Identification of a transcriptionally active peroxisome proliferator-activated receptor α-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator (original) (raw)
Abstract
Peroxisome proliferator-activated receptor α (PPARα) plays a central role in the cell-specific pleiotropic responses induced by structurally diverse synthetic chemicals designated as peroxisome proliferators. Transcriptional regulation by liganded nuclear receptors involves the participation of cofactors that form multiprotein complexes to achieve cell- and gene-specific transcription. Here we report the identification of such a transcriptionally active PPARα-interacting cofactor (PRIC) complex from rat liver nuclear extracts that interacts with full-length PPARα in the presence of ciprofibrate, a synthetic ligand, and leukotriene B4, a natural ligand. The liganded PPARα-PRIC complex enhanced transcription from a peroxisomal enoyl-CoA hydratase/l-3-hydroxyacyl-CoA dehydrogenase bifunctional enzyme gene promoter template that contains peroxisome proliferator response elements. Rat liver PRIC complex comprises some 25 polypeptides, and their identities were established by mass spectrometry and limited sequence analysis. Eighteen of these peptides contain one or more LXXLL motifs necessary for interacting with nuclear receptors. PRIC complex includes known coactivators or coactivator-binding proteins (CBP, SRC-1, PBP, PRIP, PIMT, TRAP100, SUR-2, and PGC-1), other proteins that have not previously been described in association with transcription complexes (CHD5, TOG, and MORF), and a few novel polypeptides designated PRIC300, -285, -215, -177, and -145. We describe the cDNA for PRIC285, which contains five LXXLL motifs. It interacts with PPARα and acts as a coactivator by moderately stimulating PPARα-mediated transcription in transfected cells. We conclude that liganded PPARα recruits a distinctive multiprotein complex from rat liver nuclear extracts. The composition of this complex may provide insight into the basis of tissue and species sensitivity to peroxisome proliferators.
Peroxisome proliferators such as ciprofibrate, nafenopin, Wy-14, 643, and di(2-ethylhexyl)phthalate are known to induce marked peroxisome proliferation in liver parenchymal cells in rats and mice (1, 2). Peroxisome proliferator-activated receptor α (PPARα), a member of the nuclear receptor superfamily, plays a central role in the induction of cell-specific pleiotropic responses, including the development of liver tumors by structurally diverse peroxisome proliferators (3–5). The PPAR subfamily consists of three isotypes, namely PPARα, PPARγ, and PPARβ/δ, and like all other nuclear receptors, PPARs possess a highly conserved DNA-binding domain that recognizes peroxisome proliferator response elements (PPREs) in the promoter regions of target genes (3, 6). PPARs also contain two transcriptional activation function (AF) domains, termed AF-1 in the N-terminal domain and AF-2 in the hormone-binding domain (3). After ligand binding, PPARs heterodimerize with retinoid-X receptor (RXR), and PPAR–RXR heterodimers bind to PPRE to initiate the transcriptional regulation of target genes (3, 6).
The molecular mechanisms by which nuclear receptors achieve transcriptional activation of specific genes in a tissue/cell-specific fashion remain largely unknown. Nevertheless, it is becoming increasingly clear that many cofactors interact with the liganded nuclear receptors for the successful completion of gene transcription (see refs. 7 and 8 for review). These include the p160/steroid receptor coactivator-1 (SRC-1) family with three members (SRC-1, TIF2/GRIP1/SRC2, and pCIP/ACTR/AIB1/RAC3/TRAM1/SRC-3) (see refs. 7 and 8 references therein), CREB-binding protein (CBP) (9), adenovirus E1A-binding protein p300 (10), and PPAR-binding protein PBP (TRAP220/DRIP205) (11, 12), PGC-1α (13), and PRIP/ASC-2/RAP250/TRBP/NRC (see refs. 14 and 15 for additional references). They all contain one or more conserved LXXLL (where L is leucine and X is any amino acid) signature motif, which has been found to be necessary and sufficient for ligand-dependent interactions with AF-2 domain present in the C-terminal ligand-binding domain of the nuclear receptor (16). It appears that these coactivators exert their nuclear receptor transactivation enhancing function by forming two large multiprotein complexes, one anchored by CBP/p300 and the other by PBP (7, 12, 17). The current model of transcription proposes that the initial CBP/p300-mediated complex performs the acetylation–methylation functions (7, 8, 18–20). A transition from CBP/p300-dependent to a mediator-dependent stage of transcription involves the thyroid hormone receptor-associated protein (TRAP)/vitamin D receptor-interacting protein (DRIP)/activator-recruited cofactor (ARC) complex of coactivators anchored by PBP (12, 21–23). Recently, we found that PIMT, a PRIP-binding protein with RNA methyltransferase activity, interacts with CBP/p300 as well as PBP under both in vitro and in vivo conditions, suggesting that PIMT bridges CBP/p300-anchored and PBP-anchored coactivator complexes (15, 24). This finding raises the possibility that both complexes can merge into one megacomplex in the process of transcription (15, 24).
