell permeabilization with no detectable levels of the ER markers GRP94 or TRAPα. Further solubilization of the cell pellet using higher detergent concentrations yielded release of the ER proteins GRP94 and TRAPα (ER fraction). These data, along with past reports (Lerner et al., 2003), demonstrate the efficient separation of the cytosol and ER compartments by this technique.
Additional fractionation studies were performed using an alternative approach, where rough ER vesicles were purified from cellular homogenates by buoyant density centrifugation. By immunoblot analysis, the RM fraction was enriched in the ER marker TRAPα and did not contain detectable levels of the cytosolic maker tubulin (Supplemental Figure S1D). Although this technique yields a pure rough ER membrane fraction, the nonbuoyant soluble fraction enriched with cytosolic markers such as tubulin, also contained contaminating ER, nuclear, and Golgi markers (Supplemental Figure S1D; data not shown). For these reasons, fractionation analyses will emphasize sequential detergent extraction methods to yield paired cytosol and membrane fractions, whereas the buoyant density flotation protocol will be restricted to analyses of the ER membrane fraction alone.
UPR Induction Elicits 80S Ribosome Accumulation on the Endoplasmic Reticulum
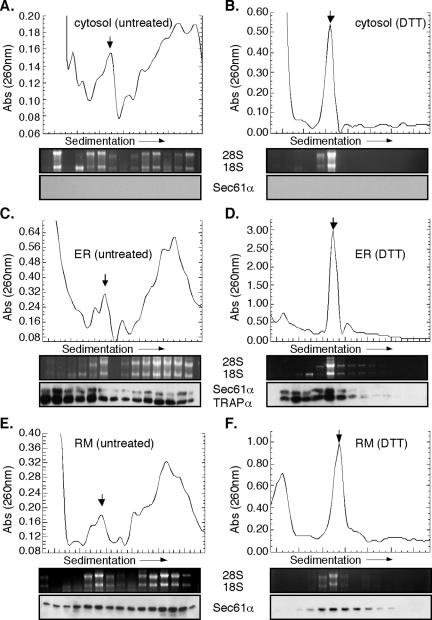
Using the experimental systems described above, the consequences of UPR induction on polyribosome structure were examined (Figure 2). In control J558L cells, cytosolic and ER-bound ribosomes were recovered predominately in the faster migrating polyribosome fractions (Figure 2, A, C, and E) (note position of 80S ribosomes, indicated by the downward-facing arrow). Immunoblot analysis demonstrated that ER-bound polyribosomes were in association with components of the protein translocation machinery, Sec61α and/or TRAPα; these proteins were absent from the cytosol-derived polyribosome fractions (Figure 2, A, C, and E). After a brief (30 min) treatment with DTT, polyribosomes from both the cytosol and ER have collapsed into 80S ribosomes, as indicated by RNA gel analysis (Figure 2, B and D). At 4 h of DTT treatment, the 80S ribosome remained as the dominant peak, as demonstrated in analyses of ribosome profiles derived from buoyant density-isolated ER membranes (Figure 2F). In addition, immunoblot analysis demonstrated that the ER-associated 80S ribosome species sedimented in association with Sec61α and/or TRAPα (Figure 2, D and F), in agreement with previous data in which inhibition of the initiation stage of protein synthesis was achieved by pactamycin addition (Potter and Nicchitta, 2002). Similar results were obtained in experiments using thapsigargin-treated NIH 3T3 cells (our unpublished data).
Figure 2.
UPR elicits polyribosome breakdown and accumulation of 80S monosomes in the cytosolic and ER fractions. Cytosol (A and B), total membrane-derived (C and D) and rough microsome-derived (E and F) fractions from control (A, C, and E) and 10 mM DTT-treated (B, D, and F) J558L cells were sedimented through 15-50% linear sucrose gradients for 3 h at 151,000 × g. Ribosomes were monitored by UV absorbance (260 nm) with the downward facing arrow denoting the 80S monosome peak. Total RNA isolated from individual gradient fractions was resolved on denaturing agarose gels. Immunoblots were performed from individual gradient fractions for translocon markers Sec61α and TRAPα.
Ribosome Partitioning to the ER Is Maintained during the UPR
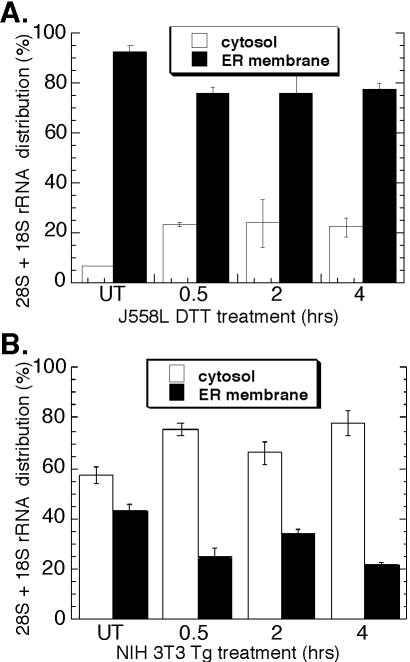
The results presented in Figure 2 indicate that ER-bound ribosomes remain membrane associated after termination. This observation contrasts with current models, which predict that ribosome release from the ER membrane is coupled to termination, and so was further examined (Blobel et al., 1979; Lingappa and Blobel, 1980; Alberts et al., 2002). In these experiments, ribosome partitioning between the cytosol and ER was assessed by analysis of the distribution of 18S and 28S ribosomal RNAs in J558L and NIH 3T3 cells. For the data reported in Figure 3A, cell cultures were incubated overnight with [3H]uridine to metabolically label rRNA. Total RNA isolated from cytosol and membrane fractions from control and DTT-treated J558L cells was resolved by denaturing RNA agarose gel electrophoresis, and the bands corresponding to the 18S and 28S rRNAs were excised and quantified by liquid scintillation spectrometry. In untreated J558L cells (t = 0 h), [3H]ribosomes were predominantly recovered (∼90%) in the ER-derived fraction, as predicted for a terminally differentiated secretory cell (Figure 3A). After UPR induction, the majority (∼75%) of the radiolabeled ribosome pool remained bound to the ER membrane and persisted throughout the course of this experiment (Figure 3A).
