Distribution and Evolution of von Willebrand/Integrin A Domains: Widely Dispersed Domains with Roles in Cell Adhesion and Elsewhere (original) (raw)
Abstract
The von Willebrand A (VWA) domain is a well-studied domain involved in cell adhesion, in extracellular matrix proteins, and in integrin receptors. A number of human diseases arise from mutations in VWA domains. We have analyzed the phylogenetic distribution of this domain and the relationships among ∼500 proteins containing this domain. Although the majority of VWA-containing proteins are extracellular, the most ancient ones, present in all eukaryotes, are all intracellular proteins involved in functions such as transcription, DNA repair, ribosomal and membrane transport, and the proteasome. A common feature seems to be involvement in multiprotein complexes. Subsequent evolution involved deployment of VWA domains by Metazoa in extracellular proteins involved in cell adhesion such as integrin β subunits (all Metazoa). Nematodes and chordates separately expanded their complements of extracellular matrix proteins containing VWA domains, whereas plants expanded their intracellular complement. Chordates developed VWA-containing integrin α subunits, collagens, and other extracellular matrix proteins (e.g., matrilins, cochlin/vitrin, and von Willebrand factor). Consideration of the known properties of VWA domains in integrins and extracellular matrix proteins allows insights into their involvement in protein–protein interactions and the roles of bound divalent cations and conformational changes. These allow inferences about similar functions in novel situations such as protease regulators (e.g., complement factors and trypsin inhibitors) and intracellular proteins (e.g., helicases, chelatases, and copines).
INTRODUCTION
The rapid accumulation of genomic sequences offers both the challenge of understanding the functions of proteins encoded by those genomes and the opportunity for drawing inferences about the evolution of functions in proteins in different phyla. Effective annotation of the genes and their products requires both analyses of sequence and structural homologies among genes and incorporation of biochemical and biological information about the proteins to make best use of the genomic information.
We have been interested in the structure, function, and evolution of proteins involved in cell adhesion and interaction (Hynes and Zhao, 2000). Annotation of these proteins represents a significant challenge because we estimate that there are >2000 such proteins encoded by mammalian genomes. Searching proteomes for conserved domains is a first step toward overcoming that challenge. Some conserved domains have been extensively studied and their presence within a protein suggests specific biological properties. In this essay, we present an analysis of a subset of proteins; those including the so-called von Willebrand A (VWA) domain (reviewed in Colombatti et al., 1993; Tuckwell, 1999). Proteins containing VWA domains are present in Eukaryota (Metazoa, fungi, plants, and protists), Eubacteria, and Archaea. VWA domains are Rossmann folds consisting of a β-sheet sandwiched by multiple α helices. Many VWA domains bind metal ions via a noncontiguous sequence motif called metal ion-dependent adhesion site (MIDAS). Frequently, VWA domain-containing proteins function in multiprotein complexes. The eponymous VWA domains of von Willebrand factor play key roles in the linkage of platelets to collagen (see below). The homologous inserted or I domains of some integrin α subunits are also involved in interactions with collagen and other ligands. Our initial interest in VWA/I domains arose from considerations of the evolution of integrin subunits because VWA domain-containing integrin α subunits seem to be restricted to chordates (Hynes and Zhao, 2000). In the course of investigating the verity and significance of this supposition, we explored the universe of VWA/I domains, and we present herein the results of that inquiry, including a number of novel proteins and new insights.
The majority of well-characterized VWA domains are found in cell adhesion and extracellular matrix (ECM) proteins (Tuckwell, 1999). In many cases, it is clear, or plausible, that they are involved in protein–protein (e.g., receptor–ligand) interactions that frequently involve divalent cations. In exploring what might be the origin of this domain, we discovered that the VWA domains most widely distributed phylogenetically are intracellular proteins present in all eukaryotic genomes sequenced thus far. The roles of the VWA domains in these intracellular proteins are not clear, but many are components of multiprotein complexes and a plausible hypothesis is that the VWA domains mediate protein–protein interactions involved in the assembly or function of these complexes. It seems that the presumptive primordial VWA domains subsequently were deployed by metazoans in extracellular protein–protein interactions, with integrin β subunits as very early representatives. Later incorporations of VWA domains also into integrin α subunits and into many ECM molecules seem to be predominantly chordate elaborations presumably related to the large expansion of ECM complexity in chordates, although other eukaryotes, notably, Caenorhabditis elegans, have also deployed VWA domains in ECM proteins. VWA domains also pop up in other surprising and interesting contexts such as ion channel subunits, the anthrax toxin receptor, and protease regulators.
In this review, we consider first the best understood VWA domains; those in integrins and von Willebrand factor (vWF) from which one can infer their likely functions. We then consider the other contexts in which one finds these domains and discuss their possible functions and potential evolution.
MATERIALS AND METHODS
As of 24 March 2002 the SMART nonredundant database (nrdb) (a merger of swissprot, swissnew, sptrembl, and sptremblnew) contained 948 proteins containing 1196 VWA domains, with new additions being deposited regularly (Letunic et al., 2002; http://smart.embl-heidelberg.de/). The survey presented in this essay is based largely on the SMART database and analysis because of the experience with integrin β subunits. The properties of integrin β subunits prompted speculation that they contained a VWA domain (Lee et al., 1995; Tozer et al., 1996; Loftus and Liddington, 1997; Tuckwell and Humphries, 1997), in spite of the fact that many domain prediction algorithms were unable to support this claim. However, the SMART analysis techniques predict the VWA domain in integrin β subunits with high confidence (Tuckwell, 1999; Ponting et al., 2000; Schultz et al., 2000). Recently, the crystal structure of the extracellular portion of the integrin αvβ3 was reported (Xiong et al., 2001), and the presence of a VWA domain in the β subunit was confirmed. Therefore, we feel that the SMART algorithm for prediction of VWA domains is currently the best available, and we have used SMART analysis as the basis for this essay. We have complemented these analyses with Interpro where helpful. In addition, we have used each human VWA domain predicted by SMART (as of March 2002) to query the Genomescan-predicted protein database at National Center for Biotechnology Information (Yeh et al., 2001;http://www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html). The National Center for Biotechnology Information genome annotation effort is ongoing, and many novel proteins detected by Genomescan have not yet been assigned to the databases included in the SMART nonredundant database. In many cases, particularly the novel collagens we report herein, the public and private human and mouse genome assemblies were also used extensively to arrive at plausible structures for uncharacterized molecules.
In addition to overall homology of domain structure and database queries with basic local alignment search tool (BLAST), we also made use of whole-molecule and individual VWA domain (extracted using SMART) alignments. Alignments, bootstrap analysis, and tree preparation were done using ClustalW, ClustalTree, and DrawTree provided by the Biology Workbench 3.2 (http://workbench.sdsc.edu/; Felsenstein, 1989) and the VectorNTI software package. Frequently, bootstrap numbers, expressed as a percentage of 1000 pseudoreplicates, are provided to give a confidence level for a given relationship (Efron et al., 1996). In the case of whole-molecule alignments, the resulting phylogenetic trees were used to provide a framework for discussion of families of molecules. In the case of the individual domain alignments, phylogenetic trees and bootstrap values were used to identify uncharacterized homologues by calculating relatedness among different domains. This type of analysis was more systematic and quantifiable than individual BLAST searches and was particularly useful in cases where the molecules in the databases were fragments or gene predictions. Due to the relatively large number of VWA domains in the database, it was sometimes necessary to create species-specific minimal sets where a single representative sequence was selected for each group of closely related sequences (usually paralogues; Table S1). This was done by first removing nearly identical sequences such as allelic variants and fragments to create a more strictly defined nonredundant set. The nonredundant sets were then aligned and subjected to bootstrap analysis and groups of closely related sequences were reduced by randomly selecting a single representative from each node with bootstrap support >94%. These minimal collections were then small enough to allow pairwise comparisons of the complement of VWA domains within a species or group.
We also investigated the occurrence of MIDAS motifs, first defined as metal ion-binding motifs in the VWA domains of integrin α subunits (Lee et al., 1995). Because metal ions play key roles in the functions of VWA domains in integrins, we scored the three noncontiguous elements of the MIDAS motif (D-x-S-x-S… . T… . D) in hand-edited alignments of all the VWA domains identified in the five completed eukaryotic genomes as well as all prokaryotic VWA domains and a subset of protist VWA domains. For the purpose of discussion, the D-x-S-x-S will be referred to as region 1 and the other conserved residues will be called T4 and D5 (Figure S1, a–c). Approximately 46% of VWA domains have a perfectly conserved MIDAS motif (see figures, red stars); others are missing one or more elements. Structural studies of integrin VWA domains and biochemical analysis of copines (Tomsig and Creutz, 2000) indicate that a perfectly conserved MIDAS motif is not required for metal ion binding. To accommodate these observations and emphasize the importance in metal binding of the region 1 D followed by spaced alcohol residues (S or T), we coined the term imperfect MIDAS (open red stars) to refer to VWA domains that lack a subset of MIDAS elements but are likely to bind metal ions. Examples of imperfect motifs include those with region 1 (D-x-S-x-S) but without one or both of T4 and D5 or those with conservative changes in region 1 (D-x-T-x-S in copines) with and without conservation of T4 and D5. It is likely that confident conclusions regarding the presence or absence of a functional MIDAS motif will require structural analysis of the VWA domain in question in both ligand-bound and -unbound states (Xiong et al., 2002). In light of these considerations, we have presented our analysis of MIDAS motifs in an Excel (Microsoft, Redmond, WA) spreadsheet (Table S3) that will allow interested readers to sort VWA domains with respect to conservation and sequence of any or all of the three MIDAS elements.
RESULTS
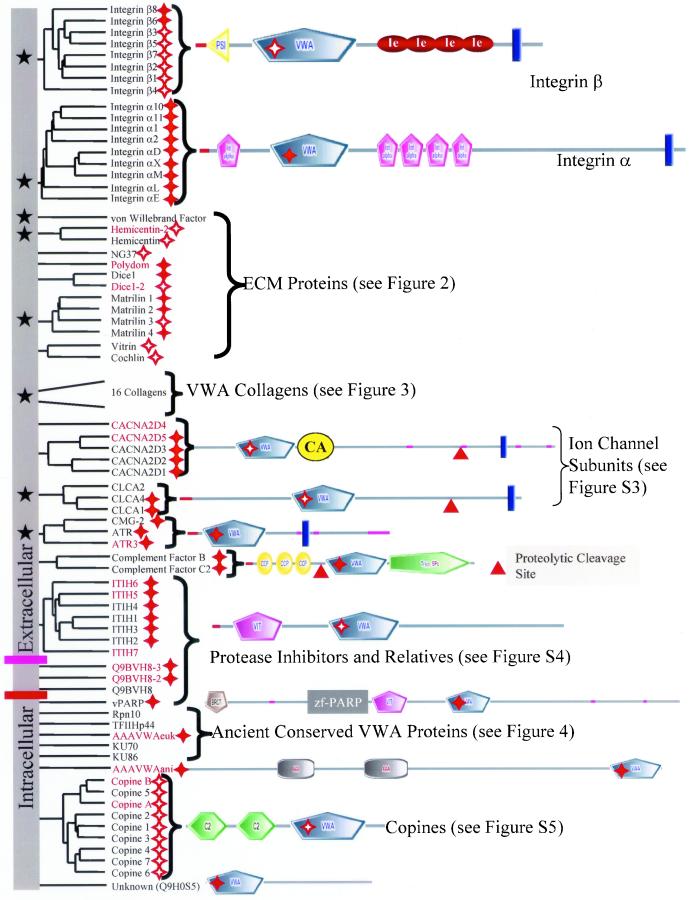
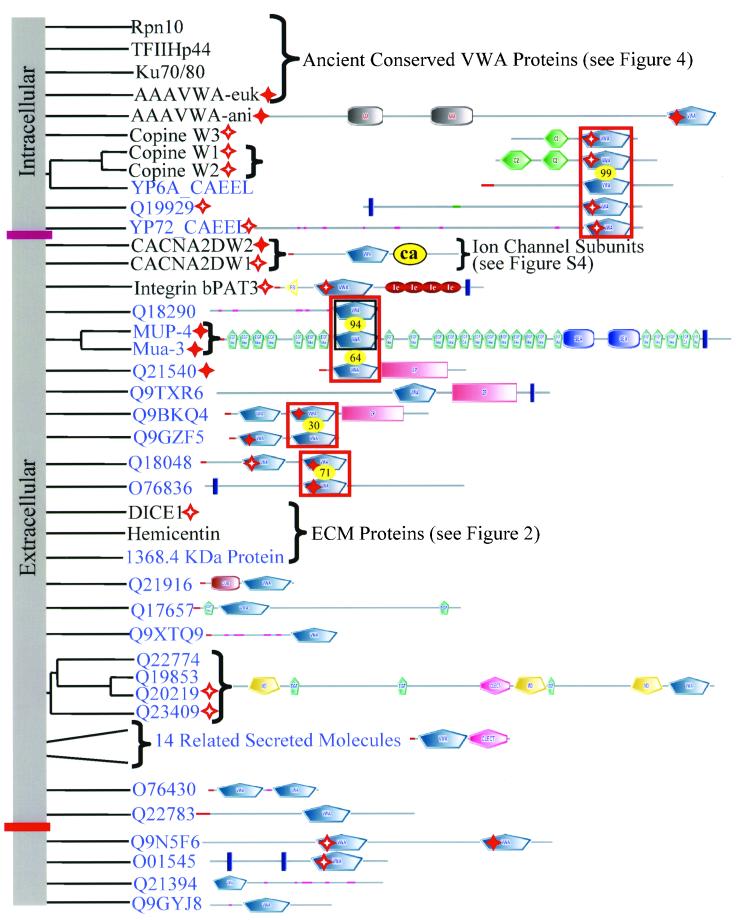
Our analysis of the human proteome uncovered 86 proteins containing 134 VWA domains (pseudogenes and splice variants excluded). Phylogenetic analysis and examination of domain architectures indicate that most of these proteins fall into 15 clearly paralogueous groups (Figure 1). These molecules can be clustered into the following categories.