In an effort to identify specific nuclear proteins that interact with PPARα, we have focused our attention on the rat liver because rat and mouse are highly sensitive to peroxisome proliferator-induced transcriptional activation of PPARα-target genes (2, 5). Rat liver nuclear extracts that contain presumably all of the cofactors for mediating PPARα-dependent transcription were used to capture the PPARα-interacting cofactor (PRIC) complex of proteins. The results presented here demonstrate that PRIC is a conglomeration of some components of both CBP/p300-anchored and PBP-anchored complexes, as well as several polypeptides that have not been previously known to be associated with transcriptional complexes. This PRIC complex also contains the bridge protein PIMT.
Materials and Methods
Plasmid Constructions.
pGEX-PPARα, pCMX-PBP, pCDNA3.1-CBP, pCDNA3.1-SRC-1 and pCDNA3.1-PRIP, GST-PPARγ, GST-RXRα, pCDNA3.1-PGC1-FLAG, and GST-ERα have been described previously (15, 24). pGL-PBE-PPRE was constructed by using the 3.3-kb promoter of rat peroxisomal L-PBE (enoyl-CoA hydratase/l-3-hydroxyacyl-CoA dehydrogenase bifunctional enzyme) gene (25).
Purification and Fractionation of Rat Liver Nuclear Extracts.
Male F-344 rats were fasted overnight to deplete liver glycogen and were killed 1 h after administering a single dose of ciprofibrate (250 mg/kg body weight) by gavage. Liver nuclei were isolated and purified by sucrose density gradient centrifugation (26), solubilized by homogenizing in buffer C [20 mM Hepes, pH 7.9/0.42 M NaCl/1.5 mM MgCl2/0.2 mM EDTA/0.2 mM EGTA/25% (vol/vol) glycerol/0.5 mM DTT/1 mM PMSF] and dialyzed for 6 h against 100 vol of buffer D [20 mM Hepes, pH 7.9/0.1 M KCl/0.2 mM EDTA/0.2 mM EGTA/20% (vol/vol) glycerol/1 mM DTT/1 mM PMSF]. The precipitated material from the dialysate was removed by centrifugation at 20,000 × g for 20 min and the supernatant fractionated on Bio-Rex-70 ion exchanger (Bio-Rad). The column was extensively washed to remove unbound proteins, and proteins bound to Bio-Rex-70 were sequentially eluted with a KOAc step gradient (0.3 M, 0.6 M, and 1.5 M) in buffer D and 1-ml fractions were collected. Fractions eluted at 0.6 M and 1.5 M were pooled and dialyzed in buffer D and this dialysate, designated purified liver nuclear extract (PLNE), was used in the identification of PPARα-interacting proteins.
Isolation of PPARα-Bound Protein Complex.
Full-length mouse PPARα cDNA was expressed as a glutathione _S_-transferase (GST)-PPARα fusion protein in Escherichia coli BL21(DE3). GST-PPARα fusion protein immobilized on beads was incubated for 12 h with 5 mg of PLNE in the presence of ligand (1 μM LTB4 or 10 μM ciprofibrate) in a 500-μl reaction mixture, which was adjusted to 180 mM KCl. After incubation, the beads were washed with 40 vol of binding buffer with 0.1% Nonidet P-40. The bound proteins were eluted with 0.2% Sarkosyl, concentrated with Microcon concentrators, and subjected to SDS/PAGE. Gels were analyzed after silver or Coomassie brilliant blue staining.
Histone Acetyltransferase (HAT) Assay.
HAT activity of the protein complex bound to GST-PPARα was assayed essentially as described (27). Briefly, the 30-μl reaction mixture contained 50 mM Hepes at pH 7.9, 10% (vol/vol) glycerol, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate, 1 μl of [14C]acetyl-CoA (57 mCi/mmol, Amersham Pharmacia; 1 mCi = 37 MBq), and 10 μl of eluted protein mix (2.5 μg of protein) and was incubated for 1 h at 30°C. The reaction products were analyzed by filter binding assays or by electrophoresis.
In Vitro Transcription Assays.
PLNE proteins bound to GST-PPARα were eluted with 0.2% Sarkosyl or 20 mM glutathione (GSH) in binding buffer, and eluted proteins were tested for the transcription initiation ability. Rat L-PBE gene promoter (3.3 kb) was used as the template for transcription assay. Typically, 25 μl of transcription reaction contained 5 μl of 5× transcription buffer [100 mM Hepes, pH 7.9/500 mM KOAc/50 mM Mg(OAc)2/25 mM EDTA/50% glycerol], 0.125 μl of 0.5 M DTT, 0.1 μl of creatine kinase (2 mg/ml), 3 μl of phosphocreatine (64 mg/ml), 100 ng of DNA template, and 15 units of RNase inhibitor and was primed with 1 μl of translated PPARα or RXR or both (28). The incubations included 4 μl of minimal nuclear extract and 6 μl of Sarkosyl- or GSH-eluted proteins that bound to GST or GST-PPARα. Total volume of the reaction mix was adjusted to 25 μl with RNase-free water. The reaction was started with the addition of NTPs containing [32P]UTP and incubated at 30°C for 1 h. The reaction was terminated with the addition of 7 vol of stop solution (0.3 M Hepes, pH 7.9/0.3 M NaOAc/0.5% SDS/2 mM EDTA/3 μg/ml tRNA) and extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, vol/vol). The radiolabeled RNA was precipitated with cold ethanol on dry ice for 15 min, and the transcription run-off products were electrophoresed on a 10% urea/TBE gel (TBE is 100 mM Tris/90 mM boric acid/1 mM EDTA, pH 8.3) and autoradiographed.