Figure 3.
Ribosome partitioning to the ER is maintained during the UPR. J558L cells were treated with 10 mM DTT (A) and NIH 3T3 cells treated with 500 nM thapsigargin (B) for the indicated times. RNA isolated from the cytosol and ER fractions was resolved by denaturing agarose gel. (A) Bands corresponding to the 28S and 18S rRNAs were excised and measured by liquid scintillation spectroscopy or (B) 28S and 18S rRNAs were measured by ethidium bromide fluorescence at 610 nm.
In Figure 3B, thapsigargin was used to elicit the UPR in NIH 3T3 cultures. In this experiment, the ribosome distribution was quantitated from the ethidium bromide fluorescence of the 18S and 28S rRNAs. In untreated NIH 3T3 cultures (t = 0 h), ribosomes are roughly evenly distributed with a slight bias toward the cytosol fraction as expected. After thapsigargin treatment, ∼20% of the cellular ribosome pool shifted into the cytosol fraction, similar to the 15% shift observed in J558L cells. These data, which reflect analyses performed in two cell types using two different methods of UPR induction, are consistent with the data presented in Figure 2 and further buttress the conclusion that the predominant fate of ER-bound ribosomes upon termination is continued association with the protein translocation machinery.
ER-bound Ribosomes Exist as “mRNA-free” or “mRNA-associated” Populations
The data presented above indicate that ER-bound ribosomes retain their membrane association after polyribosome breakdown. From this observation, we postulated that the ribosome pool would shift from a predominately mRNA-associated pool (polyribosome) to a predominately mRNA-free pool, with similar responses occurring in the cytosol. To distinguish these two ribosome populations, we used polyuridylate-Sepharose (poly(U)-Sepharose) affinity chromatography to isolate the poly(A)+ mRNA fraction from detergent-solubilized cytosol and ER fractions (Lindberg and Persson, 1972). Using this experimental approach, the relative levels of mRNA-associated ribosomes present in the ER membrane and cytosol compartments after UPR induction were evaluated.
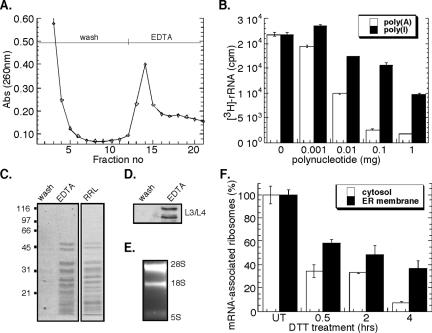
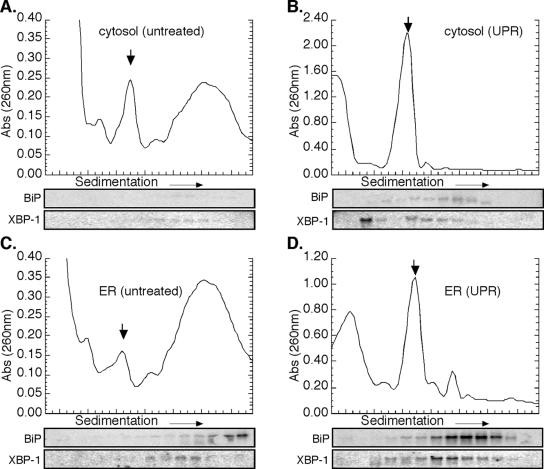
A representative poly(U)-Sepharose chromatography profile is shown in Figure 4A. After loading of a detergent-solubilized ribosome fraction, the affinity matrix was extensively washed and bound ribosomes subsequently eluted by addition of EDTA. The identity of the EDTA-releasable fraction was confirmed as ribosomes by SDS-PAGE, where the EDTA-releasable fraction displayed an identical protein profile to that seen for rabbit reticulocyte lysate-derived ribosomes (Figure 4C). Immunoblot analysis demonstrated the presence of ribosomal proteins L3 and L4 in the EDTA eluant (Figure 4D), whereas the 28S and 18S rRNAs were identified by denaturing RNA agarose gel electrophoresis (Figure 4E). In additional experiments, the resin was washed with 4 M guanidinium thiocyanate after EDTA elution. This latter eluting fraction was identified to contain predominately mRNAs by Northern blot analysis (our unpublished data).
Figure 4.
