Leptotrichia amnionii sp. nov., a Novel Bacterium Isolated from the Amniotic Fluid of a Woman after Intrauterine Fetal Demise (original) (raw)
Abstract
A novel bacterium was isolated and characterized from the amniotic fluid of a woman who experienced intrauterine fetal demise in the second trimester of pregnancy. The bacterium was a slow-growing, gram-negative anaerobic coccobacillus belonging to the genus Leptotrichia. Unlike Leptotrichia sanguinegens, the isolate did not grow in chopped-meat glucose broth or on sheep blood agar upon subculturing. The isolate was characterized by sequencing and analyzing its 16S rRNA gene. The 1,493-bp 16S ribosomal DNA sequence had only 96% homology with L. sanguinegens. Several phylogenetic analyses indicated that L. amnionii is a distinct species and most closely related to L. sanguiegens.
Molecular-based diagnostic and identification methods for fastidious or uncultivable bacteria have resulted in the recognition of many new pathogenic microorganisms (3, 16). One of the most successful methods is PCR amplification and sequencing of the bacterial 16S rRNA gene. This method has been successfully applied to environmental as well as clinical samples (6, 15). The large rRNA sequence databases at GenBank and at Ribosomal Database Project II allow for a quick comparison of 16S ribosomal DNA (rDNA) sequences and accurate identification of bacteria that are difficult to identify on the basis of phenotypic properties alone (10). The use of this method has greatly expanded the list of indigenous microbial flora of humans and has helped in recognizing the numerous opportunistic pathogens that cause infections related to severe physiological stress and immunosuppression due to chemotherapy.
Leptotrichia species are slow-growing, gram-negative anaerobic flora of the oral cavity and genital tract (5). Colonization by Leptotrichia species has been reported in over 40% of children less than a year old (19). Leptotrichia buccalis, which is considered indigenous oral flora, has been associated with endocarditis in patients with Down's syndrome (2) and bacteremia in neutropenic children and adults (14, 22). They seem to colonize permucosal implants of edentulous patients (12) and, not surprisingly, are often considered contaminants if isolated from clinical specimens.
Leptotrichia sanguinegens has recently been proposed as an agent of postpartum and neonatal bacteremia (4). It has not been identified from a healthy individual. We describe an isolate that is related to the species L. sanguinegens, but is different in its genotypic properties and nutritional requirements (4). For this isolate, we proposed the name “L. amnionii sp. nov.” (from “amnion,” the extraembryonic membrane enveloping the embryo in utero and containing the amniotic fluid), to signify its source of isolation.
CASE REPORT
A 27-year-old previously healthy, multiparous female in the second trimester of pregnancy presented to the emergency room with severe headache, neck and back pain, and a temperature of 102°F. The abdominal examination demonstrated no guarding or rebound. A purulent vaginal discharge was noted, but the physical examination was otherwise normal. Fetal heart tones were present. The patient was hospitalized. Initial laboratory values demonstrated a leukocyte count of 7,800 with the differential showing 1 metamyelocyte, 10 band forms, 85 segmented neutrophils, and 3 lymphocytes. The hemoglobin level was 11.5 g/dl, and the C-reactive protein level was 11.4 mg/dl. A urinalysis was unremarkable, showing no evidence of infection. A wet preparation of the vaginal discharge demonstrated no abnormal organisms or evidence of significant vaginal infection. A PCR test for Chlamydia trachomatis was negative. Cerebral spinal fluid evaluation was normal. The symptoms gradually resolved, and the patient was discharged. Six days later, the patient was seen in the outpatient clinic. No fetal heart tones could be heard, and an ultrasound confirmed an intrauterine fetal demise. The patient was admitted for uterine evacuation by labor induction. Prior to induction, an amniocentesis was performed. The amniotic fluid was turbid and brown in color and had a distinct foul smell. A gram stain of the amniotic fluid demonstrated gram-negative coccobacilli. A slow-growing, gram-negative anaerobic coccobacillus was recovered. Scant growth of Bacteroides fragilis and Propionibacterium acnes was observed in cultures of the placenta. The mother was given amoxicillin-clavulanic acid and had an uneventful recovery.
MATERIALS AND METHODS
Microbiology.
