Expression of the suppressor of cytokine signaling-5 (SOCS5) negatively regulates IL-4-dependent STAT6 activation and Th2 differentiation (original) (raw)
Abstract
The development of helper T (Th) cell subsets, which secrete distinct cytokines, plays an important role in determining the type of immune response. The IL-4-mediated Janus kinase–signal transducer and activator of transcription signaling pathway is crucial for mediating Th2 cell development. Notably, this pathway is selectively impaired in Th1 cells, although the molecular basis of this impairment remains unclear. We show here that during Th1 differentiation a reduction in the association of Janus kinase 1 with the IL-4 receptor (IL-4R) correlated with the appearance of the suppressor of cytokine signaling-5 (SOCS5). SOCS5 protein was preferentially expressed in committed Th1 cells and interacted with the cytoplasmic region of the IL-4Rα chain irrespective of receptor tyrosine phosphorylation. This unconventional interaction of SOCS5 protein with the IL-4R resulted in the inhibition of IL-4-mediated signal transducer and activator of transcription-6 activation. T cells from transgenic mice constitutively expressing SOCS5 exhibited a significant reduction of IL-4-mediated Th2 development. Therefore, the induced SOCS5 protein in Th1 differentiation environment may play an important role by regulating Th1 and Th2 balance.
Early events in the immune response initiate cytokine production, which in turn determine the subsequent development of helper T (Th) cell subsets (1). Th cells exhibit a characteristic cytokine profile that can divide them into at least two distinct subsets. Th1 cells produce IL-2, IFN-γ, and tumor necrosis factor β and promote cell-mediated effector responses to eliminate intracellular pathogens. Th2 cells produce IL-4, IL-5, IL-6, IL-10, and IL-13 and promote the augmentation of humoral responses as well as the immune response against parasites and nematodes. IL-4 and IL-5 are known to promote the immune response via mast cells and eosinophils and to accelerate allergic responses.
The best-defined factors that determine development of either Th1 or Th2 cells are cytokines present at the initial stage of activation through T cell receptor (TCR). IL-12, produced by activated macrophages and dendritic cells, is responsible for the development of Th1 cells, whereas IL-4 is a crucial cytokine for commitment to Th2 development (1, 2). The Janus protein kinase (Jak)/signal transducers and activators of transcription (STAT) signaling pathway is one of the major mechanisms by which cytokine receptors transduce intracellular signals. IL-12 and IL-4 activate STAT4 and STAT6, respectively, and their functional significance in helper T cell differentiation has been demonstrated in STAT4- and STAT6-deficient mice (2). IL-4-mediated STAT6 activation regulates the expression of transcription factors GATA-3, which result in the production of Th2 cytokines and chromatin remodeling at the IL-4 locus (3, 4).
Recently, it has been shown that STAT6 activation after IL-4 stimulation is selectively impaired in Th1 cells (5, 6), indicating that the status of IL-4 receptor (IL-4R)-mediated signaling during initial activation may have a significant impact on the differentiation path of the Th cell. The major signal transduction pathway initiated by IL-4 has been well documented by a series of elegant biological studies (7). However, the molecular mechanisms controlling negative regulation of IL-4 signaling during Th cell differentiation remain unclear. This article addresses the possibility that the impairment in IL-4R signaling is a consequence of a reduction of the association of Jak1 with the IL-4R, and preferential binding of suppressor of cytokine signaling-5 (SOCS5) to the IL-4R may be responsible for this impairment. The SOCS molecules are a family of cytokine-inducible Src homology 2 (SH2) domain-containing proteins and negative regulators for many cytokine-signaling pathways (8–10). However, very little is known about the role of the SOCS family in Th cell differentiation through IL-4R-mediating signaling (2). SOCS5 has been identified as a member of this gene family by sequence homology, but its function is still unknown (11). In this study, we provide evidence that SOCS5 is preferentially expressed in Th1 cells, specifically inhibits the IL-4R-mediated signaling pathway, and thereby facilitates Th1 differentiation.
Materials and Methods
Animals and Cell Lines.
The Flag-tagged SOCS5 was expressed under the control of the lck proximal promoter Eμ enhancer (12) and backcrossed with C57BL/6(B6) mice. Chicken β-actin promoter-controlled transgenic (Tg) mice were independently established and backcrossed with B6 mice. OVA-specific TCRαβ Tg mice (DO 11.10 Tg) were kindly provided by K. Murphy (Washington University, St. Louis). Six clones [D10.G4(D10), MS-SB, 24–2, 28–2, 28–4, and 23–1-8] and T cell hybridoma line 68–41 have been described (5). Th2–3, Th1–2-3, and Th1–4 are H-2d-restricted, OVA-specific cell lines established from DO 11.10 Tg mice.
Preparation of Th1/Th2 Cells and ELISA Assay.
