Genetic diversity and selection in the maize starch pathway (original) (raw)
Abstract
Maize is both phenotypically and genetically diverse. Sequence studies generally confirm the extensive genetic variability in modern maize is consistent with a lack of selection. For more than 6,000 years, Native Americans and modern breeders have exploited the tremendous genetic diversity of maize (Zea mays ssp. mays) to create the highest yielding grain crop in the world. Nonetheless, some loci have relatively low levels of genetic variation, particularly loci that have been the target of artificial selection, like c1 and tb1. However, there is limited information on how selection may affect an agronomically important pathway for any crop. These pathways may retain the signature of artificial selection and may lack genetic variation in contrast to the rest of the genome. To evaluate the impact of selection across an agronomically important pathway, we surveyed nucleotide diversity at six major genes involved in starch metabolism and found unusually low genetic diversity and strong evidence of selection. Low diversity in these critical genes suggests that a paradigm shift may be required for future maize breeding. Rather than relying solely on the diversity within maize or on transgenics, future maize breeding would perhaps benefit from the incorporation of alleles from maize's wild relatives.
Maize molecular diversity is roughly 2- to 5-fold higher than that of other domesticated grass crops (1). Tenaillon et al. (2) reported that in 25 maize individuals, one nucleotide every 28 base pairs is polymorphic, and overall nucleotide diversity is almost 1.3%. That study, the largest examination of random maize loci, found almost no evidence of selection in 21 genes along chromosome 1. Maize's closest wild relative, Z. mays ssp. parviglumis (a teosinte), often has levels of nucleotide diversity that surpass 2% (3–6). The tremendous diversity of maize and teosinte has been the raw genetic material for the radical transformation of maize into the world's highest yielding grain crop.
To date, only two loci have been identified as targets of selection in maize. Teosinte branched, tb1, is responsible for modifying apical dominance, tillering, and inflorescence position and was vital to maize domestication (7). A nucleotide diversity survey by Wang et al. (8) showed that selection was strongly directed at tb1. During the improvement of maize, kernel color was modified through the anthocyanin pathway. A nucleotide diversity survey of the c1 gene (an anthocyanin regulator) indicated the locus also was under selection (9). In both cases, surveys of nucleotide diversity were critical in linking selection pressure to specific genes in pathways. There has been little examination of selection in the maize starch metabolic pathway, which is probably the single most important pathway for grain production.
Starch production is critical to both the yield and the quality of the grain. In the maize endosperm, sucrose is converted to glucose and then into starches that normally account for 73% of the kernel's total weight (Fig. 1). Roughly three-quarters of the total starch is amylopectin, which consists of branched glucose chains that form insoluble, semicrystalline granules. The remainder of the starch is amylose, which is composed of linear chains of glucose that adopt a helical configuration within the granule (10). The chemical and structural nature of amylose and amylopectin confer specific properties related to the viscosity of starch that are important in food processing. Although amylose and amylopectin may have synergistic effects on viscosity (11), amylose is typically thought to affect the gelling of starch (12). Gelatinization is a property that controls starch firmness, because of reassociation of glucose molecules. The contribution of amylose to starch viscosity is an increase in pasting temperatures and shear stress stability (11, 13). Alternatively, amylopectin is primarily responsible for granule swelling and eventual thickening of pastes upon addition of heat (14). Starch pasting modifies the ability of foods to hold fat and protein molecules that enhance flavor and texture (15). Selection for yield and better kernel quality may have contributed to the maize domestication and improvement process.
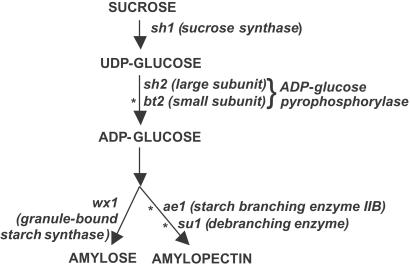
Figure 1.
A simplified pathway of starch production in maize and the position of the six sampled genes in the pathway. *, loci with strong evidence of selection (either HKA or Tajima's D test).