Protein Sequencing.
Protein bands were excised, digested with trypsin in situ, and analyzed by matrix-assisted laser-desorption/ionization reflection time-of-flight mass spectrometry (MALDI-TOF MS; PE Voyager DE-Pro) in the Analytical Services Laboratory, Northwestern University. Peptide sequences were identified by using MS-Fit (Protein Prospector, University of California, San Francisco).
PRIC285 cDNA Cloning, Northern Blotting, and in Vitro Translation.
On the basis of MALDI-TOF peptides derived from PRIC285, a full-length cDNA sequence was obtained (KIAA1769 from Kazusa, Japan) by checking human expressed sequence tags in GenBank. The sequence was confirmed and the cDNA was cloned into pcDNA3.1 and pCMV-PRIC285-FLAG. Northern blotting was performed with CLONTECH human multiple tissue and human cancer cell line blots. PRIC285 was translated in vitro by using the TNT coupled transcription–translation system (Promega).
GST Pull-Down Assays, Cell Culture, and Transfection.
GST alone and GST fusion proteins were used for GST pull-down assays by incubating with 10 μl of [35S]methionine-labeled proteins obtained by in vitro translation using the TNT coupled transcription–translation system. Immunofluorescence studies were performed with HEK293 cells transfected with pCMV-PRIC285-FLAG and PPARα expression plasmid as described (15, 24). The coactivator activity of PRIC285 was assayed by using HEK293 cells transfected for 5 h with 1.25 μg of luciferase reporter plasmid DNA, 0.75 μg of the appropriate expression plasmid DNA, and 0.5 μg of β-galactosidase expression vector pCMVB (CLONTECH) DNA by using _N-_[1-(2,3-dioleoyloxy)propyl]-N,N,N,-trimethylammonium methylsulfate (DOTAP)-mediated transfection method (Roche Molecular Biochemicals). Cell extracts were prepared 36 h after transfection and assayed for luciferase and β-galactosidase activities.
Results
Interaction of Nuclear Proteins with Liganded Immobilized PPARα.
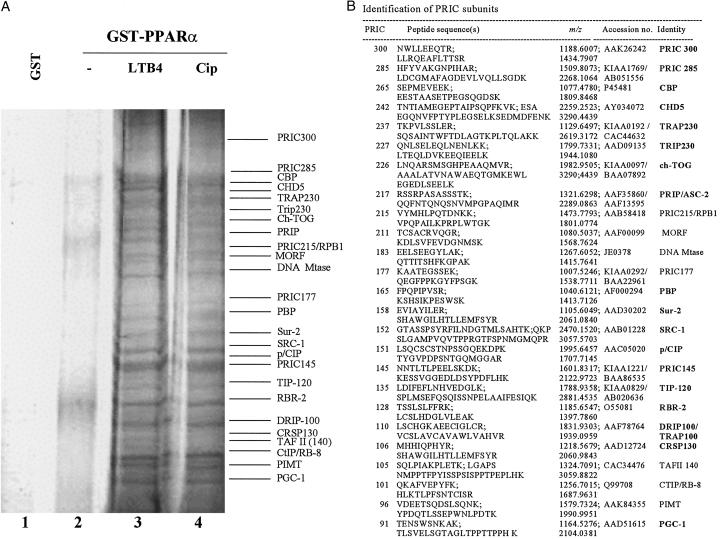
Nuclei were isolated from the livers of rats 1 h after a single intragastric dose of ciprofibrate to ensure the translocation of PPARα into the nucleus and initiation of transcriptional activity of target genes (29). Liver nuclear extract was fractionated on a Bio-Rex-70 column, and pooled fractions, designated PLNE, containing RNA polymerase II as well as PPARα and RXRα (data not shown), were incubated with a GST-fused full-length PPARα in the presence of ciprofibrate, a synthetic ligand, or LTB4, a natural ligand. PLNE is enriched in cofactors necessary for the formation of PPARα- responsive transcriptional complex in rat liver nuclei. To ensure the capture of proteins that interact with the C- terminal AF-2 domain for ligand-dependent and N-terminal AF-1 domain for ligand-independent transactivation, we chose to use the full-length PPARα. These domains interact with a variety of coactivator and corepressor proteins that in turn interact with RNA polymerase II to bring about the formation of preinitiation complex on the promoter region of the target genes (7). Proteins in PLNE that interacted with full-length PPARα, designated PRIC complex, are those that are exclusively bound in the presence of ligand, either ciprofibrate or LTB4 (Fig. 1A). An array of identical proteins (molecular mass 80–300 kDa) bound GST-PPARα in the presence either of ciprofibrate (Fig. 1A, lane 4) or of LTB4 (Fig. 1A, lane 3). Protein-sequencing data revealed that each PRIC polypeptide identified matched sequences represented by human or mouse partial cDNAs and expressed tags (GenBank) (Fig. 1B). Some of these proteins had identities to sequences of known nuclear receptor coactivators, including those isolated by using the PPAR-ligand binding domain as bait in a yeast two-hybrid screen, thus confirming the efficacy of the strategy of GST pull-down using full-length PPARα and purified rat liver nuclear extracts. These proteins include CBP, TRAP230, PBP, PRIP, SRC-1, TRAP100, CRSP130, PGC-1, and PIMT. Genes that are novel (such as PRIC300, -285, -177, and -145), or that are known but not previously described in connection with transcriptional complexes [such as CHD5 (30), TOG (31), MORF (32), and RBR-2 (33)], encode some of the other peptides of this complex. Of considerable interest is the presence of CBP, SRC-1, PBP, PRIP, and PIMT in PRIC complex because all of these have not been found collectively in the previously described TRAP/DRIP/ARC complexes (7, 12, 21, 22). Of about 25 PRIC peptides, 18 contained one or more LXXLL motifs required for interaction with nuclear receptors (Fig. 1B).