Poly(U)-Sepharose affinity chromatographic analysis of mRNA-associated ribosomes. Native J558L ribosome-mRNA complexes were affinity purified by poly(U)-Sepharose resin. (A) Chromatogram of ribosome elution by EDTA was monitored by UV absorbance (260 nm). Competitive inhibition of metabolically labeled 3H-ribosome binding using the polynucleotides poly(A) or poly(I) (B). Coomassie blue staining of SDS-PAGE profile of EDTA-eluted fractions compared with rabbit reticulocyte-derived ribosomes (C). Immunoblot analysis of L3/L4 ribosomal proteins from wash versus EDTA-eluted fractions (D). Ethidium bromide-stained denaturing agarose gel of EDTA-eluted fractions (E). J558L cells were treated with 10 mM DTT for the indicated times. The mRNA occupancy levels of cytosolic and ER-derived, metabolically labeled 3H-ribosomal fractions were determined from the EDTA-eluted fractions. mRNA (100%) association corresponds to the occupancy level obtained in untreated cells (t = 0 h) (F).
The mRNA dependence of ribosome isolation by poly(U)-Sepharose affinity chromatography was confirmed in competition studies with two purine-based polymers, polyadenylic acid [poly(A)] and polyinosinic acid [poly(I)]. If ribosome binding were mRNA dependent, addition of poly(A), and to a lesser degree poly(I), should effectively compete binding by blocking available poly(A)+ mRNA-poly(U)-Sepharose interaction sites. Conversely, if ribosome binding to poly(U)-Sepharose were direct (i.e., mRNA independent), poly(A) and poly(I) would be equally effective in blocking ribosome binding. As shown in Figure 4B, the binding of ribosomes to the poly(U) affinity matrix was substantially inhibited by the addition of excess poly(A), whereas at least 2 orders of magnitude higher concentrations of poly(I) were required to achieve similar inhibition. This indicates that ribosome association with the poly(U) matrix is occurring primarily via the 3′-poly(A) tail of mRNAs.
Using this experimental approach, ribosome loading onto mRNAs in the cytosol and ER compartments as a consequence of the UPR was evaluated. As depicted in Figure 4F, a rapid decrease in ribosome loading on cytosolic and ER-associated mRNAs was observed after a 30-min treatment with DTT. At all time periods after UPR induction, cytosolic ribosomes demonstrated a reduced degree of mRNA association relative to ER-bound ribosomes. This difference was accentuated at later time points (4 h) where the relative fraction of mRNA-associated ribosomes in the ER-bound pool was approximately fivefold greater than that observed in the cytosol pool. These data indicate that although total protein synthesis activity is dramatically reduced after UPR induction, ER-associated mRNAs remain predominately assembled in small polysomes where they presumably undergo continued, albeit much reduced, translation, whereas cytosolic polysomes disassemble to yield free ribonuclear particle (RNP) and ribosome fractions. Alternatively, UPR induction may elicit a global repartitioning of mRNAs from the cytosol to the ER. These two hypotheses were examined in the experiments described below.
mRNA Partitioning between the Cytosol and ER Compartments Is Maintained during the UPR
To determine whether the cellular partitioning of mRNAs to the ER is altered during the UPR, cytosolic and ER-derived RNA pools were isolated from thapsigargin and DTT-treated NIH 3T3 cells as well as DTT-treated J558 cells, and the fractions were analyzed by Northern blot. In additional experiments, RNA distributions were quantitated by RNA dot blot; these data are included as a Supplemental Figure S2.
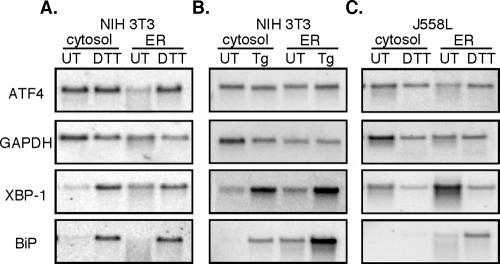
Depicted in Figure 5, A-C, are representative Northern blots in which three patterns of mRNA partitioning can be identified. In one pattern, displayed by ATF4, mRNA is recovered in both the cytosol and ER fractions of untreated (UT) cells, with partitioning favoring the cytosol (Figure 5, A and B). After UPR induction with either DTT (Figure 5A) or thapsigargin (Figure 5B), an approximately equal mRNA distribution was observed between the ER and cytosol fractions in both control and thapsigargin-treated cells. A similar mRNA partitioning pattern was observed in control and DTT-treated J558 cells (Figure 5C). In a second pattern displayed by glyceraldehyde-3-phosphate dehydrogenase (GAPDH), mRNAs were enriched in the cytosol fraction and exhibited no significant change upon UPR induction. For these mRNAs, ER stress was accompanied by a modest decrease in mRNA levels, presumably the consequence of UPR-enhanced mRNA degradation. In a third pattern exhibited by mRNAs encoding BiP and the soluble UPR transcription factor XBP-1, mRNAs were enriched on ER-bound ribosomes in both control and UPR-induced cells. As a consequence of the UPR, there was a substantial increase in BiP mRNA levels; this was not accompanied by significant changes in the relative partitioning between the free and ER-bound mRNA pools (Figure 5, A-C). These data demonstrate that individual mRNAs can display distinct and overlapping partitioning between free and membrane-bound ribosomes, a conclusion mirroring genome-wide analyses of mRNA distributions in yeast, fly, and human cells (Kopczynski et al., 1998; Diehn et al., 2000). Significantly, these data indicate that activation of the UPR program does not yield a significant global repartitioning of mRNAs between the cytosol and ER compartments.