Figure 1.
Cell adhesion and extracellular matrix molecules predominate in the diverse collection of human VWA domain-containing proteins. The domain architecture of all human VWA domain-containing proteins is indicated in this figure and the following ones as noted. Paralogous molecules are grouped together in the phylogenetic tree derived from a clustalW alignment. The groups of paralogues or unrooted individual molecules have been shuffled along the vertical axis for clarity of presentation; so there is no information in the root of the tree (vertical gray bar). Molecules with known or likely roles in cell adhesion are marked by black stars in the gray bar on the left-hand side of the figure. Perfect and imperfect MIDAS motifs are indicated by solid and hollow red stars, respectively. All molecules above the purple bar seem to be extracellular; those below the pink bar are intracellular. Signal sequences are designated by a red line and transmembrane segments by a blue bar. Proteins that were assembled or originally characterized in this study are in red, and their sequences are availablein Table S4. See Table S2 to cross-reference molecules in this figure with database identifiers. Figure S2 contains SMART or INTERPRO identifiers for the various domains shown in the diagram (see alsohttp://web.mit.edu/ccrhq/hyneslab/vwapaper/FigureS2.html). The diagrams in this and other figures are extracted largely from SMART (http://smart.embl-heidelberg.de/) but supplemented in some cases with information from other sources (see text).
Integrins
Integrin β Subunits
Integrin heterodimers are the major cell surface receptors for extracellular matrix and can also support cell-cell adhesion (Hynes, 1992). We have searched the available human genome assemblies and found no additional integrin β subunits beyond the eight already known. The same is true for the mouse genome with the exception of a molecule called pactolus (Chen et al., 1998). At the sequence level, murine pactolus is closely related to the integrin β2 subunit but it does not seem to associate with α subunits (Garrison et al., 2001) and is not detected in the human genome. We were unable to detect any credible homologues of β integrins in nonmetazoan phyla.
All integrin β subunits are predicted to have VWA domains (Tuckwell, 1999; Ponting et al., 2000; Figure 1). This prediction was recently confirmed by analyses of the crystal structure of the extracellular portion of the αvβ3 integrin heterodimer (Xiong et al., 2001), which shows a clear VWA domain in the β3 subunit. Integrin β subunits also contain an N-terminal PSI domain and repeated EGF-related domains termed I-EGF domains (Beglova et al., 2002). The integrin β VWA domain interfaces with the α subunit and ligand binds across the interface (Xiong et al., 2002). There is selectivity in α/β associations, and ligand specificity is conferred by the α/β combination. Sequences within the integrin β VWA domains are important determinants of both heterodimer and ligand specificity (Takagi et al., 1997, 2002). MIDAS motifs (at least imperfect) are present in all β subunit VWA domains (Table S3) and integrin–ligand interaction involves joint coordination of a metal ion by the integrin MIDAS and a carboxylate from the ligand (Xiong et al., 2002). As we shall see, these features are commonly observed in VWA domains, and the VWA domains of integrin β subunits are among the most ancient of those involved in cell adhesion that use these properties of the domain.
Integrin α Subunits
Nine of the 18 known human integrin α subunits have a VWA domain (Figure 1). No new α subunits (with or without VWA domains) were detected in detailed searches of the complete human and mouse genomes. The VWA-positive α subunits can be divided into two groups. Integrins α1, α2, α10, and α11 associate with integrin β1 subunit to form receptors for collagen. Integrins αM, αX, αD, αL complex with the β2 subunit, whereas αE complexes with β7, and all five are expressed on leukocytes where they mediate cell-cell adhesion. Like integrin β subunits, α subunits in general are restricted to Metazoa. VWA domain-containing α subunits, however, seem to be a chordate-specific radiation of the gene family because they have been found only in vertebrates and in the primitive chordate Halocynthia roretzi (Miyazawa et al., 2001) but are absent in the C. elegans and Drosophila melanogaster genomes. The H. roretzi α subunit is expressed on phagocytic hemocytes, perhaps suggesting that it is an orthologue of the leukocyte α subunits. Because H. roretzi contains homologues of the B and C3 components of complement (Nonaka and Azumi, 1999), it is possible that the H. roretzi VWA-containing α chain functions as a C3b receptor like αMβ2 in mammals. The β subunit VWA domains probably predate the α subunit VWA domains because β subunits are found in all metazoans, whereas the VWA domain-containing α subunits so far are known only in chordates. Recombinant VWA domains from integrin α subunits retain the ligand-binding specificity and dependence on divalent cations observed in intact heterodimers (Randi and Hogg, 1994; Ueda et al., 1994; reviewed in Shimaoka_et al._, 2002). As a result, integrin α VWA domains are understood in detail and are informative reference points for considering the functions of other VWA domains. The crystal structures of the isolated VWA domains of α1 (Nolte et al., 1999), α2 (Emsley et al., 1997), αL (Qu and Leahy, 1995), and αM (Lee et al., 1995; Baldwin et al., 1998) are known. It is clear that two α subunit VWA domain conformations (open and closed) represent high-affinity and low-affinity ligand-binding states, respectively (Shimaoka et al., 2001; Ma et al., 2002). This observation highlights the possibility that nonintegrin VWA domains may be subject to similar regulation.
All available data emphasize the importance of the VWA domain and MIDAS motif in integrin–ligand interaction (Xiong et al., 2002). The MIDAS motif is perfectly conserved in all integrin α subunit VWA domains and interaction with ligand involves mutual coordination of metal ion (Emsley et al., 2000). The β VWA domain MIDAS motif is critical for ligand binding in heterodimers that include VWA domain-containing α subunits. Mutations in the β2 and β7 VWA domain MIDAS motifs inhibit ligand binding despite being located a distance from the point of integrin-ligand contact (Bajt et al., 1995; Higgins et al., 2000). Given the high percentage of nonintegrin VWA domains with conserved MIDAS motifs (Figure S1, a–c, and Table S3), the coordination of metal ion by MIDAS and ligand may be a common feature of protein–protein interactions mediated by VWA domains.
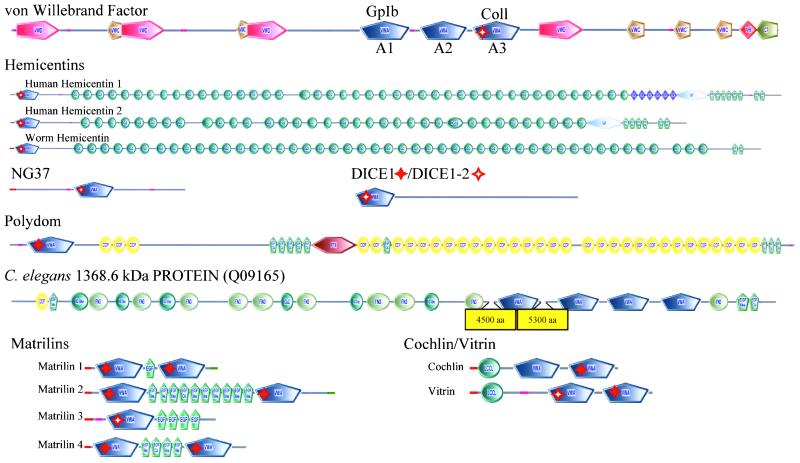
Multidomain ECM Proteins (Figure 2)
Figure 2.
Noncollagenous VWA domain-containing ECM proteins All molecules shown are human except where designated (see text for discussion of orthologues/paralogues). All these proteins are found only in Metazoa and are secreted to the extracellular matrix with the possible exceptions of NG37 and DICE (see text), and most are clearly implicated in cell adhesion. Domains are designated as in Figures 1 and S2. Perfect and imperfect MIDAS motifs are indicated by solid and hollow red stars, respectively.
von Willebrand Factor
vWF is a plasma and ECM protein that mediates adhesion of platelets to fibrillar collagen underlying injured vascular endothelium (reviewed in Sadler, 1998). It is known only in vertebrates. Mutations in the vWF gene result in von Willebrand disease (vWD), a common human bleeding disorder (reviewed in Keeney and Cumming, 2001). There are three VWA domains in vWF (referred to as A1, A2, and A3); A1 and A2 are related and neither has a MIDAS motif. At the sequence level, A3 has an imperfect MIDAS. The crystal structures of A1 and A3 are known and neither contains a coordinated metal ion and the conserved MIDAS elements in A3 are not required for function (Bienkowska et al., 1997; Emsley et al., 1998). However, the VWA domains support the vWF-mediated linkage between platelets and fibrillar collagens. The platelet receptor GPIb/IX/V binds to domain A1 (Cruz et al., 2000) and mutations in A1 cause type 2B and type 2M vWD. These are characterized, respectively, by increased or decreased platelet interaction. Domain A3 supports cation-independent vWF interaction with fibrillar collagens (Cruz et al., 1995; Bienkowska et al., 1997; Romijn et al., 2001). A missense mutation in A3 causes vWD due to defective collagen binding (Ribba et al., 2001). The function of the vWF A2 domain is less well defined but missense mutations in A2 frequently cause type 2A vWD, a dominant disorder characterized by a decrease in high-molecular-weight multimers of vWF. This is due to improper secretion of vWF or increased proteolysis of the secreted mutant protein (Keeney and Cumming, 2001) or perhaps to a defect in assembly of multimers. Recombinant vWF lacking A2 is resistant to proteolysis after denaturation, suggesting that a protease sensitivity site within the domain may be exposed in misfolded mutant protein (Lankhof et al., 1997). Thus, like those in integrins, the vWF VWA domains are well-characterized domains clearly involved in protein–protein interactions important for cell adhesion. One difference is that, in contrast with the integrin domains, the VWA domains of vWF seem to be cation-independent, perhaps related to their imperfect MIDAS motifs.
Hemicentins, NG37, and DICE1
Hemicentin is an adhesion molecule originally characterized in C. elegans, where it is secreted by body wall muscle cells and gonadal leader cells. Once secreted, hemicentin assembles into track-like matrix structures. C. elegans lacking hemicentin have defects in mechanosensory neuron development and germline cell migration (Vogel and Hedgecock, 2001). Both humans and mice have two homologues of C. elegans hemicentin but orthologues are not detectable in the D. melanogaster, yeast, and Arabidopsis thaliana genomes. Hemicentin is the only confirmed VWA domain-containing ECM protein found in both_C. elegans_ and mammals (but see DICE1 below). The domain architecture is similar between C. elegans and humans with a single VWA domain near the N terminus followed by >40 Ig domains. All hemicentin VWA domains have imperfect MIDAS motifs and are highly conserved among themselves; region 1 has the sequence D-x-T-x-S, T4 is a D, and D5 is conserved. In contrast with the C. elegans gene, mammalian hemicentins contain additional domains that are likely to be functionally important (Figure 2). Mammalian hemicentin 1 contains multiple TSP-1 domains. TSP-1 domains were originally identified in thrombospondin and are present in a wide range of proteins with roles in cell adhesion. The only other known proteins containing a combination of TSP-1 and VWA domains are proteins found in parasites from the protist kingdom of eukaryotes. These organisms cause malaria in humans and the TSP-1-VWA proteins seem to be secreted or transmembrane and function in adhesion and motility during the invasive phase of the parasitic life cycle (reviewed in Naitza et al., 1998; Figure S8). The plasmodium proteins are not closely related to any of the metazoan VWA-containing proteins. Both mammalian hemicentins also contain a G2F domain. The functions of G2F domains are unclear but they seem to be restricted to mammalian hemicentins and the metazoan nidogens, also adhesion proteins.