The CD4+ T cells from DO11.10 Tg spleen were stimulated with OVA peptide (residues 323–339) in the presence of irradiated antigen-presenting cells. The induction of Th1 and Th2 cells was controlled by the addition of either recombinant IL-12 (10 unit/ml) plus anti-IL-4 mAb (11B11) or recombinant IL-4 (100 unit/ml) plus anti-IL-12 mAb (PharMingen), respectively. Each preparation was restimulated with anti-TCR mAb for 6 h in the presence of 4 μM monensin (Sigma). Intracellular cytokine staining with FITC-labeled anti-IFN-γ Ab and phycoerythrin-labeled anti-IL-4 Ab was performed as described (13). For ELISA assay, the preactivated T cells were stimulated with anti-TCR mAb for 24 h. The cytokine concentration in the supernatant was measured with anti-IL-2, anti-IL-4, anti-IL-5, anti-IL-10, and anti-IFN-γ mAb (PharMingen) by ELISA.
Western Blot, Immunoprecipitation, and Association Analysis.
For Western analysis, antibodies against SOCS1 and SOCS3 were raised in rabbit by using a synthetic peptide. Goat anti-mouse SOCS5 C-terminus Abs were obtained from Santa Cruz Biotechnology. For immunoprecipitation, total cellular lysates were prepared in RIPA solution (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and immunoprecipitated with anti-Jak3 and anti-Jak1 Ab (Upstate Biotechnology, Lake Placid, NY), anti-IL-4R mAb (M1) (Immunex), and anti-STAT6 mAb (Sigma). To detect tyrosine phosphorylation, the blots were probed with horseradish peroxidase-conjugated anti-phosphotyrosine antibody (RC20) (Sigma). For association analysis, cells were incubated in the presence or absence of recombinant IL-4 on ice for 2 h. After chemical cross-linking with disuccinimidyl suberate, the cells was lysed by 1% Triton X-100, and the cell extracts were immunoprecipitated with either anti-γC (common γ) (TUGm3) (K. Sugamura, Tohoku Univ., Sendai, Japan) (14) or anti-IL-4R mAb. The association was detected with anti-Jak1 (Santa Cruz Biotechnology) and anti-γC (4G3) (T. R. Malek, Univ. of Miami, Miami) (15).
GST Pull-Down Analysis and Binding to Phosphorylated Receptor.
Pull-down assay was performed by using a GST fusion protein with cytoplasmic region of mouse IL-4R spanning between 258 and 426 aa. The GST fusion protein was immobilized on glutathione Sepharose beads. After incubation with cell extracts, binding protein was visualized by silver staining and probed with Abs against Lck, Fyn, Shc, and Lyn (Santa Cruz Biotechnology) and anti-IRS-1/2 Abs (Upstate Biotechnology). GST was fused to the cytoplasmic domain of IL-12Rβ1, IL-12Rβ2, or IL-4R in pGEX4T-1 (Amersham Pharmacia). These constructs were transformed into an Escherichia coli BL21 (DE3) strain or TKB1 strain, which is a tyrosine kinase derivative of BL21 (DE3) (Stratagene), and GST fusion protein was immobilized on glutathione beads. SOCS5 was transfected into 293T cells, and the cell extracts were incubated with GST fusion proteins. The precipitates were probed with anti-SOCS5 Ab.
Northern Hybridization.
Total RNA was isolated by the TRIzol reagent (GIBCO/BRL). RNA was transferred to nylon membranes, and then the membranes were hybridized with digoxigenin (DIG)-labeled riboprobes and visualized by alkaline phosphatase-leveled anti-DIG Ab (Roche Diagnostics). Probe cDNAs for CIS1 and SOCS1–SOCS6 have been described (16).
Luciferase Assay.
We generated stable transfectants of 68–41 cells expressing either IL-2R, IL-12R, or IL-4R. The transfectants were cotransfected with either the STAT6 reporter, Tpu474 (U. Schindler, Tularik, South San Francisco, CA) (17) or the STAT4 and 5 reporter, 3X BCAS-SRE (R. Goiizuka, Tokyo University of Science) in the presence of the SOCS5 expression vector (Flag SOCS5) by electroporation. The transfected cells were stimulated with either IL-4, IL-2, or IL-12. The emitted luciferase light was measured with a luminometer (Analytical Luminescience Laboratory, San Diego).
Results
Impairment of IL-4 Signaling in Th1 Cells and Failure of Jak1 to Associate with the IL-4Rα Chain.