Plant genetics and biochemistry have so far identified over 20 genes involved in starch production (10, 16). We have focused on six key genes known to play major roles in this pathway: amylose extender1 (ae1), brittle2 (bt2), shrunken1 (sh1), shrunken2 (sh2), sugary1 (su1), and waxy1 (wx1) (Fig. 1). bt2, sh1, and sh2, located upstream in the pathway, aid in the formation of glucose. High SH1 activity plays a role in better grain filling, probably by providing more glucose for ADP-glucose pyrophosphorylase (AGPase) (17, 18). sh2 and bt2 encode subunits of the AGPase enzyme, which controls the rate-limiting step in starch production and is regulated by allosteric effectors (19, 20). The enzymes coded by ae1, su1, and wx1 produce the final products of starch metabolism, amylose and amylopectin (21, 22). Mutations at the wx1 locus eliminate amylose and have been used in modern breeding to create high amylopectin maize (23). Reduction of amylopectin has been accomplished with ae1 and su1 mutants (22). We evaluated selection at these six loci by examining patterns of nucleotide diversity in maize and Z. mays ssp. parviglumis.
Materials and Methods
Sampling.
To examine diversity and selection in the starch pathway, we sequenced the six loci from inbred lines of maize that represent much of the breeding diversity available. Diversity estimates were performed by sequencing 30 maize inbred lines that included both coding and noncoding genic regions: A272, A6, B103, B14A, B37, B73, B97, CI187–2, CML254, CML258, CML333, D940Y, EP1, F2, I205, IDS28, IL101, Ki9, Ki21, Ky21, M162W, Mo17, N28Ht, NC260, NC348, Oh43, Pa91, T232, Tx601, and W153R. The entire gene and 500–2,000bp upstream of the translation start site were sampled from bt2, sh1, sh2, su1, and wx1. Approximately 8 kbp were sequenced from the 23 kbp ae1 gene, spanning some of the introns and almost all of the exons. Exon 15 and the large introns 11 and 14 were not sequenced at all.
From Z. m. ssp. parviglumis, 500–3,400 bp of each gene were amplified and cloned, and then a single clone was sequenced. The 10 accessions sampled represent much of the range and diversity of the subspecies (USDA: PI566686, PI566688, PI566691, PI331783, PI331785, PI331786, PI331787, Iltis & Cochrane Site 3, Beadle & Kato Site 5, and Benz 967). Tripsacum dactyloides (PLT457 seeds supplied courtesy of Joseph Burns, USDA/ARS, North Carolina State University) was sequenced directly from at least 1,000 bp of amplification product for each gene. Because PCR errors were a concern in these heterozygous teosinte samples, high fidelity enzyme (Pfu) was used, and statistical tests between the maize and teosinte diversity numbers were not conducted.
To clarify the origin of sweet corn, we PCR-amplified and sequenced a 1,000-bp area in the promoter and the 1,000-bp area surrounding residue 578 from seven Mexican and South American maize accessions (AYA-32, BOV-331, BOV-344, BOV-396, CUN-465, JAL-304, NAR-494). We sequenced the entire su1 locus in AYA-32 and JAL-304.
Statistics.
Nucleotide diversity, π, is the average number of nucleotide differences per site between two sequences. π was estimated by using DNASP (24). Insertions and deletions were excluded from the estimates. Tajima's test of selection (25) was also conducted by using DNASP (24).
The Hudson–Kreitman–Aguade (HKA) tests were used to evaluate selection at the loci (26). We used the HKA test to compare the six starch loci with 11 neutral loci sampled previously (2) and applied the test at two levels within our germplasm. In the first sample, we used all 30 diverse lines (set A), whereas in the second sample, we used a narrower subset of 9 lines (set B). Testing at two levels helps determine whether selection occurred across all breeding germplasm or only in a narrower subset of predominantly U.S. germplasm. In the test for all breeding germplasm (set A), the complete set of 30 diverse lines in this study and all lines from Tenaillon et al. (2) were used. For the narrow germplasm, U.S. lines were the predominant focus (set B). This set was designed to be equivalent to the Tenaillon et al. U.S. germplasm (2). Set B excluded sweet corn, popcorn, and most maize with tropical germplasm. Six of 9 lines (B73, Mo17, W153R, Ky21, Oh43, Tx601) were identical to Tenaillon et al. (2), whereas three substitutes were made (Ki9 for Mo24W, I205 for T8, and NC348 for NC258) based on the closest genetic distance between lines from SSR data (data not shown). In the narrow set, only Ki9 is from outside the U.S.