Figure 1.
Purification and identification of proteins interacting with PPARα. (A) Identification of ligand-dependent PPARα-interacting complex. SDS/PAGE analysis and silver staining of proteins bound to GST alone (lane 1), GST-PPARα in the absence of ligand (lane 2), and GST-PPARα in the presence of natural ligand LTB4 (lane 3) or synthetic ligand ciprofibrate (Cip, lane 4). (B) Mass spectrometric and limited sequence analysis data. PRIC subunit number, peptide sequence(s), experimental monoisotopic mass (m/z), SwissProt database accession number(s), and identity are included. Of the 25 PRIC peptides, 18 (bold-faced) have one or more LXXLL motifs.
HAT and Transcriptional Activity of PRIC Complex.
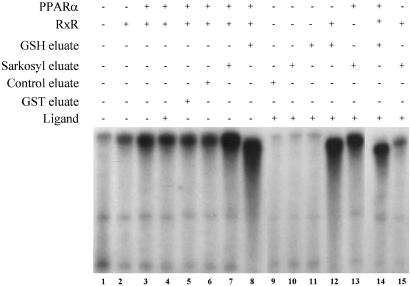
We measured the HAT activity of PRIC complex because this activity is a prerequisite for transcription. Proteins bound to GST-PPARα in the presence of ligand, when eluted by GSH or Sarkosyl, exhibited significantly higher HAT activity than those eluted from nonliganded PPARα (data not shown). This observation is consistent with the reports that some of the proteins found in PRIC complex possess intrinsic HAT activity (e.g., CBP and SRC-1). To investigate whether the proteins in PRIC act as a coactivator complex for PPARα, an in vitro transcription assay system was used to ascertain the transcriptional enhancement capability of this complex. The addition of Sarkosyl-eluted proteins (Fig. 2, lane 7; contains PRIC but no PPARα), or the GSH-eluted proteins (Fig. 2, lane 8; contains PRIC along with PPARα) that bound to GST-PPARα increased the activation of PPARα by >5-fold as compared with various controls (Fig. 2, lanes 1–6). With both GSH and Sarkosyl eluates addition of ligand did not result in substantial additional increases in transcriptional activity (Fig. 2, lanes 8 and 14), and this is attributed to the presence of traces of natural ligand in the complex. The inclusion of rat liver nuclear extract was to provide factors TFIIBs and TFIIDs and RNA polymerase II required for the transcription (34). GSH eluate, which contains PPARα in the PRIC, did not require PPARα supplement (Fig. 2, lane 12), whereas Sarkosyl eluate, which removes the PRIC components from GST-PPARα fusion and thus does not contain PPARα, appeared to depend upon PPARα supplement for transcription (Fig. 2, lanes 13 and 15). These findings suggest that an external PPARα is required to bind to PPRE present in the L-PBE promoter and that both GSH- and Sarkosyl-eluted PRIC complexes exert robust coactivator function.
Figure 2.
Purified rat nuclear PRIC potentiates PPARα transcription in a cell-free system. In vitro transcription reactions were set up as described in Materials and Methods. A 3.6-kb promoter of the rat L-PBE gene served as template for the transcription assay, with minimal nuclear extract as the source for basal transcription factors and DNA-dependent RNA polymerase II. Purified PPARα or RXR protein was added to the template DNA before the addition of nuclear extract and other components. The reactions were allowed to proceed for 60 min at 30°C, and labeled RNA transcript was extracted and separated on a urea/TBE gel and autoradiographed. GST eluate and control eluate represent, respectively, fraction bound to GST alone and to GST-PPARα in the absence of added ligand. GSH and Sarkosyl eluates represent proteins bound to GST-PPARα in the presence of ligand.
Characterization of PRIC285 cDNA.