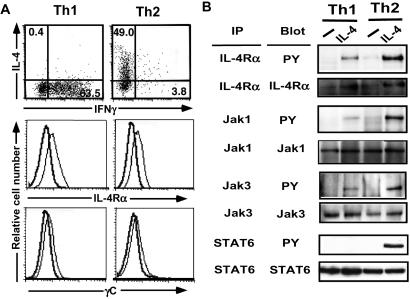
We first asked whether inactivation of IL-4 signaling occurred at an early stage of Th1 differentiation. Tyrosine phosphorylation of the IL-4Rα chain, Jak3, and STAT6 was studied in Th1 and Th2 populations prepared from DO11.10 Tg mice (Fig. 1A). Th2 cells exhibited significant tyrosine phosphorylation after IL-4 stimulation, whereas Th1 cells showed little or no phosphorylation (Fig. 1B). This result indicated that the impairment of IL-4-induced STAT6 activation occurred during Th1 differentiation. However, Th1 cells revealed marked Jak1, Jak3, and STAT5 phosphorylation after IL-2 stimulation (data not shown), demonstrating that the impairment in Th1 cells was specific for IL-4-dependent signaling. Furthermore, the signaling defect observed in Th1 cells cannot be explained by a loss of expression of IL-4R, Jak1, Jak3, and STAT6, because these expressions in the Th1 cells were comparable to that seen in the Th2 cells (Fig. 1).
Figure 1.
Impairment of IL-4-dependent signaling in Th1 cells. (A) Th1 and Th2 cells were induced from DO11.10 Tg mice, and cytokine profile was assessed by intracellular staining (Top). Surface expression of IL-4Rα (Middle) and γC (Bottom). Solid lines indicate negative control. Thin lines indicate expression of IL-4Rα or γC (Middle and Bottom). (B) Cell extracts were prepared from the Th1 and Th2 cells stimulated with IL-4. After immunoprecipitation with IL-4Rα, Jak1, Jak3, and STAT6, tyrosine phosphorylation was assessed by horseradish peroxidase-labeled RC20.
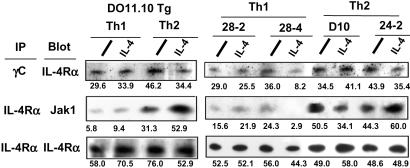
Next, we studied impairment within the conformational modification of IL-4R. The association of IL-4Rα chain with either γC chain or Jak1 was tested in Th1 and Th2 cells after incubation with or without IL-4. The dimerization of the IL-4R and the γC is an important process initiated by ligand binding, although the γC chain coprecipitated with the α chains in both Th1 and Th2 cells even in the absence of ligand binding (Fig. 2). Modest decease found in the association between the IL-4R and the γC was observed in a panel of Th1 clones but not DO11.10-derived Th1 cells. Jak1 is constitutively associated with the box1 region on the α chain, allowing phosphorylation of the α chain through activation of Jak1 and Jak3 (18). Indeed, Jak1 was coprecipitated with the α chain in Th2 cells. However, Th1 cells revealed weaker association, and this profile was consistent across the clones and DO11.10-derived cells (Fig. 2). Therefore, we focused on the impaired interaction between Jak1 and the IL-4R in Th1 cells.
Figure 2.
Impairment of the interaction between Jak1 and the IL-4R in Th1 cells. The DO11.10 Th1 and Th2 cells and Th1 and Th2 clones were incubated in the presence or absence of IL-4. The cells were extracted and immunoprecipitated with TUGm3 or M1, and association of IL-4Rα or Jak1 was analyzed by Western blotting. Data are representative of two experiments. The quantitative number by densitometry analysis is shown in each column.
SOCS5 Bound to the Box1 Region of IL-4R in Th1 Cells.
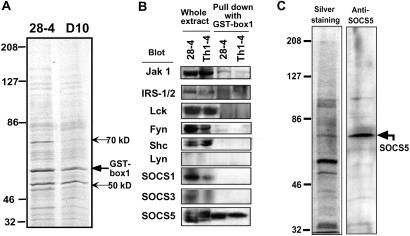
The above results raise the question regarding how the interaction between Jak1 and the IL-4R was negatively regulated in Th1 cells. We speculated this process might involve molecules that interfere with the interaction between Jak1 and the IL-4R. To determine whether other proteins could interact with the box1 region on IL-4R specifically associated with Jak1 a GST fusion protein with the cytoplasmic region of IL-4R-containing box1 was generated for pull-down assays (GST-box1). The GST-box1 protein was immobilized on Sepharose beads and mixed with cell extracts prepared from Th1 (28–4) and Th2 (D10) clones. Two binding proteins at molecular sizes around 50 and 70 kDa were preferentially precipitated from Th1 cells (Fig. 3A).
Figure 3.
Identification of IL-4R binding protein in Th1 cells. (A) Cell extracts from Th1 (28–4) and Th2 (D10) clones were incubated with the GST-box1 fusion protein coupled Sepharose beads, and the binding protein was visualized with silver staining. (B) Cell extract was prepared from two Th1 lines, 28–4 and Th1–4. The two left lanes represent whole-cell extract, and the two right lanes represent GST-box1 binding protein. Identification was performed by immune blotting of the precipitates with variety of the antibodies against signaling molecules indicated. (C) The GST-box1 binding protein in the cell extracts from 24–8 cells was silver-stained (Left) and immune-blotted with anti-SOCS5 antibody (Right) in side-by-side experiments.