Association tests were conducted by using the STRAT program (27), and population structure effects were reduced by the method of Thornsberry et al. (28).
Results
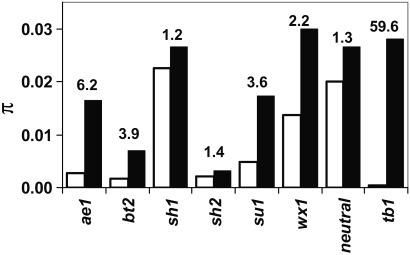
Although the starch loci exhibited a wide range in diversity, average diversity (π) in the starch loci was 2.3-fold lower than 20 random maize loci at silent sites (T test; P < 0.05; Table 1), and 4.8-fold lower at nonsynonymous sites (2). We also sampled genetic diversity in Z. mays spp. parviglumis. In nonselected loci (adh1, adh2, glb1, hm1, hm2, and te1), maize has 1.3-fold lower diversity than Z. mays ssp. parviglumis (Fig. 2, and refs. 2–5). In contrast, three of the starch loci (su1, bt2, and ae1) exhibited a dramatic 3- to 7-fold reduction in diversity (Fig. 2), which is consistent with artificial selection since domestication. Rare divergent haplotypes at ae1 and su1 are responsible for most of the diversity. If these rare divergent haplotypes were excluded, then diversity at these loci among the 30 inbreds would be almost zero.
Table 1.
Summary of maize nucleotide diversity
| Sites | Diversity, π | ||
|---|---|---|---|
| Silent | Nonsyn. | ||
| Locus | |||
| ae1 | 6,781 | 0.0029 | 0.0007 |
| bt2 | 6,098 | 0.0023 | 0.0010 |
| sh1 | 6,176 | 0.0121 | 0.0005 |
| sh2 | 6,754 | 0.0050 | 0.0013 |
| su1 | 9,378 | 0.0027 | 0.0004 |
| wx1 | 2,978 | 0.0115 | 0.0014 |
| Starch loci* | 37,330 | 0.0052 | 0.0008 |
| Random loci† | 10,908 | 0.0122 | 0.0038 |
Figure 2.
Comparison of silent diversity in maize (white bars) and its wild relative Z. mays ssp. parviglumis (black bars). For each locus, 500–2,700 silent bases were sampled. The numbers above the bars indicate the fold reduction in diversity between maize and Z. m. ssp. parviglumis. Neutral refers to the average of six nonselected genes (adh1, adh2, glb1, hm1, hm2, and te1); tb1 is the only cloned domestication gene from maize (only the highly selected promoter region is shown).
To test formally for selection, we used the HKA test (26). This test uses an outgroup, in this case, the wild-relative Tripsacum dactyloides, to compare rates of divergence between species to levels of polymorphism within species. A low level of intraspecific diversity to interspecific divergence relative to other loci suggests that selection has reduced diversity. We used the HKA test to compare starch loci with 11 neutral loci sampled previously (2), and applied the test at two levels within our germplasm. In the first sample, we used all 30 diverse lines (set A), whereas in the second sample, we used a narrower subset of 9 lines (set B) that contains no sweet, popcorn, and little tropical germplasm. Testing at two levels helps determine whether selection occurred on a broad scale across all breeding germplasm or only in the narrower germplasm base. Over the diverse maize sample, sh2 had significant HKA results (Table 2), but both maize and Z. mays ssp. parviglumis had low levels of diversity, indicating that selection may have occurred before the divergence of maize from Z. m. ssp. parviglumis (Fig. 2). bt2 and su1 had highly significant HKA tests for both germplasm sets and exhibited high levels of diversity in Z. m. ssp. parviglumis (Table 1); therefore, bt2 and su1 were likely targets of positive selection after the divergence of maize from Z. m. ssp. parviglumis. ae1 had nonsignificant HKA results for diverse germplasm (set A), but it had low diversity for all germplasm and significant HKA results for the narrow subset of germplasm (set B). Additionally, another test of selection, Tajima's D, indicated strong confirmation of selection at ae1 (D = −2.29; P < 0.01; ref. 25). Together, these significant tests suggest selection may be ongoing at ae1.
Table 2.