Figure 5.
mRNA distribution between the cytosol and ER compartments is maintained during the UPR. Northern blot analysis of ATF4, GAPDH, XBP-1, and BiP mRNAs was performed on equivalent cytosol and ER fractions derived from control (UT), 2 mM DTT, or thapsigargin-treated NIH 3T3 cells (A and B) or 10 mM DTT-treated J558L cells (C).
ER-bound Ribosomes Serve a Primary Role in mRNA Translation during the UPR
To determine whether UPR induction leads to a compartment-specific regulation of protein synthesis, mRNA translation profiles were examined in the cytosol and ER-membrane fractions of control and UPR-induced cells. In these experiments, cytosol and ER fractions from control and (4 h) DTT-treated J558L cells were prepared and separated by sucrose gradient velocity sedimentation. The distribution of ribosomes and individual mRNAs in the gradient fractions was then determined by UV-spectrometry and Northern blot analysis, respectively. To allow comparisons between the two fractions, identical cell equivalents were analyzed. For the Northern blot analyses, four mRNAs were chosen, encoding the ER-resident chaperone BiP, the soluble stress response transcription factors XBP-1 and ATF4, and the glycolytic enzyme GAPDH. BiP, XBP-1, and ATF4 mRNAs were chosen for their role in the UPR program; GAPDH mRNA was chosen because it displays overlapping distribution between the cytosol and ER (Figure 6; Lerner et al., 2003) and has previously been demonstrated to reside in polyribosomes after induction of ER stress responses (Kawai et al., 2004).
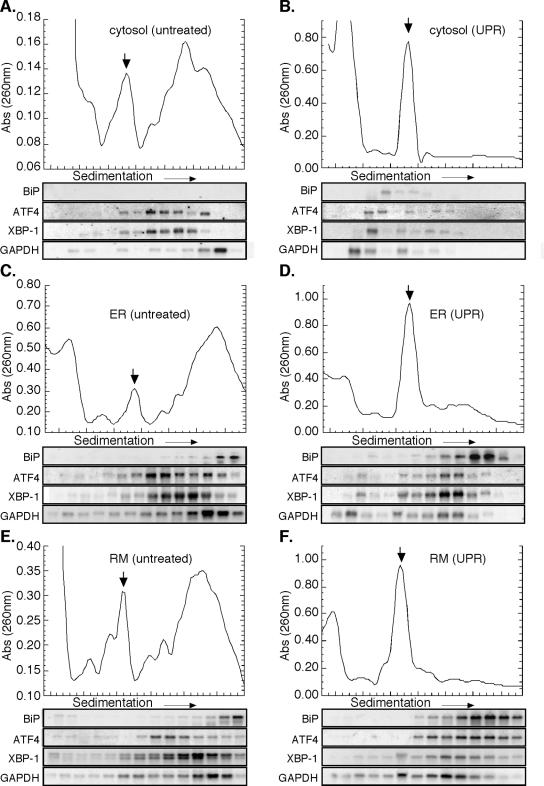
Figure 6.
ER-bound ribosomes preferentially participate in mRNA Translation during the UPR-J558L cells. Cytosol (A and B), total membrane-derived ER (C and D), and rough microsome-derived (E and F) fractions from control (A, C, and E) and DTT-treated (B, D, and F) J558L cells were sedimented through 15-50% linear sucrose gradients for 3 h at 151,000 × g. Ribosomes were monitored by UV absorbance (260 nm), and the downward facing arrow denotes the 80S monosome. RNA was isolated from individual gradient fractions and used for Northern blot analysis as indicated.
In control fractions, mRNAs residing in both the cytosol and ER compartments were assembled into polyribosomes of varying sizes (Figure 6, A, C, and E; Supplemental Figure S3). Sedimenting with the largest class of polyribosomes was the mRNA encoding BiP, which was exclusive to ER membrane fractions (Figure 6, A, C, and E). Conversely, GAPDH mRNA, which also sediments with large polyribosomes, was identified in both the cytosol and ER fractions and displayed essentially equivalent sedimentation patterns (Figure 6, A, C, and E). The compartmental distribution of the mRNAs encoding the stress response transcription factors XBP-1 and ATF4 also was determined. Although neither message is efficiently translated in untreated cells, both mRNAs were found in small polyribosome fractions of both cytosolic and ER polyribosomes and exhibited identical sedimentation patterns in the two cellular compartments. (Figure 6, A, C, and E) (Harding et al., 2000a; Calfon et al., 2002). Interestingly, the mRNA encoding the yeast homolog to XBP-1, HAC1, also has been shown to be ER-localized at steady state, indicating this may be a conserved feature of its RNA localization pathway (Chapman and Walter, 1997; Diehn et al., 2000).
After UPR induction, dramatic differences were observed in the mRNA sedimentation patterns of cytosolic and ER-bound ribosomes (Figure 6, B, D, and F; Supplemental Figure S3). Transcripts detected in the cytosol ribosome fractions (ATF4, GAPDH, and XBP-1) predominately sedimented in the ≤40S region, indicating that they were present as free messenger (m)RNPs (Figure 6B). Interestingly, small amounts of BiP mRNA were detected in the 40S region of the cytosol fraction, perhaps representing preinitiation complexes of newly exported BiP mRNAs. In contrast, ATF4, BiP, GAPDH, and XBP-1 mRNAs present in the ER compartment displayed very different sedimentation behaviors; these mRNAs were assembled into small polyribosomes (compare Figure 6D, F vs. B). BiP mRNA, which exhibited a slight UPR-dependent decrease in ribosome loading, remained in large polyribosome fractions, indicative of its relatively efficient translation during UPR induction. The mRNAs encoding the soluble proteins ATF4, XBP-1, and GAPDH exhibited more pronounced UPR-dependent shifts. Although XBP-1 and GAPDH mRNAs had minor to moderate reductions in ribosome loading, respectively, ATF4 mRNA shifted into larger polyribosomes. A similar UPR-elicited increase in ATF4 sedimentation has been reported previously, although the cellular compartmentalization of ATF4 synthesis had not been examined (Harding et al., 2000a). The data depicted in Figure 6, A-F, were quantified by PhosphorImager analysis; these data are depicted in Supplemental Figure S3. In examining the distribution of the different mRNAs, relative to the 80S ribosome peak, two patterns can be identified: mRNAs in the cytosol fraction predominate in the <80S fractions, but mRNAs present in the ER fraction are enriched in the ≥80S fraction.