To identify the binding proteins, the precipitates prepared from Th1 clones 28–4 and Th1–4 blotted with Abs against Jak1, IRS1/2, Lck, Fyn, Shc, and Lyn. We also asked whether SOCS family proteins were candidate negative regulators of IL-4R signaling, and coprecipitates were also immunoblotted with antibodies to SOCS1, SOCS3, SOCS4, and SOCS5. The blotting of whole-cell extracts showed no expression of Lyn and SOCS4, whereas the other eight molecules examined were clearly expressed (Fig. 3B Left). Among these, only Jak1 and SOCS5 were precipitated with the GST-box 1 (Fig. 3B Right). Silver staining and Western analysis of the precipitated extract showed that the molecular mass of the box1 binding protein to be about 70 kDa corresponded to the molecular mass of endogenous SOCS5 (Fig. 3C).
Preferential Expression of SOCS5 in Th1 Cells.
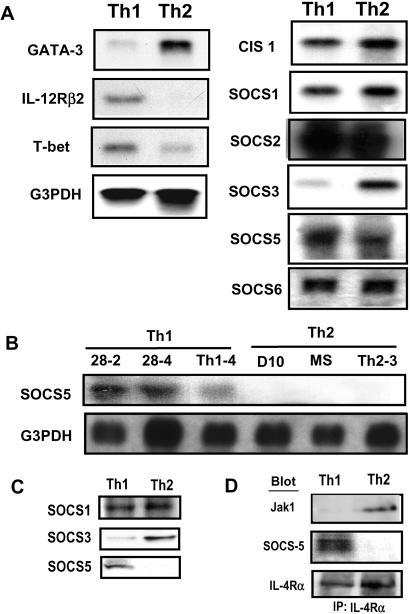
Because the 70-kDa protein was apparent in the pull-downed extract from Th1 cells, it is conceivable that SOCS5 is preferentially expressed in Th1 cells. To test this possibility, mRNA expression of CIS-1, SOCS1, SOCS2, SOCS3, SOCS5, and SOCS6 was examined within the DO11.10 Th1 and Th2 cells. Differential expression of IL-12Rβ2, T-bet, and GATA-3 confirmed the differentiation profiles of Th1 and Th2 cells. Both Th1 and Th2 cells showed comparable mRNA expression of CIS-1, SOCS1, SOCS2, and SOCS6 (Fig. 4A). The SOCS5 mRNA expression in Th1 cells was higher than that in Th2 cells although the difference was modest in DO11.10-derived cells (Fig. 4A). The fully committed cell lines exhibited the selective expression of SOCS5 mRNA. All three Th1 clones, but not Th2 clones, examined here showed SOCS5 expression, (Fig. 4B). Interestingly, SOCS3 exhibited the reversed expression pattern being exclusively expressed in Th2 but not Th1 cells (Fig. 4A).
Figure 4.
Selective expression of SOCS5 in Th1 cells. (A) Total RNA was isolated from DO11.10 Th1 and Th2 cells. The mRNA expressions of GATA-3, IL-12Rβ2, T-bet, CIS1, SOCS1–6, and G3PDH were detected by digoxigenin-labeled RNA probe. (B) Total RNA was isolated from a panel of Th1 and Th2 clones, and the expressions of SOCS5 and G3PDH were detected as described in A. (C) Protein expression of SOCS5 in DO11.10 Th1 and Th2 cells was detected by anti-SOCS5 Ab. (D) The DO11.10 Th1 and Th2 cell extracts were immunoprecipitated with anti-IL-4Rα mAb and blotted with anti-Jak1, SOCS5, or mIL-4Rα Abs.
We next examined the protein expression of SOCS1, SOCS3, and SOCS5 in Th1 and Th2 cells. The SOCS5 protein was detected in Th1 but not Th2 cells, whereas the SOCS3 protein was found only in Th2 cells (Fig. 4C). On the other hand, SOCS1 protein was equally expressed in both Th1 and Th2 cells. Together, our protein and mRNA data provide evidence that SOCS3 and SOCS5 are differentially expressed in Th1 and Th2 cells, and expression of SOCS5 protein was preferentially induced during Th1 differentiation.
These results raised the question of whether the endogenous IL-4Rα chain preferentially associated with SOCS5 or Jak1 in Th1 cells. Thus, we carried out immunoprecipitation by anti-IL-4R mAb in DO11.10 Th1 and Th2 cells (Fig. 4D). SOCS5 protein was preferentially coprecipitated with the endogenous α chain in Th1 cells. In this circumstance, Th1 cells again showed a lack of Jak1 association with the IL-4R (Fig. 4D). Thus, the association of Jak1 with the IL-4Rα chain was impaired when SOCS5 protein bound to the α chain.