The VWA domain of a potentially secreted protein, termed NG37 in human and G7c in mouse, is closely related to the VWA domain of hemicentin. The residues at the MIDAS positions are identical with those in all three hemicentins, suggesting that they may have similar divalent cation-binding properties. NG37 and G7c were identified in genomic sequencing of the major histocompatibility complex class III region (Snoek et al., 1998, 2000). The close sequence relationship among the VWA domains of NG37 and hemicentins, coupled with the predicted signal sequence make it seem likely that NG37/G7c plays some role in cell adhesion.
Kumanovics and Lindahl (2001) and our results also suggest a relationship between the VWA domains of NG37/G7c and the domain found in DICE1, a protein encoded by a gene located in a human tumor suppressor locus (Wieland et al., 1999). A Genomescan-predicted protein on the X chromosome is a paralogue of Dice1 (see Table S4 for sequence). Mice have orthologues of both human DICE1 and DICE1-2, and single orthologues are present in the genomes of_C. elegans_ and D. melanogaster (Wieland et al., 2001). Orthologues were not detected in Saccharomyces cerevisiae and A. thaliana. In all cases, the DICE VWA domains are located at the N terminus of the proteins. MIDAS motifs are perfectly conserved except for C. elegans where S substitutes for T4. The Dice proteins are being considered herein as potential ECM proteins because of their VWA domain similarities with the hemicentin/NG37 molecules. However, signal sequences are not predicted in any DICE orthologue and the mammalian molecules have a DEAD box motif that is found in RNA helicases (reviewed in Tanner and Linder, 2001). Because the DEAD box is not conserved in the C. elegans and D. melanogaster orthologues, we have weighted the relationship with hemicentin/NG37 but inclusion in the ECM group is tentative.
Polydom
Polydom is a secreted protein originally identified in mice in a screen for molecules containing EGF domains (Gilges et al., 2000). The domain architecture consists of an N-terminal VWA domain and a central PTX domain; the remainder of the protein is made up of a large number of repeated CCP and EGF modules (Figure 2). The human orthologue of polydom was previously uncharacterized and has a similar domain architecture (see Table S4 for sequence). The VWA domain of polydom contains a conserved MIDAS motif, suggesting a potential role for divalent cations in polydom function. Included in this list of enormous ECM molecules with proven or likely roles in cell adhesion is a predicted nematode protein of >1300 kDa (Figure 2). This molecule has four VWA domains (all lacking MIDAS motifs), three EGF-like, six Ig-like, one CCP, and 10 Fn3 domains. Its function, like that of polydom, is unknown but their domain composition, including VWA domains, strongly suggests roles in cell adhesion.
Matrilins
The matrilins are a family of fibril-forming vertebrate ECM proteins with four paralogues in the human genome (reviewed in Deak et al., 1999). With the exception of matrilin 3, which has a single VWA domain in all orthologues examined, matrilins contain two VWA domains that flank a variable number of EGF domains (Figure 2). Matrilins 1 and 3 are expressed in cartilage and matrilins 2 and 4 have a widespread distribution (Deak et al., 1999). The matrilins form oligomers (Wu and Eyre, 1998), and both VWA domains of matrilin 1 play a role in oligomerization (Chen et al., 1999). All matrilin VWA domains have MIDAS motifs (imperfect in the matrilin 3 VWA domain) and mutation of the MIDAS motif in both matrilin 1 VWA domains blocks filamentous network formation, suggesting that cation binding by the VWA domains of matrilins may be required for function (Chen_et al._, 1999). Matrilin-1 supports integrin α1β1-mediated adhesion and spreading of chondrocytes (Makihira_et al._, 1999). Two groups observed normal development in mice lacking matrilin-1 despite abnormal type II collagen fibrillogenesis (Aszodi et al., 1999; Huang et al., 1999). Two different recessive mutations in the exon encoding the VWA domain of matrilin-3 found in unrelated families cause the EDM5 form of multiple epiphyseal dysplasia (Chapman et al., 2001). These mutations result in single amino acid changes of V194D or R121W. These residues are conserved in all matrilin family members, are not part of the MIDAS motif, and the disease-causing mechanism is unknown. Like the VWA collagens, matrilins seem to be a chordate invention.
Cochlin and Vitrin
Cochlin and vitrin are proteins containing a single LCCL domain followed by two VWA domains (Figure 2). Cochlin is expressed by fibrocytes in the inner ear, localized to extracellular spaces, and missense mutations in the LCCL domain lead to the autosomal-dominant hearing disorder DFNA9 (Robertson et al., 2001). Vitrin was isolated from the vitreous of the bovine eye (Mayne et al., 1999). The LCCL is a domain found in Limulus Factor C, cochlins, and lgl1. The C-terminal VWA domains of cochlin and vitrin form a clade with those of matrilins (51% bootstrap support). Whole-molecule alignments also support the relationship between cochlin/vitrin and matrilins. The C-terminal VWA domains of both cochlin and vitrin have perfectly conserved MIDAS motifs. The functions of the VWA domains in these molecules are unknown but, given their extracellular localization and sequence similarities, they are likely similar to the roles in matrilins and VWA collagens (see below). All seem most likely to comprise ECM molecules and many, perhaps all, their VWA domains are involved in protein–protein interactions.
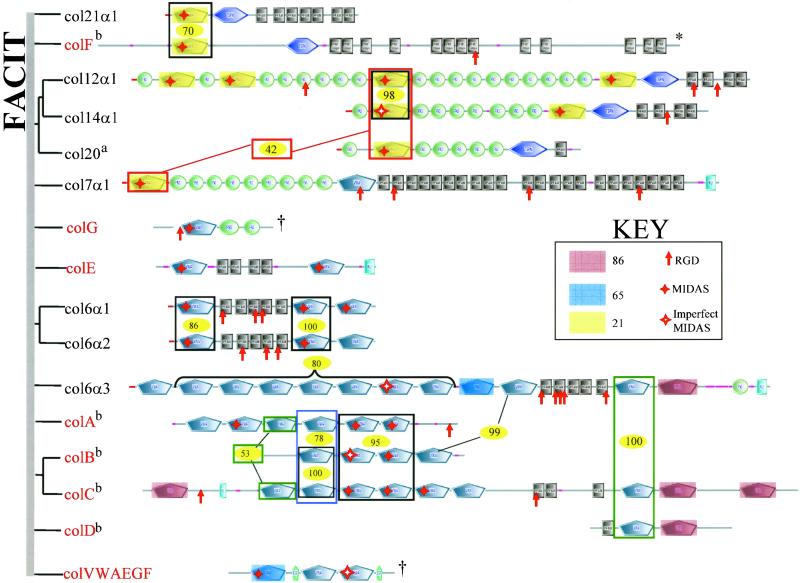
VWA Collagens (Figure3)
Figure 3.
Sixteen unconventional mammalian collagens contain VWA domains. The figure shows the predicted structures of the eight known unconventional collagens and eight related novel proteins (red), all containing VWA domains. The eight novel proteins have been given letter designations for the purpose of discussion, and their sequences are available in Table S4. Appropriate names should be given after cDNA analysis. On the left is a phylogenetic tree showing only the most confidently clustered groups of paralogues. The vertical gray bar replaces the root of the tree. The VWA domains of these collagens comprise a clade and are more closely related among themselves than with any other VWA domains. They also show familial relationships within the group, shown herein by shading and boxes designating the degree of similarity with percentage bootstrap scores for individual groups shown (yellow ovals). Perfect and imperfect MIDAS motifs are indicated by solid and hollow red stars, respectively. Also marked are all occurrences of RGD motifs (red arrows). Domain structures shown were predicted by SMART except for the collagen motifs (black boxes, each of which represents 20 G-X-Y repeats predicted by Pfam). SMART does not predict VWA 2 or the KU domain in collagen 7α1 but the domains are predicted by Pfam so they were added to the figure. The asterisk (*) indicates that colF is the mouse orthologue used to show C-terminal collagen motifs (see text). The dagger (†) indicates these molecules lack predicted collagen motifs but are included because of close homology of their VWA domains with those of known collagens, as is also true for collagens A–D (see text). aThe human orthologue of chicken collagen 20 is KIAA1510 identified in a full-length cDNA sequencing project (Nagase et al., 2000). bCurrent assemblies a bit ambiguous (see text).
The collagens are a large family of extracellular matrix molecules defined by the presence of repeating (G-X-Y) sequences that form a triple helix (Ricard-Blum et al., 2000). Sixteen collagens also contain 57 of the 134 VWA domains found in the human proteome. Of these, 25 have perfectly conserved MIDAS motifs (Figure 3, solid red stars); four imperfect MIDAS motifs are also indicated (Figure 3, hollow red stars). Eight of the 16 collagens are well characterized and have been studied experimentally; the other eight are novel genes detected in this study (sequences available in Table S4). These 16 collagens can be considered in several related groups (Figure 3).
Collagens 12, 14, 20, and 21 (Fitzgerald and Bateman, 2001; Tuckwell, 2002) are fibril-associated collagen with interrupted triple helix (FACIT) collagens that associate with fibrillar collagen via their C-terminal collagenous domains. Their N-terminal segments containing the VWA domains extend out from the surface of the collagen fibrils and are suggested/known to bind other ECM components. The newly predicted collagen F seems to fall into this same group. The gene predicted in the human genome assembly lacks C-terminal collagen repeats, possibly due to a gap in the sequence assembly near the 3′ end of the gene. To present a more likely picture of this molecule, we have shown the mouse orthologue of collagen F. Both the human and the mouse sequences are available in Table S4. These five FACIT collagens share a related set (clade) of VWA domains (Figure 3, highlights in yellow) as well as the universal presence of a TSPN domain just N-terminal of their C-terminal collagenous segments. Collagens 12, 14, and 20 also share tandem arrays of FN3 repeats interspersed with a closely related set of VWA domains (Figure 3). Both collagen 12 and collagen 14 are cell adhesion collagens and both have RGD sequences.
Collagen 7 shares many of these features and homologies of FACIT collagens but differs from them in having a C-terminal KU domain that forms a globular noncollagenous domain in collagen 7. Collagen 7 is a homotrimer with a cross-like structure. The short arms of the cross are formed by the three globular N-terminal noncollagenous domains consisting of the VWA and FN3 domains. The long arm is the collagen triple-helical region and a globular C-terminal domain. In the extracellular space, homotrimers associate in an antiparallel manner via the C-terminal globular domains. The N-terminal globular domains bind a variety of ECM/BM components, including collagens 1 and 4 and laminins 5 and 6. This bivalent ECM-binding feature of collagen 7 results in linkage of epidermal basal lamina and dermal ECM. Mutations in collagen 7 are involved in many types of epidermolysis bullosa, a human skin disease characterized by separation of dermal and epidermal layers (Pulkkinen and Uitto, 1999). The new predicted protein colG is included here because of its homologies, although it has no predicted collagen repeats and may not be a true collagen.
Another large group of collagens includes collagen 6 and related proteins (Figure 3) Collagen 6 (α1, α2, and α3 chains) is a heterotrimer that forms filamentous structures flanked by globular domains containing the VWA domains. These globular domains can associate with themselves or with other ECM molecules. The monomeric form of collagen 6 is a heterotrimer consisting of 6α1, 6α2, and 6α3 chains. Antiparallel association of collagen 6 monomers forms dimers and the dimers laterally associate to form tetramers. The tetramers associate end-to-end to form a beaded filament. Collagen 6 interacts with a wide range of ECM components (reviewed in Ricard-Blum_et al._, 2000). The VWA domains of col6α3 interact with fibrillar collagen 1 (Bonaldo et al., 1990) and the triple-helical part of collagen 6 interacts with the A1 VWA domain of vWF (Hoylaerts et al., 1997). The new collagen E is similar in overall structure to col6α1 and col6α2 but is not clearly related at the sequence level. Three new collagens (A, B, and C) are in a cluster on chromosome 3 in human, and their orthologues are on chromosome 9 in mouse. The gene predictions are a little ambiguous in the current genome assemblies, and the structures shown in Figure 3represent a best estimate based on both the human and mouse assemblies (public and Celera). Collagens A–D show close homologies in domain arrangements and similarities among VWA domains with collagen 6α3 (Figure 3). Although the gene predictions shown represent open reading frames (ORFs) without stop codons, their domain organizations have some puzzling features such as lack of collagen repeats in collagens A and B and the anomalous position of a VWA-Ku domain pair at the N terminus of collagen C. Whether these represent genome assembly problems or pseudogenes will have to await further information, but it seems likely that there may exist a family of collagen subunits related to collagen 6α3.
