The dynamic organization of gene-regulatory machinery in nuclear microenvironments (original) (raw)
Abstract
Nuclear components are functionally linked with the dynamic temporal and spatial compartmentalization, sorting and integration of regulatory information to facilitate its selective use. For example, the subnuclear targeting of transcription factors to punctate sites in the interphase nucleus mechanistically couples chromatin remodelling and the execution of signalling cascades that mediate gene expression with the combinatorial assembly of the regulatory machinery for biological control. In addition, a mitotic cycle of selective partitioning and sequential restoration of the transcriptional machinery provides a basis for the reassembly of regulatory complexes to render progeny cells competent for phenotypic gene expression. When this intranuclear targeting and localization of regulatory proteins is compromised, diseases, such as cancer, can occur. A detailed understanding of this process will provide further options for diagnosis and treatment.
Keywords: chromatin organization, gene regulation, intranuclear transport, nuclear microenvironments, transcriptional control
Introduction
Advances in experimental strategies have provided essential tools for studying the nuclear structure–function interrelationship. New protocols for image analysis (for example, intranuclear informatics) have indicated that regulatory proteins in the nucleus have a characteristic architectural signature (Young et al, 2004). Biochemical and cell-biological approaches have also shown that nuclear regulatory proteins (for example, RUNX transcription factors; Zeng et al, 1997; Zaidi et al, 2001) are directed to intranuclear sites by unique subnuclear targeting motifs (for example, the nuclear-matrix-targeting signal or NMTS). Similarly, biophysical and structural properties of proteins support the study of combinatorial control at the molecular level (Taatjes et al, 2004). Here, we address the dynamic organization of proteins in nuclear microenvironments for the combinatorial assembly and activities of the gene-regulatory machinery under biological conditions, and consider the consequences of the perturbations that occur during human disease.
Organizing nuclear microenvironments
Physiologically responsive gene expression in vivo requires the temporal and spatial assembly and activities of the transcriptional regulatory machinery. The architectural organization of nucleic acids and regulatory proteins supports an interrelationship between nuclear structure and gene expression. Biochemical in situ immunofluorescence and genetic approaches have identified numerous levels of control in transcriptional regulation, which include the following: the convergence of several regulatory signals at promoter elements; the integration of regulatory information between independent promoter domains; the selective use of redundant regulatory pathways; thresholds for transcriptional initiation or suppression through limiting amounts of promoter elements and regulatory factors; mechanisms that render promoters of cell growth and phenotypic genes competent for protein–DNA and protein–protein interactions in response to regulatory cues; the combinatorial organization of focally placed regulatory complexes at the promoter sites of target genes and in subnuclear domains where the transcriptional machinery is assembled; and the intranuclear trafficking of regulatory proteins to transcriptionally active foci.
The biological response of cells to diverse regulatory cues requires the optimal temporal and spatial representation of nucleic acids and proteins. These threshold concentrations of the genetic-regulatory machinery are supported by the presence of architecturally associated macromolecular complexes in punctuate nuclear microenvironments. Nuclear microenvironments can be functionally defined as DNA-based (for example, chromosome territories) or protein-based (for example, subnuclear domains). The compartmentalization of multifunctional protein complexes into nuclear microenvironments is illustrated by the focal organization of promyelocytic leukaemia (PML) bodies (Dyck et al, 1994), RUNX sites (Zeng et al, 1997; Zaidi et al, 2001), the nucleolus (Olson et al, 2002) and chromosomes (Ma et al, 1999), as well as by the intranuclear distribution of sites for replication (Cook, 1999), DNA repair (Scully & Livingston, 2000), transcription (Verschure et al, 1999), steroid and polypeptide responsiveness (Nagaich et al, 2004), and RNA processing (Misteli & Spector, 1999; Smith et al, 1999; Wagner et al, 2003). This architectural organization could allow the optimal representation of nucleic acids and regulatory proteins that are required for protein–protein and protein–DNA interactions to organize and assemble regulatory complexes (Fig 1).
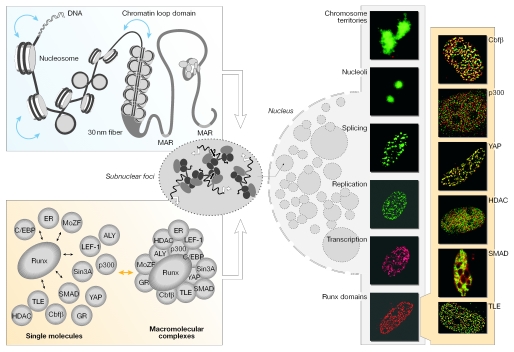
Figure 1.