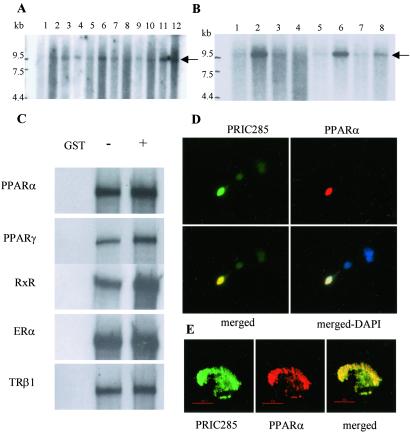
cDNA and genomic sequence analysis of all four novel PRIC peptides, PRIC300, -285, -177, and -145, showed that all except PRIC177 contained one or more LXXLL motifs (data not presented). In this study we chose to report the characterization of PRIC285. The full-length cDNA encoding this protein is confirmed on the basis of amino acid sequences (underlined in Fig. 3) derived from cognate polypeptides in the GST-PPARα complex. PRIC285 contains five LXXLL motifs found in nuclear receptor coactivators and also revealed the presence of UvrD-helicase motif found in the REP family of helicases (35). The full-length human PRIC285 cDNA has an ORF of 6,240 nucleotides that encodes a 2,080-aa protein with an estimated molecular mass of 285 kDa. The gene encoding human PRIC285 is localized on chromosome 20q13.3 and consists of 18 exons (data not presented). Exon 7, the longest of all, encodes 1,238 aa and has three LXXLL motifs. A computer search of the database revealed no protein that has significant overall similarity to PRIC285. Nevertheless, PRIC285 has a region between amino acids 1584 and 1991 with substantial similarity to UvrD/REP helicase (35). To determine the size of PRIC285 protein, we performed in vitro transcription and translation and found that the synthesized a protein has an estimated molecular mass of 285 kDa (data not presented). Northern analysis revealed a prominent ≈9.5-kb PRIC285 mRNA transcript in various human tissues such as skeletal muscle, colon, spleen, liver, kidney, lung, peripheral blood, and placenta (Fig. 4A), and in certain cancer cell lines such as HeLa cells, colorectal adenocarcinoma, and melanoma cells (Fig. 4B).
Figure 3.
Deduced human PRIC285 amino acid sequence with five LXXLL motifs (boldfaced). Two peptide sequences found by mass spectrometry are underlined. The putative UvrD/helicase domain is between amino acids 1584 and 1991.
Figure 4.
Northern blotting, binding of PRIC285 to PPARα, and colocalization. (A) Northern analysis of human multiple tissue blot (CLONTECH). Lanes 1–12 represent, respectively, brain, heart, skeletal muscle, colon, thymus, spleen, kidney, liver, small intestine, placenta, lung, and leukocytes. Arrow indicates PRIC285 mRNA. (B) Northern analysis of human cancer cell line blot (CLONTECH) probed with a partial PRIC285 cDNA probe. Lanes 1–8 represent, respectively, HL-60 cells line, HeLa cell S3, chronic myelogenous leukemia (K-562), lymphoblastic leukemia MOLT-4, Burkitt's lymphoma Raji, colorectal adenocarcinoma SW480, lung carcinoma A-549, and melanoma G-361. Arrow indicates PRIC285 mRNA. (C) [35S]Methionine-labeled full-length PRIC285 generated by in vitro translation was incubated with GSH-Sepharose beads bound with purified _E. coli_-expressed GST-PPARα or GST. PRIC285 bound to PPARα in the absence of ligand, and the addition of ligand increased the binding ≈2-fold. PRIC285 also interacts with PPARγ, RXRα, ERα (estrogen receptor α), and TRβ1 (thyroid hormone receptor β1). (D and E) Colocalization of PRIC285 and PPARα in the nucleus as visualized by conventional fluorescence microscopy (D) and by DELTAVISION deconvolution microscopy (E). PRIC285 was expressed transiently in HEK293 cells by using three FLAG epitopes linked to the C terminus, and it was visualized by using anti-FLAG antibodies (green). PPARα was also transiently expressed in these cells and localized by using anti-PPARα antibodies (red). Merging of the PRIC285 and PPARα localization images of the same cell reveals overlapping localization (yellow) of these two proteins. 4×,6-Diamidino-2-phenylindole (DAPI) staining shows nuclei in D (Lower Right).
Interaction of PRIC285 with PPARα and Other Nuclear Receptors.
To determine whether PRIC285 directly interacts with PPARα, we evaluated the in vitro binding by using bacterially generated GST-PPARα fusion protein and in vitro translated PRIC285. As shown in Fig. 4C, PRIC285 interacted with PPARα but not with GST. The addition of ligand, either LTB4 or ciprofibrate, slightly enhanced the interaction of PPARα with PRIC285. We also examined the interaction of PRIC285 with PPARγ, TRβ1, ERα, and RXRα and found that it exhibits ligand-dependent interaction (Fig. 4C). The in vivo interaction of PRIC285 with PPARα has been determined by using immunofluorescence microscopy mainly to ascertain whether they are colocalized in the nucleus. A plasmid containing three FLAG epitopes linked to the N-terminal portion of PRIC285 was cotransfected into HEK293 cells along with PPARα plasmid. Immunofluorescence with anti-FLAG revealed that the expressed PRIC285 protein is localized in the nucleus (Fig. 4D, green). As expected, PPARα localization using anti-PPARα antibodies revealed the nuclear localization of this receptor (Fig. 4D, red). When PRIC285 and PPARα images were merged in cells expressing both proteins, an appreciable degree of overlapping localization of these two molecules was discerned (Fig. 4D, yellow), and this colocalization was more striking in high-resolution images of the nucleus (Fig. 4E).