These experiments shown in Figure 6 also were performed in DTT-treated NIH 3T3 cells, with similar results. Data describing the mRNA translation profiles for ATF4 and XBP-1 in control and DTT-treated fibroblasts is shown in Figure 7. As seen in J558L cells, BiP mRNA was enriched in the heavy polyribosome fraction derived from the ER but not the cytosol fractions derived from NIH 3T3 cells (Figure 7, A and C). mRNAs encoding XBP-1 were present at very low, but detectable, levels in both polyribosome fractions (Figure 7, A and C). After addition of DTT to NIH 3T3 cells, extensive polyribosome breakdown was observed (Figure 7, B and D). Again similar to observations obtained in J558L cells, DTT-elicited UPR induction in NIH 3T3 cells was accompanied by a pronounced accumulation of XBP-1 mRNA in the <40S portion of the cytosol extract and the enhanced appearance of XBP-1 mRNA in the small ER-associated polyribosome fraction (Figure 7, B and D).
Figure 7.
ER-bound ribosomes preferentially participate in mRNA translation during the UPR-NIH 3T3 cells. Cytosol (A and B) and total membrane-derived ER (C and D) fractions from control (A and C) and DTT-treated (B and D) NIH 3T3 cells were sedimented through 15-50% linear sucrose gradients for 3 h at 151,000 × g. Ribosomes were monitored by UV absorbance (260 nm), and the downward facing arrow denotes the 80S monosome. RNA was isolated from individual gradient fractions and used for Northern blot analysis as indicated.
In total, these data indicate that protein synthesis is preferentially compartmentalized to the ER as a consequence of UPR induction. Additionally, the similarities in the overall ribosome loading patterns of the mRNAs encoding the soluble cytosolic proteins XBP-1, ATF4, and GAPDH suggest that enhanced compartmentalization of mRNA translation to the ER is a global regulatory feature of protein synthesis during the UPR.
Protein Synthesis Partitions to the ER during the UPR
To determine whether the findings presented in Figures 6 and 7 were globally representative of mRNAs translation profiles, we examined ribosome loading onto poly(A)+ mRNA in the cytosol and ER after UPR induction. Global mRNA levels from individual gradient fractions were determined by oligod(T)-primed reverse transcription. In this assay, mRNA was measured as a function of incorporated radiolabeled nucleotide precursor ([α-32P]dCTP) into cDNA products.
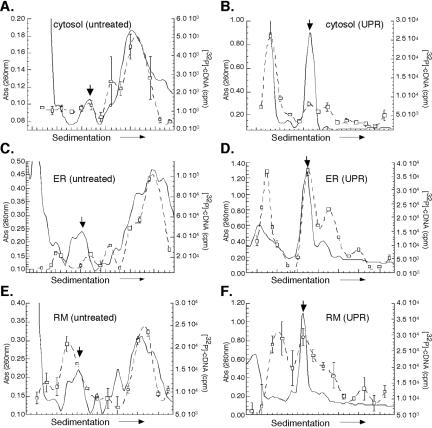
In control J558L cells, mRNAs are highly enriched in the polyribosome fractions prepared from the cytosol and ER compartments (Figure 8, A, C, and E). At 4 h post-UPR induction, ribosomes from both the cytosol and ER are predominately recovered as 80S monosomes (Figure 8, B, D, and F). However, examination of the mRNA distribution in the two compartments identified striking differences. In the cytosol fractions, the majority of the mRNAs were recovered in the lighter sucrose gradient fractions (<40S), indicating that they were not ribosome associated (Figure 8B). The sedimentation behavior of the nonribosome-associated mRNA fraction (i.e., <40S) was verified by comparison with EDTA-treated fractions (our unpublished data). In marked contrast, ER-associated transcripts were recovered predominantly in ribosome/polyribosome-containing fractions (≥80S) and to a lesser degree in nonribosomal fractions (<40S) (Figure 8, D and F). These data extend the analyses presented in Figures 6 and 7 and further document the finding that UPR induction elicits pronounced changes in the mRNA translation profiles of cytosolic and ER ribosomes.
Figure 8.
Protein synthesis is compartmentalized to the ER during the UPR. Cytosol (A and B), total membrane-derived ER (C and D), and rough microsome-derived (E and F) fractions from control (A, C, and E) and 10 mM DTT-treated (B, D, and F) J558L cells were sedimented through 15-50% linear sucrose gradients for 3 h at 151,000 × g. Ribosomes were monitored by UV absorbance (260 nm), and the downward facing arrow denotes the 80S monosome. Ribosome distribution is depicted by the solid line. The mRNA content of individual gradient fractions was measured by the quantitative incorporation of [α-32P]dCTP radiolabeled nucleotide precursor into cDNA products using an oligo(dT)20-primed reverse transcription reaction. Reactions were sampled in triplicate; mean values with error bars representing the SD values are shown. The mRNA distribution is represented by the dotted line/open squares.
