Differentiation of Closely Related Carnobacterium Food Isolates Based on 16S-23S Ribosomal DNA Intergenic Spacer Region Polymorphism (original) (raw)
Abstract
A novel strategy for identification of Carnobacterium food isolates based on restriction fragment length polymorphism (RFLP) of PCR-amplified 16S-23S ribosomal intergenic spacer regions (ISRs) was developed. PCR amplification from all Carnobacterium strains studied always yielded three ISR amplicons, which were designated the small ISR (S-ISR), the medium ISR (M-ISR), and the large ISR (L-ISR). The lengths of these ISRs varied from one species to another. Carnobacterium divergens NCDO 2763T and C. mobile DSM 4849T generated one major S-ISR band (ca. 400 bp) and minor M-ISR and L-ISR bands (ca. 500 and ca. 600 bp, respectively). The ISRs amplified from C. gallinarum NCFB 2766T and C. piscicola NCDO 2762T were larger (S-ISR, ca. 600 bp; M-ISR, ca. 700 bp; and L-ISR, ca. 800 bp). The L-ISR contained two tDNAs coding for tRNAIle and tRNAAla genes. The M-ISR included one tRNAAla gene, and the S-ISR did not contain a tDNA gene. The RFLP scheme devised involves estimation of variable PCR product sizes together with _Hin_fI, _Taq_I, and _Hin_dIII restriction analysis. Forty-two isolates yielded four unique band patterns that correctly resolved these isolates into four Carnobacterium species. This method is very suitable for rapid, low-cost identification of a wide variety of Carnobacterium species without sequencing.
Lactic acid bacteria (LAB) have been the focus of extensive research due to their value in the food-processing industry (14, 27). One of the most recent taxonomic additions to the LAB group is the genus Carnobacterium. Members of this genus are widespread in nature, and the habitats of these organisms range from foods, such as poultry, meat, cheese, and seafood, to fish intestines and anoxic Antarctic lake water (2, 10, 16, 26, 29). The four species of Carnobacterium isolated from food are Carnobacterium divergens, C. piscicola, C. gallinarum, and C. mobile (6). Most research on the genus Carnobacterium has been focused on the production of bacteriocins and the regulation of metabolic enzymes (9, 28). In the last few years, many ways to differentiate Carnobacterium species have been developed. The phylogenetic interrelationships of species in the genus Carnobacterium have been investigated by using numerical taxonomic matrices and sequencing of the 16S rrn segments encoding mature rRNAs (19, 41). The Carnobacterium species form a phylogenetically coherent group, which is quite distinct from all other LAB. However, the number of polymorphic sites in the 16S rRNA of Carnobacterium species is rather low, since some species have the same sequence (the sequences of C. piscicola and C. gallinarum are 98% identical) and other species exhibit very high degrees of sequence similarity (the sequences of C. divergens and C. mobile are 96% similar) (41). Thus, it is difficult to define specific 16S RNA rrn sequences that can be used for differentiation of these closely related species. Carnobacteria have been identified at the genus level by nucleic acid hybridization by using 16S rRNA-targeted genus-specific probes (32). Species-specific primers have been designed by using the domains that exhibit low homology in the 16S ribosomal DNA (rDNA) sequences of species of Carnobacterium (1, 4). However, the PCR primers used in these studies were not specific enough to differentiate Carnobacterium spp. from other genera of LAB. Data obtained with the randomly amplified polymorphic DNA PCR technique enable only C. divergens to be differentiated from other Carnobacterium species (20).
In view of this, a study of the more variable sequences in the rRNA operon of phylogenetically closely related Carnobacterium species could be useful. In most prokaryotes, the ribosomal genes form an operon with the order 5′-16S-23S-5S-3′, and the genes are separated by two intergenic spacer regions (ISRs). ISRs, especially those located between the 16S and 23S rDNAs, are thought to be under less evolutionary pressure and, therefore, to provide greater genetic variation than rRNA coding regions (13, 17). Thus, the 16S-23S ISR is an important tool for developing DNA-based typing methods because it varies in length and sequence from one species to another (11, 31, 37, 40). The heterogeneity of rRNA operons is mainly due to the presence of several functional units within the operons, such as tRNA genes (18, 43). Therefore, the genetic variations in ISRs are not only interstrain variations but also intercistronic variations (7). Analysis of the sequences of the ribosomal operons (rrn) is a method of choice for determining phylogenetic relationships among organisms. Even so, the high expense makes sequencing unacceptable for general use. This leads to problems in the development of simpler analysis methods, such as PCR-restriction fragment length polymorphism (RFLP) analysis of the 16S-23S ISR.
The aim of the present study was to investigate the suitability of the 16S-23S ISR in the four closely related Carnobacterium species that are of interest to the food industry, C. divergens, C. piscicola, C. gallinarum, and C. mobile, for establishing a rapid PCR-RFLP-based identification scheme. The first goal was to design universal primers for genus-specific amplification and evaluation of the ISR length polymorphism in these species. Restriction analysis of the ISRs was used to develop a reliable diagnostic algorithm for identification of a broad spectrum of Carnobacterium species with one to three endonucleases. The degree of interspecies divergence and the intraspecies discriminatory power of ISR sequences were investigated by using 42 strains belonging to four species. 16S-23S ISRs of the type strains of four Carnobacterium species were sequenced to examine the variation found in wider phylogenetic gaps.
MATERIALS AND METHODS
Bacterial strains.
The strains of the genus Carnobacterium used in this study are listed in Table 1. C. divergens NCDO 2763, C. piscicola NCDO 2762, C. gallinarum NCFB 2766, and C. mobile DSM 4849 are type strains. Thirty-eight Carnobacterium strains isolated from different habitats were tested in order to evaluate the molecular differentiation and degree of intraspecific diversity in the genus. LAB other than Carnobacterium were chosen in order to improve our PCR strategy. The bacteria tested in this study represented 175 strains (32 reference strains and 143 isolates) belonging to 40 defined species in seven genera (Table 2). The strains were stored as 15% (wt/vol) glycerol stock cultures at −80°C in MRS medium. Cultures were grown at 30°C for 24 to 36 h in brain heart infusion medium, M17, or MRS medium (Biokar). Carnobacterium strains were identified by phenotypic tests as described by Montel et al. (30).
TABLE 1.