PRIC285 Potentiates the Transcriptional Activity of PPARα in Mammalian Cells.
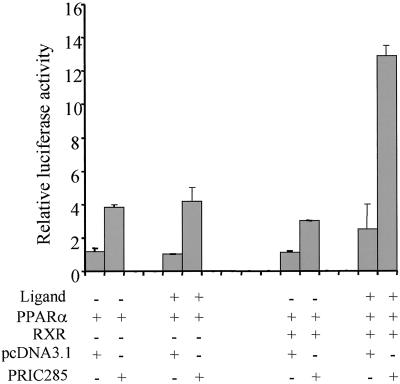
We examined the functional relevance of the binding of PRIC285 with PPARα by transiently coexpressing PRIC285 and PPARα with the reporter luciferase gene, PPRE-TK-LUC in HEK293. PRIC285 coactivates a PPRE in the presence of PPARα even in the absence of exogenous ligand, implying the presence of natural ligands in the cells and medium (Fig. 5).
Figure 5.
PRIC285 functions as a coactivator for PPARα. HEK293 cells were cotransfected with 1.5 μg of reporter construct PPRE-TK-LUC, 0.25 μg of PCMV-PPARα, 0.25 μg of PCMV-RXR, 0.5 μg of PCMV-PRIC285, and 0.5 μg of pcDNA3.1 as indicated. Transfections without the indicated plasmid were compensated by adding the same amount of pcDNA3.1. The activity obtained on transfection of PPRE-TK-LUC without exogenous PRIC285 was taken as 1. Results are the mean of three independent transfections.
Discussion
In this study we used rat liver nuclear extract to identify specific nuclear proteins that interact with PPARα for the main reason that rat liver is highly sensitive to PPARα ligand-mediated inductive effects, including the activation of several target genes, peroxisome proliferation, and the development of liver tumors (1, 2, 5, 36). We have used a full-length PPARα in GST pull-down assays to capture a portrait of proteins that interact not only with AF-2 domain in a ligand-dependent fashion but also with proteins that interact with other parts of the receptor, including the AF-1 domain. The use of C-terminal AF-2 activation domain of nuclear receptors resulted mostly in the identification of PBP-anchored TRAP/DRIP protein complexes (12, 21, 37, 38). As pointed out before, the nuclear receptor coactivators presumably exist in two complexes, one with the p160 family anchored by CBP/p300 and the other known variously as TRAP/DRIP/ARC complex anchored by PBP (7). Although there is a debate as to whether these two complexes form sequentially (39), or combinatorially (40), emerging evidence points to proteins that facilitate the linkage of these two complexes even if transiently (15, 41). One such protein is PRIP/ASC-2/RAP250/TRBP/NRC, which links CBP/p300 to TRAP/DRIP130 (41), and the other is PIMT, an RNA methyltransferase, which interacts directly with CBP/p300 and PBP (15, 24).
Consistent with this bridging function is our finding that the PRIC complex consists of components of both p160/CBP/p300 complex and PBP-anchored complex (Fig. 1). First, of the 25 or so peptides found in PRIC, the identification of SRC-1, p/CIP, and the anchor CBP, all of which have HAT activity, establishes that PPARα clearly associates with so-called platform proteins (7, 8). Second, the complex contains PBP, TRAP230, TRAP/DRIP100, and TRAP/DRIP/CRISP130, which are found in the PBP-anchored multiprotein complex that links to the basal transcription machinery (12). Third, PRIC reveals the presence of linker proteins PRIP and PIMT, which have recently been shown to link the p160/CBP complex with PBP-anchored complex (15, 41). Thus, it is more than likely that by virtue of PRIP and PIMT and possibly other linker proteins, the two complexes may merge into one megacomplex on DNA template at least in rat liver. This complex can then facilitate signal transduction passing through a specific set of proteins of this complex to account for the cell/species-specific PPARα-mediated transcription. The identification of one composite complex with PPARα does not exclude the possibility that it can arise from both sequential and combinatorial assembly of components. The presence in PRIC complex of CBP, SRC-1, p/CIP, PBP, PRIP, and SUR-2 [a member of the mammalian mediator complex (42)], and of HAT activity together with the transcriptional enhancement capability of PRIC establish the authenticity of the PPARα transcriptional complex we isolated.
