The evolution of molecular biology (original) (raw)
Summary
Biology's various affairs with holism and reductionism, and their contribution to understanding life at the molecular level
In 1865, Gregor Mendel discovered the laws of heritability and turned biology into an exact science, finally on a par with physics and chemistry. Although the scientific community did not immediately realize the importance of his discovery—it had to be 'rediscovered' around 1900 by Correns, deVries and Tschermak—it came as a relief for a science in crisis. Many biologists in the twentieth century were tired of the purely descriptive nature of their science, with its systematic taxonomy and comparative studies. Charles Darwin's theory of evolution had already provided a first glimpse at the larger mechanisms at work in the living world. Scientists therefore felt that it was time to move from a descriptive science to one that unravels functional relationships—the annual 'Cold Spring Harbor Symposia on Quantitative Biology' was a clear testimonial to that desire. Mendel's four basic laws of genetics, formulated from meticulous experimentation, sparked a revolution in biology as they finally provided biologists with a rational basis to quantify observations and investigate cause–effect relationships.
To relate observed effects to the events that caused them is one of mankind's strong mental abilities. By understanding their relationship, it allows us to remember recurrent events and estimate their likelihood and reproducibility. This usually works well if a cause and its effect are linked by a short chain of events, but the challenge increases with complexity. Living organisms in their natural environment are probably the most complex entities to study, and causes and effects are not usually linked in single linear chains of causalities but rather in large multi-dimensional and interconnected meshworks. To unravel and understand these meshworks, it is therefore important first to study simple systems, in which the chains of near-causalities are relatively short and can be subdivided into single causalities, which are reproducible and thus comparable with what we call the 'laws of nature' in physics. Most of the important 'rules of nature', such as the 'genetic code', 'protein biosynthesis at ribosomes' or the 'operon' are examples of such chains. This new understanding has sparked a fresh debate about reductionist versus holistic approaches to biological research, which has implications for the public view and acceptance of biology and of its application in medicine and the economy.
The laws of genetics as formulated by Mendel were comparable with the basic laws of thermodynamics and therefore attracted many physicists to biological research. The German physicist Max Delbrück, for instance, who spent his earlier research career in astronomy and quantum physics, moved to biology in the late 1930s to study the basic rules of inheritance in the simplest organisms available, namely bacterial viruses (bacteriophages) and their hosts. As the political situation in Nazi Germany worsened, he left for the California Institute of Technology (Caltech; Pasadena, CA, USA), where he and Emory L. Ellis established the standard methods for this field (Ellis & Delbrück, 1939). The author of the present paper is no exception. Trained as a physicist around the middle of the last century, he changed to studying biophysics and biology. This followed the lead of his thesis advisor, Jean J. Weigle, who quit his position as a professor of experimental physics in Geneva, Switzerland, to join Max Delbrück's phage group at Caltech as a research fellow.
Living organisms in their natural environment are probably the most complex entities to study, and causes and effects are not usually linked in single linear chains of causalities...
It is interesting to note that at that time chemistry and, in particular, biochemistry were not yet ready to participate in this newly emerging field, which would later become known as molecular biology. The predominant understanding of chemistry was concentrated on other aspects; it had reached a new peak as a research field and everybody was convinced that the future belonged to chemistry. The main laws of mass action and thermodynamics were established and solidly anchored in every chemist. The specificity of organic substances was well explained by stoichiometry and the steric arrangement of atoms. This knowledge resulted in many important discoveries and products—such as fertilizers, pesticides, plastics and explosives—with a huge impact on society, agriculture, medicine, consumer products and the military, to name a few. Chemistry was then predominantly focused on synthesizing new molecules; biological polymers therefore had little chance to be recognized as the conservative carrier of genetic information. The understanding of the role of enzymes as a catalyst of chemical reactions enforced this general belief; the catalogue of identified and characterized enzymes grew daily and gave rise to a euphoria comparable with that of the heyday of genomics.