Carnobacterium strains used in this study
| Species | Straina | Source (reference) |
|---|---|---|
| C. divergens | NCDO 2763T | Meat (30) |
| C. divergens | INRA 541 | Meat (30) |
| C. divergens | INRA 695 | Meat (30) |
| C. divergens | INRA 507 | Meat (30) |
| C. divergens | INRA 548 | Meat (30) |
| C. divergens | INRA 733 | Meat (30) |
| C. divergens | INRA 508 | Meat (30) |
| Identified as C. piscicola | This study | |
| C. divergens | INRA 524 | Meat (30) |
| C. divergens | INRA 586 | Meat (30) |
| Identified as C. piscicola | This study | |
| C. divergens | INRA 687 | Meat (30) |
| C. divergens | INRA 515 | Meat (30) |
| Identified as C. piscicola | This study | |
| C. divergens | INRA 337 | Meat (30) |
| C. divergens | ENITIAA V41 | Fish (34) |
| C. mobile | DSM 4849T | Poultry |
| C. mobile | CIP 103159 | Poultry |
| C. piscicola | NCDO 2762T | |
| C. piscicola | IFREMER 644 | Smoked salmon (23) |
| C. piscicola | IFREMER 665 | Smoked salmon (23) |
| C. piscicola | IFREMER 666 | Smoked salmon (23) |
| C. piscicola | IFREMER 692 | Smoked salmon (23) |
| C. piscicola | IFREMER 694 | Smoked salmon (23) |
| C. piscicola | IFREMER 668 | Smoked salmon (23) |
| C. piscicola | INRA 545 | Meat (30) |
| Identified as C. divergens | This study | |
| C. piscicola | INRA 725 | Meat (30) |
| C. piscicola | INRA 722 | Meat (30) |
| Identified as C. divergens | This study | |
| C. piscicola | INRA 501 | Meat (30) |
| C. piscicola | INRA 527 | Meat (30) |
| C. piscicola | INRA 572 | Meat (30) |
| Identified as C. divergens | This study | |
| C. piscicola | INRA 543 | Meat (30) |
| C. piscicola | INRA 528 | Meat (30) |
| C. piscicola | ENITIAA V1 | Trout (34) |
| C. piscicola | INRA 336 | Meat (30) |
| C. piscicola | INRA 338 | Meat (30) |
| C. piscicola | INRA 526 | Meat (30) |
| C. piscicola | ENSAIA 1 | Cheese (29) |
| C. piscicola | ENSAIA 7 | Cheese (29) |
| C. divergens | ENSAIA 13 | Cheese (29) |
| Identified as C. piscicola | This study | |
| C. piscicola | ENSAIA 16 | Cheese (29) |
| C. piscicola | ENSAIA 29 | Cheese (29) |
| C. piscicola | ENSAIA 32 | Cheese (29) |
| C. gallinarum | NCDO 2766T | Poultry (6) |
| C. gallinarum | INRA 680 | Poultry (6) |
TABLE 2.
Numbers and lengths of PCR-amplified 16S-23S ISRs obtained with primers 16S/4 and 23S/7 from 175 strains of LAB isolated from food
| Species or subspecies | Reference strain(s) | No. of isolates | No. of ISR PCR products | Length(s) of PCR product(s) containing ISR (bp) | |
|---|---|---|---|---|---|
| Carnobacterium | divergens | NCDO 2763T | 14 | 3 | 400, 500, 600 |
| Carnobacterium | piscicola | NCDO 2762T | 24 | 3 | 600, 700, 800 |
| Carnobacterium | gallinarum | NCDO 2766T | 2 | 3 | 600, 700, 800 |
| Carnobacterium | mobile | DSM 4849T | 2 | 3 | 400, 500, 600 |
| Enterococcus | faecium | ATCC 19434 | 2 | 2 | 550, 650 |
| Enterococcus | faecalis | CIP 103015, JH 22 | 2 | 2 | 450, 600 |
| Lactobacillus | sakei | ATCC 15531, ATCC 15521, ATCC 3453 | 12 | 2 | 450, 650 |
| Lactobacillus | acidophilus | ATCC 4356 | 5 | 2 | 450, 650 |
| Lactobacillus | alimentarius | DSM 20181 | 8 | 2 | 450, 650 |
| Lactobacillus | brevis | CIP 102806T, ATCC 3548 | 17 | 2 | 450, 650 |
| Lactobacillus | buchneri | ATCC 9460 | 2 | 2 | 450, 650 |
| Lactobacillus | curvatus | DSM 20019, CIP 102986T | 7 | 2 | 450, 650 |
| Lactobacillus | delbrueckuii subsp. bulgaricus | CIP 101027 | 2 | 2 | 450, 650 |
| Lactobacillus | delbrueckuii subsp. lactis | CIP 101028 | 2 | 2 | 450, 650 |
| Lactobacillus | farciminis | DSM 20180 | 3 | 2 | 450, 650 |
| Lactobacillus | kefir | ATCC 35411 | 6 | 2 | 450, 650 |
| Lactobacillus | helveticus | CIP 103146T | 4 | 2 | 450, 650 |
| Lactobacillus | kefiranofaciens | ATCC 43761 | 2 | 2 | 450, 650 |
| Lactobacillus | parakefiri | CIP 104242T | 2 | 2 | 450, 650 |
| Lactobacillus | kefirgranum | CIP 104241T | 2 | 2 | 450, 650 |
| Lactobacillus | casei | ATCC 11578T | 4 | 2 | 450, 650 |
| Lactobacillus | pentosus | ATCC 8041 | 2 | 2 | 450, 650 |
| Lactobacillus | plantarum | ATCC 14917, NCDO 343 | 19 | 2 | 450, 650 |
| Lactobacillus | graminis | CIP 105164T | 1 | 2 | 450, 650 |
| Lactococcus | lactis subsp. lactis | ATCC 11454 | 3 | 1 | 500 |
| Lactococcus | lactis subsp. cremoris | CNRZ 117 | 3 | 1 | 500 |
| Leuconostoc | mesenteroides subsp. mesenteroides | DSM 20200, DSM 20240, DSM 20241 | 8 | 1 | 600 |
| Leuconostoc | mesenteroides subsp. dextranicum | DSM 20484 | 4 | 1 | 600 |
| Leuconostoc | pseudomesenteroides | DSM 20193 | 2 | 1 | 600 |
| Leuconostoc | lactis | DSM 20192 | 2 | 1 | 600 |
| Weissella | confusa | DSM 20186 | 2 | 3 | 450, 550, 650 |
| Streptococcus | thermophilus | CNRZ 216 | 5 | 1 | 550 |
Escherichia coli INV αF′ (Invitrogen), which was used for cloning procedures, was grown in Luria-Bertani broth (0.1% tryptone, 0.1% sodium chloride, 0.05% yeast extract; pH 7.0) for 16 h at 37°C. Transformed E. coli was grown on Luria-Bertani agar plates containing 100 μg of ampicillin/ml, 0.5 mM isopropyl-β-d-thiogalactopyranoside, and 80 μg of 5-bromo-4-chloro-3-indolyl-β-galactopyranoside/ml.
PCR primers for ISR amplification.
The ISRs were amplified with primers 16S/p2 (5′-CTTGTACACACCGCCCGTC-3′) and 23S/p10 (5′-CCTTTCCCTCACGGTACTG-3′), which anneal to positions 1388 to 1406 of the 16S rRNA gene and to positions 456 to 474 of the 23S rRNA gene (E. coli numbering; GenBank accession number V00331), respectively. The resultant PCR products contained the complete 16S-23S ribosomal ISR and parts of the flanking rDNAs (ca. 150 bp of 16S rDNA and 474 bp of 23S rDNA).
