Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line (original) (raw)
Abstract
An HEK293S cell line resistant to ricin was prepared by mutagenesis by using ethyl methanesulfonate. It was shown to lack _N_-acetylglucosaminyltransferase I (GnTI) activity, and consequently unable to synthesize complex N-glycans. The tetracycline-inducible opsin expression system was assembled into this GnTI− HEK293S cell line. Stable cell lines were isolated that gave tetracycline/sodium butyrate-inducible expression of the WT opsin gene at levels comparable with those observed in the parent tetracycline-inducible HEK293S cell line. Analysis of the N-glycan in rhodopsin expressed by the HEK293S GnTI− stable cell line showed it to be Man5GlcNAc2. In a larger-scale expression experiment (1.1 liter) a WT opsin production level of 6 mg/liter was obtained. Further, the toxic constitutively active rhodopsin mutant, E113Q/E134Q/M257Y, previously shown to require inducible expression, has now been expressed in an HEK293S GNTI−-inducible cell line at levels comparable with those obtained with WT rhodopsin.
Except for WT rhodopsin, G protein-coupled receptors are available from natural sources only in extremely small amounts. Structure–function studies involving, in particular, NMR spectroscopy and crystallization for 3D structural information require in the order of many milligrams of the receptor proteins. Recently, the development of expression systems based on mammalian stable cell lines has allowed preparation of recombinant rhodopsin and a variety of its mutants in amounts suitable for NMR studies (1, 2). Further, toxicity problems encountered in the development of stable cell lines for expression of certain proteins have been overcome by the development of a tetracycline-inducible expression system (3). For the potentially important application to crystallization of integral membrane proteins, such as rhodopsin mutants and the G protein-coupled receptors, the methods so far developed have the potential drawback that the complex N-glycans in the expressed proteins are extremely heterogeneous (see below). Thus, the aim of the work herein reported has been to develop an expression system in which N-glycan formation is restricted and homogeneous. With this aim, we undertook to construct an HEK293S mutant cell line that is defective for production of complex N-glycans. Such cell lines can be prepared by virtue of resistance to lectins and this approach has been used for preparation of Chinese hamster ovary cell lines with deficient N-glycosylation (4). Indeed, one class of Chinese hamster ovary mutants, designated Lec1, was shown to be _N_-acetylglucosaminyltransferase-I (GnTI) negative (5). Here, we report on the preparation of a ricin-resistant (ricinR) HEK293 mutant cell line that has been characterized to be GnTI negative. The tetracycline-inducible opsin expression system (3) has been assembled into the above-mentioned GnTI− HEK293 cell line. Here we describe the application of this cell line in the expression of WT rhodopsin and the constitutively active rhodopsin mutant, E113Q/E134Q/M257Y in 3–6 mg of the proteins per liter of cell culture. The N-glycans in both the expressed proteins have been characterized to be homogeneous Man5GlcNAc2.¶
Materials and Methods
The materials and buffers were used as described (3) except for the following. Ricin (RCA II) was from EY Laboratories. Triton X-100 and ethyl methanesulfonate (EMS) were from Sigma. [35S]Methionine (Translabel) and DMEM deficient for L-glutamine, L-cysteine, and L-cystine were from ICN. The polyclonal antibody anti-TetR was from MoBiTec (Marco Island, FL). 3H-labeled rainbow protein standards were from Amersham Pharmacia. The plasmid pRVSTag (6) was a gift from J. Nathans (Johns Hopkins University, Baltimore).
Arthrobacter ureafaciens sialidase (Glyko, Novato, CA) at 2 units/ml was used for N-glycan profiling. Trichoderma reesei α-1,2-mannosidase at 30 μg/ml was prepared from Pichia pastoris (7). APTS (8-Amino-1,3,6-pyrenetrisulfonic acid) was from Molecular Probes. The N-glycan mixture for the GnTI activity assay was prepared from bovine RNase B (Glyko).
Chemical Mutagenesis of HEK293S Cells and Isolation of Mutants Resistant to Ricin.
HEK293S cell monolayers were grown in DMEM/F12 supplemented with FBS (10%), penicillin (100 units/ml), streptomycin (100 μg/ml), and glutamine (292 μg/ml). Cells in exponential growth phase were trypsinized and plated at a density of 2 × 105 cells per ml in 70-cm2 T flasks. After incubation for 20 h the growth medium was replaced with fresh medium (13 ml) containing EMS (150 μg/ml). Cells were grown in the presence of the mutagen for 20 h, washed twice with PBS, trypsinized, and then stored in three aliquots (each containing ≈8 × 105 cells) at −70°C. One aliquot of frozen mutagenized cells was thawed and grown for 5 days to ≈70% confluence. These cells were then split 1:10 and the aliquot incubated for a further 2 days. The medium was then replaced with fresh medium containing ricin (1 ng/ml, 10 ng/ml, 100 ng/ml, or 1 μg/ml). Cells were refed with fresh medium containing appropriate levels of ricin every 4 days until colonies formed.
Identification of HEK293S Cell Lines with Altered N-Glycosylation.