However, the most important question—how heritable information is stored in the cell—remained open. DNA was not considered seriously: how could a heteropolymer of only four different monomers explain the high specificities of genes? Proteins, with their 20 different amino acids, seemed much more suitable. Furthermore, the specific structure of proteins or DNA as a linear sequence of amino acids or nucleotides had not even been suspected, let alone elucidated. Even biologists were instilled with this bias. This went so far that they sought all kinds of alternative explanations for the results of their experiments, such as Oswald Avery's research demonstrating the important role of DNA in the genetic transformation of bacteria (Avery et al, 1944; other examples in Kellenberger, 1995).
It was up to a physicist to provide the first clue to this enigma. Erwin Schrödinger, in a few lines from his famous book What is Life? (Schrödinger, 1943), suggested a genetic code similar to the Morse code, then still widely used for the transmission of telegrams. His idea went unnoticed and was only rediscovered several years later when magnetic tape recorders became available for general use. Suddenly, Schrödinger's proposal of information being represented by a specific, linear sequence of only a few elements was appreciated. The experiments of Alfred Hershey and Martha Chase further supported the idea that DNA is the carrier of genetic information (Hershey & Chase, 1951), although they provided less convincing evidence than Avery's work. Once James Watson and Francis Crick solved the structure of DNA on the implicit basis of such a code (Watson & Crick, 1953), the precise nature of how it stores and copies information was abundantly discussed in theoretical terms (Asimov, 1963; Gamow, 1954), before it was eventually proven by experiments (Nirenberg et al, 1965; Nishimura et al, 1964).
As biological macromolecules—nucleic acids and proteins—became the topic of investigation, molecular biology and biochemistry found a common denominator. Now, the biochemists stood whole-heartedly behind 'molecular biology', newly defined as the biochemistry related to DNA and its expression into proteins (Rheinberger, 1997), and focused their collective experience on investigating how genetic information is stored, transmitted and translated into phenotypes. Biochemists had long learned to examine reactions by dividing them into the smallest individual steps in chemical terms. Under given conditions, such as pH, temperature and salt concentrations, these elementary steps are absolutely reproducible. Known as 'Descartes' clockwork', such a one-dimensional, linear chain of elementary steps leading from cause to effect was for a long time the basic understanding of technical and scientific processes, including biological ones. As the name indicates, Descartes used the mechanical clock as a model in which every cog induces the movement of the next in a reproducible and predictable way. This model is still applicable to the majority of biochemical reactions. This reductionist approach to molecular biology proved to be extremely successful initially and helped to unravel many of the basic molecular and cellular processes. However, some biologists started realizing quite early that the immense complexity of living organisms could not be explained solely on the basis of a clockwork mechanism.
Electronics and computer science, particularly the development of integrated circuits and software with feedback controls, imposed a new view; the linear network was no longer adequate to explain complex systems. Only meshworks with two or even more dimensions could account for these new features (Fig 1). Such a meshwork comprises individual, reproducible causalities as its basic elements, which interact with and influence each other (Kellenberger, 1997; R. Ernst, personal communication). A causal chain in a multi-dimensional meshwork is a linear sequence of causalities within the larger meshwork, but at every point where two or more causalities join, deviations from the chain to other parts of the meshwork are possible (Figs 2,3). This reduces the probability that a given cause leads to only one effect; instead, side effects or completely changed effects are possible as well. In contrast to Descartes' clockwork, meshworks in complex systems can only give probabilities about the outcome of a cause; they do not describe predictable and reproducible linear cause–effect relationships. One speaks now of 'near-causalities' rather than causal chains.
Figure 1.