In order to improve the specificity of 16S-23S ISR PCR amplification, the PCR products were used as templates in a second PCR (nested PCR) with primers 16S/p4 (5′-GCTGGATCACCTCCTTTCT-3′) and 23S/p7 (5′-GGTACTTAGATGTTTCAGTTC-3), which anneal to positions 1526 to 1542 of the 16S rRNA gene and positions 207 to 189 of the 23S rRNA gene (E. coli numbering), respectively. Primer 23S/p7 was designed by using sequences of the 23S gene conserved in various bacteria (11). The DNA that was PCR amplified with primers 16S/p4 and 23S/p7 contained the complete 16S-23S ribosomal ISR and parts of the flanking rDNAs (ca. 17 bp of 16S rDNA and 207 bp of 23S rDNA). The tRNAAla primer (5′-TAGCTCAGCTGGGAGAGC-3′) was designed by using a conserved sequence of the tDNAAla gene located in 16S-23S ISR of Oenococcus oeni (22). The PCR products obtained after amplification with the tRNAAla and 23S/p10 primers were used for ISR-RFLP analysis.
DNA isolation and PCR.
Chromosomal DNA was isolated as previously described by Tudor et al. (39). The resultant DNA samples were used for PCR amplification (ca. 50 ng of DNA per PCR mixture) performed with a PTC-100 thermocycler (MJ Research). The PCR mixture (total volume, 50 μl) contained 5 μl of 10× PCR buffer, 2 μl of a mixture containing each deoxynucleoside triphosphate at a concentration of 0.25 mM, each primer at a concentration of 0.3 μM, 2.5 mM MgCl2, and 1 U of Taq DNA polymerase (Appligene). The amplification reaction consisted of a 60-s denaturation step at 94°C, a 75-s annealing step at 60°C, and a 75-s extension step at 72°C. The first cycle was preceded by incubation for 5 min at 94°C. After 30 cycles, there was a final 7-min extension at 72°C. Negative controls containing no DNA template were included in parallel. PCR products were purified with a QIA-quick PCR purification kit (Qiagen) used according to the manufacturer's protocol.
Restriction enzyme analysis and computer-assisted analysis of rDNA restriction patterns.
The following enzymes were used: _Alu_I, _Rsa_I, _Hha_I, _Hin_fI, _Hin_dIII, and _Taq_I. Digestion was performed in a 25-μl (final volume) mixture at the optimal temperature according to the manufacturer's protocols (BioLabs). The total digested products were separated by electrophoresis in a 2% (wt/vol) agarose gel. PCR-amplified products were separated by horizontal electrophoresis in 1.5% (wt/vol) agarose gels in Tris-borate-EDTA buffer. Gels were stained with 0.5 μg of ethidium bromide per ml and visualized with UV light. Gel images were digitized with a charged-coupled device video camera (Sony) and stored as TIFF files. These files were converted, normalized with the molecular size markers in a 100-bp DNA ladder (BioLabs), and analyzed with Bio Profile software (Vilbert Lourmat). For ISR-RFLP analysis, a band-matching algorithm was used to calculate pairwise similarity matrices with the Dice coefficient (15). A band-matching tolerance of 5% was chosen.
16S-23S ISR DNA cloning and sequencing.
Clone libraries of the carnobacterial PCR-amplified 16S-23S rDNA ISRs obtained with primers 16S/p4 and 23S/p7 were constructed by using a pCRR2.1. TA cloning kit (Invitrogen). From each clone library, 30 white colonies were picked randomly. Clones were screened by PCR for the presence of rDNA ISR inserts and for the sizes of inserts by restriction mapping. For each strain, three independent clones of each 16S-23S ISR identified in Carnobacterium were selected and sequenced. Double-stranded DNAs from the recombinant plasmids of the positive clones obtained in the screening assays described above were purified by using a QIAprep Spin Miniprep kit (Qiagen). The nucleotide sequences of the cloned 16S-23S ISRs were determined by the dideoxynucleotide chain termination method (35) with an ABI 370 automated sequencer by using a Taq DyeDeoxy terminator cycle sequencing kit (Perkin-Elmer). The M13 primers flanking the multiple cloning site of pCRR2.1 DNA were used to sequence both DNA strands.
DNA analysis.
Sequences were submitted to the National Center for Biotechnology Information (Bethesda, Md.) for similarity searches with the GenBank database. The computer program CLUSTALW (38) was used for sequence alignment, and the BLAST 2 program (36) was used to represent ISR similarities for sequences which did not include 16S or 23S rDNA.
Nucleotide sequence accession numbers.
The 16S-23S ISR rDNA sequences determined in this study have been deposited in the EMBL, GenBank, and DDBJ nucleotide databases under the following accession numbers: AF374286, AF374287, AF374288, AF374289, AF 374290, AF374291, AF374292, AF374293, AF374294, AF374295, AF374296, and AF374297.
RESULTS
Length polymorphism of the carnobacterial 16S-23S ISR.
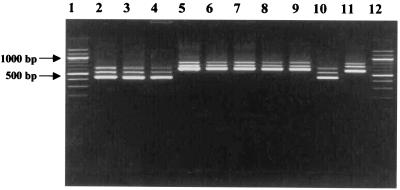
PCR amplification with primers 16S/p2 and 23S/p10 designed by using the flanking terminal sequences of the 16S and 23S genes was performed with chromosomal DNA isolated from 42 Carnobacterium strains. Amplification yielded nearly identical band patterns containing three fragments_._ To verify that amplified products were specific to the 16S-23S ISR region, a second PCR was performed. Nested PCR amplification of these fragments with primers 16S/p4 and 23S/p7 resulted in similar profiles. PCR products from each Carnobacterium strain always contained three ISR amplicons, which were designated the small ISR (S-ISR), the medium ISR (M-ISR), and the large ISR (L-ISR) spacer region, and their lengths varied from one species to another (Fig. 1). C. divergens NCDO 2763T and C. mobile DSM 4849T generated a major S-ISR band (ca. 400 bp) and minor M-ISR and L-ISR bands (ca. 500 and ca. 600 bp, respectively). The ISRs amplified from C. gallinarum NCFB 2766T and C. piscicola NCDO 2762T were larger (S-ISR, ca. 600 bp; M-ISR, ca. 700; and L-ISR, ca. 800 bp). Extensive investigations of a large number of Carnobacterium strains gave similar results (data not shown). Polymorphism in the length of the 16S-23S amplicon for different Carnobacterium species was evident, and two groups were distinguished. The first group, designated group A, included C. divergens and C. mobile. The second group (group B) contained all strains of C. piscicola and C. gallinarum. The three different 16S-23S ISR PCR products obtained for each strain indicated that there are at least three types of rrn operons in Carnobacterium species. The variations in ISR length observed could be due in part to variations in the number and type of tRNA sequences found in this region. The 16S-23S ISRs were PCR amplified from LAB related to Carnobacterium. The LAB strains used in this study included reference strains and independent isolates which are geographically and temporally distinct (Table 2). 16S-23S ISR PCR amplification with primers 16S/4 and 23S/7 of 175 LAB strains resulted in very different patterns (in terms of the size and number of bands). Two ISR fragments that were approximately 450 and 650 bp long were amplified for all Lactobacillus strains. A single amplified fragment was obtained for Lactococcus (ca. 500 bp), Leuconostoc (ca. 600 bp), and Streptococcus thermophilus (ca. 550 bp) strains. Polymorphism in the lengths of ISRs was found for strains of Enterococcus. Two bands at ca. 450 and 600 bp were observed for Enterococcus faecalis, and two fragments at ca. 550 and 650 bp were found for Enterococcus faecium. The variation in PCR amplicon number and length was useful for differentiation of the Carnobacterium strains at the genus level in the LAB group (Table 2).