It is remarkable that of the 25 PRIC peptides, 18 have one or more LXXLL motifs, and 11 of these (CBP, TRAP230, TRIP230, PRIP, PBP, Sur-2, SRC-1, p/CIP, DRIP/TRAP100, CRSP130, and PGC-1) (Fig. 1) have been previously described in association with nuclear receptors or coactivator complexes (7, 15, 43). Three of the remaining 7 proteins with LXXLLs, namely TOG (31), CHD5 (30), and TIP120 (44), appear to belong to the SWI2/SNF2 family (45), 1, RBR-2, is a member of the retinoblastoma gene family (33), and the remaining 3 are novel proteins (PRIC300, -285, and -145). SWI2/SNF2 proteins, some of which are helicases, play a role in transcription control, DNA repair, and chromatin remodeling (45). TIP120, a TATA-binding protein-interacting protein, is known to cause transcriptional stimulation by all three eukaryotic RNA polymerases (44). Therefore, the presence in PRIC complex of TOG, CHD5, and TIP120 strongly suggests a role for these proteins, possibly in promoter-specific transcription. In this study, we have shown that PRIC285, which has an UvrD helicase-like domain in its C-terminal region (amino acids 1584–1991), acts as a nuclear receptor coactivator and interacts with other nuclear receptors. We have also found that TOG, a microtubule- and tubulin-interacting protein (46), also interacts with PPARα in the GST pull-down assays (unpublished data), which is consistent with the presence of five LXXLL motifs in this protein (31). Further studies are needed to ascertain the coactivator functions of these newly identified proteins in PRIC complex that contain the LXXLL motif necessary for the interaction with nuclear receptors (16).
In the PPARα-interacting cofactor complex, we found only 7 proteins without LXXLL motifs, implying that these may not directly interact with PPARα but serve as accessory proteins that interact with coactivators. These include a rat homologue of human MORF, which is a histone acetyltransferase (32), DNA Mtase, a DNA (cytosine-5) methyltransferase (47), TAFII140 (48), CTIP/RB-8 (a retinoblastoma-binding protein) (49), PIMT, an RNA methyltransferase (15, 24, 50), PRIC215, and PRIC177 (Fig. 1). Elucidation of the functional role of these proteins both singly and collectively is necessary to fully appreciate receptor-specific transduction of signal through the assembled transcriptional complex. The composition of rat liver PRIC complex should serve as a prototype for discerning clues rearding cell- and species-specific differences in peroxisome proliferator-induced pleiotropic responses.
Acknowledgments
We thank Dr. Tom Lukas for generous advise on MALDI-TOF and Wen-Qing Cao and Travis Harrison for assistance with deconvolution microscopy. This work was supported by National Institutes of Health Grants GM23750 (to J.K.R.) and CA84472 (to M.S.R.), by a Department of Veterans Affairs Merit Review Grant (to A.V.Y.), and by the Joseph L. Mayberry, Sr., Endowment Fund.
Abbreviations
PPAR
peroxisome proliferator-activated receptor
PPRE
peroxisome proliferator-activated response element
AF
activation function
L-PBE
enoyl-CoA hydratase/l-3-hydroxyacyl-CoA dehydrogenase bifunctional enzyme
SRC-1
steroid receptor coactivator-1
PBP
PPAR-binding protein
CREB
cAMP response element-binding protein
CBP
CREB-binding protein
TRAP
thyroid hormone receptor-associated protein(s)
DRIP
vitamin D receptor-interacting protein(s)
ARC
activator-recruited cofactor
PRIC
PPARα-interacting cofactor
PRIP
PPAR-interacting protein
PIMT
PRIP-interacting protein with methyltransferase domain
RXR
retinoid-X receptor for 9-_cis_-retinoic acid
LTB4
leukotriene B4
HAT
histone acetyltransferase
GST
glutathione _S_-transferase
GSH
glutathione
MALDI-TOF
matrix-assisted laser-desorption/ionization reflection time-of-flight
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF517673).