Unconventional Interaction of SOCS5 with the IL-4Rα Chain.
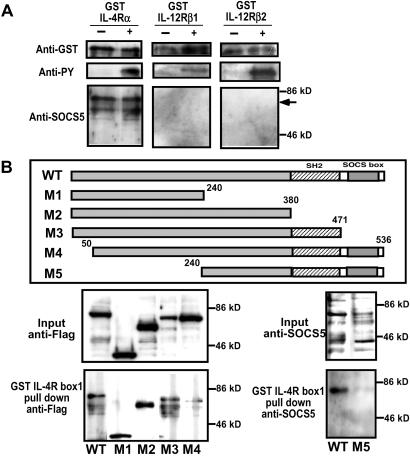
Interestingly, tyrosine phosphorylation of the IL-4R induced by ligand binding did not appear necessary for the SOCS5 interaction. To confirm this possibility, the binding of SOCS5 to bacterially expressed cytoplasmic IL-4R was examined with or without tyrosine phosphorylation of IL-4R. SOCS5 was clearly coprecipitated with the cytoplasmic IL-4R irrespective of tyrosine phosphorylation (Fig. 5A), indicating that the binding of SOCS5 to the IL-4R did not require a tyrosine-based interaction. We also examined the binding of SOCS5 to the cytoplasmic region of other cytokine receptors, IL-12R β1 and β2 chain, with or without tyrosine phosphorylation. SOCS5 precipitated with control IL-4R, but not with the IL-12R β1 and β2 chain (Fig. 5A), suggesting that SOCS5 specifically binds to the cytoplasmic region of the IL-4Rα chain. Because the interaction between SOCS5 and the IL-4Rα chain was not tyrosine-based, we speculated that the binding site to the IL-4R box1 region was not located at the SH2 domain. In fact, a series of SOCS5 deletion mutants exhibited that the deletion of SH2, M1, and M2 did not affect this interaction. However, the deletion of N-terminal side (M4 and M5) impaired the binding (Fig. 5B), indicating that the first 50 aa of SOCS5 protein are critical for the interaction with the box1 region on IL-4R.
Figure 5.
Interaction of SOCS5 protein with the IL-4R. (A) The GST fusion protein of cytoplasmic IL-4Rα, IL-12Rβ1, or IL-12Rβ2 chain was bacterially expressed with or without tyrosine phosphorylation. The GST fusion proteins were tyrosine-phosphorylated in TKB1 E. coli strain. After incubation with the cell extracts of HEK293 cells that SOCS5 was transiently expressed, the GST fusion proteins were precipitated with glutathione beads and blotted with anti-GST, anti-phosphotyrosine (anti-PY), and anti-SOCS5. (B) A series of SOCS5 deletion mutants (M1–M5) were incubated with GST-IL-4R box1. The mutants M1–M4 were tagged with Flag protein and probed with anti-Flag mAb. The mutant M5 was probed by anti-SOCS5 Ab.
Constitutive Expression of SOCS5 Inhibited IL-4-Dependent STAT6 Activation and Th2 Differentiation.
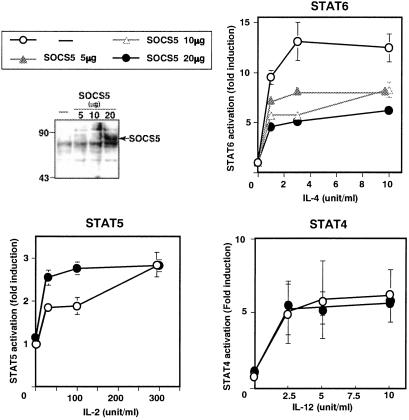
The SOCS5 protein expressed in Th1 cells is capable of associating with IL-4R, which raised the possibility that the binding inhibits IL-4-mediated STAT6 activation. To address this possibility, a STAT6 reporter construct was transiently expressed in a T cell hybridoma, 68–41 cells with or without SOCS5 cDNA expression vector. The transfected SOCS5 protein was clearly expressed at detectable levels (Fig. 6). The overexpression of SOCS5 reduced IL-4-mediated STAT6 activation to half of the maximum response (Fig. 6). In contrast, SOCS5 expression revealed no inhibition on the IL-2-dependent STAT5 activation as well as the IL-12-mediated STAT4 activation.
Figure 6.
SOCS5 inhibited IL-4-mediated STAT6 activation. STAT4, STAT5, and STAT6 reporter were transiently transfected with various concentrations of Flag-tagged SOCS5 into 68–41 cells expressing either IL-4R, IL-2R, or IL-12R. SOCS5 protein expression was probed by anti-Flag mAb. After either IL-4, IL-2, or IL-12 stimulation for 6 h, luciferase activity was measured. Data represent fold induction against the luciferase light unit of no stimulation.