Levels of nuclear organization. The linear placement of DNA-regulatory elements in gene promoters constitutes the primary level of nuclear organization. The distance between these regulatory sites is intricately regulated by the packaging of DNA into nucleosomes and higher order chromatin structures (left, upper panel). Scaffolding nuclear proteins, such as RUNX, provide structural platforms for the assembly of multiprotein supercomplexes to facilitate the combinatorial control of gene expression (left, bottom panel). Genes and macromolecular regulatory complexes together give rise to dynamic nuclear microenvironments in the nucleus. RUNX bodies are nuclear microenvironments that contain various co-regulatory proteins that are involved in gene activation, as well as repression, chromatin remodelling and cellular signalling (immunofluorescence images on the right, shaded yellow). RUNX was visualized using the Alexa 488 secondary antibody in all images and the proteins were detected using Alexa 568 fluorochrome-conjugated secondary antibodies, as indicated.
Remodelling nuclear microenvironments
Nuclear microenvironments are often dynamic in nature to accommodate temporal macromolecular interactions. For example, the S-phase-specific transcriptional regulation of histone gene clusters reflects the dynamic nature of regulatory nuclear microenvironments. In proliferating cells, histone gene expression requires the recruitment of transcription factors to organize S-phase-specific nuclear microenvironments. These punctate intranuclear sites contain HiNF-P, which is a transcription factor that binds to the cell-cycle regulatory sites of the histone genes (Mitra et al, 2003), NPAT, which is an HiNF-P co-regulatory protein (Zhao et al, 1998; Ma et al, 2000), and Cajal bodies (Shopland et al, 2001). In addition, PML bodies also associate with histone genes in the S phase (Wang et al, 2004). Therefore, histone-gene-related nuclear microenvironments facilitate the combinatorial activity of proteins with diverse functions, which are obligatory for S-phase-specific histone gene expression. Similarly, Ikaros proteins offer a conceptually analogous example of dynamic nuclear microenvironments during lymphoid differentiation. Ikaros first localizes diffusely in the nucleoplasm and then accumulates at centromeric loci during lymphoid differentiation. This movement coincides with the relocation and inactivation of Ikaros target genes (Brown et al, 1999). Overall, the dynamic remodelling of nuclear microenvironments allows numerous protein–protein and protein–DNA interactions, and organizes functional intranuclear domains where transcription takes place.
Nuclear microenvironments also contain scaffolding proteins that recruit co-regulatory factors for the combinatorial control of proliferation and differentiation. Scaffolding proteins, such as RUNX and acute lymphoblastic leukaemia 1 (ALL1), provide a model for understanding the assembly of the gene-regulatory machinery in the three-dimensional context of nuclear organization (Fig 1). The dual recognition of the RUNX transcription factor by both promoter elements and co-regulatory proteins modulates the structural and functional properties of target genes by facilitating the assembly of complexes that form nuclear microenvironments. Sequence-specific interactions with promoter elements place RUNX proteins at strategic sites where they act as scaffolds for protein–protein interactions to organize the regulatory machineries. These include machineries for histone modifications and chromatin remodelling, which increase the competency for transcription-factor binding and establish genomic conformations that mediate crosstalk between proximal and upstream promoter regions (Stein et al, 2003). At the same time, regulatory cues from signalling pathways that activate or suppress gene expression in a physiologically responsive manner are integrated (Zaidi et al, 2002, 2004). In addition, the RUNX proteins are post-translationally modified to influence their activity further (Lian et al, 2004). Similarly, the complexity of the ALL1 regulatory protein that assembles as a supercomplex of basal transcription factors, chromatin-remodelling factors and histone-modifying proteins, which are architecturally associated and focally organized, illustrates the potential influence of leukaemia-related chromosomal translocations on gene expression (Nakamura et al, 2002). Therefore, transcription factors that function as scaffolds and interact with co-regulatory proteins provide a platform for the remodelling of the architecturally organized regulatory machinery in the nucleus.