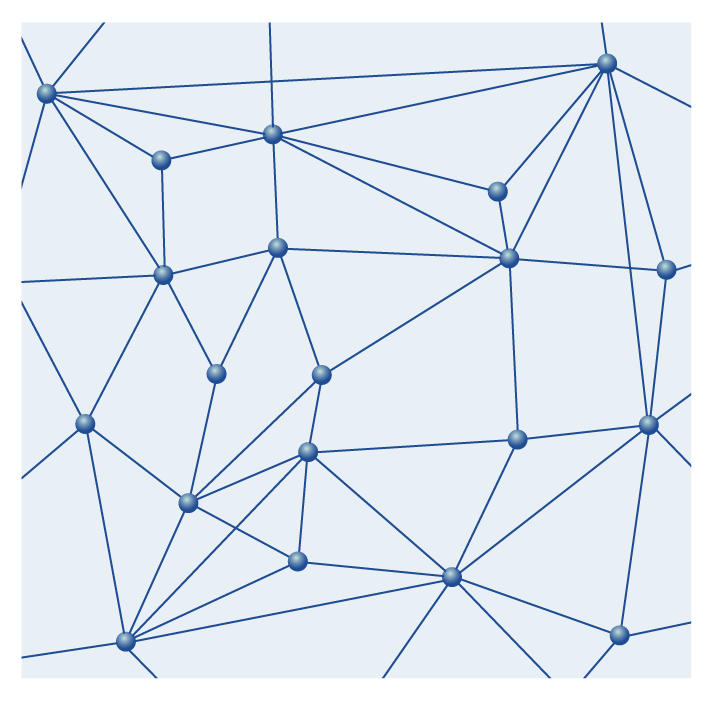
An area of a two-dimensional network in which all single, reproducible causalities are known, here represented by the straight lines between junctions. They obviously depend on the physicochemical conditions, such as pH, temperature or pressure, which also determine the direction of a reaction.
Figure 2.
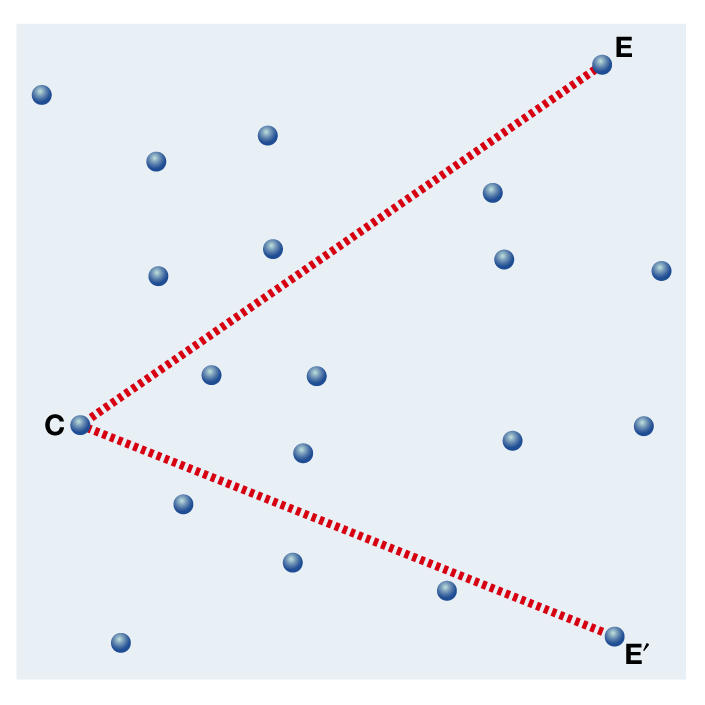
A near-causality in a network is represented by the straight connections (red) between the cause C, and the effect E. As deviations are possible at each junction, a cause can also lead to a side effect E′ (see also Fig 3).
Figure 3.