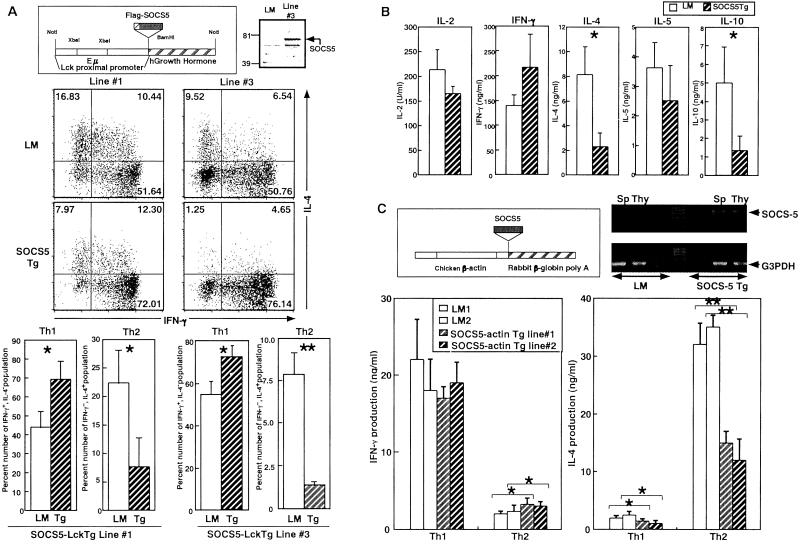
Activation of STAT6 pathway has a crucial process for Th2 differentiation. Thus, we next examined whether the inhibitory function by SOCS5 for the IL-4 signaling is sufficient for a negative regulation of Th2 development. Because SOCS5 showed no expression in naïve T cells, we attempt to establish Tg mice constitutively expressing SOCS5 in T cells by two distinct promoters: one is the Lck proximal promoter and Eμ enhancer(SOCS5-LckTg), and the other is chicken β-actin promoter (SOCS5-actin Tg). In SOCS5-Lck Tg, line 3 showed detectable SOCS5 protein expression in thymus and spleen (Fig. 7A), but lines 1 and 2 expressed very faint SOCS5 protein (data not shown). Th1 and Th2 cells were induced in unskewed conditions, and the proportions of IL-4- and IFN-γ-producing cells were assessed after 7 days. Lines 1 and 3 were in a different generation of backcross with B6, thus we compared the proportion rates between littermate (LM) and Tg mice in the same line. T cells from high-expression line 3 revealed five times less Th2 development compared with that of LM mice (Fig. 7A). The inhibition of Th2 development was consistent even in the low-expression lines 1 and 2 (Fig. 7A). Furthermore, the forced expression of SOCS5 also reduced production levels of Th2 cytokines, IL-4, IL-5, and IL-10 (Fig. 7B).
Figure 7.
The forced expression of SOCS5 inhibited Th2 development. (A) SOCS5 protein expression in thymocytes from SOCS5-Lck Tg line 3 was assessed by anti-SOCS5 Ab. T cells from SOCS5-Lck Tg were stimulated with anti-TCR plus anti-CD28 mAb. Lines 1 and 3 were in second and fourth generation with backcross into B6, respectively. After 7 days, IFN-γ- and IL-4-producing cells were analyzed by intracellular cytokine staining. The number represents the percentage of IFN-γ- and IL-4-producing cells. The mean ± SD shown was obtained from three independent mice in LM. *, P < 0.05 and **, P < 0.01 versus control LM. (B) T cells from control and line 3 were stimulated as described in A and restimulated with anti-TCR mAb for 24 h. Cytokine productions were analyzed by ELISA. The mean ± SD was obtained from three independent experiments. *, P < 0.05 versus LM. (C) SOCS5 mRNA expression in thymocytes and spleen from SOCS5-actin Tg lines 1 and 2 was assessed by RT-PCR. T cells were stimulated with anti-CD3 (100 μg/ml) in Th1 (IL-12 + anti-IL-4 mAb) and Th2 (IL-4 + anti-IL-12mAb) skewing condition. The mean ± SD shown was obtained from three independent mice in same LM. *, P < 0.05 and **, P < 0.01 versus control LM.
T cells from SOCS5-actin Tg 1 and 2 lines revealed significant SOCS5 mRNA expression (Fig. 7C) but not a detectable level of SOCS5 protein. The CD4+ T cells were preactivated in the presence of either IL-12 or IL-4 that skewed Th1 or Th2 differentiation, respectively. SOCS5 clearly inhibited IL-4-induced Th2 development, reducing it to half that of control LM (Fig. 7C). On the other hand, in Th1 skewing conditions, the expressed SOCS5 did not alter Th1 development. These results clearly demonstrated that the expression of SOCS5 at an early developmental stage can result in the impairment of Th2 differentiation.