It seems very likely that the VWA domains of these collagens are involved in protein–protein interactions with other matrix proteins and possibly with cells. This elaboration of VWA collagens seems to be chordate- or vertebrate- specific.
Other Extracellular VWA Domain Proteins
We turn next to a different class of VWA-domain proteins where the presence of VWA domains comes as something of a surprise (Figure 1).
Calcium Channel α2δ Family
Voltage-gated calcium channels are a complex of five proteins: α1, β1, γ, α2, and δ. The α2 and δ subunits result from proteolytic processing of a single gene product (De Jongh et al., 1990). The α2 subunit consists of ∼950 N-terminal amino acids and contains a VWA and a cache domain (Figure 1). In most cases, at least an imperfect MIDAS motif is conserved (Figure S3). Cache domains are only found in these molecules and in a family of prokaryotic chemotaxis receptors (Anantharaman and Aravind, 2000). The remainder of the molecule comprises the δ subunit. The α2 and δ subunits are disulfide-linked and Brickley et al. (1995) conclude that the δ subunit contains transmembrane domains and that the α2 subunit is entirely extracellular. When coexpressed with the pore-forming α1 subunit, the α2δ complex regulates various functional properties of the channel complex (reviewed in Hobom_et al._, 2000). The α2δ gene family has orthologues in the D. melanogaster and C. elegans_genomes, but none are detectable in A. thaliana or yeast. There are five paralogues in human, two in C. elegans, and four in D. melanogaster (Figure S4). In human, three paralogues were characterized previously (CACNA2D1-3), a fourth was deposited in the database from a full-length cDNA sequencing project and is the most divergent of the five. The fifth is a Genomescan-predicted protein identified in this study (see Table S4 for sequence). Both C. elegans orthologues were targeted with RNAi and no mutant phenotype was observed (Maeda et al., 2001). Mutant phenotypes have not been described for any of the_Drosophila genes. Because the annotation of several of these molecules is uninformative in the databases, we have provided tentative names based on orthologous relationships with human molecules (Figure S3).
CLCA Family of Putative Chloride Channel Subunits
The first member of the CLCA family (Pauli et al., 2000) was characterized in bovine aortic endothelial cells as a protein involved in attachment of melanoma cells to lung endothelium and was named lung-specific endothelial adhesion molecule (Lu-ECAM-1; Zhu et al., 1991). Lu-ECAM-1 was found to encode a protein 88% identical to a putative calcium-activated chloride channel from bovine trachea (Elble et al., 1997). Expression of calcium-activated chloride channel in Xenopus oocytes resulted in the appearance of anion-selective conductance (Cunningham_et al._, 1995). In the human genome, four paralogues are located in a cluster on chromosome 1p22. The transcript encoding the human orthologue of bovine Lu-ECAM-1 (hCLCA3) contains several stop codons and results in the production of a secreted variant corresponding to the N-terminal 262 amino acids (without the VWA domain) of the other family members (Gruber and Pauli, 1999). Human CLCA1 and CLCA4 have conserved MIDAS motifs. Orthologues of the CLCA family are not detectable in the C. elegans and_D. melanogaster_ genomes but a possible homologue is present in the Xenopus expressed sequence tag (EST) collection (GB# AW767641), suggesting that the CLCA family is a chordate invention. It is likely that CLCA proteins do not form channels by themselves. According to SMART predictions, CLCA family members consist of a central VWA domain and a single transmembrane domain near the C terminus (Figure 1). Published transmembrane predictions for these molecules (Cunningham et al., 1995; Gruber et al., 1999) require that the VWA domain spans the plasma membrane, a situation unlikely to occur based on VWA domain structural studies. The biochemical data presented byGruber et al. (1999), however, are entirely consistent with the model of CLCAs derived from SMART analysis: a type I transmembrane protein with an extracellular VWA domain (Figure 1). Reminiscent of the α2δ proteins, CLCAs are expressed as 125-kDa precursor proteins that are subsequently processed into 90- and 35-kDa subunits (reviewed in Pauli et al., 2000). It seems to us more likely that the CLCAs are accessory molecules for chloride channels, analogous to the α2δ subunits of calcium channels. The roles of the VWA domain in both types of molecules are unknown but could involve modulation of channel activity by binding other proteins or divalent cations via the VWA domains.
Anthrax Toxin Receptor Family
There are three members of the anthrax toxin receptor family, and the two that have been studied experimentally are type I transmembrane proteins with a single extracellular VWA domain. The third is a Genomescan-predicted protein that we have termed anthrax toxin receptor (ATR) 3 (Figure 1; see Table S4 for sequence). The ATR is the cellular receptor for the anthrax protective antigen and facilitates entry of the toxin into cells. The VWA domain of ATR mediates interaction with protective antigen and the binding is dependent on divalent cations (Bradley et al., 2001). The murine orthologue of this gene, TEM-8, was identified as a gene up-regulated in colon tumor endothelium (St Croix_et al._, 2000). The second member of this family, CMG-2, was identified in a similar screen for genes up-regulated during capillary morphogenesis (Bell et al., 2001). The normal cellular ligand for the anthrax receptor is unknown but a recombinant fragment of CMG-2 was shown in a solid-phase assay to bind collagen IV, laminin and, to a lesser extent, fibronectin (Bell et al., 2001). All three molecules have conserved MIDAS motifs and the cytoplasmic domain of ATR can be alternatively spliced (Bradley_et al._, 2001). Potential homologues of these molecules are present in the chicken and zebrafish EST databases. The observations discussed above suggest that these proteins are a family of vertebrate ECM receptors expressed by endothelial cells.
Complement Factors
Complement factors B and C2 both contain three CCP or Sushi domains, a single VWA domain with a conserved MIDAS motif, and a trypsin-type serine protease domain (Figure 1). Orthologues of these molecules are found from echinoderms to chordates (Smith et al., 1998) but are not found in_D. melanogaster_ and C. elegans, suggesting that they may be a deuterostome-specific invention. During complement activation, the CCP domains are cleaved off, resulting in the formation of an active protease that cleaves and activates complement C3. Complement C2 is in the classical pathway and complement factor B is in the alternative pathway. The interactions of C2 with C4 and of factor B with C3b are both dependent on Mg2+-binding sites within the VWA domains, and the VWA domain of factor B has been shown to mediate the binding of C3 (Tuckwell et al., 1997), consistent with the common inferred function of VWA domains as magnesium-dependent protein interaction domains. Up to this point, all of the molecules discussed have extracellular VWA domains, and most seem likely to play roles in cell adhesion or in protein–protein interactions among ECM molecules. However, VWA domains, like FN3 and Ig domains (in titin, etc.) can also be found in intracellular molecules. The next group of proteins provides a useful transition to consideration of intracellular VWA domain-containing proteins because some are extracellular, one is intracellular, and some have not been characterized.
Trypsin Inhibitors and Their Relatives
Three classes of human proteins contain a combination of VIT and VWA domains: seven inter-α-trypsin inhibitor heavy chains (ITIH), three members of the novel Q9BVH8 family, and a single poly(ADP-ribose) polymerase [vault poly(ADP-ribose) polymerase, vPARP; Figures 1 and S4]. The VWA domains of these proteins represent a discrete clade in a comparison with other human VWA domains (25% bootstrap support). The most closely related domains outside this group are those of the calcium channel α2δ subunits. The function of the VIT domain (vault protein inter-α-trypsin domain) is not known. It is found only in chordates with the exception of a single bacterial protein predicted in Cyanobacterium anabaena(NP_488452), which clusters with vPARP and the Q9BVH8 family with 100% bootstrap support (Figure S4). With the exception of the Mg chelatases discussed below, this molecule is the only clear VWA domain-containing orthologue shared between bacteria and eukaryotes and may represent a recent horizontal gene transfer event.
The four characterized ITIH family members are extracellular molecules found in complexes with the kunitz-type serine protease inhibitor bikunin (Bost et al., 1998), which confers the protease inhibitor function of the complex. The function of the heavy chains is unclear, but they are covalently bound to hyaluronic acid and may play a role in ECM binding and/or stability (reviewed in Bost et al., 1998). ITIH1–4 are well characterized and reviewed in Salier_et al._ (1996). The three new paralogues are fragments in the database or Genomescan-predicted proteins. The newly assembled protein sequences are available in Table S4. The VWA domains of ITIH proteins cluster with 100% bootstrap support. MIDAS motifs are conserved in all ITIH family members except for ITIH7.
Three proteins (Q9BVH8-1–3) also have the domain architecture VIT-VWA but are separate from the ITIH family (Figure S4). These molecules are poorly characterized and the conclusion that they contain the VIT-VWA domain arrangement is based on SMART analysis of the corresponding Genomescan-predicted proteins and clustalW alignments (see Figure S4 for details). These molecules are more closely related to vPARP (see below) than to the ITIH family, but they lack the BRCT domain found in vPARP and form their own subfamily of VIT-VWA proteins. The MIDAS motif is conserved in two of the three. Nothing is known about their subcellular localization.
The final protein that contains VIT and VWA domains is vPARP (Figure1), the 193-kDa component of cytoplasmic vault ribonucleoprotein complexes (Jean et al., 1999). Poly(ADP-ribose) polymerase is a eukaryotic enzyme but vPARP (Figure 1) seems to be restricted to mammals. We have identified a predicted protein from the mouse golden path genome assembly, and the mouse orthologue of vPARP and its sequence are available in Table S4. The vault complex is involved in DNA repair, nuclear transport, and multidrug resistance (Kickhoefer_et al._, 1999). The VWA domains in both human and mouse vPARP have conserved MIDAS motifs, implicating coordinated metal ions in their function. The function of the vPARP VWA domain is unknown, but vPARP is part of a multiprotein complex (Kickhoefer et al., 1999), raising the possibility that the VWA domain of vPARP plays a role in complex assembly.
Intracellular Proteins: The Primordial Group?
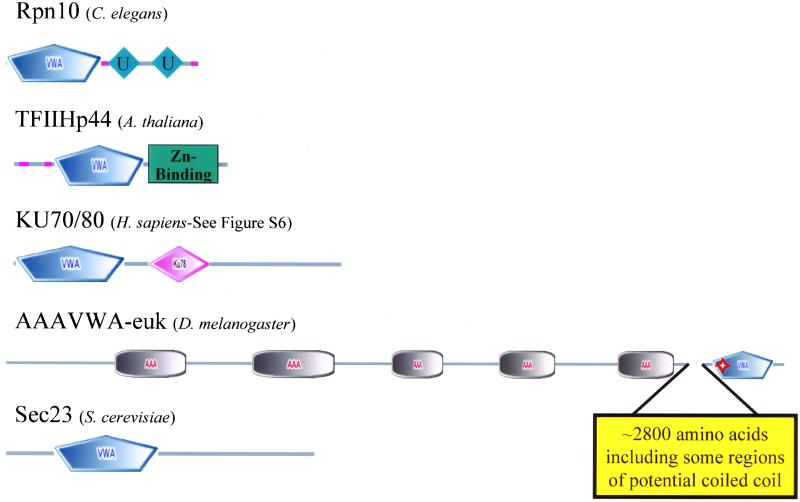
One of the more surprising results of our survey (at least to us) was the presence of a set of intracellular VWA domain-containing proteins (Figure 4). Intracellular VWA domains have been noted previously (Ponting et al., 1999). As we discuss below, these proteins are more broadly distributed phylogenetically than the metazoan extracellular proteins discussed above, suggesting that the intracellular proteins may be the most ancient.
Figure 4.