FIG. 1.
Electrophoresis in a 1.5% agarose gel of PCR-amplified 16S-23S ISRs of C. divergens, C. mobile, C. gallinarum, and C. piscicola. Lanes 1 and 12, molecular weight marker (100-bp DNA ladder); lane 2, C. divergens NCDO 2763T; lane 3, C. divergens INRA 541; lane 4, C. divergens V41; lane 5, C. piscicola NCDO 2762T; lane 6, C. piscicola V1; lane 7, C. piscicola SF 644; lane 8, C. gallinarum NCDO 2766T; lane 9, C. gallinarum INRA 680; lane 10, C. mobile DSM 4849T; lane 11, C. piscicola INRA 501.
Nucleotide sequence analysis of the 16S-23S rDNA ISR.
The polymorphism of Carnobacterium 16S-23S ISRs was investigated by sequencing the corresponding three amplicons of type strains C. divergens NCDO 2763, C. mobile DSM 4849, C. gallinarum NCFB 2766, and C. piscicola NCDO 2762. For each strain, the three ISR amplicons cloned in recombinant plasmid pCRR2.1 were screened by PCR amplification. Three independent clones containing the insert rDNA corresponding to each of three PCR ISR amplicons were identified and sequenced. Anticipated errors of PCR and sequencing reactions were avoided by sequencing both strands from each cloned fragment obtained from separate PCR experiments. Sequences at the 3′ end of the 16S coding region and at the 5′ end of the 23S coding region were recognized by comparison with previously published sequences in the GenBank database. The sequence alignments for S-ISR, M-ISR, and L-ISR are shown in Fig. 2.
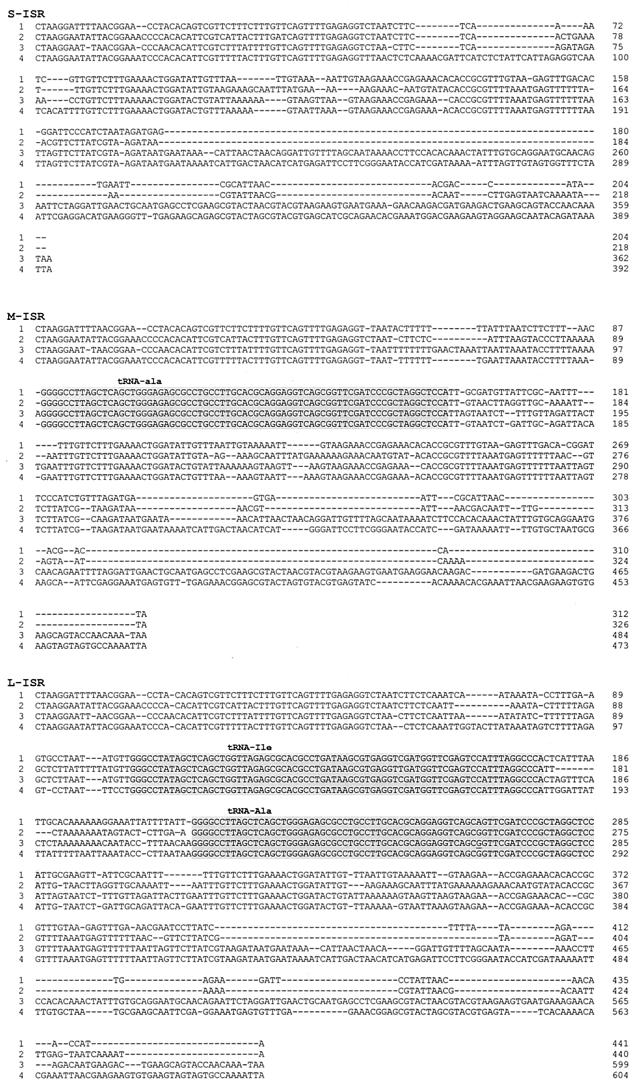
FIG. 2.
Alignment of the nucleotide sequences of the Carnobacterium 16S-23S ISRs. Line 1, C. mobile; line 2, C. divergens; line 3, C. gallinarum; line 4, C. piscicola. GenBank accession numbers of the complete sequences are listed in Materials and Methods. The tRNAs are indicated by grey boxes.
Sequence analysis of the S-ISRs of the four Carnobacterium species revealed four different sequences. Two short sequences, 204 and 218 bp long, were found for C. mobile and C. divergens, respectively. Two longer S-ISR sequences (391 and 362 bp) were amplified from C. piscicola and C. gallinarum, respectively. These four S-ISRs showed 40.7% homology and displayed multiple microheterogeneities. The unexpected lengths of the C. piscicola and C. gallinarum sequences are explained by the presence of one short insertion in the C. piscicola S-ISR (ca. 25 bp long, located at positions 68 to 93) and one long insertion in both S-ISRs (located at positions 184 to 340 and 212 to 370, respectively). In the strains examined, none of the S-ISR fragments contained a tDNA gene. A 103-bp fragment located at the 3′ end of the four S-ISRs exhibited 83 to 88% identity with the S-ISR sequences of Enterococcus muntidii, Enterococcus faecalis, Lactobacillus pentosus, and Lactobacillus plantarum (GenBank accession numbers X87188, AJ301836, U97141, and U97139).
Sequence analysis of the M-ISRs amplified from the type strains also revealed four different sequences, with 50.8% similarity. The 16S-23S M-ISRs were found to contain 326, 312, 473, and 484 bp in C. divergens, C. mobile, C. piscicola, and C. gallinarum, respectively. In all strains, the M-ISR invariably contained an insertion with a gene encoding tRNAAla (anticodon, UGC) beginning at position 90 or 99 and ending at position 162 or 171. The central region was flanked by sequences that were identical or almost identical to those of the corresponding S-ISRs. The M-ISRs of the four Carnobacterium species exhibited 36% identity with the ISR sequences of Enterococcus durans and Enterococcus faecium (GenBank accession numbers X87177 and AF082294, respectively). The M-ISRs of Carnobacterium species are organized like the ISRs of Enterococcus, Lactococcus, Leuconostoc, and Streptococcus, which contain a unique tRNAAla gene (21, 22, 31, 33, 44).