RicinR colonies were expanded to 10-cm dishes. Cells split 1:5 were transiently transfected the next day by using 30 μg of a mixture (10:1 wt/wt) of two plasmids (pMT4 and pRSVTag, respectively) by the Ca2PO4 precipitation procedure (3). Plasmid pRSVTag (6) carries a gene encoding SV40 large tumor (T) antigen that promotes pMT4 replication in HEK293S cells (6), whereas plasmid pMT4 carries the WT opsin gene (8). Transfection was performed as described (1) but after removal of the DNA mixture, cells were incubated for 48 h to allow transient expression of the opsin gene. Cells were harvested, treated with 11-_cis_-retinal (5 μM), and solubilized by using buffer B (3) in the dark. Rhodopsin was purified by using mAb 1D4-Sepharose immunoaffinity chromatography (1, 8). Purified rhodopsin was subjected to SDS/PAGE (10%) and the bands were visualized by silver stain. The N-glycans in rhodopsin expressed transiently in the ricin-resistant cell lines were also analyzed by N-glycan-profiling experiments (see below).
GnTI Activity Assays.
Preparation of HEK293S cell lysates.
HEK293S and HEK293S GnTI− cells were grown in 15-cm dishes.
On reaching confluence, cells were harvested and washed twice with 10 ml each of PBS buffer, and whole-cell pellets were frozen in liquid nitrogen in aliquots of ≈3 × 107 cells. Frozen cell pellets were resuspended by addition of 200 μl of PBS containing 1% Triton X-100 and protease inhibitors (without EDTA; Roche Molecular Biochemicals). The cells were lysed by four cycles of freeze (liquid N2)/thaw (0°C). Insoluble material was removed by centrifugation (14,000 × g, 10 min at 4°C). Protein concentration was determined by using the bicinchonic acid procedure (9).
Preparation of APTS-labeled Man5GlcNAc2.
Bovine RNaseB N-glycans [Man (5–9)GlcNAc2] (30 nmol) were treated with 50 mM APTS (1,000-fold excess) in a reaction mixture (60 μl) containing citric acid (0.6 M), NaCNBH3 (0.5 M), and DMSO (50% vol/vol). Incubation was for 4 h at 55°C followed by 16 h at 37°C. The reaction was quenched by addition of 5 vol of water. Excess APTS was removed by Sephadex G-10 gel filtration chromatography over a 25 cm × 1 cm i.d. column. Elution was performed with water at a flow rate of 0.4 ml/min. Fractions (1.2 ml) were monitored by using fluorescence spectroscopy (λem = 520 nm). The fractions containing the APTS-glycan conjugate were pooled. Water was removed by evaporation (SpeedVac) and the residual APTS-labeled N-glycans were digested for 16 h by using 3 milliunits/ml of α-1,2-mannosidase in a reaction containing 20 mM sodium acetate (pH 5.0). The mixture was then desalted by gel-filtration chromatography with a 4-ml Sephadex G-10 spin column. Mannosidase was removed by precipitation by addition of methanol to 60% (vol/vol). The supernatant was then recovered and evaporated to dryness (SpeedVac). The quantity of recovered Man5GlcNAc2-APTS was estimated by spectrophotometry at 454 nm, assuming a molar extinction coefficient equal to the one of unconjugated APTS (ɛ = 18,000 M−1⋅cm−1), as no molar extinction coefficient for glycan-conjugated APTS is available.
GnTI assay conditions.
The reaction mixture (6 μl final volume) contained 50 mM Mes (pH 6.9), Complete EDTA-free protease inhibitor mixture at the concentration specified by the manufacturer, 250 μM swainsonine (class II mannosidase inhibitor), 5 mM MnCl2, 5 mM MgCl2, ≈3 nmol Man5GlcNAc2-APTS, and 30 nmol UDP-GlcNAc. The reaction was initiated by addition of 1 μl of cell extract (≈25 μg of protein), and incubation was at 37°C. Aliquots (0.5 μl) of the reaction mixture were removed at different time points and the reaction was terminated by dilution (125-fold) in ice-cold water. Samples were further diluted 10-fold with water and a 1-μl aliquot was mixed with 1 μl of formamide and 0.5 μl of rhodamine-labeled oligonucleotide internal standard (Genescan 500, Applied Biosystems). Samples were applied to a polyacrylamide sequencing gel and electrophoresis was as described in the following section.
Analysis of N-Glycan in Rhodopsin.
The procedure has been described in detail (10). Briefly, purified rhodopsin samples (5 μg) were bound to a poly(vinylidene difluoride) membrane in a microtiter plate format (Millipore). The N-glycans were released from the bound protein by treatment with PNGaseF and then labeled with APTS. Excess APTS was removed by gel filtration with Sephadex G-10. The labeled N-glycan mixture was supplemented with Genescan 500 rhodamine-labeled internal standard. The samples were then subjected to PAGE by using an Applied Biosystems 377A gel-based DNA-sequencer. The applied voltage was 3,500 V, and the gel temperature was maintained at 23°C by using an external cooling bath. All exoglycosidase digestions were performed in 10 μl of 20 mM NaAc, pH 5. Data analysis was performed by using the GENESCAN 3.1 software (Applied Biosystems). A rhodamine-oligonucleotide standard used in all lanes on the same gel was used for alignment with the lane containing an APTS-labeled malto-oligosaccharide standard. The peaks corresponding to the rhodamine-labeled internal standards for alignment are omitted from the data shown.