VWA domain-containing proteins common to all eukaryotes are intracellular and found in multiprotein complexes. The five proteins shown are found in all eukaryotes, and we show examples from Metazoa, A. thaliana, and yeast. VWA domains are predicted in orthologues from each of the five. The VWA domain is a particular form of a Rossmann fold and some of the Rossmann folds in orthologues of Ku70/80 and Sec23 do not give predictions for the VWA domain subclass. The VWA domains of TFIIHp44 and Rpn10 are closely related. Both TFIIHp44 and Ku70/80 exhibit helicase activity. A common feature of these proteins is that they are subunits of multiprotein complexes. Given the role of VWA domains in protein–protein interactions, a possible role for the VWA domains in these proteins is in mediating assembly of these complexes. Whatever the role of the VWA domains, these intracellular molecules seem to be the most ancient eukaryotic examples of this protein domain from which other (mostly extracellular) VWA proteins presumably evolved. Molecules that may also be included in this group but have been lost in some taxa include copines (Figure S5). Perfect and imperfect MIDAS motifs are indicated by solid and hollow red stars, respectively. See text for further discussion of these molecules and their relatives.
Rpn10-26S Proteasome Regulatory Subunit
The 26S proteasome is a complex of proteins involved in degradation of ubiquitin-tagged proteins (reviewed in Voges et al., 1999). The subunit of the complex that recognizes chains of ubiquitin is Rpn10 (or S5a, PSD4). A single orthologue of this molecule is encoded by all completed eukaryotic genomes. In addition to the N-terminal VWA domain, Rpn10 proteins contain ubiquitin-interacting motifs that are involved in recognition of multiubiquitin. Yeast cells deficient in Rpn10 are viable, suggesting that Rpn10 is not the only multiubiquitin-binding protein in cells. However, the VWA domain in Rpn10 may play a role in efficient 26S complex function (Fu et al., 2001). MIDAS motifs are not found in any Rpn10 (Table S3). Region 1 of the yeast Rpn10 VWA domain has the sequence DxSxY, and Fu_et al._ (2001) demonstrated a requirement for an acidic residue in the D position by using several functional assays, suggesting that the intact VWA domain is required for 26S proteasome function. The VWA domain can also mediate interaction with Id1, a transcription regulator that is itself regulated by ubiquitin-mediated proteolysis (Anand et al., 1997; Bounpheng et al., 1999).
TFIIHp44
TFIIH is a multiprotein complex that is one of the five general transcription factors that binds the RNA polymerase II holoenzyme (Orphanides et al., 1996; Myer and Young, 1998). The p44 subunit of TFIIH is the human orthologue of yeast SSL1 (Humbert et al., 1994). Orthologues of these genes are also found in all completed eukaryotic genomes and all these proteins have VWA domains (Figure 4). TFIIHp44 functions as a DNA helicase in RNA polymerase II transcription initiation and DNA repair, and its transcriptional activity is dependent on its C-terminal Zn-binding domains (Fribourg et al., 2000). The function of the VWA domain is unclear, but it may be involved in complex assembly. MIDAS motifs are not conserved except for the fly, which has an imperfect MIDAS motif. During the course of these analyses, we performed pairwise comparisons of isolated VWA domains from each species considered. One outcome of this work was the identification of a strong relationship between TFIIHp44 and Rpn10. When the complete VWA domain collections of eukaryotes are compared, the VWA domains of TFIIHp44 and Rpn10 form a clade with >90% bootstrap support. This relationship is supported by Aravind and Ponting (1998) who recognized them as homologues. Both proteins function in intracellular multiprotein complexes so these results may suggest an analogous role for each in their respective complexes.
Ku70/80 DNA Helicase Family
In humans, the lupus autoantigens Ku70 and Ku86 form heterodimers, bind DNA, and are involved in repair of double-stranded breaks in DNA (Walker et al., 2001, and references therein). SMART predicts that human Ku70 and Ku86 have VWA domains. The Ku DNA helicases have orthologues in all completed eukaryotic genomes and, therefore, seem to be a pan-eukaryotic VWA domain-containing protein (Figure 4). The VWA domain, however, is not consistently predicted in many of these orthologues (Figure S6). The structure of the human Ku70/Ku86 heterodimer has been solved and both subunits contain a Rossmann fold, but whether this should be considered a true VWA domain is unclear. The VWA domains/Rossmann folds are probably not involved in dimer formation but may support interactions with additional molecules (Walker_et al._, 2001).
ATPases Associated with Diverse Cellular Activities (AAA)-VWA Proteins
Two proteins in the human genome contain a combination of AAA and VWA domains. AAA domains are found in all branches of life and have ATPase activity (Patel and Latterich, 1998). There is growing evidence that AAA proteins are important in assembly and disassembly of macromolecular complexes (Maurizi and Li, 2001). Both of the AAA-VWA proteins in the human genome were previously uncharacterized. The first is a 5000 amino acid protein common to all completed eukaryotic genomes (AAAVWA-euk; Figure 4). The yeast orthologue is required for cell viability (Winzeler et al., 1999) and coprecipitates with a protein complex thought to be the transport intermediate of 60S ribosomal subunits (Bassler et al., 2001). These proteins have multiple AAA domains at the N terminus and a single VWA domain near the C terminus. Human and_C. elegans_ AAAVWA-euk have perfect and the other orthologues have imperfect MIDAS motifs, suggesting that divalent cations may play a role in their function. In the databases, the yeast, D. melanogaster, and A. thaliana orthologues are full length. The human and C. elegans are fragments and concatenation of adjacent Genomescan-predicted proteins is required to generate full-length sequence. These sequences are available in Table S4. The second AAA-VWA domain protein in the human genome has orthologues in D. melanogaster and C. elegans but is not detectable in yeast and A. thaliana, suggesting it may be a metazoan invention (AAAVWA-ani; Figures 1, 5, and S7). These molecules have two AAA domains and a single C-terminal VWA domain. Perfect MIDAS motifs are found in the human and D. melanogaster orthologues, and the C. elegans molecule has an imperfect motif, suggesting that cations may be important in their function. The human protein is a fragment in the database but concatenation of adjacent Genomescan-predicted proteins results in an intact molecule that is >30% identical to the _C. elegans_orthologue over 1800 amino acids. The Drosophila protein is also a fragment in the database, and two adjacent predicted genes in Flybase were concatenated to generate a full-length molecule. The sequences of these assembled molecules are available in Table S4. The function of these proteins is unknown.
Figure 5.
C. elegans has a large number of novel VWA domain-containing ECM proteins. The domain architecture of all C. elegans VWA domain-containing proteins is indicated. Paralogous molecules are grouped together in the phylogenetic tree derived from a clustalW alignment. The groups of paralogues or unrooted individual molecules have been shuffled along the vertical axis for clarity of presentation; so there is no information in the root of the tree (vertical gray bar). The molecules in blue lack close homologues in all other completed genomes. Note the novel domain associations in many of these proteins. All molecules below the purple bar seem to be extracellular or membrane-associated; the localizations of those below the red bar are unclear. The VWA domains of the C. elegans copines are closely related to the VWA domains of three uncharacterized molecules and these relationships are indicated by a red box. Other relationships are also indicated by boxes and the bootstrap numbers are provided in yellow ovals. Perfect and imperfect MIDAS motifs are indicated by solid and hollow red stars, respectively. See Table S2 to cross-reference molecules in this figure with database identifiers.
The only other proteins that contain the combination of AAA and VWA domains are the D subunits of the protoporphyrin IX Mg chelatase complexes. These proteins are known as chloroplast Mg chelatase D (ChlD) in A. thaliana (Figure 6) and bacterial Mg chelatase D (BchD) in bacteria. They are also found in archaeal genomes, and it may be appropriate to name these molecules archaeal Mg chelatase D (AchD). The protoporphyrin IX enzymatic complex is essential for chlorophyll biosynthesis and is likely to be common to all photosynthetic organisms. Mg chelatases are complexes of three subunits that function to incorporate Mg into protoporphyrin IX in an ATP-dependent mechanism (Walker and Willows, 1997). Bacterial cobalt chelatases are also three subunit complexes (Schubert et al., 1999) and one subunit, COBT, has a VWA domain but not an AAA domain. At least an imperfect MIDAS is present in 18/19 chelatases subunits examined and in 11 of the examples the MIDAS motif is perfectly conserved. In the cobalt chelatases, a G replaces the T4 in all three available examples, possibly reflecting different cation preferences of the chelatases. In any case, it is likely that the MIDAS motif in most chelatase D subunits has the potential to coordinate a cation. The functional significance of this coordination may be in presentation of ion to the protoporphyrin IX or in complex assembly.
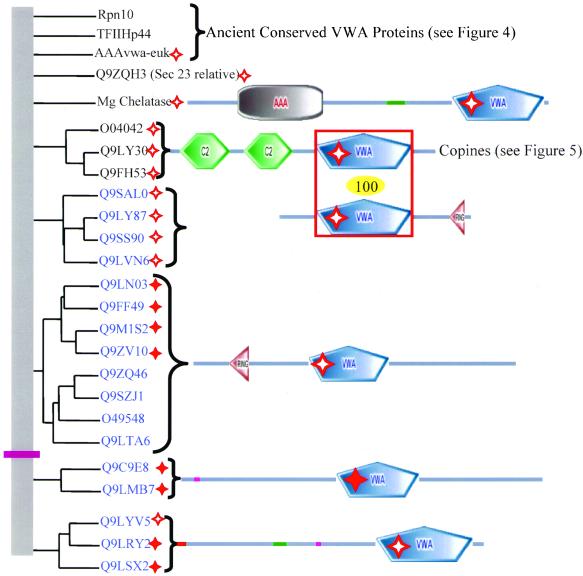
Figure 6.
A. thaliana has additional intracellular VWA domain proteins. The domain architecture of all_A. thaliana_ VWA domain-containing proteins is indicated. Paralogous molecules are grouped together in the phylogenetic tree derived from a clustalW alignment. The groups of paralogues or unrooted individual molecules have been shuffled along the vertical axis for clarity of presentation; so there is no information in the root of the tree (vertical gray bar). The molecules in blue lack homologues in completed fungal and metazoan genomes. All molecules above the purple bar seem to be intracellular. No information is available for the molecules below the purple bar although one (Q9LSX2) has a predicted signal sequence (indicated by red bar), suggesting that that group of three molecules might be secreted. The VWA domains of the copines are closely related to the VWA domains of the VWA-RING proteins (Q9LVN6lacks the RING domain). The relationship is indicated by the red box and the number in the yellow oval. Perfect and imperfect MIDAS motifs are indicated by solid and hollow red stars, respectively. See Table S2 to cross-reference molecules in this figure with database identifiers.
Sec-23
The S. cerevisiae protein Sec-23 has orthologues in all completed eukaryotic genomes, but only the S. cerevisiae molecule is predicted to have a VWA domain. Sec-23 is part of a multiprotein complex involved in transport of vesicles from the Golgi to the endoplasmic reticulum. It is possible that the Sec-23 VWA domain prediction is a false positive because the e-value is high (1.01) and all three MIDAS regions are unconserved.Q9ZQH3 is an uncharacterized A. thaliana molecule whose VWA domain is similar to yeast Sec23 (63% bootstrap support). This molecule, however, is distinct from the _A. thaliana_orthologues of Sec-23, all of which lack a predicted VWA domain. These five intracellular VWA proteins found in all eukaryotes (Figure 4) could be viewed as the primordial set of VWA proteins. All are involved in multiprotein complexes, and it is tempting to speculate that the role of VWA domains is in the protein–protein interactions contributing to these complexes. In most cases, divalent cations are key to the structure and or function of these complexes, suggesting another potential role for the VWA domains, although most lack MIDAS motifs.
Copines
The copines are phospholipid-binding proteins, originally identified in Paramecium (Creutz_et al._, 1998). There are nine family members in human (three groups of three) and three orthologues in both C. elegans and A. thaliana, but none are detectable in D. melanogaster or S. cerevisiae(Figure S5). Two of the human copines (A and B) are previously uncharacterized Genomescan-predicted proteins and their sequences are available in Table S4. It seems that the ancestral organism common to_Homo sapiens_, C. elegans, and A. thaliana had a single copine that independently expanded into three in C. elegans and A. thaliana and nine in H. sapiens. The phylogenetic distribution suggests that copines have been lost from some eukaryotic phyla (Figure S5). Each copine contains two C2 domains followed by a single VWA domain except for copine W3, which has a single C2 domain. Three additional C. elegans molecules (Figure5) have VWA domains that are closely related to the copines but all lack C2 domains. No functional properties have been assigned to the VWA domains of copines. They contain a functional MIDAS motif based on preferential binding to magnesium and manganese (Tomsig and Creutz, 2000) despite the fact that the MIDAS motif is not perfectly conserved. In all 15 cases, region 1 of the copine MIDAS consists of the sequence D-x-T-x-S. The_Paramecium_ sequence is D-x-T-x-Q at this location. The C2 domains mediate calcium-dependent phospholipid binding (Davletov and Sudhof, 1993) and support oligomerization (Tomsig and Creutz, 2000). In_A. thaliana_, copines may play a role in growth regulation. BONZAI1 mutants that lack copine 1 (Figure S5) produce miniature plants when grown at 22°C (Hua et al., 2001). Mutations in the same gene described by another group result in abnormal cell death in response to low-humidity conditions (Jambunathan et al., 2001).