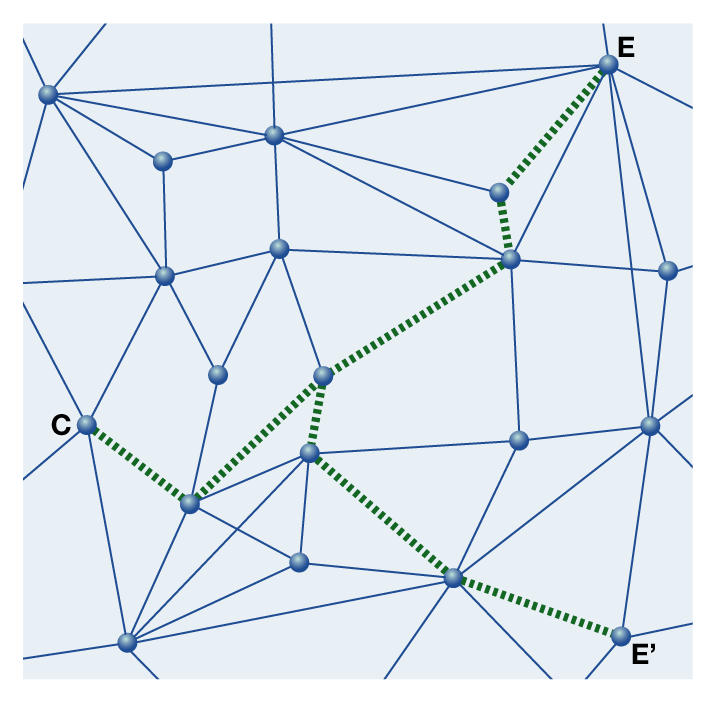
Analysing the individual causalities that link causes and effects (green) allows scientists to make better and more reliable predictions about the probability of effects and side effects.
The idea of near-causality has had an important impact on the philosophy of science and technology. In these complex meshworks, the endpoint of a chain—the effect—was eventually unstable and not reproducible. Consequently, scientists have to first learn more about the nature of the individual causalities that form a causal chain within a given meshwork and the regulatory functions that lead to deviations from this chain, which is what fundamental research actually does. This became clear shortly after molecular biology had become one of the 'exact sciences', and biologists soon realized that living systems with their manifold regulatory and feedback mechanisms are far too complex to be understood on the basis of Descartes' clockwork. In the early years of molecular biology, many biologists strongly believed that the genetic code would be different for higher organisms, particularly for humans. Even now, after the universality of the genetic code has been proven, the uncritical extrapolation of results obtained from microorganisms to higher organisms may lead to mistakes.
Some research areas, such as medical, epidemiological or environmental research, still use the idea of linear causal chains even if they understand that they are, in fact, near-causalities. The reason is that we have to act, to cure the patient or protect the environment from dangers, even if we understand only a few, if any, individual causalities within the larger meshwork. We cannot wait until research has completed its investigations and reached a complete understanding; we have to accept the probability of failure or deleterious side effects, even accepting that only fundamental experimental research can establish and prove genuine reproducible causalities. The more that are characterized and understood, the higher the probability of a reproducible outcome from a given cause (Fig 3). Furthermore, to understand complexity, we have to expand the idea of meshworks beyond the three dimensions of space and the fourth dimension of time. A fifth dimension would represent the increasing complexity of living organisms. The simple systems, such as microorganisms, are located at the bottom, whereas increasing numbers of regulatory meshes are added higher up the axis of complexity. Frequently, these are in themselves increasingly complex; for example, the regulatory network of hormones that gets more complicated the more complex the organism.
Around 1968, a popular movement, even among scientists, alleged that near-causalities could be successfully investigated without going along the cumbersome path of reducing them to genuine causalities, followed by their combination to complete chains. This approach is based on the ideas of the biologist Ludwig von Bertalanffy (von Bertalanffy, 1949), who later created 'systems theory'. Holism is the art of treating complex systems as a whole and not reconstituting them from their individual components. However, for reasons that I discuss below, attempts to materialize such an integrated or 'holistic' approach did not give rise to new methods and results. Instead, one again uses the old methods with a large number of experimental repeats and a statistical analysis. Nevertheless, the concept of holism helped to counterbalance increasing specialization in many academic disciplines, and gave rise to what is now called inter- and transdisciplinary research. This necessary development led to the recognition of many new near-causalities, in fields and disciplines in which even those had been completely lacking.