Construction of Tetracycline-Inducible Cell Lines.
The HEK293S GnTI− cell line was maintained as described (1, 3), and cell growth was in the absence of ricin. Cells were transfected with plasmid pCDNA6-TR and blasticidin-resistant colonies were obtained as described (3). Individual blasticidin-resistant colonies were expanded and screened for tetR expression by analyzing solubilized cell extracts by SDS/PAGE followed by immunodetection with an anti-TetR polyclonal antibody. One anti-TetR immunopositive cell line was chosen and expanded for use in all subsequent experiments. This cell line was transfected with pACMVtetO-rho or pACMVtetO-rho (E113Q/E134Q/M257Y) (3). The HEK293S (GnTI−) TetR cell line was found to be hypersensitive to Geneticin, and this drug was used at 200–250 μg/ml during the selection process. After 7 days, Geneticin-resistant cells were collected by trypsinization. Purified clones were isolated from this pool of cells by limiting dilution cloning, for which it was necessary to use medium supplemented (30%) with conditioned medium (see below). Cell lines exhibiting tetracycline-inducible opsin expression were identified by immunodetection.
Conditioned medium for limited dilution cloning was prepared as follows. WT HEK293S cells were grown to 90% confluence in dishes. The medium was replaced with new medium and incubation was continued for a further 24 h. This conditioned medium was then collected and filtered (0.2-μm membrane) before storage at −20°C.
[35S]Methionine Pulse–Chase in Examination of Recombinant Rhodopsin Maturation in Stable Cell HEK293S Cell Lines.
HEK293S stable cell lines containing the inducible opsin gene were grown to 90% confluence in medium containing Geneticin (200 μg/ml) by using 10-cm dishes. The growth medium was then replaced with fresh medium containing tetracycline (2 μg/ml) and sodium butyrate (5 mM) without Geneticin. The next day the cells were grown for 1 h in the same DMEM induction medium but without methionine and cysteine and supplemented with 10% FBS that had been dialyzed with a 1-kDa exclusion membrane. The medium was then replaced with the same medium (5 ml) containing [35S]methionine (100 μCi Translabel; 1 Ci = 37 GBq). After incubation for 40 min (pulse time) the medium was replaced with DMEM containing unlabeled methionine (60 μg/ml) and supplemented with nondialyzed FBS but no tetracycline or sodium butyrate. After further incubations for specified time intervals, cells were pelleted and frozen immediately. Cells were solubilized in buffer B (1%) and opsin was purified by immunoaffinity chromatography. Purified opsin samples were examined by SDS/PAGE (10%) followed by autoradiography. The gels were soaked in Fluoro-Hance (Research Products International, Mount Prospect, IL) for 30 min and dried. Autoradiography was by exposure to Biomax film for 48 h.
Growth of 293S Cell Line Suspension Cultures in a Bioreactor.
HEK293S GnTI− stable cell lines harboring the inducible opsin genes were expanded in medium containing Geneticin (G418) (200 μg/ml). Cells from six 15-cm dishes were used to inoculate 1.1 liters of medium in a bioreactor (3).
Results
Construction of a RicinR HEK293S Cell Line Performing Limited N-Glycosylation.
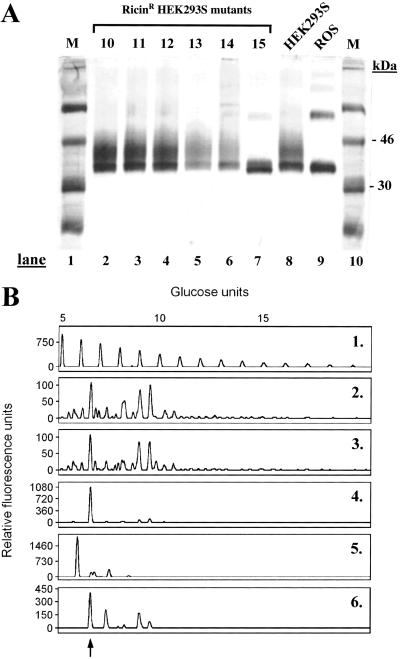
HEK293S cells growing exponentially were treated with the mutagen EMS and then incubated in the presence of ricin as described in Materials and Methods. After incubation for 2–3 weeks, ricinR colonies formed in growth media containing ricin at concentrations of 1 or 10 ng/ml. No colonies were detected in dishes containing the higher concentrations, 100 ng/ml and 1 μg/ml, of ricin. The frequency of ricinR colony formation at 10 ng/ml ricin concentration was 1–2 times per 10-cm dish, whereas the frequency of ricinR colony formation at 1 ng/ml ricin concentration was 10–20 times higher. Isolated ricinR HEK293S cell colonies were collected by using cloning rings and expanded in the absence of ricin. On reaching confluence in 10-cm dishes, ricinR HEK293S cell lines were transiently transfected (Materials and Methods) by using a plasmid containing the WT opsin gene. Rhodopsin was prepared from 14 ricinR cell lines obtained in the presence of 1 ng/ml ricin and one ricinR cell line obtained in medium containing 10 ng/ml ricin. Purified rhodopsin was analyzed by SDS/PAGE followed by visualization by silver stain (Fig. 1A). Rhodopsin prepared from transiently transfected WT HEK293S cells (Fig. 1A, lane 8) migrated at about 34 kDa with a trailing smear. This SDS/PAGE pattern is characteristic of rhodopsin purified from transiently transfected COS-1 cells (8). The SDS/PAGE migration pattern of rhodopsin prepared from all of the HEK293S ricinR cell lines obtained in the presence of 1 ng/ml ricin (Fig. 1A, lanes 2–6) was similar to that prepared from the WT HEK293S cell line (lane 8). However, rhodopsin prepared from the single HEK293S ricinR cell line obtained in the presence of 10 ng/ml ricin (Fig. 1A, lane 7) migrated as a slightly faster doublet without a trailing smear. This mobility resembles that of rhodopsin prepared from bovine rod outer segments (ROS) (Fig. 1A, lane 9).