Cell-biological and biochemical evidence points to a dynamic, but stable, organization of nuclear microenvironments to accommodate the temporal and spatial requirements for the regulation of gene expression. In this model, scaffolding proteins use a hierarchy of targeting sequences to assemble regulatory complexes at punctate sites in the nucleus. These motifs include the nuclear-localization signal for nuclear import, the DNA-binding domain for interaction with promoter regulatory elements and the subnuclear-targeting sequence for intranuclear trafficking. The interaction of scaffolding proteins with numerous co-regulatory proteins that have diverse functions then results in the assembly and organization of functional nuclear microenvironments (Fig 2). This model for the organization and remodelling of nuclear microenvironments is compatible with the self-organization model of nuclear assembly based on live-cell microscopy (Misteli, 2001). However, the distinction between the self-assembly and the self-organization of regulatory complexes and subnuclear compartments is essentially semantic. Nuclear microenvironments arise from transient and long-term components with both in vivo and in vitro support for dynamic changes that provide competency for physiological responsiveness. A definitive and comprehensive characterization of punctately organized transcription sites has yet to be performed. Experimental approaches that are based on native concentrations of endogenous proteins and, where possible, in vivo genetic models should definitively address the biologically relevant mechanisms that contribute to the organization of regulatory mechanisms at sites in the nucleus.
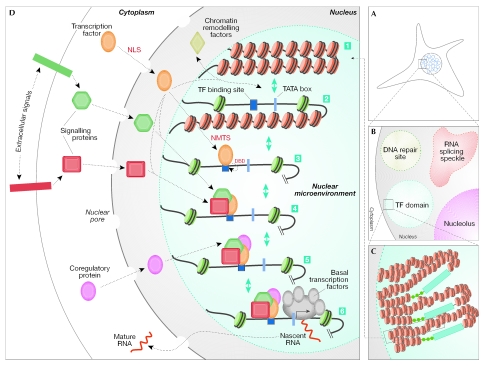
Figure 2.
Biogenesis and regulatory activities of nuclear microenvironments that support biological control. The multistep organization and assembly of nuclear microenvironments that integrate extracellular and intracellular regulatory signals to establish transcriptional competency is shown. (A) The mammalian nucleus is functionally compartmentalized into punctate subnuclear domains (blue circles). (B) An enlarged area of the mammalian nucleus shows four nuclear microenvironments with distinct functions: the nucleolus (purple circle) is the site of ribosomal RNA synthesis; RNA-splicing speckles (shown in red), such as SC35, are involved in RNA splicing/processing; various proteins accumulate at sites of DNA damage to organize distinct nuclear domains for DNA repair (green circle); and transcription factors are present in distinct foci in the nucleus, the subnuclear placement of which often correlates with gene regulation (light-blue circle). (C) A transcription domain can contain several target genes (shown here in repressed or closed chromatin conformations) for a transcription factor to amplify the representation of the regulatory machinery, thereby facilitating the combinatorial control of transcriptional regulation. (D) This composite schematic for the control of gene expression in the context of nuclear architecture incorporates various regulatory parameters that are operative in vivo, although not necessarily in a specific temporal sequence. (1) Transcriptional repression is mediated by nucleosomes that restrict the access of promoter elements to regulatory proteins. Inactive genes show the closed chromatin conformation (red circles). (2) To accommodate cellular response to specific extracellular stimuli, chromatin modifications, which are mediated by macromolecular chromatin-remodelling complexes, render the open conformation to the gene-promoter region; this results in the accessibility of promoter domains to the tissue-restricted, as well as the basal, transcription machinery. (3) Transcription factors that are synthesized in the cytoplasm translocate into the nucleus through a nuclear-localization signal (NLS), are targeted to the subnuclear domains by the intranuclear-targeting signal (NMTS) and bind DNA through the DNA-binding domain (DBD). (4) Various physiological cues, with positive (green hexagon) or negative (red square) regulatory potential, are transmitted to the nucleus through signalling proteins; these proteins interact with transcription factors and use their subnuclear targeting signals, as well as DBDs, to converge at the target genes. (5) Other regulatory proteins (purple circle) also converge at independent, or similar, promoter regions of the target genes, which results in the combinatorial control of gene expression. (6) Finally, the basal transcription machinery is recruited to the promoter, which leads to the activation of genes and mRNA synthesis (shown as nascent RNA). The nascent RNA is then exported from the nucleus, after various steps of RNA splicing and processing that are carried out in RNA-splicing speckles, as mature RNA.