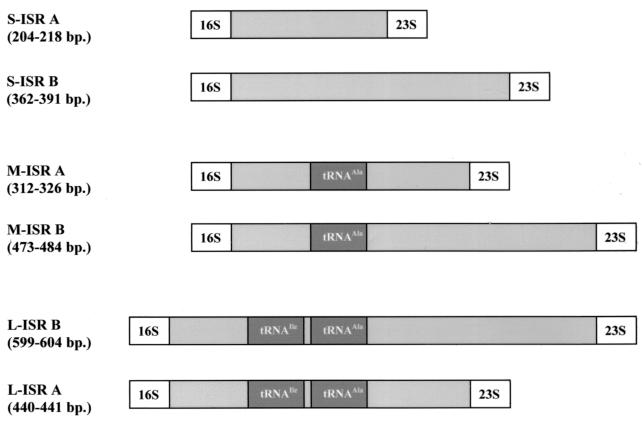
Sequence analysis of the L-ISRs of the type strains revealed four different sequences. The L-ISR amplicons of C. piscicola and C. gallinarum consisted of 604 and 599 nucleotides, respectively. In contrast, C. divergens and C. mobile had shorter versions (440 and 441 bp). All L-ISRs contained two tDNA genes, coding for tRNAIle (located at position 104, 106, or 111; anticodon, GAU) and tRNAAla (located at position 204, 214, or 221), which were 73 to 74 bp long. The difference in length between groups A and B was attributed to an approximately 149-bp deletion in C. mobile and C. divergens, located between the end of the tDNAAla gene and the start of the 23S gene. The four carnobacterial L-ISRs showed 56.5% similarity and 36.7% identity with the L-ISR sequences of Lactobacillus graminis, Lactobacillus curvatus, and Lactobacillus sakei (GenBank accession numbers U97136, U97135, and U97137). The L-ISRs of Carnobacterium species are organized like those of the Lactobacillus species (3). They are composed of the corresponding S-ISRs and M-ISRs, interrupted by sequences (which are 213 to 237 bp long) containing two tDNA genes (Fig. 3).
FIG. 3.
Schematic representation of S-ISR, M-ISR, and L-ISR of 16S-23S rDNA in the genus Carnobacterium. Group A is the group containing C. divergens and C. mobile; group B is the group containing C. gallinarum and C. piscicola. The positions of both tRNAs (tRNAIle and tRNAAla) in M-ISR and L-ISR are indicated.
The primary structures of the two tDNA genes found in M-ISR and L-ISR were similar in the four Carnobacterium species. We detected only two microheterogeneities in both tDNA genes. The tDNAIle of C. divergens NCDO 2763T differed by a single T-C substitution at position 154. At the tDNAAla level, only one G-A heterogeneity was suspected in the L-ISR of C. mobile DSM 4849T (at position 264). The results of a BLAST search of tRNAs are summarized in Table 3. The highest levels of similarity of carnobacterial tDNAAla and tDNAIle genes were found with Bacillus, Lactobacillus, and Listeria strains.
TABLE 3.
Sequence similarities of tRNAs found in 16S-23S ISRs of Carnobacterium species
| ISR | tRNA | Organism(s) | Matching organisms | % Similaritya |
|---|---|---|---|---|
| M-ISR | tRNAAla (UGC) | C. mobile, C. gallinarum, C. piscicola, C. divergens | Bacillus megaterium | 100 (0/73) |
| Bacillus pseudomycoides | 100 (0/73) | |||
| Bacillus mycoides | 100 (0/73) | |||
| Bacillus thuringiensis | 100 (0/73) | |||
| Streptococcus difficile | 98 (1/73) | |||
| Lactococcus lactis subsp. lactis | 98 (1/73) | |||
| L-ISR | tRNAIi (GAu) | C. mobile, C. gallinarum, C. piscicola | Lactobacillus sakei | 100 (0/74) |
| Bacillus sporothermodurans | 100 (0/74) | |||
| Bacillus pseudomycoides | 100 (0/74) | |||
| Listeria innocua | 100 (0/74) | |||
| Listeria monocytogenes | 100 (0/74) | |||
| Lactobacillus graminis | 100 (0/74) | |||
| Lactobacillus curvatus | 100 (0/74) | |||
| Bacillus subtilis | 97 (2/74) | |||
| C. divergens | Lactobacillus sakei | 98 (1/74) | ||
| Bacillus sporothermodurans | 98 (1/74) | |||
| Bacillus megaterium | 95 (3/74) | |||
| tRNAAla (UGC) | C. mobile | Bacillus megaterium | 98 (1/73) | |
| Alcanivorax borkumensis | 98 (1/73) | |||
| C. divergens, C. gallinarum, C. piscicola | Bacillus megaterium | 100 (0/73) | ||
| Bacillus cereus | 100 (0/73) | |||
| Alcanivorax borkumensis | 100 (0/73) | |||
| Lactococcus lactis subsp. lactis | 98 (1/73) |
PCR-based restriction analysis.
The variation in 16S-23S ISR length among the four Carnobacterium species was not substantial. The limitations of the ISR PCR method are evident, and this leads to problems in the development of simple sequence analysis methods, such as PCR-RFLP. Since PCR amplification with primers 16S/p4 and 23S/p7 yields three ISR amplicons of different lengths, it was difficult to obtain clear patterns in a PCR-RFLP analysis. The L-ISR and M-ISR sequence analysis showed that there is polymorphism in the region located between tRNAAla and 23S DNA. The gene encoding tRNAAla contains an 18-nucleotide sequence which is conserved in all the tRNAAla sequences compared, making this region a suitable target for performing PCR inside the ISR. Primer 23S/p10 is therefore the recommended primer, as it is the primer most likely to detect all copies of the spacer region and hence to determine spacer and sequence variation (11). The tRNAAla gene is located the same distance from 23S rRNA in M-ISR and L-ISR (Fig. 3). For this reason, PCR performed with primers tRNAAla and 23S/p10 allowed amplification of two amplicons of the same length, which appeared as a single DNA fragment at about 740 bp for group A organisms (C. divergens and C. mobile) and at about 900 bp for group B organisms (C. gallinarum and C. piscicola). These amplicons included polymorphism of the two targets (M-ISR and L-ISR). The PCR products derived from each strain were digested separately with six endonucleases, _Alu_I, _Rsa_I, _Hha_I, _Hin_fI, _Hin_dIII, and _Taq_I. These restriction enzymes were selected as first-line enzymes that, together with information about amplicon sizes and sequence polymorphism of the 16S-23S ISR, would produce the most discriminatory RFLP patterns. Digestion with _Alu_I, _Hha_I, and _Rsa_I generated two genotypes, corresponding to group A (C. divergens and C. mobile) and group B (C. gallinarum and C. piscicola) (data not shown). The two groups required further analysis with additional endonucleases for accurate identification.