Figure 1.
Characterization of rhodopsin and its N-glycans prepared from ricinR HEK293S cell lines. (A) HEK293S ricinR cell lines expressing the opsin gene by transient transfection were used for purification of rhodopsin (Materials and Methods). Rhodopsin was characterized by SDS/PAGE (10%) followed by visualization of proteins by silver stain. Lanes 1 and 10 (M) (molecular weight standards); lanes 2–6, rhodopsin purified from HEK293S ricinR cell lines obtained by using 1 ng/ml ricin; lane 7, rhodopsin purified from a ricin cell line obtained by using 10 ng/ml ricin; lane 8, rhodopsin from WT HEK293S cells; lane 9, rhodopsin from bovine ROS. (See text for further details.) (B) Rhodopsin was prepared as described above. N-glycans removed by treatment with PNGaseF were analyzed as in Materials and Methods. Profile 1, profile of molecular weight standard (malto-oligosaccharides) and the corresponding number of glucose units indicated above the profile. N-glycan profiles of rhodopsin purified from HEK293S (profile 2), one HEK293S ricinR cell line at 1 ng/ml ricin concentration (profile 3), the HEK293S ricinR cell line at 10 ng/ml ricin concentration (profile 4), and bovine ROS (profile 5). The profile of RNase B N-glycans is in profile 6; the peak corresponding to Man5GlcNAc2 is indicated by an arrow. The peaks show relative fluorescence intensity corresponding to eluting N-glycans.
Analysis of N-Glycans Prepared from Rhodopsin Mutants by Transient Transfection of RicinR HEK293S Cell Lines.
N-glycans were released from rhodopsin mutants by treatment with PNGaseF, labeled with the fluorescent dye, APTS (Materials and Methods), and separated by PAGE. The results are shown in Fig. 1B. Profile 1 shows the mobilities of malto-oligosaccharide molecular weight standards. Rhodopsin purified from the WT HEK293S cell line (Fig. 1B, profile 2) carries heterogeneous N-glycans as indicated by numerous peaks that correspond to N-glycans of various size and monosaccharide content.
Rhodopsin prepared from all other HEK293S ricinR cell lines similarly obtained displayed a very similar N-glycan profile as WT HEK293S rhodopsin (data not shown). An example of _N_-glycan from rhodopsin prepared from ricinR cell lines obtained by using 1 ng/ml ricin concentration is shown in Fig. 1B, profile 3. Profile 4 is analysis of rhodopsin N-glycan prepared from the HEK293S ricinR cell line obtained at 10 ng/ml ricin concentration. In this case, one N-glycan predominates and the single peak corresponds to 7 monosaccharide units. This peak has the same mobility as that of the Man5GlcNAc2 peak in the N-glycan mixture prepared from RNase B (Fig. 1B, profile 6). The N-glycan profile of ROS rhodopsin is shown in lane 5.
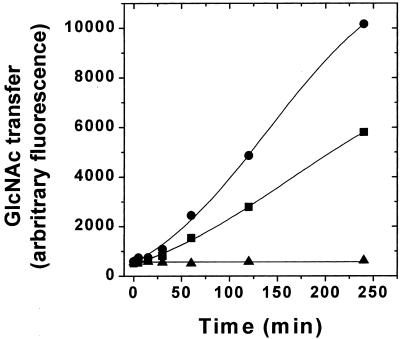
HEK293S RicinR-Resistant Mutant Has Undetectable GnTI Activity.