Intranuclear trafficking of regulatory proteins
Mechanisms are emerging for the selective trafficking of proteins to nuclear microenvironments to organize functional regulatory complexes in the interphase nucleus. The discovery of the NMTS that is responsible for the architectural association of osteoblast, myeloid and lymphoid RUNX transcription factors has allowed the examination of mechanisms that direct regulatory proteins to transcriptionally active subnuclear domains (Zeng et al, 1997; Zaidi et al, 2001). The NMTS functions autonomously and is necessary, as well as sufficient, to direct the transcriptionally active RUNX factors to nuclear matrix-associated sites where gene expression occurs. The biological role of Runx intranuclear trafficking has been established in vivo by mouse models that express Runx proteins with impaired subnuclear targeting. These mice show similar phenotypes to _Runx_-null mice, which indicates that the assembly and organization of Runx-containing macromolecular complexes at subnuclear sites is intimately linked to their biological activity (North et al, 1999; Choi et al, 2001).
In addition to organizing and remodelling regulatory microenvironments in the interphase nucleus, the post-mitotic restoration of nuclear organization and the assembly of regulatory complexes are required to render progeny cells competent for phenotypic gene expression. A mitotic cycle of selective partitioning and sequential restoration of the regulatory machinery for RNA synthesis (for example, RUNX foci) and processing (for example, SC35 speckles), correlates with the ordered use of the regulatory machinery that is involved in the various steps of transcriptional control (Zaidi et al, 2003). Several other proteins, such as ALL1 (Ennas et al, 1997) and upstream binding factor 1 (UBF1; Gebrane-Younes et al, 1997), show similar mitotic transport. The mechanisms that regulate the association of regulatory proteins with mitotic chromosomes and/or the mitotic apparatus are only now beginning to be clarified. A viable possibility is that chromosomal foci contain co-regulatory proteins and several target genes that are poised for postmitotic transcription. Further studies are required to provide mechanistic insight. However, such trafficking of proteins seems to be crucial for phenotypic gene expression and provides a basis for understanding the post-mitotic events that ensure the fidelity of gene expression in progeny cells.
Catastrophic perturbations in nuclear microenvironments
Compromised architectural organization of regulatory complexes in the nucleus results in perturbed gene expression (Fig 3). For example, a functional linkage between the deregulated nuclear import of regulatory proteins and a disease state is reflected by impaired fibroblast growth factor (FGF) signalling and tumorigenesis (Stachowiak et al, 2003), the insulin-like growth factor 1 (IGF1) signalling pathway in haematopoiesis and leukaemogenesis (Baserga et al, 2003), α-thalassaemia retardation syndrome X-linked (ATRX) signalling in neurological abnormalities (Berube et al, 2002), anaphase-promoting complex (APC)/β-catenin signalling in colon cancer (Clevers, 2004) and the detection of translocation-fusion proteins in myeloid leukaemias (Spencer & Davie, 2000; Zelent et al, 2001).
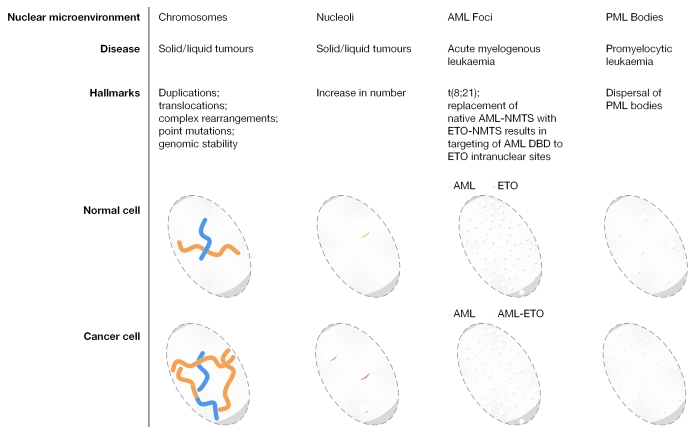
Figure 3.
The catastrophic consequences of perturbations in nuclear microenvironments. Examples of nuclear microenvironments that undergo alterations under pathological conditions are shown. Chromosomes undergo extensive changes, including translocations, duplications and mutations, in a broad range of cancers. Similarly, the number of nucleoli, which are the sites of ribosomal gene expression, is increased in tumours. The (8;21) translocation in acute myeloid leukaemia (AML) results in the misrouting of RUNX1 to AML–eight twenty one (ETO) subnuclear sites (yellow circles). Similarly, promyelocytic leukaemia (PML) bodies increase in number and decrease in size in PML patients.