Discussion
Signaling mediated through IL-4R is known to be essential for the development of Th2 cells. In this article, we investigated the functional impairment of IL-4-dependent STAT6 activation in Th1 cells (5, 6). This impairment was caused by a functional alteration rather than a down-regulation in the expression of the molecules involved in the IL-4-dependent Jak-STAT cascade. Our results indicate that the defect may occur at a step relatively proximal to the IL-4R recognition process. This defect is likely responsible for an alteration in the interaction between Jak1 and the α chain, which is relatively weak in Th1 cells (Fig. 2). We isolated SOCS5 as a molecule that can interact with the cytoplasmic domain of the IL-4Rα chain. SOCS5 has been isolated based on structural similarities with other SOCS proteins such as the central SH2 domain and a SOCS box at the C terminus (11), but its biological target has not been previously identified. Our data suggest that SOCS5 preferentially interacts with the IL-4R and has the functional effect of impairing IL-4-induced STAT6 activation (Figs. 5 and 6). These results lead us to propose that the IL-4Rα chain is a specific target of SOCS5.
It is noteworthy that SOCS5 can interact with the IL-4R without tyrosine phosphorylation. This characteristic would be unique to SOCS5, because other SOCS members interact with phosphorylated Jaks or receptors through the SH2 region (19–21). We speculate that SOCS5 binding results in the reduction of the Jak1 association based on the evidence that the impaired IL-4 signaling coincides with the appearance of SOCS5 (Figs. 2 and 4). SOCS5 specifically binds to the region adjacent to Box1 of the IL-4Rα chain, leading to the possibility that the SOCS5 binding may inhibit the interaction between Jak1 and the IL-4R. Of course, it is still a possibility that other molecules may alter the conformation of the IL-4R or may inhibit IL-4R-mediated signaling.
The expression profiles and the function of the SOCS molecules during Th cell differentiation are not fully understood. Our results clearly showed that CIS-1, SOCS1, SOCS2, and SOCS6 were also inducible during Th cell differentiation, but there was no difference in the expression levels between Th1 and Th2 cells, except SOCS3 and SOCS5 (Fig. 4). A cytokine environment that favors Th1 commitment may induce SOCS5 expression, leading to a concurrent suppression of IL-4-dependent STAT6 activation. Recent reports have indicated that SOCS1 preferentially expressed in Th1 cells derived from the HEL-specific TCR Tg mouse (22). However, SOCS1 act as a potent inhibitor for the IL-4-dependent STAT6 activation (23–25). Therefore, Th1 selective expression of SOCS1 may elucidate Th1-specific impairment in the IL-4 signaling. Also the IL-6-mediated inhibition of Th1 differentiation accounts for SOCS1 expression induced by IL-6, indicating that SOCS1 can be expressed in Th2 cells (26). Our data showed that the SOCS1 expression levels are almost identical in Th1 and Th2 cells (Fig. 4A). SOCS1 is known to inhibit a wide range of cytokine-mediated signaling (8, 10, 27). Thus, the role of SOCS1 as specific negative regulator in Th2 differentiation still remains unclear. Furthermore, T cells from Tg mice expressing either CIS-1, SOCS2, or SOCS3 revealed no effect on STAT6 activation and Th2 differentiation (28) (data not shown). Therefore, CIS-1, SOCS2, and SOCS3 may not play a role as a negative regulator of Th2 development via the inhibition of STAT6 activation.
The data from Tg expression of SOCS5 clearly indicate that SOCS5 plays a role as a specific negative regulator for Th2 differentiation. We have generated eight Tg lines, and seven lines showed lower expression of SOCS5 protein compared with that in committed Th1 cells. Even under these circumstances, the constitutive expression of SOCS5 in naïve T cells consistently resulted in the reduction of Th2 development (Fig. 7). This inhibition was restored by the addition of excess amount of IL-4, 100 times excess of that secreted from naïve T cells (data not shown), thus we speculate that the inhibition of Th2 development is a result of the blockade of IL-4 signaling by SOCS5. However, inhibitory activity of SOCS5 for the IL-4-mediated STAT6 activation was marginal, thus it remains a possibility that other SOCS family members, such as SOCS1, may participate in the inhibition of the IL-4 signaling in Th1 cells.
Acknowledgments
This work was supported by a Grant in Aid for Scientific Research on Priority Areas of the Ministry of Education, Culture, Sports, Science, and Technology (Japan). Y.S. and A.M. are research fellows of the Japan Society for the Promotion of Science.