DISCUSSION
In this communication, we examined the cellular processes governing the partitioning of ribosomes and mRNAs between the two primary protein synthesis compartments of the cell, the cytoplasm, and the ER. Using the unfolded protein response as means to physiologically modulate mRNA translation and gene transcription, we report a novel translational control mechanism whereby the ER serves as the preferred site for the synthesis of both cytosolic and signal sequence-bearing proteins. After UPR induction, ER-associated mRNAs were enriched in the small polyribosome fraction, whereas cytosolic mRNAs accumulated as free, nonribosome-associated RNPs. Additionally, UPR-elicited polyribosome remodeling was not accompanied by a repartitioning of mRNAs between cellular compartments. As a result, mRNAs that underwent translation on both cytosolic and ER-bound ribosomes in the homeostatic condition displayed ER-restricted loading into polyribosomes in response to UPR induction. Together, these data indicate that the observed compartmental segregation of protein synthesis is not uniquely confined to UPR target mRNAs; rather, the capacity to differentially regulate protein synthesis in the cytosol and ER compartments allows the ER to serve as a privileged site of protein synthesis. As a key and perhaps essential element of this process, ER-bound ribosomes were observed to remain in continued association with the ER membrane after termination, thereby providing the ER compartment with a stable and abundant source of protein synthesis capacity.
It is well established that ribosomes engaged in the synthesis of secretory/membrane proteins are trafficked to the endoplasmic reticulum via the SRP pathway (Walter and Johnson, 1994). Less well established is the mechanism whereby ER-bound ribosomes and mRNAs recycle from the ER membrane to the cytosol (Potter et al., 2001). Current models propose that the dissociation of ER-bound ribosomes is functionally linked to termination with the individual subunits departing the ER membrane to join a common cytosolic pool (Blobel et al., 1979; Walter and Johnson, 1994; Alberts et al., 2002). In contrast, data presented in this report indicate that the physiological induction of polyribosome breakdown yields the accumulation of membrane-bound, 80S ribosomes. In previous reports, we examined the coupling of termination and ribosome release from the ER by pharmacologically inhibiting the initiation reaction of protein synthesis (Seiser and Nicchitta, 2000; Potter and Nicchitta, 2002). Under these conditions, ER-bound polyribosomes underwent efficient termination yet remained in stable association with the ER translocon (Seiser and Nicchitta, 2000; Potter and Nicchitta, 2002). On the basis of past and current studies, we now conclude that the physiological process of ribosome dissociation from the ER membrane is regulated independently of the termination reaction. This latter finding helps to refocus future studies on a series of fundamental and as yet unanswered questions first proposed by Palade (1975): is initiation of mRNA translation restricted to the cytosol compartment, with ribosome traffic to the ER occurring as a cotranslational event, or can both ER-bound and free ribosomes function to initiate de novo translation? Second, what regulates the duration of ribosome attachment to the ER?
From the viewpoint of existing models, the observation that mRNAs encoding cytosolic proteins can be partitioned to, and translated on, ER-bound ribosomes is unexpected (Alberts et al., 2002; Blobel and Dobberstein, 1975). Nonetheless, rigorous examinations of mRNA partitioning between free and membrane-bound ribosomes, conducted in multiple cell types and tissues, in lower and higher eukaryotes and by multiple fractionation techniques, also have concluded that the mechanism of mRNA partitioning in the eukaryotic cell is complex (Mechler and Rabbitts, 1981; Mueckler and Pitot, 1981; Kopczynski et al., 1998; Diehn et al., 2000; Lerner et al., 2003). Little is known regarding the mechanism for how such complex mRNA partitioning patterns are conferred. It is reasonable to consider that encoded localization information, presumably present within the untranslated regions, and/or particular cohorts of RNA binding proteins contribute significantly to this phenomenon. In this context, it is also possible that a given mRNA species may comprise multiple subpopulations, differing in RNA binding protein composition and thus in relative partitioning between the cytosol and ER compartments. Regardless of the mechanism, the data presented herein demonstrate that ER-associated, rather than cytosolic, mRNAs serve as the preferred templates for translation during the UPR.
Our analyses of mRNA recruitment by ribosomes present in the cytosol and ER compartments of UPR-induced cells identified a clear distinction between the two compartments; ER-bound ribosomes continued to function in protein synthesis and thus are recruited, albeit inefficiently, into small polyribosomes, whereas cytosolic ribosomes display dramatically reduced function and a much reduced steady-state association with mRNAs. These data indicate that the translation activities of cytosolic and ER-bound ribosomes are differentially regulated during the UPR. A potential key to this puzzle is the regulation of the eIF2 phosphorylation cycle, which has a well-established role in the global modulation of protein synthesis rates during cell stress. There exist four eIF2a kinases functioning in the cellular response to stress; HRI, the heme-repressed kinase, functions during erythroid precursor differentiation; GCN2, the general control non-repressed kinase, provides a cellular response to the accumulation of uncharged tRNAs; PKR, or protein kinase repressor, responds to double-stranded (viral) RNA; and PERK, the ER-resident PKR-like kinase, senses the accumulation of unfolded proteins in the ER (reviewed in Rutkowski and Kaufman, 2004). In addition to the eIF2α-directed kinases, at least two phosphatase activities are known to act on eIF2α, GADD34/PP1 and CreP (Novoa et al., 2001; Jousse et al., 2003). The activity of GADD34/PP1 is known to reside on the ER (Brush et al., 2003) and thus in the context of an ER-restricted translational control response, it would seem likely that the regulation and/or expression of PERK and GADD34/PP1 would contribute significantly to differential regulation of the relative translation capacity of the cytosol and ER compartments. In this view, compartment-specific regulation of dephospho-eIF2 levels would allow for disparate translation activities between the cytosol and ER during stress, when eIF2a availability is limiting. How this might occur is currently under investigation.