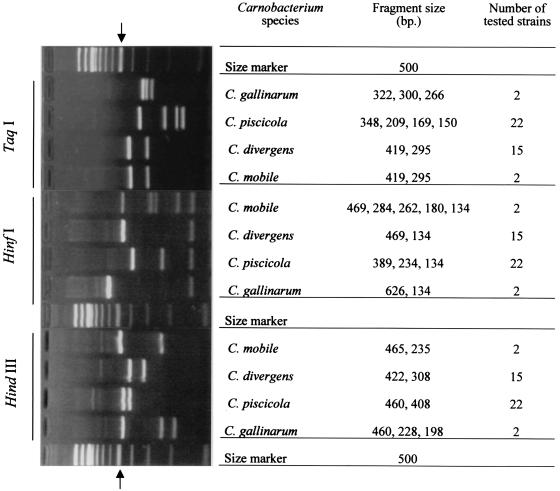
The observed individual _Taq_I, _Hin_fI, and _Hin_dIII RFLP patterns of reference strains are shown in Fig. 4. _Taq_I digestion revealed three genotypes with well-resolved bands. In this case, polymorphism in the closely related organisms C. piscicola and C. gallinarum was found. The three genotypes produced the following patterns: four bands at 150, 169, 209, and 348 bp for C. piscicola, three bands at 266, 300, and 322 bp for C. gallinarum and two bands at 295 and 419 bp for C. divergens and C. mobile.
FIG. 4.
Gel electrophoresis of PCR-amplified 16S-23S ISR fragments of the Carnobacterium species digested with _Taq_I, _Hin_fI, and _Hin_dIII. Algorithms of RFLP pattern species are presented by molecular size. The position of the 500-bp fragment of the 100-bp DNA ladder is indicated by arrows.
The four Carnobacterium species were finally distinguished by digestion of the ISR amplicon with _Hin_fI and _Hin_dIII. _Hin_fI digestion revealed four genotypes with two to five bands. Two bands at 134 and 469 bp were distinguished for C. divergens, two bands at 134 and 626 bp were distinguished for C. gallinarum, three bands at 134, 234, and 389 bp were distinguished for C. piscicola, and five bands at 134, 180, 262, 284, and 469 bp were distinguished for C. mobile.
The results showed that the total length of the _Hin_fI DNA fragments was much more than 740 bp only for C. mobile (1,329 bp). On the basis of the alignment of the M-ISR and L-ISR sequences of C. mobile, we detected a microheterogeneity at the _Hin_fI restriction site. In M-ISR a _Hin_fI site was located at position 267. In L-ISR, two _Hin_fI sites located at positions 393 and 419 were identified (Fig. 2). These results suggested that when the tRNAAla and 23S/p10 primers were used, the amplicons corresponding to the 5′ part of 23S rDNA and the 3′ extremity of M-ISR or L-ISR contained two and three _Hin_fI sites, respectively. Digestion of M-ISR yielded three fragments (134, 180, and 469 bp), and restriction of L-ISR resulted in four fragments (27, 180, 262, and 284 bp). The small 27-bp fragment was not detected on the gel. This explains why the total length of the _Hin_fI DNA fragments from C. mobile is about twice the length of the ISR amplicon. _Hin_dIII restriction resulted in two or three fragments, as follows: for C. mobile, 465 and 235 bp; for C. divergens, 422 and 308 bp; for C. gallinarum, 460, 228, and 198 bp; and for C. piscicola, 460 and 408 bp.
The RFLP patterns resolved 42 isolates into the following four species: C. mobile, C. divergens, C. gallinarum, and C. piscicola. _Hin_fI and _Hin_dIII restriction produced genotypes whose sizes could be easily estimated and analyzed with Bio Profile software.
According to the spacer PCR-RFLP method, 12 isolates belonging to C. divergens and 14 isolates belonging to C. piscicola displayed monomorphic restriction patterns with all endonucleases tested and are phylogenetically related to C. divergens NCDO 2763T and C. piscicola NCDO 2762T, respectively.
During phenotypic identification of our Carnobacterium collection, some strains appeared to be incorrectly identified. This was confirmed by the 16S-23S ISR-RFLP method. A group of three isolates (INRA 508, INRA 586, and INRA 515) previously identified as C. divergens exhibited C. piscicola phenotypic and PCR-RFLP profiles. These isolates were reclassified as C. piscicola.
The four isolates deposited as C. piscicola (INRA 545, INRA 572, INRA 722, and ENSAIA 13) produced patterns that were in full agreement with the patterns obtained for C. divergens. These isolates were reclassified as C. divergens.
DISCUSSION
In this study, we sought to develop a new molecular method for differentiating the four closely related Carnobacterium species isolated from food. We found that our method is useful for identifying all strains at the species level accurately and reliably and is simple enough for use even in routine laboratories.
The conventional methods for identifying Carnobacterium strains based on phenotypic tests are time-consuming, and interpretation of their results is often ambiguous. Currently, the widely accepted strategy formulated to improve methods of Carnobacterium strain identification includes sequence analysis of the 16S rRNA gene and construction of genus- and/or species-specific oligonucleotide probes for use with a multiplex and multistep PCR (1, 4, 8, 32, 42). Each technique has several advantages and disadvantages. The small number of polymorphic positions in the 16S rDNA obviates the need for nucleotide sequencing. The species-specific PCR primers, designed from 16S rDNA, were not specific enough to differentiate Carnobacterium spp. from other bacterial strains (4). They revealed 100% matches with members of several bacterial genera, including Vagococcus spp., Enterococcus spp., and Listeria spp. The specific primer for C. piscicola (Cpg) anneals to both C. piscicola and C. gallinarum because these species exhibit >98% homology in their 16S rDNA sequences.