Unknown Conserved in Humans and Flies (Q9H0S5/Q9VPY0)
The final human VWA protein, Q9H0S5, is an uncharacterized molecule with a clear MIDAS domain and widespread expression based on EST frequency. Q9VPY0 is the Drosophila orthologue of this gene with several embryonic ESTs (TIGR Drosophila gene index TC71868; Adams et al., 2000; Figure S7). Q9VPY0does not have a conserved MIDAS motif. Both molecules consist of an N-terminal VWA domain followed by ∼400 amino acids of sequence that lacks obvious distinguishing features, including a signal sequence. The human and D. melanogaster molecules are ∼33% identical over their entire length. Orthologues of these molecules are not detectable in the C. elegans, A. thaliana, or S. cerevisiae genomes. This is the only VWA domain-containing protein found in humans and D. melanogaster but not in the C. elegans genome.
Additional C. elegans VWA Proteins
It has been noted (Hutter et al., 2000) that C. elegans contains a novel set of ECM and adhesion genes, and the same can be said for VWA-domain proteins (Figure 5, marked in blue). This genome encodes a large number of C-type lectin proteins, several of which also contain a VWA domain. This combination of domains is not seen in any other organism, and many of the proteins contain other domains as well or instead (Figure 5; CUB, ZP, and EGF). These all seem to reflect separate evolution of VWA-domain proteins in C. elegans. It remains unclear why this species has elaborated such a plethora of apparent adhesion proteins; perhaps it has something to do with cuticle formation or with the highly reproducible arrangements of cells in this organism, providing additional zip codes not required in less determinative organisms.
The C. elegans proteins mup-4 and mua-3 are VWA domain-containing transmembrane receptors for extracellular matrix (Figure 5). Mup-4/mua-3 have functional and molecular similarities with the mammalian integrin β4, suggesting that the molecules may represent an example of convergent evolution (Bercher et al., 2001; Hong et al., 2001). Like all integrin β subunits, mup-4 and mua-3 have extracellular VWA domains with conserved MIDAS motifs and EGF modules and, like integrin β4, their cytoplasmic domains link to intermediate filaments. In support of this idea, D. melanogaster do not have intermediate filaments and lack a homologue of either integrin β4 or mup-4 (Bercher et al., 2001).
Additional A. thaliana VWA Proteins
In contrast with the elaboration of novel extracellular VWA domain proteins in C. elegans, A. thaliana seems to have elaborated the intracellular portion of the VWA domain-containing protein set (Figure 6). There are no integrins or ECM proteins in the plant set of VWA proteins.
Two groups of A. thaliana proteins contain a combination of VWA and ring finger domains. Ring finger domains frequently have E3 ubiquitin-protein ligase activity. Rice and A. thaliana are the only sequenced organisms containing this combination, suggesting that it is a plant-specific domain architecture. One group of three paralogues has VWA-ring arrangement. A fourth paralogue, Q9LVN6, lacks the ring domain but the VWA domain is closely related to the other three. The VWA domains of these molecules are also closely related (100% bootstrap support) to the A. thaliana copine VWA domains, including the D-x-T-x-S sequence at region 1 of the MIDAS (Figure 6).
The second group, which seems to be unrelated to the first, has eight paralogues and the architecture ring-VWA. One-half of these have conserved MIDAS domains (Figure 6). The VWA domains of these molecules are related to the human Q9BVH8 family of VIT-VWA molecules (70% bootstrap support) and a group of bacterial proteins (NP_442565 and relatives; 92% bootstrap support).
With the possible exception of two groups of homologues, one with two members and the other with three members (Figure 6, below purple line), all A. thaliana VWA domain proteins seem to be intracellular. No functional information is available for the molecules below the purple line, but all have at least imperfect MIDAS motifs and one has a predicted signal sequence, suggesting that it might be secreted.
Prokaryotic VWA Proteins
In general, the archaeal and bacterial VWA domain-containing proteins are not well characterized. Of the 148 prokaryotic VWA domains in the databases, 90 have the words hypothetical, putative, ORF, or unknown in their descriptions. More than 80% of prokaryotic VWA domains have at least an imperfect MIDAS motif, indicating that divalent cation coordination may play a role in protein function (Figure S1, a–c, and Table S3).
In Archaea, there are 16 VWA domain proteins from nine different species (Table S2). Five of these are Mg chelatases based on sequence homology. The remaining proteins are all described as hypothetical or ORF. Within this list of unknown proteins, there are two groups of clear homologues; one group contains four members, the other contains two. All archaeal genomes sequenced to date have at least one VWA protein, but there are no universally present VWA domain proteins and the only molecules clearly related to eukaryotic VWA domain proteins are the Mg chelatases (see below).
In bacteria, there are 132 VWA domain proteins from 49 different species (Table S2). Examples of intracellular, secreted, and membrane-associated proteins can be found in this list. There are 11 Mg and four Co chelatases in the list. Eight molecules are orthologues of norD, a protein required for nitric oxide reductase activity (Bartnikas_et al._, 1997). The streptococcal opacity factors, named for their ability to cloud mammalian serum, contain VWA domains and bind fibronectin (Katerov et al., 2000). The TadG molecules mediate nonspecific adherence of bacterial cells to surfaces and contain VWA domains (Kachlany et al., 2000).
One-half of bacterial genomes (28/53) lack VWA domain proteins and, when present, most prokaryotic VWA domain proteins are not clear orthologues of one another. The number of VWA domain proteins in a proteome is usually one to four but in some cases, the number can be much higher. For example, Rhizobium loti has 12 VWA domain proteins (1 Mg chelatase and 11 uncharacterized molecules) but a closely related species, R. meliloti, has only three. The absence of VWA domains from many bacterial genomes supports the idea that the most ancient VWA proteins are intracellular eukaryotic molecules and suggests that they are not required for prokaryotic existence. Instead, horizontal gene transfer events may be responsible for the presence of some of the VWA domains in prokaryotes. We have already mentioned the VIT-VWA protein in the cyanobacterium_Anabaena_ (Figure S4) that seems to be a clear example of transfer from eukaryotes to prokaryotes.
The Mg and Co chelatases discussed earlier may be an exception to this generality. These AAA-VWA proteins play essential roles in protoporphyrin IX biosynthesis and occur in chloroplasts, bacteria, and Archaea. Photosynthesis (and Mg chelatases) originated in prokaryotes (Xiong and Bauer, 2002) and was acquired by eukaryotes through an endosymbiotic event (Gray, 1999). Therefore, it is plausible that these proteins originated in prokaryotes and were incorporated into plants. Whether other AAA-VWA proteins in eukaryotes represent a subsequent or separate evolution is unclear.
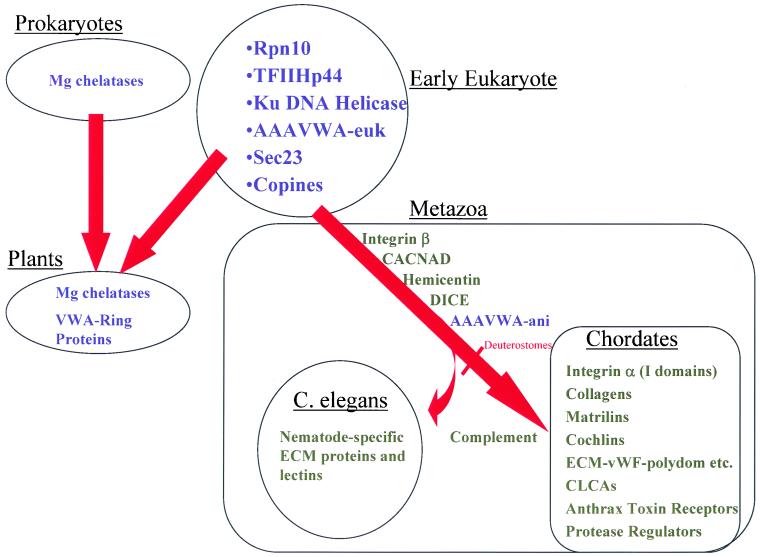
CONCLUSIONS
In considering the phylogenetic distribution of VWA domains (Figure 7), it is easiest to start from a few simplifying statements. First, all VWA proteins from S. cerevisiae and D. melanogaster (Figure S7) have orthologues in the human (and mouse) genomes as do many A. thaliana VWA proteins (Figure 6). Second, plants, nematodes, and chordates have each separately evolved characteristic sets of VWA-containing proteins. In plants, the radiation occurred in intracellular proteins, whereas in nematodes and chordates there were separate large expansions in the sets of extracellular, largely adhesive proteins, Third, if one considers the small set of VWA proteins common to all eukaryotes, they are all intracellular proteins involved in fundamental cellular functions (e.g., DNA repair, transcription, ribosomal transport, and protein degradation). This suggests that this domain originally evolved from a Rossmann fold, perhaps more than once, acquiring specialized functions, apparently related to multiprotein assemblies and perhaps involving divalent cations. The common presence of VWA domains in Mg and Co chelatases in_A. thaliana_ and prokaryotes could imply that this was a separate evolution from the Rossmann fold and a lateral transfer from prokaryotes to early photosynthetic eukaryotes.
Figure 7.
Phylogenetic distribution of VWA domain-containing proteins in the organisms with completed genomes is summarized in the diagram. Intracellular proteins are in blue; those with extracellular VWA domains are black. The most ancient VWA domain-containing proteins all seem to be intracellular (see list under early eukaryote). S. cerevisiae and S. pombe have all these genes except copines. With the exception of AAAVWA-ani, metazoan-specific VWA domains are extracellular. Mg chelatases may be of prokaryotic origin with subsequent transfer to plants in chloroplasts. They might also represent the ancestors of AAAVWA-euk, although, at the sequence level, they do not seem closely related. In present-day fungi and plants, it seems that all VWA domain proteins remain intracellular. In contrast, there have been significant expansions in extracellular proteins in Metazoa, a few being widely distributed (e.g., integrin β subunits) and others restricted as in chordates and C. elegans.
However, by far the majority of VWA domains now occur in extracellular proteins, notably integrin subunits and ECM proteins that are metazoan- and, in many cases, chordate-specific developments. These are the best understood VWA domains, and it is clear that their role is to mediate protein–protein interactions frequently involving a key role for divalent cations coordinated by the VWA domain (often by a so-called MIDAS motif). The most ancient of these extracellular VWA domains seem to be those residing in integrin β subunits from sponges to humans. Incorporation into integrin α subunits came much later, perhaps as late as chordates. A case can be made that some VWA domains in ECM proteins were an early event (e.g., hemicentin) but that most VWA-containing matrix proteins appeared much later, in chordates (VWA collagens, matrilins, cochlin/vitrin, polydom, and vWF itself) or as separate evolutions in bizarre ECM or membrane proteins in C. elegans and Plasmodium. Other manifestations of VWA domains include membrane proteins such as the anthrax toxin receptor family, subunits of ion channels (probably modulators) and modulators of protease cascades (complement factors and trypsin inhibitors). Some of these proteins have been shown to be involved in cell adhesion and others in protein–protein interactions, and it seems likely that this is also true of the others.
The VWA domains of the integrin α and β subunits and those present in vWF are the best studied, and there is abundant evidence for the importance of these domains in protein-ligand binding. Knowledge about these domains should form the framework for VWA domain research in the future. A plausible hypothesis is that many VWA domains will support protein–protein interaction by joint coordination of a metal ion at the MIDAS (Emsley et al., 2000; Xiong et al., 2002), and a systematic effort to identify motifs that can support these interactions would be instructive. Another common feature of the VWA domains of integrins and vWF is that they undergo significant conformational changes on ligand-binding, which seem to propagate to other parts of these proteins, altering their shape and function (Sadler, 1998; Liddington, 2002; Shimaoka et al., 2002). A very appealing hypothesis is that this property may be a general feature of VWA domains, coupling binding of associated proteins to long-distance conformational changes in large multidomain proteins and multiprotein complexes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Mary Connolly and Aaron Cook for technical assistance and Arjan van der Flier, Chris Liu, and Sunny Wong for critical reading of the manuscript. This work was supported by National Cancer Institute grant RO1CA17007 and the Howard Hughes Medical Institute. R.O.H. is an Investigator of Howard Hughes Medical Institute.