The importance of the subnuclear localization of transcription factors for biological control is indicated by the compromised subnuclear organization and activity of the RUNX1/acute myeloid leukaemia 1 (AML1) haematopoietic regulatory protein in AML patients. The chromosomal translocation in a significant proportion of AML patients results in a chimeric protein acute myelogenous leukaemia–eight twenty one (AML–ETO) translocation. This fusion protein has several subnuclear targeting signals and organizes into nuclear microenvironments that are distinct from those of wild-type RUNX1 (McNeil et al, 1999; Barseguian et al, 2002). Therefore, the pathology of AML involves the subnuclear misrouting of RUNX1, which results from the loss of the carboxyl terminus and a gain-of-function by the fusion protein. Similarly, the chimeric promyelocytic leukaemia–retinoic-acid receptor (PML–RAR) fusion protein results in the dispersal of PML bodies, and leads to apoptosis and stress responses. The treatment of PML patients with all-trans retinoic acid results in remission from leukaemia accompanied by the restoration of PML bodies (Zelent et al, 2001). Crosstalk between nuclear microenvironments that results in modifications to the organization of regulatory complexes is illustrated by the AML–ETO-dependent dispersal of PML bodies in a manner that is analogous to the intranuclear distribution of PML–RAR foci (McNeil et al 2000).
Proliferative diseases, such as cancer, are traditionally treated by blocking proliferation in a non-selective manner. However, a new generation of therapeutics is now emerging that selectively targets regulatory processes. Combinatorial activity and architectural association of the regulatory machinery provides an added dimension to drug specificity and selectivity. Several parameters of nuclear organization are being used for the treatment of cancer. For example, intercalating agents and derivatives of butyrate allow the manipulation of DNA and chromatin structure. Similarly, kinase inhibitors intercept and modulate the integration of regulatory signals in nuclear microenvironments. Intranuclear trafficking offers mechanistic insight into the assembly and organization of nuclear microenvironments for the combinatorial control of gene expression, and can be exploited to modulate gene expression under pathological conditions.
Nuclear microenvironments in biological control
Evidence for focal thresholds of genes and regulatory factors that optimize informative encounters to support transcription, replication and repair has emerged from the interrogation of fundamental parameters of biological control. Additional insight into the regulation of gene expression should be forthcoming from further exploration of the mechanisms and underlying parameters that mediate the multistep dynamic positioning and combinatorial association of regulatory macromolecules in the nucleus. Despite compelling support for the physiological relevance of intranuclear regulatory domains, there is a requirement to define the rate-limiting parameters of mechanisms that mediate the temporal and spatial components of nuclear structure–gene expression interrelationships. The rules governing organization of the regulatory machinery in the three-dimensional context of nuclear architecture are being functionally enhanced by the combined application of molecular, cellular and in vivo genetic approaches. It is realistic to expect that mechanistic explanations for the dynamic organization of the gene-regulatory machinery in nuclear microenvironments, which are compromised in many diseases, including cancer, can provide a platform for new approaches to diagnosis and treatment.
Acknowledgments
The studies from our laboratory that are presented in this review were partly supported by grants PO1 AR48818, PO1 CA82834 and 5 P30 DK32520 from the National Institutes of Health (NIH). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The authors thank E. Bronstein and J. Rask for their editorial assistance with the preparation of this manuscript.