The principal goal of our study was to investigate the level of ISR polymorphism and thereby assess the utility of this target for Carnobacterium species identification. In this study we demonstrated that the ISR of the genus Carnobacterium exhibits considerable variation in length, which is useful for distinguishing two Carnobacterium groups from other LAB. Importantly, the sequence divergence includes a reasonable number of insertion and deletion sites. Besides its greater variability, two additional advantages of this target can be pointed out. Unlike a 16S rDNA-based PCR, in which the genus-specific primers are separated by a long stretch of target sequence, an ISR-based PCR should result in a smaller PCR product and in more efficient and sensitive target amplification. In addition, the ISR has the potential to be used for environmentally significant species and strain differentiation. Hence, ISR sequences have been proposed as a useful supplement when 16S rDNA shows insufficient diversity to differentiate recently diverged species (11). The ISR sequence does not code for a final product, but it has an important processing function in forming pre-RNAs. As a consequence, there is presumably some functional selective pressure for conservation of this region. This assumption is consistent with the stability of species-specific ISR signatures found with high reproducibility in different strains. As shown with other taxa, the evolutionary rate of the ISR is higher than that of 16S rRNA, and rearrangements in the central region are relatively recent. We showed that the number of tDNA genes found in carnobacterial 16S-23S ISRs varied from zero to two. It is, therefore, surprising that the Carnobacterium species have at least three classes of spacer regions and form three types of rrn operons (rrn S [rrn S], rrn Medium [rrn M], and rrn Large [rrn L]). The rrn S and rrn L operons were similar to those found in Lactobacillus species, which are organized as follows: one rrn S operon without tDNA and one rrn L operon with two tDNA genes (tRNAIle and tRNAAla). The rrn L operon was also present in a variety of bacterial taxa, including proteobacteria, cytophagas, and gram-positive bacteria with low DNA G+C contents (7, 17, 24, 25, 40). This suggests that this ISR type may be widespread among bacteria and may have been present in the common ancestor of the domain Bacteria (5). Two tDNAs found in carnobacterial L-ISR are homologous; i.e., they originated from the same ancestral organism. The phylogenetic relationship based on tRNAIle (GAU)-tDNAAla (UGC) sequences found in L-ISR was readily comparable to current bacterial taxonomy based on 16S rDNA sequence data. The majority of LAB, including members of the genera Lactococcus, Streptococcus, and Leuconostoc, have one type of rrn operon with a tRNAAla gene which is similar to the M-ISR of Carnobacterium species (21, 33, 44). Staphylococcus aureus (12) has three types of rrn operons with structures identical to those of Carnobacterium species. The genus Carnobacterium is phylogenetically related to the genus Enterococcus (6). In contrast, Enterococcus species contain two types of rrn operons, which correspond to the carnobacterial rrn S and rrn M operons (31). As described previously for all other bacteria, the sequences of small, medium, and large Carnobacterium ISRs are related to the sequences of tRNA genes inserted between the common sequences. The sequence variability of C. divergens, C. mobile, C. gallinarum, and C. piscicola can be considered an example of the higher resolution of ISR data at the species level. The resolution of the PCR-RFLP methods depended on the part of the rrn operon which was analyzed. The data obtained by RFLP analyses of part of the rrn M and rrn L operons showed that C. divergens, C. mobile, C. gallinarum, and C. piscicola belong to the same genomic group. Concerning the intraspecies stability of ISR-RFLP patterns, we found that the spacer-based method was successfully evaluated with expanded groups of strains belonging to Carnobacterium species. The closely related species in groups A and B clustered with the same or similar _Alu_I, _Rsa_I, and _Hha_I genotypes. In contrast, very different profiles were found with _Taq_I, _Hin_fI, and _Hin_dIII. The four species clustered in four genotypes. The identification of distinct C. divergens, C. mobile, C. gallinarum, and C. piscicola groups, accurately defined by unique spacer RFLP genotypes, correlates perfectly with a previous numerical phenetic study of the genus Carnobacterium (19). The congruence between the results of analyses of rRNA sequences and the RFLP patterns of the 16S-23S-ISR obtained in the present study indicates that these molecular chronometers are well synchronized.
In conclusion, 16S-23S ISR PCR-RFLP analysis is a promising new method for reliable and rapid differentiation of Carnobacterium species.
REFERENCES
- 1.Barakat, R. K., M. W. Griffiths, and L. J. Harris. 2000. Isolation and characterization of Carnobacterium, Lactococcus and Enterococcus spp. from cooked, modified atmosphere packaged refrigerated poultry meat. Int. J. Food Microbiol. 62**:**83-94. [DOI] [PubMed] [Google Scholar]
- 2.Baya, A. M., A. E. Toranzo, B. Lupiani, T. Li, B. S. Robertson, and F. M. Hetrick. 1991. Biochemical and serological characterization of Carnobacterium spp. isolated from farmed and natural populations of striped bass and catfish. Appl. Environ. Microbiol. 57**:**3114-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthier, F., and S. D. Ehrlich. 1998. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 161**:**97-106. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, J. L., A. S. Moore, R. A. Patchett, M. D. Collins, and R. G. Kroll. 1992. Use of the polymerase chain reaction and oligonucleotide probes for rapid detection and identification of Carnobacterium species from meat. J. Appl. Bacteriol. 72**:**294-301. [DOI] [PubMed] [Google Scholar]
- 5.Chun, J., A. Huq, and R. R. Colwell. 1999. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl. Environ. Microbiol. 65**:**2202-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, M. D., J. A. Farrow, B. Phillips, S. Ferusu, and D. Jones. 1987. Classification of Lactobacillus divergens, Lactobacillus piscicola and some catalase-negative, asporogenous, rod-shaped bacteria from poultry in a new genus, Carnobacterium. Int. J. Syst. Bacteriol. 37**:**310-316. [Google Scholar]
- 7.Condon, C., C. Squires, and L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59**:**623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connil, N., X. Dousset, B. Onno, M. F. Pilet, M. F. Breuil, and M. C. Montel. 1998. Enumeration of Carnobacterium divergens V41, Carnobacterium piscicola V1 and Lactobacillus brevis LB 62 by in situ hybridization-flow cytometry. Lett. Appl. Microbiol. 27**:**302-306. [DOI] [PubMed] [Google Scholar]
- 9.Coombs, J., and J. E. Breanchley. 1999. Biochemical and phylogenetic analyses of a cold-active β-galactosidase from the lactic acid bacterium Carnobacterium piscicola BA. Appl. Environ. Microbiol. 65**:**5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franzmann, P. D., P. Hopfl, N. Weiss, and B. J. Tindall. 1991. Psychrotrophic lactic acid-producing bacteria from anoxic waters in Ace Lake, Antarctica; Carnobacterium funditum and Carnobacterium alterfunditum sp. nov. Arch. Microbiol. 156**:**255-262. [DOI] [PubMed] [Google Scholar]