Footnotes
D⃞
Online version of this article contains supplemental material.
*
Corresponding author. E-mail address: rohynes@mit.edu.
REFERENCES
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Anand G, Yin X, Shahidi AK, Grove L, Prochownik EV. Novel regulation of the helix-loop-helix protein Id1 by S5a, a subunit of the 26 S proteasome. J Biol Chem. 1997;272:19140–19151. doi: 10.1074/jbc.272.31.19140. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Cachea signaling domain common to animal Ca(2+)-channel subunits, and a class of prokaryotic chemotaxis receptors. Trends Biochem Sci. 2000;25:535–537. doi: 10.1016/s0968-0004(00)01672-8. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszodi A, Bateman JF, Hirsch E, Baranyi M, Hunziker EB, Hauser N, Bosze Z, Fassler R. Normal skeletal development of mice lacking matrilin 1: redundant function of matrilins in cartilage? Mol Cell Biol. 1999;19:7841–7845. doi: 10.1128/mcb.19.11.7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajt ML, Goodman T, McGuire SL. B 2 (CD18) mutations abolish ligand recognition by I domain integrins LFA-1 (aLb2, CD11a/CD18) and MAC-1 (aMb2, CD11b/CD18) J Biol Chem. 1995;270:94–98. doi: 10.1074/jbc.270.1.94. [DOI] [PubMed] [Google Scholar]
- Baldwin ET, et al. Cation binding to the integrin CD11bI domain and activation model assessment. Structure. 1998;6:923–35. doi: 10.1016/s0969-2126(98)00093-8. [DOI] [PubMed] [Google Scholar]
- Bartnikas TB, Tosques IE, Laratta WP, Shi J, Shapleigh JP. Characterization of the nitric oxide reductase-encoding region in Rhodobacter sphoeroides2.4.3. J Bacteriol. 1997;179:3534–3540. doi: 10.1128/jb.179.11.3534-3540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Beglova N, Blacklow SC, Takagi J, Springer TA. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nat Struct Biol. 2002;9:282–287. doi: 10.1038/nsb779. [DOI] [PubMed] [Google Scholar]
- Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, Davis GE. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci. 2001;114:2755–2773. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- Bercher M, Wahl J, Vogel BE, Lu C, Hedgecock EM, Hall DH, Plenefisch JD. mua-3, a gene required for mechanical tissue integrity in Caenorhabditis elegans, encodes a novel transmembrane protein of epithelial attachment complexes. J Cell Biol. 2001;154:415–426. doi: 10.1083/jcb.200103035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska J, Cruz M, Atiemo A, Handin R, Liddington R. The von Willebrand factor A3 domain does not contain a metal ion-dependent adhesion site motif. J Biol Chem. 1997;272:25162–25167. doi: 10.1074/jbc.272.40.25162. [DOI] [PubMed] [Google Scholar]
- Bonaldo P, Russo V, Bucciotti F, Doliana R, Colombatti A. Structural and functional features of the a3 chain indicate a bridging role for chicken collagen VI in connective tissues. Biochemistry. 1990;29:1245–1254. doi: 10.1021/bi00457a021. [DOI] [PubMed] [Google Scholar]
- Bost F, Diarra-Mehrpour M, Martin JP. Inter-α-trypsin inhibitor proteoglycan family-a group of proteins binding and stabilizing the extracellular matrix. Eur J Biochem. 1998;252:339–346. doi: 10.1046/j.1432-1327.1998.2520339.x. [DOI] [PubMed] [Google Scholar]
- Bounpheng MA, Dimas JJ, Dodds SG, Christy BA. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- Brickley K, Campbell V, Berrow N, Leach R, Norman RI, Wray D, Dolphin AC, Baldwin SA. Use of site-directed antibodies to probe the topography of the α2 subunit of voltage-gated Ca2+channels. FEBS Lett. 1995;364:129–133. doi: 10.1016/0014-5793(95)00371-f. [DOI] [PubMed] [Google Scholar]
- Chapman KL, Mortier GR, Chapman K, Loughlin J, Grant ME, Briggs MD. Mutations in the region encoding the von Willebrand factor A domain of matrilin-3 are associated with multiple epiphyseal dysplasia. Nat Genet. 2001;28:393–396. doi: 10.1038/ng573. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhang Y, Johnson DM, Goetinck PF. Assembly of a novel cartilage matrix protein filamentous network: molecular basis of differential requirement of von Willebrand factor A domains. Mol Biol Cell. 1999;10:2149–2162. doi: 10.1091/mbc.10.7.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Garrison S, Weis JJ, Weis JH. Identification of pactolus, an integrin β subunit-like cell-surface protein preferentially expressed by cells of the bone marrow. J Biol Chem. 1998;273:8711–8718. doi: 10.1074/jbc.273.15.8711. [DOI] [PubMed] [Google Scholar]
- Colombatti A, Bonaldo P, Doliana R. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix. 1993;13:297–306. doi: 10.1016/s0934-8832(11)80025-9. [DOI] [PubMed] [Google Scholar]
- Creutz CE, Tomsig JL, Snyder SL, Gautier MC, Skouri F, Beisson J, Cohen J. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J Biol Chem. 1998;273:1393–1402. doi: 10.1074/jbc.273.3.1393. [DOI] [PubMed] [Google Scholar]
- Cruz MA, Diacovo TG, Emsley J, Liddington R, Handin RI. Mapping the glycoprotein Ib-binding site in the von willebrand factor A1 domain. J Biol Chem. 2000;275:19098–19105. doi: 10.1074/jbc.M002292200. [DOI] [PubMed] [Google Scholar]
- Cruz MA, Yuan H, Lee JR, Wise RJ, Handin RI. Interaction of the von Willebrand factor (vWF) with collagen. Localization of the primary collagen-binding site by analysis of recombinant vWF a domain polypeptides. J Biol Chem. 1995;270:10822–10827. doi: 10.1074/jbc.270.18.10822. [DOI] [PubMed] [Google Scholar]
- Cunningham SA, Awayda MS, Bubien JK, Ismailov II, Arrate MP, Berdiev BK, Benos DJ, Fuller CM. Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem. 1995;270:31016–31026. doi: 10.1074/jbc.270.52.31016. [DOI] [PubMed] [Google Scholar]
- Davletov BA, Sudhof TC. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- De Jongh KS, Warner C, Catterall WA. Subunits of purified calcium channels. α2 and δ are encoded by the same gene. J Biol Chem. 1990;265:14738–14741. [PubMed] [Google Scholar]
- Deak F, Wagener R, Kiss I, Paulsson M. The matrilins: a novel family of oligomeric extracellular matrix proteins. Matrix Biol. 1999;18:55–64. doi: 10.1016/s0945-053x(98)00006-7. [DOI] [PubMed] [Google Scholar]
- Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci USA. 1996;93:13429–13434. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble RC, Widom J, Gruber AD, Abdel-Ghany M, Levine R, Goodwin A, Cheng HC, Pauli BU. Cloning and characterization of lung-endothelial cell adhesion molecule-1 suggest it is an endothelial chloride channel. J Biol Chem. 1997;272:27853–27861. doi: 10.1074/jbc.272.44.27853. [DOI] [PubMed] [Google Scholar]
- Emsley J, Cruz M, Handin R, Liddington R. Crystal structure of the von Willebrand Factor A1 domain and implications for the binding of platelet glycoprotein Ib. J Biol Chem. 1998;273:10396–10401. doi: 10.1074/jbc.273.17.10396. [DOI] [PubMed] [Google Scholar]
- Emsley J, King SL, Bergelson JM, Liddington RC. Crystal structure of the I domain from integrin α2β1. J Biol Chem. 1997;272:28512–28517. doi: 10.1074/jbc.272.45.28512. [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP–Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fitzgerald J, Bateman JF. A new FACIT of the collagen family: COL21A1. FEBS Lett. 2001;505:275–280. doi: 10.1016/s0014-5793(01)02754-5. [DOI] [PubMed] [Google Scholar]
- Fribourg S, Kellenberger E, Rogniaux H, Poterszman A, Van Dorsselaer A, Thierry JC, Egly JM, Moras D, Kieffer B. Structural characterization of the cysteine-rich domain of TFIIH p44 subunit. J Biol Chem. 2000;275:31963–31971. doi: 10.1074/jbc.M004960200. [DOI] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison S, Hojgaard A, Patillo D, Weis JJ, Weis JH. Functional characterization of pactolus, a β-integrin-like protein preferentially expressed by neutrophils. J Biol Chem. 2001;276:35500–35511. doi: 10.1074/jbc.M104369200. [DOI] [PubMed] [Google Scholar]
- Gilges D, Vinit MA, Callebaut I, Coulombel L, Cacheux V, Romeo PH, Vigon I. Polydom: a secreted protein with pentraxin, complement control protein, epidermal growth factor and von Willebrand factor A domains. Biochem J. 2000;352:49–59. [PMC free article] [PubMed] [Google Scholar]
- Gray MW. Evolution of organellar genomes. Curr Opin Genet Dev. 1999;9:678–687. doi: 10.1016/s0959-437x(99)00030-1. [DOI] [PubMed] [Google Scholar]
- Gruber AD, Pauli BU. Molecular cloning and biochemical characterization of a truncated, secreted member of the human family of Ca2+-activated Cl−channels. Biochim Biophys Acta. 1999;1444:418–423. doi: 10.1016/s0167-4781(99)00008-1. [DOI] [PubMed] [Google Scholar]
- Gruber AD, Schreur KD, Ji HL, Fuller CM, Pauli BU. Molecular cloning and transmembrane structure of hCLCA2 from human lung, trachea, and mammary gland. Am J Physiol. 1999;276:C1261–C1270. doi: 10.1152/ajpcell.1999.276.6.C1261. [DOI] [PubMed] [Google Scholar]
- Higgins JM, Cernadas M, Tan K, Irie A, Wang J, Takada Y, Brenner MB. The role of α and β chains in ligand recognition by β7 integrins. J Biol Chem. 2000;275:25652–25664. doi: 10.1074/jbc.M001228200. [DOI] [PubMed] [Google Scholar]
- Hobom M, Dai S, Marais E, Lacinova L, Hofmann F, Klugbauer N. Neuronal distribution and functional characterization of the calcium channel α2δ-2 subunit. Eur J Neurosci. 2000;12:1217–1226. doi: 10.1046/j.1460-9568.2000.01009.x. [DOI] [PubMed] [Google Scholar]
- Hong L, Elbl T, Ward J, Franzini-Armstrong C, Rybicka KK, Gatewood BK, Baillie DL, Bucher EA. MUP-4 is a novel transmembrane protein with functions in epithelial cell adhesion in Caenorhabditis elegans. J Cell Biol. 2001;154:403–414. doi: 10.1083/jcb.200007075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoylaerts MF, Yamamoto H, Nuyts K, Vreys I, Deckmyn H, Vermylen J. von Willebrand factor binds to native collagen VI primarily via its A1 domain. Biochem J. 1997;324:185–191. doi: 10.1042/bj3240185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Grisafi P, Cheng SH, Fink GR. Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 2001;15:2263–2272. doi: 10.1101/gad.918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Birk DE, Goetinck PF. Mice lacking matrilin-1 (cartilage matrix protein) have alterations in type II collagen fibrillogenesis and fibril organization. Dev Dyn. 1999;216:434–441. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<434::AID-DVDY11>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Humbert S, van Vuuren H, Lutz Y, Hoeijmakers JH, Egly JM, Moncollin V. p44 and p34 subunits of the BTF2/TFIIH transcription factor have homologies with SSL1, a yeast protein involved in DNA repair. EMBO J. 1994;13:2393–2398. doi: 10.1002/j.1460-2075.1994.tb06523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter H, et al. Conservation and novelty in the evolution of cell adhesion and extracellular matrix genes. Science. 2000;287:989–994. doi: 10.1126/science.287.5455.989. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150:F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- Jambunathan N, Siani JM, McNellis TW. A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell. 2001;13:2225–2240. doi: 10.1105/tpc.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean L, Risler JL, Nagase T, Coulouarn C, Nomura N, Salier JP. The nuclear protein PH5P of the inter-α-inhibitor superfamily: a missing link between poly(ADP-ribose)polymerase and the inter-α-inhibitor family and a novel actor of DNA repair? FEBS Lett. 1999;446:6–8. doi: 10.1016/s0014-5793(99)00173-8. [DOI] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, Figurski DH. Nonspecific adherence by Actinobacillus actinomycetemcomitansrequires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katerov V, Lindgren PE, Totolian AA, Schalen C. Streptococcal opacity factor: a family of bifunctional proteins with lipoproteinase and fibronectin-binding activities. Curr Microbiol. 2000;40:149–156. doi: 10.1007/s002849910031. [DOI] [PubMed] [Google Scholar]