References
- Barseguian K, Lutterbach B, Hiebert SW, Nickerson J, Lian JB, Stein JL, van Wijnen AJ, Stein GS (2002) Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proc Natl Acad Sci USA 99: 15434–15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga R, Peruzzi F, Reiss K (2003) The IGF-1 receptor in cancer biology. Int J Cancer 107: 873–877 [DOI] [PubMed] [Google Scholar]
- Berube NG, Jagla M, Smeenk C, De Repentigny Y, Kothary R, Picketts DJ (2002) Neurodevelopmental defects resulting from ATRX overexpression in transgenic mice. Hum Mol Genet 11: 253–261 [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG (1999) Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell 3: 207–217 [DOI] [PubMed] [Google Scholar]
- Choi J-Y et al. (2001) Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA 98: 8650–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2004) Wnt breakers in colon cancer. Cancer Cell 5: 5–6 [DOI] [PubMed] [Google Scholar]
- Cook PR (1999) The organization of replication and transcription. Science 284: 1790–1795 [DOI] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WH, Chen JD, Kakizuka A, Evans RM (1994) A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76: 333–343 [DOI] [PubMed] [Google Scholar]
- Ennas MG, Sorio C, Greim R, Nieddu M, Scarpa A, Orlandini S, Croce CM, Fey GH, Marschalek R (1997) The human ALL-1/MLL/HRX antigen is predominantly localized in the nucleus of resting and proliferating peripheral blood mononuclear cells. Cancer Res 57: 2035–2041 [PubMed] [Google Scholar]
- Gebrane-Younes J, Fomproix N, Hernandez-Verdun D (1997) When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci 110: 2429–2440 [DOI] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS (2004) Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr 14: 1–41 [PubMed] [Google Scholar]
- Ma H, Siegel AJ, Berezney R (1999) Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol 146: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW (2000) Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev 14: 2298–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil S, Zeng C, Harrington KS, Hiebert S, Lian JB, Stein JL, van Wijnen AJ, Stein GS (1999) The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFa2 transcription factor. Proc Natl Acad Sci USA 96: 14882–14887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil S, Javed A, Harrington KS, Lian JB, Stein JL, van Wijnen AJ, Stein GS (2000) Leukemia-associated AML1/ETO (8;21) chromosomal translocation protein increases the cellular representation of PML bodies. J Cell Biochem 79: 103–112 [PubMed] [Google Scholar]
- Misteli T (2001) The concept of self-organization in cellular architecture. J Cell Biol 155: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL (1999) RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3: 697–705 [DOI] [PubMed] [Google Scholar]
- Mitra P, Xie R, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, Harper JW, Stein JL, van Wijnen AJ, Stein GS (2003) Identification of HiNF-P, a key regulator of cell cycle-controlled histone H4 genes at the onset of S phase. Mol Cell Biol 23: 8110–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Rayasam GV, Martinez ED, Becker M, Qiu Y, Johnson TA, Elbi C, Fletcher TM, John S, Hager GL (2004) Subnuclear trafficking and gene targeting by steroid receptors. Ann NY Acad Sci 1024: 213–220 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 10: 1119–1128 [DOI] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA (1999) Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126: 2563–2575 [DOI] [PubMed] [Google Scholar]
- Olson MO, Hingorani K, Szebeni A (2002) Conventional and nonconventional roles of the nucleolus. Int Rev Cytol 219: 199–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Livingston DM (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408: 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB (2001) Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell 12: 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB (1999) Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol 144: 617–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer VA, Davie JR (2000) Signal transduction pathways and chromatin structure in cancer cells. J Cell Biochem 35(Suppl): 27–35 [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Fang X, Myers JM, Dunham SM, Berezney R, Maher PA, Stachowiak EK (2003) Integrative nuclear FGFR1 signaling (INFS) as a part of a universal 'feed-forward-and-gate' signaling module that controls cell growth and differentiation. J Cell Biochem 90: 662–691 [DOI] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi J-Y, Stein JL, Lian JB, Javed A (2003) Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol 13: 584–592 [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Marr MT, Tjian R (2004) Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol 5: 403–410 [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van Der Kraan I, Manders EM, van Driel R (1999) Spatial relationship between transcription sites and chromosome territories. J Cell Biol 147: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Chiosea S, Nickerson JA (2003) The spatial targeting and nuclear matrix binding domains of SRm160. Proc Natl Acad Sci USA 100: 3269–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D (2004) Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol 164: 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Zaidi SK, Furcinitti PS, Javed A, van Wijnen AJ, Stein JL, Lian JB, Stein GS (2004) Quantitative signature for architectural organization of regulatory factors using intranuclear informatics. J Cell Sci 117: 4889–4896 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Choi J-Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS (2001) A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci 114: 3093–3102 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, Lian JB (2002) Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proc Natl Acad Sci USA 99: 8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SH, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS (2003) Mitotic partitioning and selective reorganization of tissue specific transcription factors in progeny cells. Proc Natl Acad Sci USA 100: 14852–14857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS (2004) Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J 23: 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelent A, Guidez F, Melnick A, Waxman S, Licht JD (2001) Translocations of the RARα gene in acute promyelocytic leukemia. Oncogene 20: 7186–7203 [DOI] [PubMed] [Google Scholar]
- Zeng C et al. (1997) Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA 94: 6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dynlacht B, Imai T, Hori T, Harlow E (1998) Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev 12: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]