On the basis of the data presented in this report, we suggest that the global suppression of protein synthesis and polyribosome breakdown elicited by UPR-induced phosphorylation of eIF2α serves two primary functions: 1) the ER is relieved of the bulk influx of newly synthesized polypeptides, which would exacerbate stress-related disruptions in protein folding efficiency; and 2) as a consequence of polyribosome breakdown, ribosome loading onto individual mRNAs is dramatically reduced and a substantial fraction of the (ER) ribosomes become available for the de novo translation of stress response mRNAs (Rutkowski and Kaufman, 2004). In this view, polyribosome breakdown is an essential component of the UPR because it makes available ribosomes that otherwise would be assembled in polyribosomes. In the absence of such a response, newly transcribed mRNAs would either compete for access to a limited ribosome pool or ribosome biogenesis would need to increase in proportion to the magnitude of the transcriptional response. The observations reported here indicate that the UPR program includes an essential process of “mRNA remodeling” as ribosomes redistribute between polyribosome complexes. These data also suggest that the compartmental regulation of translation is of key significance to UPR gene expression. In this context, we postulate that restricting the translation of mRNAs encoding UPR effector proteins to the two-dimensional geography of the ER membrane provides a kinetic advantage to the assembly of translation complexes in the three-dimensional cytosol.
Supplementary Material
[Supplemental Material]
Acknowledgments
We thank members of the Nicchitta laboratory, particularly Brook Pyhtila, Rachel Lerner, Jeff Baker, and Tianli Zheng for helpful comments and criticisms and Matt Brush and the Shenolikar laboratory for providing us with antibodies recognizing phospho-eIF2α. This work was supported by National Institutes of Health Grants DK-47897 (to C.V.N.), GM61970 (to J.W.B.), AI46451 and CA79907 (to J.D.K.), and American Heart Association predoctoral fellowship 0515333U (to S.B.S.).
Abbreviations used: DTT, dithiothreitol; eIF2, eukaryotic initiation factor 2; ER, endoplasmic reticulum; PERK, RNA-activated protein kinase-like endoplasmic reticulum kinase; poly(A), polyadenylic acid; poly(I), polyinosinic acid; poly(U), polyuridylic acid; RNP, ribonuclear particle; SRP, signal recognition particle.
References
- Adam, S. A., Marr, R. S., and Gerace, L. (1990). Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111, 807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). Molecular Biology of the Cell, New York: Garland Science.
- Aza-Blanc, P., Cooper, C. L., Wagner, K., Batalov, S., Deveraux, Q. L., and Cooke, M. P. (2003). Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell 12, 627-637. [DOI] [PubMed] [Google Scholar]
- Blobel, G., and Dobberstein, B. (1975a). Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent Ig light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67, 835-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel, G., and Dobberstein, B. (1975b). Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 67, 852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel, G., Walter, P., Chang, C. N., Goldman, B. M., Erickson, A. H., and Lingappa, V. R. (1979). Translocation of proteins across membranes: the signal hypothesis and beyond. Symp. Soc. Exp. Biol. 33, 9-36. [PubMed] [Google Scholar]
- Braakman, I., Helenius, J., and Helenius, A. (1992a). Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 11, 1717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman, I., Helenius, J., and Helenius, A. (1992b). Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature 356, 260-262. [DOI] [PubMed] [Google Scholar]
- Brostrom, C. O., and Brostrom, M. A. (1998). Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid Res. Mol. Biol. 58, 79-125. [DOI] [PubMed] [Google Scholar]
- Brush, M. H., Weiser, D. C., Shenolikar, S. (2003). Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1α to the endoplasmic reticulum and promotes dephosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 23, 1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon, M., Zeng, H., Urano, F., Till, J. H., Hubbard, S. R., Harding, H. P., Clark, S. G., and Ron, D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92-96. [DOI] [PubMed] [Google Scholar]
- Chapman, R. E., and Walter, P. (1997). Translational attenuation mediated by an mRNA intron. Curr. Biol. 7, 850-859. [DOI] [PubMed] [Google Scholar]
- Choi, S. B., Wang, C., Muench, D. G., Ozawa, K., Franceschi, V. R., Wu, Y., and Okita, T. W. (2000). Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407, 765-767. [DOI] [PubMed] [Google Scholar]
- Diehn, M., Eisen, M. B., Botstein, D., and Brown, P. O. (2000). Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 25, 58-62. [DOI] [PubMed] [Google Scholar]
- Gass, J. N., Gifford, N. M., and Brewer, J. W. (2002). Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277, 49047-49054. [DOI] [PubMed] [Google Scholar]
- Gilmore, R., and Blobel, G. (1983). Transient involvement of signal recognition particle and its receptor in the microsomal membrane prior to protein translocation. Cell 35, 677-685. [DOI] [PubMed] [Google Scholar]
- Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., and Ron, D. (2000a). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099-1108. [DOI] [PubMed] [Google Scholar]
- Harding, H. P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000b). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897-904. [DOI] [PubMed] [Google Scholar]
- Harding, H. P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271-274. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A. G. (2000). Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Translational Control of Gene Expression, ed. N. Sonenberg, J.W.B. Hershey, and M. B. Mathews [eds.], Cold Spring Harbor, NY: Cold Spring Harbor Press, 185-243.