Abbreviations
γC
common γ
Jak
Janus kinase
STAT
signal transducer and activator of transcription
SOCS
suppressor of cytokine signaling
TCR
T cell receptor
Tg
transgenic
Th
helper T
IL-4R
IL-4 receptor
SH2
Src homology 2
LM
littermate
References
- 1.O'Garra A. Immunity. 1998;8:275–283. doi: 10.1016/s1074-7613(00)80533-6. [DOI] [PubMed] [Google Scholar]
- 2.Murphy K M, Ouyang W, Farrar J D, Yang J, Ranganath S H, Asnagli H, Afkarian M, Murphy T L. Annu Rev Immunol. 2000;18:451–494. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S M, Radbruch A, Murphy K M. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 4.Lee H-J, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O'Garra A, Arai N. J Exp Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubo M, Ransom J, Webb D, Hashimoto Y, Tada T, Nakayama T. EMBO J. 1997;16:4007–4020. doi: 10.1093/emboj/16.13.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Paul W E. J Exp Med. 1998;187:1305–1313. doi: 10.1084/jem.187.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelms K, Keegan A D, Zamorano J, Ryan J, Paul W E. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 8.Hilton D J. Cell Mol Life Sci. 1999;55:1568–1577. doi: 10.1007/s000180050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naka T, Fujimoto M, Kishimoto T. Trends Biochem Sci. 1999;24:394–398. doi: 10.1016/s0968-0004(99)01454-1. [DOI] [PubMed] [Google Scholar]
- 10.Yasukawa H, Sasaki A, Yoshimura A. Annu Rev Immunol. 2000;18:143–164. doi: 10.1146/annurev.immunol.18.1.143. [DOI] [PubMed] [Google Scholar]
- 11.Hilton D J, Richardson R T, Alexander W S, Viney E M, Willson T A, Sprigg N S, Starr R, Nicholson S E, Metcaf D, Nicola N A. Proc Natl Acad Sci USA. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iritani B M, Forbush K A, Farrar M A, Perlmutter R M. EMBO J. 1997;16:7019–7031. doi: 10.1093/emboj/16.23.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubo M, Yamashita M, Abe R, Tada T, Okumura K, Ransom J T, Nakayama T. J Immunol. 1999;163:2432–2442. [PubMed] [Google Scholar]
- 14.Kondo M, Takeshita T, Ishi N, Nakamura M, Watanabe S, Arai K, Sugamura K. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 15.He Y-W, Adkins B, Furse R K, Malek T R. J Immunol. 1995;154:1596–1605. [PubMed] [Google Scholar]
- 16.Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, et al. Biochem Biophys Res Commun. 1997;239:439–446. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- 17.Mikita T, Campbell D, Wu P, Williamson K, Schindler U. Mol Cell Biol. 1996;16:5811–5820. doi: 10.1128/mcb.16.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard W J, O'Shea J O. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 19.Cohney S, Asanden D, Cacalano N A, Yoshimura A, Mui A M, Migone T S, Johnston J A. Mol Cell Biol. 1999;19:4980–4988. doi: 10.1128/mcb.19.7.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichlson S E, Willson T A, Farley A, Starr R, Zhang J G, Baca M, Alexander W S, Metcaf D, Hilton D J, Nicola N A. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle J N, Yoshimura A. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egwuagu C E, Yu C R, Zhang M, Mahdi R M, Kim S J, Gery I. J Immunol. 2002;168:3181–3187. doi: 10.4049/jimmunol.168.7.3181. [DOI] [PubMed] [Google Scholar]
- 23.Dickensheets H L, Venkataraman C, Schindler U, Donnelly R P. Proc Natl Acad Sci USA. 1999;96:10800–10805. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losman J A, Chen X P, Hilton D J, Rothman P. J Immunol. 1999;162:3770–3774. [PMC free article] [PubMed] [Google Scholar]
- 25.Naka T, Matsumoto T, Narazaki M, Fujimoto M, Morita Y, Ohsawa Y, Saito H, Nagasawa T, Uchiyama Y, Kishimoto T. Proc Natl Acad Sci USA. 1998;95:15577–15582. doi: 10.1073/pnas.95.26.15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diehl S, Anguita J, Hoffmeyer A, Zapton T, Ihle J N, Fikrig E, Rincon M. Immunity. 2000;13:805–815. doi: 10.1016/s1074-7613(00)00078-9. [DOI] [PubMed] [Google Scholar]
- 27.Naka T, Tsutsui H, Fujimoto M, Kawazoe Y, Kohzaki H, Morita Y, Nakagawa R, Narazaki M, Adachi K, Yoshimoto T, et al. Immunity. 2001;14:535–545. doi: 10.1016/s1074-7613(01)00132-7. [DOI] [PubMed] [Google Scholar]
- 28.Matsumoto A, Seki Y, Kubo M, Ohtsuka S, Suzuki A, Hayashi I, Tsuji K, Nakahara T, Okabe M, Yamada S, Yoshimura A. Mol Cell Biol. 1999;19:6396–6407. doi: 10.1128/mcb.19.9.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]