GnTI activity was measured by monitoring the transfer of GlcNAc from the donor (UDP-GlcNAc) to a fluorescently tagged acceptor (Man5GlcNAc2-APTS). The product (GlcNAcMan5GlcNAc2-APTS) was separated from the acceptor by PAGE as described in Materials and Methods. GnTI activity assays were performed by using Triton X-100-solubilized cell extracts (11) prepared from WT HEK293S cells or the HEK293S ricinR mutant described in the preceding section. The results are shown in Fig. 2. The WT HEK293S cell extract contains GnTI activity that catalyzes conversion of Man5GlcNAc2-APTS to GlcNAcMan5GlcNAc2-APTS in a time-dependent manner. No GnTI− activity could be detected in the HEK293S ricinR mutant. Combination of equal parts of the two cell extracts results in a GnTI activity that is approximately half of the WT value. This result demonstrates that the solubilized cell extract prepared from the HEK293S ricinR mutant does not contain an inhibitor of GnTI. Thus, the HEK293S ricinR mutant is completely deficient for GnTI activity, and this defect most likely arises from mutations in both GnTI alleles. The HEK293S ricinR mutant deficient in GnTI activity will be hereafter referred to as HEK293S GnTI−.
Figure 2.
Assay of GnTI enzyme activity in the cell extracts. Triton X-100-solubilized cell extracts (Materials and Methods) were prepared from HEK293S WT or the HEK293S GnTI− (ricinR) cell line obtained by using 10 ng/ml ricin. The reaction mixtures contained 23.7 μg of HEK293S WT protein, 25.8 μg of HEK293S GnTI− (ricinR) protein, or 24.7 μg of protein of a mixture (1:1) of each lysate. GnTI enzymatic activity was measured by monitoring transfer of GlcNAc from UDP-GlcNAc to a fluorescently tagged acceptor (Man5GlcNAc2-APTS) as described in Materials and Methods. Time-dependent formation of the product (GlcNAcMan5GlcNAc2-APTS) was detected by analysis of samples by PAGE by using an ABI377 sequencer system (Materials and Methods). Samples analyzed were prepared from HEK293S (circles), HEK293S GnTI− (ricinR) (triangles), or a 1:1 mixture of each lysate (squares).
Construction of an HEK293S GnTI− Stable Cell Line for Tetracycline-Inducible Opsin Expression.
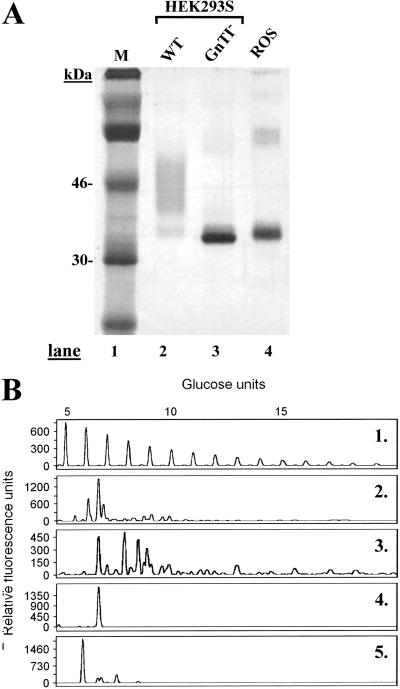
Construction of an HEK293S GnTI− cell line expressing stably the tetR gene was done by transfection with plasmid pCDNA6-TR as described in Materials and Methods (3). An HEK293S GnTI− cell line expressing tetR at high levels was identified. This cell line was expanded and then transfected with plasmid pACMVtetO-rho (Materials and Methods). Stable cell lines were derived from colonies arising from single cells by using limiting dilution cloning. One cell line displaying high levels of tetracycline-inducible WT opsin gene expression was identified and used in all subsequent experiments. Rhodopsin as expressed by this cell line was analyzed by SDS/PAGE and visualized by silver stain (Fig. 3A). Rhodopsin prepared after inducible opsin gene expression by using the WT HEK293S cell line and ROS rhodopsin served as markers for comparison. Rhodopsin prepared from the WT HEK293S cell line migrated as a smear, whereas rhodopsin prepared from the HEK293S GnTI− stable cell line migrated predominantly as a single species (Fig. 3A, lanes 2 and 3). The protein from the GnTI− cell line migrates slightly faster than ROS rhodopsin (Fig. 3A, lane 4).
Figure 3.
Characterization of rhodopsin and rhodopsin N-glycans prepared from HEK293S and HEK293S-GnTI− stable cell lines. The cell lines were induced for 2 days by using tetracycline and sodium butyrate. (A) Rhodopsin was purified and examined by SDS/PAGE (10%) followed by silver stain. Lane 1, M (molecular weight standards); lane 2, rhodopsin from inducible HEK293S; lane 3, rhodopsin from HEK293S GnTI−, and lane 4, rhodopsin from ROS. (B) Characterization of rhodopsin N-glycan composition. Profile 1, mobilities of molecular weight standards (malto-oligosaccharides) used for calibration. Profiles 2 and 3, N-glycans from rhodopsin samples prepared from inducible HEK293S cell line (before and after treatment with sialidase, respectively); profile 4, rhodopsin from HEK293S GnTI−; and profile 5, ROS rhodopsin.
The N-glycans in the rhodopsin samples above were then analyzed. As seen in profiles 2 and 3, rhodopsin made by the WT HEK293S stable cell line has extremely heterogeneous glycans, which is further demonstrated after removal of sialic acid residues by treatment with sialidase (profile 3). In contrast, rhodopsin prepared from the HEK293S-GnTI−-inducible cell line gives a sharp band indicating a single glycan species (Fig. 3B, lane 4). The mobility of this species corresponds to Man5GlcNAc2. The N-glycan in bovine ROS rhodopsin carries mainly GlcNAc2Man3GlcNAc (12) (Fig. 3B, lane 5). The mobility of this glycan relative to that from HEK293S-GnTI− corresponds to one less sugar unit.