- Jousse, C., Oyadomari, S., Lu, P., Zhang, Y., Harding, H. P., and Ron, D. (2003). Inhibition of constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163, 767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, T., Fan, J., Mazan-Mamczarz, K., and Gorospe, M. (2004). Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell. Biol. 24, 6773-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski, C. C., Noordermeer, J. N., Serano, T. L., Chen, W. Y., Pendleton, J. D., Lewis, S., Goodman, C. S., and Rubin, G. M. (1998). A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 95, 9973-9978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi, Y., Segal, M., Normington, K., Gething, M., and Sambrook, J. (1988). The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332, 462-464. [DOI] [PubMed] [Google Scholar]
- Lerner, R. S., Seiser, R. M., Zheng, T., Lager, P. J., Reedy, M. C., Keene, J. D., and Nicchitta, C. V. (2003). Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9, 1123-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, U., and Persson, T. (1972). Isolation of mRNA from KB-cells by affinity chromatography on polyuridylic acid covalently linked to Sepharose. Eur. J. Biochem. 31, 246-254. [DOI] [PubMed] [Google Scholar]
- Lingappa, V. R., and Blobel, G. (1980). Early events in the biosynthesis of secretory and membrane proteins: the signal hypothesis. Recent Prog. Horm. Res. 36, 451-475. [DOI] [PubMed] [Google Scholar]
- Lodish, H. F., and Kong, N. (1993). The secretory pathway is normal in DTT-treated cells, but disulfide-bonded proteins are reduced and reversibly retained in the endoplasmic reticulum. J. Biol. Chem. 268, 20598-20605. [PubMed] [Google Scholar]
- Lytton, J., Westlin, M., and Hanley, M. R. (1991). Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 266, 17067-17071. [PubMed] [Google Scholar]
- Mechler, B., and Rabbitts, T. H. (1981). Membrane-bound ribosomes of myeloma cells. IV. mRNA complexity of free and membrane-bound polysomes. J. Cell Biol. 88, 29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechler, B., and Vassalli, P. (1975). Membrane-bound ribosomes of myeloma cells. III. The role of the messenger RNA and the nascent polypeptide chain in the binding of ribosomes to membranes. J. Cell Biol. 67, 25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler, M. M., and Pitot, H. C. (1981). Structure and function of rat liver polysome populations. I. Complexity, frequency distribution, and degree of uniqueness of free and membrane-bound polysomal polyadenylate-containing RNA populations. J. Cell Biol. 90, 495-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta, C. V. (2002). A platform for compartmentalized protein synthesis: Protein translation and translocation in the ER. Curr. Opin. Cell Biol. 14, 412-416. [DOI] [PubMed] [Google Scholar]
- Novoa, I., Zeng, H., Harding, H. P., and Ron, D. (2001). Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2a. J. Cell Biol. 153, 1011-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, T., Yoshida, H., Akazawa, R., Negishi, M., and Mori, K. (2002). Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366, 585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade, G. (1975). Intracellular aspects of the process of protein synthesis. Science 189, 347-358. [DOI] [PubMed] [Google Scholar]
- Potter, M. D., and Nicchitta, C. V. (2000). Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J. Biol. Chem. 275, 33828-33835. [DOI] [PubMed] [Google Scholar]
- Potter, M. D., and Nicchitta, C. V. (2002). Endoplasmic reticulum-bound ribosomes reside in stable association with the translocon following termination of protein synthesis. J. Biol. Chem. 277, 23314-23320. [DOI] [PubMed] [Google Scholar]
- Potter, M. D., Seiser, R. M., and Nicchitta, C. V. (2001). Ribosome exchange revisited: a mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 11, 112-115. [DOI] [PubMed] [Google Scholar]
- Rutkowski, D. T., and Kaufman, R. J. (2004). A trip to the ER: coping with stress. Trends Cell Biol. 14, 20-28. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Press.
- Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R. J. (2001). Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165-1176. [DOI] [PubMed] [Google Scholar]
- Seiser, R. M., and Nicchitta, C. V. (2000). The fate of membrane-bound ribosomes following the termination of protein synthesis. J. Biol. Chem. 275, 33820-33827. [DOI] [PubMed] [Google Scholar]
- Shen, X., et al. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893-903. [DOI] [PubMed] [Google Scholar]
- Travers, K. J., Patil, C. K., Wodicka, L., Lockhart, D. J., Weissman, J. S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249-258. [DOI] [PubMed] [Google Scholar]
- Walter, P., Ibrahimi, I., and Blobel, G. (1981). Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to invitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 91, 545-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P., and Johnson, A. E. (1994). Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10, 87-119. [DOI] [PubMed] [Google Scholar]
- Washida, H., Sugino, A., Messing, J., Esen, A., and Okita, T. W. (2004). Asymmetric localization of seed storage protein RNAs to distinct subdomains of the endoplasmic reticulum in developing maize endosperm cells. Plant Cell Physiol. 45,: 1830-1837. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]