HKA tests of selection
| Silent sites | Diverse germplasm set A lines | Narrow germplasm set B lines | |||||
|---|---|---|---|---|---|---|---|
| Ratio* | f (P < 0.05), %† | _P_all‡ | Ratio* | f (P < 0.05), %† | _P_all‡ | ||
| Locus | |||||||
| ae1 | 2,216 | 0.32 | 0 | 0.109 | 0.07 | 18 | 0.023 |
| bt2 | 999 | 0.01 | 100 | <0.0001 | 0.01 | 91 | <0.0001 |
| sh1 | 1,630 | 0.21 | 0 | 0.844 | 0.23 | 0 | 0.815 |
| sh2 | 1,485 | 0.08 | 18 | 0.002 | 0.09 | 9 | 0.595 |
| su1 | 2,345 | 0.04 | 91 | <0.0001 | 0.06 | 18 | <0.0001 |
| wx1 | 417 | 0.13 | 0 | 0.712 | 0.16 | 0 | 0.967 |
| Starch loci§ | 8,951 | 0.13 | 35 | 0.10 | 23 | ||
| Random loci¶ | 4,567 | 0.22 | 2 | 0.19 | 2 |
Because of the close proximity of bt2 and su1 to the domestication locus tga, we were concerned about a possible hitchhiking effect. The uncloned tga locus is 3.3 cM from bt2 and 4 cM from su1 (29). To determine whether low diversity of bt2 and su1 was the product of selection on the neighboring tga locus, we characterized diversity in nr (nitrate reductase; ref. 30), the closest known and cloned gene to tga. nr diversity (πsilent = 0.008; πnonsyn. = 0.003) was roughly 3-fold higher than su1 and bt2. Thus, a hitchhiking effect because of selection on tga is not responsible for the su1 and bt2 low diversity. Selection has been shown to impact only parts of a single gene, as in the domestication gene tb1 (8). Therefore, it is not surprising that a hitchhiking effect does not extend 3 cM in a species where linkage disequilibrium decays rapidly (2, 31).
Our survey of su1 discovered a polymorphism unique to all sampled U.S. sweet corns. This polymorphism converted tryptophan to arginine at conserved residue 578. Association tests between the su1 polymorphism and the sweet phenotype were significant (P < 0.001), even while controlling for population structure (27, 28). The amino acid change was also one of two identified in the molecular and biochemical study by Dinges et al. (32). Sweet maize cultivars from central and South America did not carry the tryptophan-to-arginine mutation. Rather, the Mexican sample had a 1.3-kbp transposable element in exon 1 that disrupts normal translation of the gene. Further investigation of the South American samples is needed to determine the mutation responsible for the sweet phenotype.
Discussion
Previous studies of random maize loci have shown departures from neutrality are rare (2), whereas selection was prevalent across genes in the starch production pathway. bt2, su1, and ae1 have average levels of diversity in Z. mays ssp. parviglumis, but low levels of diversity in maize, consistent with artificial selection during maize domestication and improvement. The significant HKA results for bt2 and su1 indicate that this selection probably occurred before the dispersal of maize germplasm throughout the world, whereas at ae1 the HKA test with narrow germplasm (set B) and Tajima's test suggest selection is ongoing. It is striking that at least half of these starch loci exhibit strong evidence of selection, although random loci in maize show almost no proof of selection.
Why was there selection in this pathway? Given the position of ae1, bt2, and su1 in the starch pathway, we propose that selection, both historically and currently, has been for increased yield and different amylopectin qualities. Starch (unlike protein) is often lacking in hunter-gatherer diets in the tropics and subtropics (33), so it would be reasonable that the early cultivators of maize focused on improving their yield of starch. Native American and modern breeders have boosted yield and starch in maize several-fold over its wild relative, and genes like bt2 may have had an important role. Grain quality is also critical, as evinced in wheat for gluten levels and rice for stickiness. In maize, the ratio of amylose to amylopectin, as well as amylopectin branch chain length is important for altering starch gelatinization and pasting properties that could affect everything from porridge to tortilla texture (11, 34, 35, ‖). Indeed, increased amylopectin improves the textural properties of tortillas, making them softer (∥). Obviously, the relationship between amylose and amylopectin in the starch granule is a complex one. However, because of the substantial consequence to food processing, it is quite probable that people selected for quality traits very early in the process of maize improvement. If starch gelatinization had been an important target for selection, then we should have seen selection evidence at wx1. Instead, we saw strong selection at the starch branching (ae1) and debranching enzymes (su1), which suggests that amylopectin structure and, therefore, pasting properties were the key targets of selection. The exact nature of this selection will not be understood until a wide range of teosinte starch alleles are examined in maize genetic backgrounds and are combined with subsistence archaeological studies. The identified genes and alleles will provide the background necessary for further dissection of this pathway.