While 'holism' and 'transdisciplinarity' became the new catchwords both in academic circles and in the general public, reductionism has made a recent comeback in a new form. It proposes that experimental results obtained from simple organisms—that is, for relatively short chains of causalities—can be extrapolated to higher organisms, in which the corresponding chains are believed to be much longer, in particular through the addition of 'regulatory meshes'. This 'reversed' reductionism is often justified to save time and effort; sometimes it is not. A case in point is the generation of transgenic organisms. Even if the same method of genetic engineering is used to transfer aliquots of the same sample of DNA into dozens of identical organisms, the results obtained can vary by factors of more than ten. As we do not understand most of the individual causalities, the procedure—a near-causality—is not reproducible. Among these transgenic organisms, the researcher selects those that produce most of the desired product and at the same time behave 'normally'; that is, similarly to the parent strain. It is obvious that the way the new gene is integrated into the host genome has an enormous influence, and this is some-thing that we need to investigate thoroughly. However, for gene technology firms it is less expensive to make a commercial product by selection rather than by first fully understanding the process of gene transfer.
In the early years of molecular biology, many biologists strongly believed that the genetic code would be different for higher organisms, particularly for humans
Just as physicists had a strong influence on molecular biology in its early days, physics is again providing new insights and clues for biological research. Among the many revolutionary developments in physics, particle-wave dualism and the uncertainty principle, first described by Werner Heisenberg and Nils Bohr, have become very important ideas, frequently quoted and extrapolated by biologists and biochemists. In essence, they state that the means to explore an object or event also modifies it at the same time. For the sub-microscopic world of atoms and molecules, it means that the physical properties of a system—for example, its energy—are uncertain; in particular, these properties depend on the type of observation that is performed on that system, defined by the experimental set-up. In the fashionable field of nano-biotechnology, which examines biological structures at the level of atoms, these relations are directly relevant. At larger dimensions, the interactions between an experimental set-up and biological matter give rise to modifications too; for example, bleaching during fluorescence microscopy or the electron-beam-induced alteration—such as burning or carbonization—of a specimen.
The harm caused to a specimen by electrons in electron microscopy is already well investigated, and students have learned to overcome these imposed obstacles. Biochemists have frequently ridiculed these attempts by claiming that electron microscopists only observe artefacts. They forget too easily that even worse 'artefacts' are produced when cells are broken up for in vitro experiments. The physical event of destroying the boundary of a cell necessarily dilutes its content by at least two times, with all the effects due to changes of ionic composition—which is obviously different inside the cell to the medium present outside—and of the chemical equilibria, as summarized in Kellenberger & Wunderli (1995).
Today's economic situation is due to the steady increase of new capital from increasing global trade that has to be reinvested in new enterprises and ideas. During the mid-1990s, science and technology became predestined venues for investing money. This 'new economy', a fusion of neo-liberalism and globalization, valued enterprises on the basis of their ideas and potential market success rather than on existing assets. New applications of science, such as biotechnology, computer science or nanotechnology, therefore promised enormous returns on the invested capital with the result that much of it flowed from companies that produced normal consumer goods to high-tech start-up firms.
In addition, many scientists, particularly in the life sciences, were flattered by the enormous and sudden interest in their work. They also found it much easier during the early days of this new economy to raise money from private investors compared with the gruelling work of writing grants for public funding. Furthermore, many governments, pressured by the economy to reduce their activities in favour of private enterprises, turned the tables and increasingly requested that scientists obtain funding from private sources. This obviously ignores the experience that, in the long run, creativity and innovation come from fundamental research that is mainly judged by peers and supported by the public, and not by the market or the expectation of huge profits.
The new economy and the lure of the enormous amount of money that suddenly flowed into applied science and technology has created new problems, among them the increasing abuse of the phrase 'molecular biology' and its various 'offspring'. The more that start-up enterprises compete for capital to fund their research, the more they have to use advertising and public relations to attract the interest of investors. This has led to the abundant use of 'molecular' as an adjective and the redefinition of words in the public realm, to the extent that they are becoming hollow words. 'Genomics', which is actually the sequencing of genomes, and 'proteomics', another word for protein biochemistry, have instigated several years of euphoria with feedbacks and backlashes on science. The most momentous backlash was the discovery that more than 90% of the human genome does not code for genes, something that was not found in primitive microorganisms, in which all—or nearly all—of the DNA is used for coding proteins. This apparently non-functional DNA quickly acquired the name 'junk DNA', although some scientists doubted such a complete lack of biological function and postulated explanatory hypotheses (Scherrer, 1989). It took more than ten years of research to demonstrate that some of these large 'uncoding regions' have regulatory functions (Gibbs, 2003).