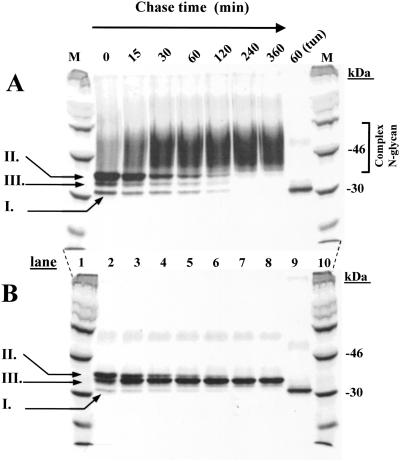
Analysis of Rhodopsin N-Glycan Maturation in HEK293S Stable Cell Lines by Pulse–Chase.
WT HEK293S and HEK293S-GnTI−-inducible cell lines were treated with tetracycline and sodium butyrate to induce opsin gene expression and then incubated in the medium containing [35S]methionine (Materials and Methods) for 40 min. Cells were then incubated in the presence of unlabeled methionine for specified times (Fig. 4) and the opsins were purified (Materials and Methods). Opsin made by WT HEK293S cells (Fig. 4A) appears first (0- and 15-min chase) as three distinct species (Fig. 4A, lanes 2 and 3) with partial conversion to the complex form as seen by the increasing smear. The three initial protein bands presumably are nonglycosylated (species I), a major species (species II) containing the initial N-glycan, GlcNAc3Man5GlcNAc2, and species III of intermediate size, presumably a trimming intermediate. Increasing chase times (Fig. 4A, lanes 4–8) show conversion of the species above to the complex form (40- to 60-kDa smear), the process being complete within 4 h. In a separate experiment, shown in Fig. 4A, lane 9, opsin was prepared from a tunicamycin-treated cell line. This nonglycosylated opsin corresponded to species I described above. As seen in Fig. 4B, lane 2, the HEK293SGnT1− opsin initially formed the same pattern as described above for WT HEK293S cells, namely species I–III, but with no trailing smear. Species II is converted to species III, which then accumulates. Probably, conversion of species II to species III represents trimming of the Man9GlcNAc2 form to the Man5GlcNAc2 form.
Figure 4.
Monitoring of opsin N-glycan formation and processing by pulse–chase. (A) Inducible HEK293S cell line. Opsin expression was induced, the total mixture pulsed with [35S]methionine for 40 min, and chased with cold methionine for times indicated. 35S-methionine-labeled opsin was purified from DM-solubilized cell extracts by using rho-1D4-Sepharose. Samples were examined by SDS/PAGE (lanes 2–9) and gels were dried before autoradiography. 14C-labeled markers (M) are in lanes 1 and 10. (B) The protocol was the same as in A except that the HEK293S GnTI− cell line was used. For details, see the text.
Large-Scale Growth of the HEK293S-GnTI− Cell Line for High-Level Inducible Expression of WT Rhodopsin.
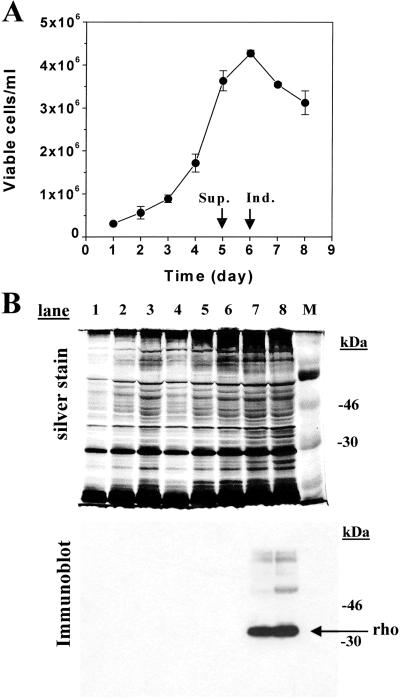
The HEK293S GnTI− cell line expressing inducibly the wild-type opsin gene was grown in suspension by using a 1.1-liter bioreactor culture. The growth profile is shown in Fig. 5A. The maximum viable cell count reached on day 6 is 4 × 106 cells per ml. Samples were collected throughout the growth curve and analyzed by SDS/PAGE (10%). Proteins were detected by silver stain (Fig. 5B Upper), whereas opsin was specifically detected by immunodetection by using mAb rho-1D4 (Fig. 5B Lower). Opsin was only detected after addition of tetracycline and sodium butyrate to the growth medium (Fig. 5B Lower, lanes 7 and 8) and migrates predominantly as a sharp band. Slower-migrating species presumably correspond to the opsin dimer and trimer. After harvest of cells and constitution with 11-_cis_-retinal to rhodopsin, the expression level of the opsin was calculated to be 6 mg/liter, comparable with the expression level of opsin in the HEK293S tetracycline-inducible cells (3).