In some varieties of maize, there has been selection for low-starch, high-sugar phenotypes, which are popular because of their sweet taste. Examples of maize races with the sweet phenotype are found throughout the Americas. The su1 mutants give rise to the sweet corn phenotype because the mutants accumulate sucrose, and su1 was one of the first loci described genetically (36). Our data clarify the origin of sweet corn, for which there were two competing hypotheses. One theory argues a single origin from a Peruvian race (Chullpi; ref. 37), whereas others propose independent origins from recurring mutations (38). Our discovery of two independent su1 mutations suggests that there have been at least two independent origins of sweet corn.
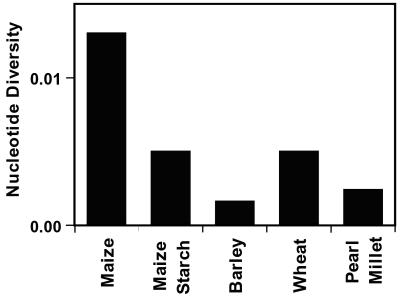
The reduction of diversity in starch loci is dramatic, and should motivate a paradigm shift for maize breeding. Maize has high levels of diversity at most loci (2), and often has 2–5 times the diversity of other grass crops (Fig. 3). This tremendous variation has allowed maize to respond to selection for industrial farming in the last century. However, limited diversity in starch and perhaps other critical pathways may preclude current breeding practices from reaching their full potential. Selection for yield may arguably be a constant throughout the millennia, while selective pressures on quality differ as cultural preferences change. Hence, useful variation, especially for grain quality, needs to be generated for these pathways. Perhaps the most efficient way to introduce potentially useful diversity into maize is to introgress or transform the abundant allelic variation present in teosinte for selected genomic regions or genes. This approach has been successful in tomato (39), and it could provide the allelic variation necessary to further increase yield and provide a much wider range of kernel qualities.
Figure 3.
Comparison of nucleotide diversity in maize and various grass crops. Maize data are the average diversity at random loci from Tenaillon et al. (2), the maize starch average is from the six genes examined here. The other grasses are diversity averages from published studies (see review in ref. 1); however, they are often based on a limited number of loci.
Acknowledgments
We thank Martha James for her wonderful suggestions on the possible targets of selection. We thank Major Goodman and John Doebley for their help in choosing and acquiring germplasm. We thank Michael Purugganan and William Tracy for useful discussions. We are grateful to the College of Agriculture and Life Sciences Genome Research Laboratory at North Carolina State University for assistance with sequencing. This research was supported by National Science Foundation Grant DBI-9872631 and by the U.S. Department of Agriculture/Agricultural Research Service.
Abbreviation
HKA
Hudson–Kreitman–Aguade
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY146786–AY146817).
‖
Yeggy, H., Zelaya, N., Suhendro, E. L., McDonough, C. M. & Rooney, L. W., American Association of Cereal Chemists 84th Annual Meeting, Oct. 31–Nov. 3, 1999, Seattle, abstr. 271.