References
- 1.Reddy J K, Krishnakantha T P. Science. 1975;190:787–789. doi: 10.1126/science.1198095. [DOI] [PubMed] [Google Scholar]
- 2.Reddy J K, Chu R. Ann NY Acad Sci. 1996;804:176–201. doi: 10.1111/j.1749-6632.1996.tb18616.x. [DOI] [PubMed] [Google Scholar]
- 3.Issemann J, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 4.Lee S S-T, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez F J, Peters J M, Cattley R C. J Natl Cancer Inst. 1998;90:1702–1709. doi: 10.1093/jnci/90.22.1702. [DOI] [PubMed] [Google Scholar]
- 6.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenfeld M G, Glass C K. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 8.McKenna N J, O'Malley B W. Cell. 2002;108:467–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 9.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y, Qi C, Jain S, Rao M S, Reddy J K. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 12.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman B M. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Kan L, Qi C, Kanwar Y S, Yeldandi A V, Rao M S, Reddy J K. J Biol Chem. 2000;275:13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 15.Misra P, Qi C, Yu S, Shah S H, Cao W-Q, Rao M S, Thimmapaya B, Zhu Y-J, Reddy J K. J Biol Chem. 2002;277:20011–20019. doi: 10.1074/jbc.M201739200. [DOI] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 18.Vo N, Goodman R H. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 19.Chen D, Ma H, Hong H, Koh S S, Huang S-M, Schurter B T, Aswad D W, Stallcup M R. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 20.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson B M, Montminy M, Evans R M. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 21.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Nature (London) 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 22.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Nature (London) 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y W, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway J W, Conaway R C, Kornberg R D. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Qi C, Cao W-Q, Yeldandi A V, Rao M S, Reddy J K. Proc Natl Acad Sci USA. 2001;98:10380–10385. doi: 10.1073/pnas.181347498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Marcus S L, Sajjadi F H, Alvares K, Reddy J K, Subramani S, Rachubinski R, Capone J P. Proc Natl Acad Sci USA. 1992;89:7541–7545. doi: 10.1073/pnas.89.16.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Widnell C C, Tata J R. Biochem J. 1964;92:313–317. doi: 10.1042/bj0920313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownell J E, Allis C D. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guermah M, Tao Y, Roeder R G. Mol Cell Biol. 2001;21:6882–6894. doi: 10.1128/MCB.21.20.6882-6894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy J K, Goel S K, Nemali M R, Carrino J J, Laffler T G, Reddy M K, Sperbeck S J, Osumi T, Hashimoto T, Lalwani N D, Rao M S. Proc Natl Acad Sci USA. 1986;83:1747–1751. doi: 10.1073/pnas.83.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster E F, Stoger R. Mamm Genome. 2002;13:117–119. doi: 10.1007/s00335-001-3042-6. [DOI] [PubMed] [Google Scholar]
- 31.Charrasse S, Mazel M, Taviaux S, Berta P, Chow T, Larroque C. Eur J Biochem. 1995;234:406–413. doi: 10.1111/j.1432-1033.1995.406_b.x. [DOI] [PubMed] [Google Scholar]
- 32.Champagne N, Bertos N R, Pelletier N, Wang A H, Vezmar M, Yang Y, Heng H H, Yang X-J. J Biol Chem. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- 33.Sawada Y, Nomura H, Endo Y, Umeki K, Fujita T, Ohtaki S, Fujinaga K. Biochem Biophys Acta. 1997;1361:20–27. doi: 10.1016/s0925-4439(97)00037-9. [DOI] [PubMed] [Google Scholar]
- 34.Dvir A, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng W, Hsieh J, Brendza K M, Lohman T M. J Mol Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 36.Reddy J K, Hignite C E, Azarnoff D L. Nature (London) 1980;283:397–398. doi: 10.1038/283397a0. [DOI] [PubMed] [Google Scholar]
- 37.Kang Y K, Guermah M, Yuan C-X, Roeder R G. Proc Natl Acad Sci USA. 2002;99:2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanagisawa J, Kitagawa H, Yanagida M, Wada O, Ogawa S, Nakagomi M, Oishi H, Yamamoto Y, Nagasawa H, McMahon S B, et al. Mol Cell. 2002;9:553–562. doi: 10.1016/s1097-2765(02)00478-1. [DOI] [PubMed] [Google Scholar]
- 39.Margeat E, Poujol N, Boulahtouf A, Chen Y, Muller J D, Gratton E, Cavailles V, Royer C A. J Mol Biol. 2001;306:433–442. doi: 10.1006/jmbi.2000.4418. [DOI] [PubMed] [Google Scholar]
- 40.Shang Y, Hu X, DiRenzo J, Lazar M A, Brown M. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 41.Ko L, Cardona G R, Chin W W. Proc Natl Acad Sci USA. 2000;97:6212–6217. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer T G, Martin M E D, Lees E, Ricciardo R P, Berk A J. Nature (London) 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 43.Chang K-H, Chen Y, Chen T-T, Chou W-H, Chen P-L, Ma Y-Y, Yang-Feng T L, Leng X, Tsai M-J, O'Malley B W, Lee W H. Proc Natl Acad Sci USA. 1997;94:9040–9045. doi: 10.1073/pnas.94.17.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makino Y, Yogosawa S, Kayukawa K, Coin F, Egly J M, Wang Z, Roeder R G, Yamamoto K, Muramatsu M, Tamura T. Mol Cell Biol. 1999;19:7951–7960. doi: 10.1128/mcb.19.12.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emerson B M. Cell. 2002;109:267–270. doi: 10.1016/s0092-8674(02)00740-7. [DOI] [PubMed] [Google Scholar]
- 46.Spittle C, Charrasse S, Larroque C, Cassineris L. J Biol Chem. 2000;275:20748–20753. doi: 10.1074/jbc.M002597200. [DOI] [PubMed] [Google Scholar]
- 47.Kimura H, Takeda T, Tanaka S, Ogawa T, Shiota K. Biochem Biophys Res Commun. 1998;253:495–501. doi: 10.1006/bbrc.1998.9802. [DOI] [PubMed] [Google Scholar]
- 48.Gangloff Y-G, Pointud J-C, Thuault S, Carre L, Romier C, Muratoglu S, Brand M, Tora L, Couderc J-L, Davidson I. Mol Cell Biol. 2001;21:5109–5121. doi: 10.1128/MCB.21.15.5109-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaeper U, Subramaniam T, Lim L, Boyd J M, Chinnadurai G. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 50.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Mol Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]