- 11.Gürtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142**:**3-16. [DOI] [PubMed] [Google Scholar]
- 12.Gürtler, V., and H. D. Barrie. 1995. Typing of Staphylococcus aureus strain by PCR-amplification of variable-length 16S-23S rDNA spacer regions: characterization of spacer sequences. Microbiology 141**:**1255-1265. [DOI] [PubMed] [Google Scholar]
- 13.Hain, T., N. Ward-Rainey, R. M. Kroppenstedt, E. Stackebrandt, and F. A. Rainey. 1997. Discrimination of Streptomyces albidoflavus strains based on the size and the number of 16S-23S ribosomal DNA intergenic spacers. Int. J. Syst. Bacteriol. 47**:**202-206. [DOI] [PubMed] [Google Scholar]
- 14.Hammes, W. P., and P. S. Tichaczek. 1994. The potential of lactic acid bacteria for the production of safe and wholesome food. Z. Lebensm. Unters. Forsch. 198**:**193-201. [DOI] [PubMed] [Google Scholar]
- 15.Heyndrickx, M., L. Vauterin, P. Vandamme, K. Kesters, and P. D. Vos. 1996. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J. Microbiol. Methods 26**:**247-259. [Google Scholar]
- 16.Hitchener, L. J., A. F. Egan, and P. J. Rogers. 1982. Characteristics of lactic acid bacteria isolated from vacuum-packaged beef. J. Appl. Bacteriol. 52**:**31-37. [DOI] [PubMed] [Google Scholar]
- 17.Jagoueix, S., J. M. Bove, and M. Carnier. 1997. Comparison of the 16S/23S ribosomal intergenic regions of Candidatus Liberobacter asiaticum and Candidatus Liberobacter africanum, the two species associated with citrus huanglongbing (greening) disease. Int. J. Syst. Bacteriol. 47**:**224-227. [DOI] [PubMed] [Google Scholar]
- 18.Krawiec, S., and M. Riley. 1990. Organization of the bacterial chromosome. Microbiol. Rev. 54**:**502-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, S., and L. N. Manchester. 2000. Numerical phenetic study of the genus Carnobacterium. Antonie Leeuwenhoek 78**:**73-85. [DOI] [PubMed] [Google Scholar]
- 20.Lai, S., H. Shojaei, and L. N. Manchester. 2000. The differentiation of Carnobacterium divergens using the random amplification of polymorphic DNA polymerase chain reaction technique. Lett. Appl. Microbiol. 30**:**448-452. [DOI] [PubMed] [Google Scholar]
- 21.Leblond-Bourget, N., H. Philippe, I. Mangin, and B. Decaris. 1996. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int. J. Syst. Bacteriol. 46**:**102-111. [DOI] [PubMed] [Google Scholar]
- 22.Le Jeune, C., and A. Lonvaud-Funel. 1997. Sequence of DNA 16S/23S spacer region of Leuconostoc oenos (Oenococcus oeni): application to strain differentiation. Res. Microbiol. 148**:**79-86. [DOI] [PubMed] [Google Scholar]
- 23.Leroi, F., J. J. Joffraud, F. Chevalier, and M. Cardinal. 1998. Study of the microbial ecology of cold-smoked salmon during storage at 8°C. Int. J. Food Microbiol. 39**:**111-121. [DOI] [PubMed] [Google Scholar]
- 24.Luz, S. P., F. Rodriguez-Valera, R. Lan, and P. S. Reeves. 1998. Variation of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J. Bacteriol. 180**:**2144-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manceau, C., and A. Horvais. 1997. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl. Environ. Microbiol. 63**:**498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauguin, S., and G. Novel. 1994. Characterization of lactic acid bacteria isolated from seafood. J. Appl. Bacteriol. 76**:**616-625. [Google Scholar]
- 27.McKay, L. L., and K. A. Baldwin. 1990. Applications for biotechnology: present and future improvement in lactic acid bacteria. FEMS Microbiol. Rev. 87**:**3-14. [DOI] [PubMed] [Google Scholar]
- 28.Metivier, A., M. F. Pilet, X. Dousset, O. Sorokine, P. Anglade, M. Zagorec, J. C. Piard, D. Marion, Y. Cenantiempo, and C. Fremaux. 1998. Divercin V41, a new bacteriocin with two disulfide bonds produced by Carnobacterium divergens V41: primary structure and genomic organization. Microbiology 144**:**2837-2844. [DOI] [PubMed] [Google Scholar]
- 29.Millière, J. B., M. Michel, F. Mathieu, and G. Lefebvre. 1994. Presence of Carnobacterium spp. in French surface mould-ripened soft-cheese. J. Appl. Bacteriol. 76**:**264-269. [Google Scholar]
- 30.Montel, M.-C., R. Talon, J. Fournaud, and M.-C. Champomier. 1991. A simplified key for identifying homofermentative Lactobacillus and Carnobacterium spp. from meat. J. Appl. Bacteriol. 70**:**469-472. [DOI] [PubMed] [Google Scholar]
- 31.Naïmi, A., G. Beck, and C. Branlant. 1997. Primary and secondary structures of rRNA spacer regions in Enterococcus. Microbiology 143**:**823-834. [DOI] [PubMed] [Google Scholar]
- 32.Nissen, H., A. Holck, and R. H. Dainty. 1994. Identification of Carnobacterium spp. and Leuconostoc spp. in meat genus-specific 16S rRNA probes. Lett. Appl. Microbiol. 19**:**165-168. [DOI] [PubMed] [Google Scholar]
- 33.Nour, M., A. Naïmi, G. Beck, and C. Branlant. 1995. 16S-23S and 23S-5S intergenic spacer region of Streptococcus thermophilus and Streptococcus salivarius, primary and secondary structure. Curr. Microbiol. 31**:**270-278. [DOI] [PubMed] [Google Scholar]
- 34.Pilet, M.-F., X. Dousset, R. Barré, G. Novel, M. Desmazeaud, and J. C. Piard. 1995. Evidence for two bacteriocins produced by Carnobacterium piscicola and Carnobacterium divergens isolated from fish and active against Listeria monocytogenes. J. Food Prot. 58**:**256-262. [DOI] [PubMed] [Google Scholar]
- 35.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74**:**5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephen, A. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilsala-Timisjarvi, A., and T. Alatossava. 1997. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequence for identifying different dairy and probiotic lactic acid bacteria by PCR. Int. J. Food Microbiol. 35**:**49-56. [DOI] [PubMed] [Google Scholar]
- 38.Tompson, J. D., G. Desmond, I. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:**4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tudor, J. J., L. Marri, P. J. Piggot, and L. Daneo-Moore. 1990. Size of the Streptococcus mutans GS-5 chromosome as determined by pulsed-field gel electrophoresis. Infect. Immun. 58:838-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Gissen, J. W., R. M. Haring, and B. A. M. van der Zeijst. 1994. Comparison of the 23S ribosomal RNA genes and the spacer region between the 16S and 23S rRNA genes of the closely related Mycobacterium avium and Mycobacterium paratuberculosis and the fast-growing Mycobacterium phlei. Microbiology 140**:**1103-1108. [DOI] [PubMed] [Google Scholar]
- 41.Wallbanks, S., A. J. Martnez-Murcia, J. L. Fryer, B. A. Phillips, and M. D. Collins. 1990. 16S rRNA sequence determination for members of the genus Carnobacterium and related lactic acid bacteria and description of Vagococcus salmoninarium sp. nov. Int. J. Syst. Bacteriol. 40**:**224-230. [DOI] [PubMed] [Google Scholar]
- 42.Yost, C. K., and F. M. Nattress. 2000. The use of multiplex PCR reactions to characterize populations of lactic acid bacteria associated with meat spoilage. Lett. Appl. Microbiol. 31**:**129-133. [DOI] [PubMed] [Google Scholar]
- 43.Young, R. A., R. Macklis, and J. A. Steitz. 1979. Sequence of the 16S-23S spacer region in two ribosomal RNA operons of Escherichia coli. J. Biol. Chem. 254**:**3264-3271. [PubMed] [Google Scholar]
- 44.Zavaleta, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1996. 16S-23S rDNA intergenic sequences indicate that Leuconostoc oenos is phylogenetically homogeneous. Microbiology 142**:**2105-2114. [DOI] [PubMed] [Google Scholar]