References
- 1.Buckler E S, IV, Thornsberry J M, Kresovich S. Genet Res. 2001;77:213–218. doi: 10.1017/s0016672301005158. [DOI] [PubMed] [Google Scholar]
- 2.Tenaillon M I, Sawkins M C, Long A D, Gaut R L, Doebley J F, Gaut B S. Proc Natl Acad Sci USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyre-Walker A, Gaut R L, Hilton H, Feldman D L, Gaut B S. Proc Natl Acad Sci USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White S E, Doebley J F. Genetics. 1999;153:1455–1462. doi: 10.1093/genetics/153.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilton H, Gaut B S. Genetics. 1998;150:863–872. doi: 10.1093/genetics/150.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goloubinoff P, Pääbo S, Wilson A C. Proc Natl Acad Sci USA. 1993;90:1997–2001. doi: 10.1073/pnas.90.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doebley J, Stec A, Hubbard L. Nature (London) 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 8.Wang R L, Stec A, Hey J, Lukens L, Doebley J. Nature (London) 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- 9.Hanson M A, Gaut B S, Stec A O, Fuerstenberg S I, Goodman M M, Coe E H, Doebley J F. Genetics. 1996;143:1395–1407. doi: 10.1093/genetics/143.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers A M, Morell M K, James M G, Ball S G. Plant Physiol. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jane J-L, Chen J-F. Cereal Chem. 1992;69:60–65. [Google Scholar]
- 12.Ott M, Hester E. Cereal Chem. 1965;42:476–484. [Google Scholar]
- 13.Miller S, Derby R I, Trimbo H B. Cereal Chem. 1973;50:271–280. [Google Scholar]
- 14.Tester R F, Morrison W R. Cereal Chem. 1990;67:551–557. [Google Scholar]
- 15.Johnson L A, Baumel C P, Hardy C L, White P J. Identifying Valuable Corn Quality Traits for Starch Production. Ames: Iowa State Univ. Press; 1999. [Google Scholar]
- 16.Nelson O, Pan D. Annu Rev Plant Phys Plant Mol Biol. 1995;46:475–496. [Google Scholar]
- 17.Chourey P S, Nelson O E. Biochem Genet. 1976;14:1041–1055. doi: 10.1007/BF00485135. [DOI] [PubMed] [Google Scholar]
- 18.Liang J S, Zhang J H, Cao X Z. Physiol Plant. 2001;112:470–477. doi: 10.1034/j.1399-3054.2001.1120403.x. [DOI] [PubMed] [Google Scholar]
- 19.Hannah L C, Nelson O E. Biochem Genet. 1976;14:547–560. doi: 10.1007/BF00485834. [DOI] [PubMed] [Google Scholar]
- 20.Greene T W, Hannah L C. Physiol Plant. 1998;103:574–580. [Google Scholar]
- 21.Fisher M B, Boyer C. Plant Physiol. 1983;72:813–816. doi: 10.1104/pp.72.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James M G, Robertson D S, Myers A M. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai C. Biochem Genet. 1974;11:83–96. doi: 10.1007/BF00485766. [DOI] [PubMed] [Google Scholar]
- 24.Rozas J, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 25.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson R R, Kreitman M, Aguade M. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard J K, Stephens M, Rosenberg N A, Donnelly P. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thornsberry J M, Goodman M M, Doebley J, Kresovich S, Nielsen D, Buckler E S, IV. Nat Genet. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- 29.Dorweiler J, Stec A, Kermicle J, Doebley J. Science. 1993;262:233–235. doi: 10.1126/science.262.5131.233. [DOI] [PubMed] [Google Scholar]
- 30.Gowri G, Campbell W H. Plant Physiol. 1989;90:792–798. doi: 10.1104/pp.90.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remington D L, Thornsberry J M, Matsuoka Y, Wilson L M, Whitt S R, Doebley J, Kresovich S, Goodman M M, Buckler E S. Proc Natl Acad Sci USA. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinges J R, Colleoni C, Myers A M, James M G. Plant Physiol. 2001;125:1406–1418. doi: 10.1104/pp.125.3.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piperno D R, Pearsall D M. The Origins of Agriculture in the Lowland Neotropics. San Diego: Academic; 1998. [Google Scholar]
- 34.Jane J, Chen Y Y, Lee L F, McPherson A E, Wong K S, Radosavljevic M, Kasemsuwan T. Cereal Chem. 1999;76:629–637. [Google Scholar]
- 35.Klucinec J D, Thompson D B. Cereal Chem. 2002;79:24–35. [Google Scholar]
- 36.Correns C. Bibl Bot. 1901;53:1–161. [Google Scholar]
- 37.Mangelsdorf P C. Corn: Its Origin, Evolution and Improvement. Cambridge, MA: Harvard Univ. Press; 1974. [Google Scholar]
- 38.Tracy W F. In: Specialty Corns. Hallauer A R, editor. Boca Raton, FL: CRC; 2001. pp. 155–197. [Google Scholar]
- 39.Tanksley S D, McCouch S R. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 40.Sokal R R, Rohlf F J. Biometry. New York: Freeman; 1995. [Google Scholar]