Figure 5.
Growth and characterization of rhodopsin from inducible HEK293S GnTI− cell line prepared in 1.1-liter suspension culture by using a bioreactor. The cell line was grown in suspension culture in a bioreactor (Materials and Methods). (A) Viable cell count obtained by microscopic examination of samples removed from the bioreactor at 24-h intervals. The culture was supplemented (Sup.) and opsin expression was induced (Ind.) as indicated by arrows and described in Materials and Methods in ref. 3. (B) Samples of cells collected throughout growth of the culture were solubilized buffer B, and the proteins were separated by SDS/PAGE (10%) and visualized by silver stain. Proteins were transferred from gels to nitrocellulose by electroblotting and opsin was visualized by immunoblotting with anti-rhodopsin mAb rho-1D4. The protein band corresponding to rhodopsin is indicated by the arrow. Lanes 1–8 are samples from days 1–8.
Purification and Characterization of Rhodopsin Containing Man5GlcNAc2 N-Glycan.
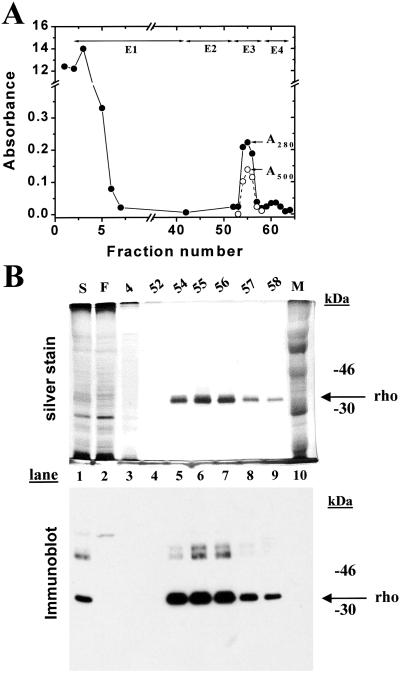
One-third portion of the harvested cells from the bioreactor run above was treated with 11-_cis_-retinal (8, 13) to measure rhodopsin concentration. The cells were solubilized by using nonylglucoside (2%) as in Materials and Methods (3). The supernatant from the cell extract was treated with Sepharose-rho-1D4 and after loading the beads into a column, elutions were performed, the profile being shown in Fig. 6A. Rhodopsin eluted in elution volumes 2, 3, and 4 in salt-free buffer containing the epitope peptide, E3. A small amount of _A_280 absorbing material eluted in high salt shown in Fig. 6A. Analysis of the eluted proteins by SDS gel is shown in Fig. 6B. Rhodopsin migrated mainly as a single species and the _A_280/_A_500 nm ratio of fraction 3 was 1.6, corresponding to that of pure rhodopsin. Recovery of rhodopsin was 1.5 mg from about 2 mg (75%) of rhodopsin present in the solubilized extract.
Figure 6.
Purification of WT rhodopsin containing only the Man5GlcNAc2 N-glycan. HEK293S GnTI− cells were collected from the bioreactor after induction of the opsin gene for 2 days (Fig. 5). A portion of the cells (one-third) were resuspended in buffer A and treated with 11-_cis_-retinal. For solubilization nonylglucoside was added to produce the composition of buffer G. Elution buffers were E1, buffer H; E2, buffer I; E3, buffer J; E4, buffer K. Absorption at 280 nm (closed circles) and 500 nm (open circles) was recorded as indicated. (Inset) SDS/PAGE (10% gel) examination of selected fractions eluted from the column as visualized by silver stain and immunoblot after electroblotting to nitrocellulose. M, molecular weight standards; S, solubilized cell extract; F, flowthrough (proteins that do not bind to rho-1D4-Sepharose). Migration of rhodopsin (rho) is indicated by an arrow.
Purification of the Rhodopsin Mutant, E113Q/E134Q/M257Y, Containing Only the Man5GlcNAc2 N-Glycan.
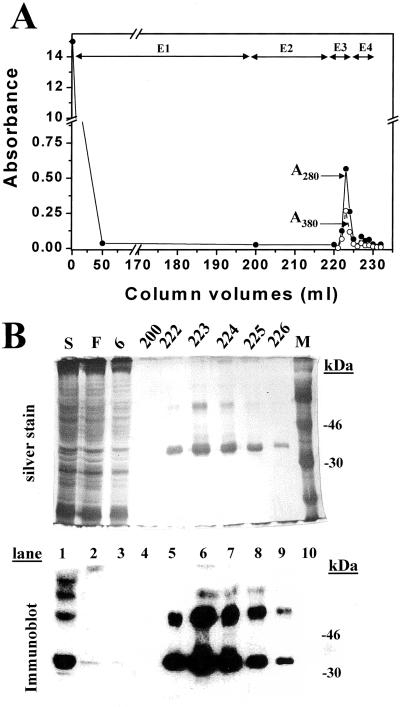
An HEK293S GnTI− cell line was constructed for tetracycline-inducible expression of the rhodopsin mutant, E113Q/E134Q/M257Y. This cell line was grown in a 1.1-liter bioreactor culture, and opsin expression was induced by addition of tetracycline and sodium butyrate (3). One-third portion of the cells was treated with 11-_cis_-retinal and the cells solubilized by using 1% DM. The rhodopsin mutant protein was purified by rho-1D4-Sepharose chromatography (Fig. 7A). The successive elutions used are described in the Fig. 7 legend. The total yield of rhodopsin obtained was close to 1 mg. SDS/PAGE followed by silver staining and immunoblotting of rhodopsin fractions demonstrated that N-glycosylation was uniform (Fig. 7B).