- Keeney S, Cumming AM. The molecular biology of von Willebrand disease. Clin Lab Hematol. 2001;23:209–230. doi: 10.1046/j.1365-2257.2001.00400.x. [DOI] [PubMed] [Google Scholar]
- Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumanovics A, Lindahl KF. G7c in the lung tumor susceptibility (Lts) region of the MHC class III region encodes a von Willebrand factor type A domain protein. Immunogenetics. 2001;53:64–68. doi: 10.1007/s002510100297. [DOI] [PubMed] [Google Scholar]
- Lankhof H, Damas C, Schiphorst ME, Ijsseldijk MJ, Bracke M, Furlan M, Tsai HM, de Groot PG, Sixma JJ, Vink T. von Willebrand factor without the A2 domain is resistant to proteolysis. Thromb Hemost. 1997;77:1008–1013. [PubMed] [Google Scholar]
- Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC. Will the real integrin please stand up? Structure. 2002;10:605–607. doi: 10.1016/s0969-2126(02)00766-9. [DOI] [PubMed] [Google Scholar]
- Loftus JC, Liddington RC. New insights into integrin-ligand interaction. J Clin Invest. 1997;100:S77–S81. [PubMed] [Google Scholar]
- Ma Q, Shimaoka M, Lu C, Jing H, Carman CV, Springer TA. Activation-induced conformational changes in the I domain region of lymphocyte function-associated antigen 1. J Biol Chem. 2002;277:10638–10641. doi: 10.1074/jbc.M112417200. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegansby high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Makihira S, et al. Enhancement of cell adhesion and spreading by a cartilage-specific noncollagenous protein, cartilage matrix protein (CMP/Matrilin-1), via integrin α1β1. J Biol Chem. 1999;274:11417–11423. doi: 10.1074/jbc.274.16.11417. [DOI] [PubMed] [Google Scholar]
- Maurizi MR, Li CH. AAA proteins; in search of a common molecular basis. EMBO Rep. 2001;2:980–985. doi: 10.1093/embo-reports/kve229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne R, Ren ZX, Liu J, Cook T, Carson M, Narayana S. VIT-1: the second member of a new branch of the von Willebrand factor A domain superfamily. Biochem Soc Trans. 1999;27:832–835. doi: 10.1042/bst0270832. [DOI] [PubMed] [Google Scholar]
- Miyazawa S, Azumi K, Nonaka M. Cloning and characterization of integrin α subunits from the solitary ascidian Halocynthia roretzi. J Immunol. 2001;166:1710–1715. doi: 10.4049/jimmunol.166.3.1710. [DOI] [PubMed] [Google Scholar]
- Monaco C, Negrini M, Sozzi G, Veronese ML, Vorechovsky I, Godwin AK, Croce CM. Molecular cloning and characterization of LOH11CR2A, a new gene within a refined minimal region of LOH at 11q23. Genomics. 1997;46:217–222. doi: 10.1006/geno.1997.5036. [DOI] [PubMed] [Google Scholar]
- Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Ishikawa K, Hirosawa M, Ohara O. Prediction of the coding sequences of unidentified human genes. XVII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:143–150. doi: 10.1093/dnares/7.2.143. [DOI] [PubMed] [Google Scholar]
- Naitza T, Spano F, Robson KJH, Crisanti A. The thrombospondin-related protein family of apicomplexan parasites: the gears of the cell invasion machinery. Parasitol Today. 1998;14:479–484. doi: 10.1016/s0169-4758(98)01346-5. [DOI] [PubMed] [Google Scholar]
- Nolte M, Pepinsky RB, Venyaminov S, Koteliansky V, Gotwals PJ, Karpusas M. Crystal structure of the α1β1 integrin I-domain: insights into integrin I-domain function. FEBS Lett. 1999;452:379–385. doi: 10.1016/s0014-5793(99)00666-3. [DOI] [PubMed] [Google Scholar]
- Nonaka M, Azumi K. Opsonic complement system of the solitary ascidian Halocynthia roretzi. Dev Comp Immunol. 1999;23:421–417. doi: 10.1016/s0145-305x(99)00021-x. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- Pauli BU, Abdel-Ghany M, Cheng HC, Gruber AD, Archibald HA, Elble RC. Molecular characteristics and functional diversity of CLCA family members. Clin Exp Pharmacol Physiol. 2000;27:901–905. doi: 10.1046/j.1440-1681.2000.03358.x. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. Eukaryotic signaling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J Mol Biol. 1999;289:729–745. doi: 10.1006/jmbi.1999.2827. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Schultz J, Copley RR, Andrade MA, Bork P. Evolution of domain families. Adv Protein Chem. 2000;54:185–244. doi: 10.1016/s0065-3233(00)54007-8. [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Uitto J. Mutation analysis and molecular genetics of epidermolysis bullosa. Matrix Biol. 1999;18:29–42. doi: 10.1016/s0945-053x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- Qu A, Leahy DJ. Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, αLβ2) integrin. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi AM, Hogg N. I domain of β2 integrin lymphocyte function-associated antigen-1 contains a binding site for ligand intercellular adhesion molecule-1. J Biol Chem. 1994;269:12395–12398. [PubMed] [Google Scholar]
- Ribba AS, Loisel I, Lavergne JM, Juhan-Vague I, Obert B, Cherel G, Meyer D, Girma JP. Ser968Thr mutation within the A3 domain of von Willebrand factor (VWF) in two related patients leads to a defective binding of VWF to collagen. Thromb Hemost. 2001;86:848–854. [PubMed] [Google Scholar]
- Ricard-Blum AS, Dublet B, van der Rest M. Unconventional Collagens Types VI, VII, VIII, IX, X, XIV, XIX. Oxford, United Kingdom: Oxford University Press; 2000. [Google Scholar]
- Robertson NG, Resendes BL, Lin JS, Lee C, Aster JC, Adams JC, Morton CC. Inner ear localization of mRNA and protein products of COCH, mutated in the sensorineural deafness and vestibular disorder, DFNA9. Hum Mol Genet. 2001;10:2493–2500. doi: 10.1093/hmg/10.22.2493. [DOI] [PubMed] [Google Scholar]
- Romijn RA, Bouma B, Wuyster W, Gros P, Kroon J, Sixma JJ, Huizinga EG. Identification of the collagen-binding site of the von Willebrand factor A3-domain. J Biol Chem. 2001;276:9985–9991. doi: 10.1074/jbc.M006548200. [DOI] [PubMed] [Google Scholar]
- Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- Salier JP, Rouet P, Raguenez G, Daveau M. The inter-α-inhibitor family: from structure to regulation. Biochem J. 1996;315:1–9. doi: 10.1042/bj3150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Raux E, Wilson KS, Warren MJ. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38:10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka M, Lu C, Palframan RT, von Andrian UH, McCormack A, Takagi J, Springer TA. Reversibly locking a protein fold in an active conformation with a disulfide bond: integrin αL I domains with high affinity and antagonist activity in vivo. Proc Natl Acad Sci USA. 2001;98:6009–6014. doi: 10.1073/pnas.101130498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biol Mol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- Smith LC, Shih CS, Dachenhausen SG. Coelomocytes express SpBf, a homologue of factor B, the second component in the sea urchin complement system. J Immunol. 1998;161:6784–6793. [PubMed] [Google Scholar]
- Snoek M, Albertella MR, van Kooij M, Wixon J, van Vugt H, de Groot K, Campbell RD. G7c, a novel gene in the mouse and human major histocompatibility complex class III region, possibly controlling lung tumor susceptibility. Immunogenetics. 2000;51:383–386. doi: 10.1007/s002510050634. [DOI] [PubMed] [Google Scholar]
- Snoek M, Teuscher C, van Vugt H. Molecular analysis of the major MHC recombinational hot spot located within the G7c gene of the murine class III region that is involved in disease susceptibility. J Immunol. 1998;160:266–272. [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Takagi J, DeBottis DP, Erickson HP, Springer TA. The role of the specificity-determining loop of the integrin β subunit I-like domain in autonomous expression, association with the α subunit, and ligand binding. Biochemistry. 2002;41:4339–4347. doi: 10.1021/bi016047u. [DOI] [PubMed] [Google Scholar]
- Takagi J, Kamata T, Meredith J, Puzon-McLaughlin W, Takada Y. Changing ligand specificities of αvβ1 and αvβ3 integrins by swapping a short diverse sequence of the β subunit. J Biol Chem. 1997;272:19794–19800. doi: 10.1074/jbc.272.32.19794. [DOI] [PubMed] [Google Scholar]
- Tanner NK, Linder P. DExD/H box RNA helicases. from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Tomsig JL, Creutz CE. Biochemical characterization of copine: a ubiquitous Ca2+-dependent, phospholipid-binding protein. Biochemistry. 2000;39:16163–16175. doi: 10.1021/bi0019949. [DOI] [PubMed] [Google Scholar]
- Tozer EC, Liddington RC, Sutcliffe MJ, Smeeton AH, Loftus JC. Ligand binding to integrin αIIββ3 is dependent on a MIDAS-like domain in the β3 subunit. J Biol Chem. 1996;271:21978–84. doi: 10.1074/jbc.271.36.21978. [DOI] [PubMed] [Google Scholar]
- Tuckwell D. Evolution of von Willebrand factor A (VWA) domains. Biochem Soc Trans. 1999;27:835–840. doi: 10.1042/bst0270835. [DOI] [PubMed] [Google Scholar]
- Tuckwell D. Identification and analysis of collagen α1(XXI), a novel member of the FACIT collagen family. Matrix Biol. 2002;21:63–66. doi: 10.1016/s0945-053x(01)00176-7. [DOI] [PubMed] [Google Scholar]
- Tuckwell DS, Humphries MJ. A structure prediction for the ligand-binding region of the integrin β subunit: evidence for the presence of a von Willebrand factor A domain. FEBS Lett. 1997;400:297–303. doi: 10.1016/s0014-5793(96)01368-3. [DOI] [PubMed] [Google Scholar]
- Tuckwell DS, Xu Y, Newham P, Humphries MJ, Volanakis JE. Surface loops adjacent to the cation-binding site of the complement factor B von Willebrand factor type A module determine C3b binding specificity. Biochemistry. 1997;36:6605–6613. doi: 10.1021/bi963155l. [DOI] [PubMed] [Google Scholar]
- Ueda T, Rieu P, Brayer J, Arnaout MA. Identification of the complement iC3b binding site in the β2 integrin CR3 (CD11b/CD18) Proc Natl Acad Sci USA. 1994;91:10680–10684. doi: 10.1073/pnas.91.22.10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel BE, Hedgecock EM. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development. 2001;128:883–894. doi: 10.1242/dev.128.6.883. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Walker CJ, Willows RD. Mechanism and regulation of Mg-chelatase. Biochem J. 1997;327:321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;12:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wieland I, Arden KC, Michels D, Klein-Hitpass L, Bohm M, Viars CS, Weidle UH. Isolation of DICE1: a gene frequently affected by LOH and downregulated in lung carcinomas. Oncogene. 1999;18:4530–4537. doi: 10.1038/sj.onc.1202806. [DOI] [PubMed] [Google Scholar]
- Wieland I, Ropke A, Stumm M, Sell C, Weidle UH, Wieacker PF. Molecular characterization of the DICE1 (DDX26) tumor suppressor gene in lung carcinoma cells. Oncol Res. 2001;12:491–500. doi: 10.3727/096504001108747503. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiaegenome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Eyre DR. Matrilin-3 forms disulfide-linked oligomers with matrilin-1 in bovine epiphyseal cartilage. J Biol Chem. 1998;273:17433–17438. doi: 10.1074/jbc.273.28.17433. [DOI] [PubMed] [Google Scholar]
- Xiong J, Bauer CE. Complex Evolution of Photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 2002;53:503–521. doi: 10.1146/annurev.arplant.53.100301.135212. [DOI] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Yeh RF, Lim LP, Burge CB. Computational inference of homologous gene structures in the human genome. Genome Res. 2001;11:803–816. doi: 10.1101/gr.175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu DZ, Cheng CF, Pauli BU. Mediation of lung metastasis of murine melanomas by a lung-specific endothelial cell adhesion molecule. Proc Natl Acad Sci USA. 1991;88:9568–9572. doi: 10.1073/pnas.88.21.9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.