...the concept of holism helped to counterbalance increasing specialization in many academic disciplines, and gave rise to what is now called inter- and transdisciplinary research
Thus, to attract funding, other innovative words and phrases have to be invented, the latest being 'systems biology', a return to von Bertalanffy. Systems biology promises to speed up the process of research by bypassing the classical methods and using a holistic approach. Instead of starting with the least complex organisms and progressively adding regulatory devices to finally reach the most complex, presumably human, organism, its proponents hope once more to harness the power of modern supercomputing to explain near-causal relationships in highly complex organisms (Kitano, 2002). When considering a 'whole' (holistic) system, a final effect is caused by a multitude of causes and modifying factors; similarly, a single cause might give rise to multiple effects. The human brain is not particularly well equipped for such a multifactorial analysis; computers can do this much better. This is illustrated by the computer's success in climate and environmental research. Time will tell if it will also help us to study complex biological systems; it may well speed up the application of biological knowledge for developing new products. But the advancement of fundamental knowledge about the functions of individual causalities of networks will more likely come from the classical reductionist approach, as exemplified by the breakthrough of molecular genetics half a century ago, when biology was already rather holistic. It promises again the discovery of new, fundamental laws and rules of nature.

Acknowledgments
I am indebted to J. Leisi for his important clarifications of the considerations in physics and to V. Bonifas for improving the manuscript.
References
- Asimov I (1963) The Genetic Code. Signet Science Library, New York, USA [Google Scholar]
- Avery OT, MacLeod CM, McCarthy M (1944) Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med 79: 137–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis EL, Delbrück M (1939) The growth of bacteriophage. J Gen Physiol 22: 365–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamow G (1954) Possible relation between deoxyribonucleic acid and protein structures. Nature 173: 318 [Google Scholar]
- Gibbs WW (2003) The unseen genome: gems among the junk. Sci Am 289: 46–53 [PubMed] [Google Scholar]
- Hershey AD, Chase M (1951) Independent functions of viral protein and nucleic acid in the growth of bacteriophage. J Gen Physiol 36: 35–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger E (1995) History of phage research as viewed by a European. FEMS Microbiol Rev 17: 7–24 [Google Scholar]
- Kellenberger E (1997) in Chaos in der Wissenschaft (ed. Onori P), 169–183. Des Kantons Basel–Landschaft, Liestal, Germany [Google Scholar]
- Kellenberger E, Wunderli H (1995) Electron microscopic studies on intracellular phage development—history and perspectives. Micron 26: 213–245 [DOI] [PubMed] [Google Scholar]
- Kitano H (2002) Systems biology: a brief overview. Science 295: 1662–1664 [DOI] [PubMed] [Google Scholar]
- Nirenberg M, Leder P, Bernfield M, Brimacomb R, Trupin J, Tottman F, O'Neal C (1965) RNA codewords and protein synthesis. VII. On the general nature of the RNA code. Proc Natl Acad Sci USA 53: 1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Jones DS, Khorana HG (1964) The _in vitro_synthesis of a copolypeptide containing two amino acids in alternating sequence dependent upon a DNA-like polymer containing two nucleotides in alternating sequence. J Mol Biol 13: 302–324 [DOI] [PubMed] [Google Scholar]
- Rheinberger HJ (1997) Toward a History of Epistemic Things. Synthesizing Proteins in the Test Tube. Stanford University Press, Stanford, California, USA [Google Scholar]
- Scherrer K (1989) A unified matrix hypothesis of DNA-directed morphogenesis, protodynamism and growth control. Biosci Rep 9: 157–188 [DOI] [PubMed] [Google Scholar]
- Schrödinger E (1943) What is Life? Cambridge University Press, Cambridge, UK [Google Scholar]
- von Bertalanffy L (1949) Das Biologische Weltbild. A. Francke, Bern, Switzerland [Google Scholar]
- Watson JD, Crick FHC (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171: 737–738 [DOI] [PubMed] [Google Scholar]