Figure 7.
Purification of rhodopsin mutant E113Q/E134Q/M257Y from the HEK293S GnTI− stable cell line. Column profile showing _A_280 and _A_500. Fractions (1.5 ml) were collected and washes were E1 (buffer C), E2 (buffer D, pH 7.2), E3 as E2 + 100 μM 9-mer rhodopsin elution peptide, and E4 as E1 + 100 μM elution peptide. (Inset) SDS/PAGE (10%) analysis of selected column fractions with detection either by silver stain or by immunoaffinity with rho-1D4. S, solubilized cell lysate; F, column flowthrough; M, molecular weight markers.
Discussion
The potential importance of the work described in this report derives from two main considerations. First, N-glycosylation at essentially conserved sites is a structural feature of the majority of all GPCR. Second, because of the extremely restricted availability of GPCR from natural sources, further advances in structure–function work can only result from development of suitable expression systems, in particular, for potential crystallization attempts. Thus, the nature and extent of N-glycosylation during expression become critically important. Two alternative possibilities for controlling N-glycan synthesis arise. The first is to abolish N-glycosylation altogether, either by mutagenic removal of the consensus tripeptide sequence sites (14) or by preventing N-glycosylation in the presence of tunicamycin (3). However, we are working with the two plausible assumptions. One, that the heterogeneous complex N-glycans produced in the WT HEK293S cell lines are not acceptable for crystallization attempts on the expressed proteins and, two, that some glycosylation, restricted and homogeneous, such as that present in ROS rhodopsin may actually be essential in stabilization of the folded expressed proteins and, consequently, for attempts at their crystallization.
In the light of the considerations above for future structural work with rhodopsin mutants and, in particular, GPCR, the main accomplishment of the work now reported is the development of a system for large-scale expression that integrates the elements of induction by tetracycline into a stable cell line that does not allow the formation of complex and heterogeneous N-glycans.
Acknowledgments
We thank Professor U. L. RajBhandary of our Biology Department for valuable discussions. We appreciate the assistance of Ms. Judy Carlin in the preparation of this manuscript. This work was supported by National Institutes of Health Grant GM28289 (to H.G.K.).
Abbreviations
GnTI
_N_-acetylglucosaminyltransferase-I
APTS
8-amino-1,3,6-pyrenetrisulfonic acid
EMS
ethyl methanesulfonate
ROS
rod outer segments
DM
_n_-dodecyl-β-d-maltoside
ricinR
ricin-resistant
Footnotes
¶
This is paper 53 in the series “Structure–Function in Rhodopsin.” The preceding paper is ref. 3.
References
- 1.Reeves P J, Thurmond R L, Khorana H G. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves P J, Klein-Seetharaman J, Getmanova E V, Eilers M, Loewen M C, Smith S O, Khorana H G. Biochem Soc Trans. 1999;27:950–955. doi: 10.1042/bst0270950. [DOI] [PubMed] [Google Scholar]
- 3.Reeves P J, Kim J-M, Khorana H G. Proc Natl Acad Sci USA. 2002;99:13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley P, Sallustio S, Kragg S S, Dunn B. Somat Cell Mol Genet. 1990;16:211–223. doi: 10.1007/BF01233357. [DOI] [PubMed] [Google Scholar]
- 5.Stanley P, Chaney W. Mol Cell Biol. 1985;5:1204–1211. doi: 10.1128/mcb.5.6.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathans J. Biochemistry. 1990;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- 7.Maras M, Callewaert N, Piens K, Claeyssens M, Martinet W, Dewaele S, Contreras H, Dewerte I, Penttila M, Contreras R. J Biotechnol. 2000;77:255–263. doi: 10.1016/s0168-1656(99)00222-9. [DOI] [PubMed] [Google Scholar]
- 8.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 10.Callewaert N, Geysens S, Molemans F, Contreras R. Glycobiology. 2001;11:275–281. doi: 10.1093/glycob/11.4.275. [DOI] [PubMed] [Google Scholar]
- 11.Vischer P, Hughes R C. Eur J Biochem. 1981;117:275–284. doi: 10.1111/j.1432-1033.1981.tb06334.x. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda M N, Papermaster D S, Hargrave P A. Methods Enzymol. 1982;81:214–223. doi: 10.1016/s0076-6879(82)81034-3. [DOI] [PubMed] [Google Scholar]
- 13.Reeves P J, Hwa J, Khorana H G. Proc Natl Acad Sci USA. 1999;96:1927–1931. doi: 10.1073/pnas.96.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaushal S, Ridge K D, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]