The amniotic fluid and the placenta tissue specimens were cultured on blood and chocolate agar under both aerobic and anaerobic conditions at 37°C. The template DNA for 16S rDNA PCR was prepared from a few colonies that were isolated on the prereduced blood agar incubated anaerobically. The DNA was extracted with a Qiagen DNA extraction kit (Qiagen, Inc., Valencia, Calif.). Broad-range prokaryotic PCR primers (23) and nested sequencing primers (17, 23) were used to amplify and sequence the 16S gene rRNA. The methodology has been described previously (17).
Phylogenetic analysis.
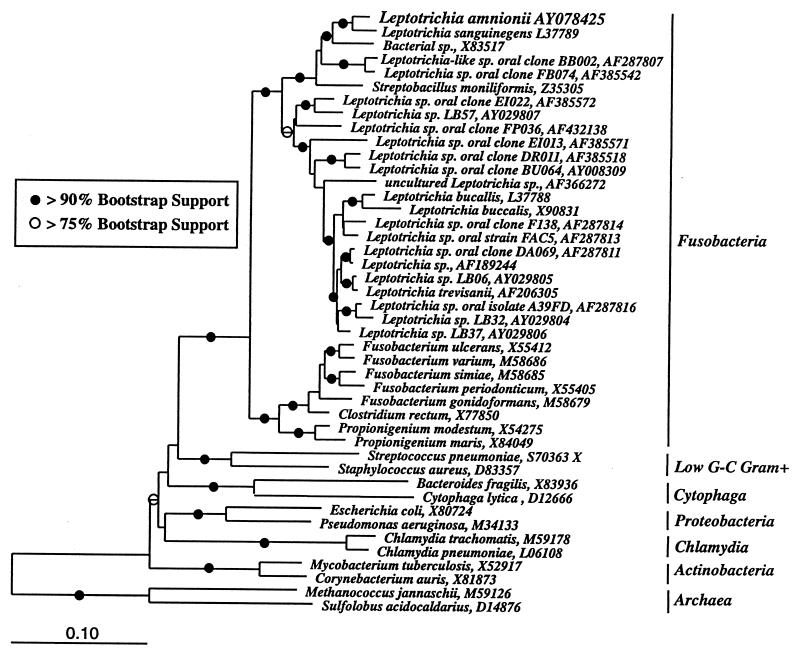
The rDNA sequence of the L. amnionii sp. nov. was aligned with a database of archaeal, bacterial, and eucaryal SSU rRNA sequences (ca. 10,000 sequences in total) by using the ARB software package (18). Both BLAST analysis and the parsimony insertion tool of ARB tentatively placed the L. amnionii sequence within the bacterial division of Fusobacteria. Consequently, a subset of the ARB alignment, which included the Leptotrichia species of the division Fusobacteria (including the species of the genus Leptotrichia), as well as members of other, outlying bacterial divisions, was selected for phylogenetic analysis. Both full-length data sets and sequence alignments minimized by the use of the Lane mask (8) were analyzed. The sequences of Methanococcus jannaschii and Sulfolobus acidocaldarius were selected as out-groups for phylogenetic analysis. The dendrogram presented in Fig. 2 was constructed by evolutionary distance analysis (neighbor joining with Olsen correction) with the ARB package (18). The robustness of this tree was assessed by bootstrap resampling (>100 replicates) of evolutionary distance trees by using weighted least-squares mean analysis with Kimura two-parameter or maximum-likelihood correction of evolutionary distances (PAUP* version 4.0b2) (20). Parsimony and maximum-likelihood analyses (ARB or PAUP*) provided results that were substantially similar to those of the evolutionary distance algorithm.
FIG. 2.
Evolutionary distance dendrogram of selected leptotrichial and fusobacterial 16S rRNA sequences, including that of the _Leptotrichia_-like sp. isolate. Two archaeal species, Methanococcus jannaschii (M59126) and Sulfolobus acidocaldarius (D14876), were chosen as out-groups for phylogenetic analysis. Sequences are identified by species name and GenBank accession number. Branch points supported by >90% bootstrap values are indicated by solid circles. Open circles represent branch points with bootstrap values in the range 75 to 89%. Branch points without circles were not resolved (bootstrap values in the range <75%) as specific groups by this analysis. The bar at the bottom indicates the number of nucleotide changes per site.
Nucleotide sequence accession number.
The 16S rRNA sequence of the L. amnionii sp. nov. was deposited in GenBank and given accession no. AY078425.
RESULTS AND DISCUSSION
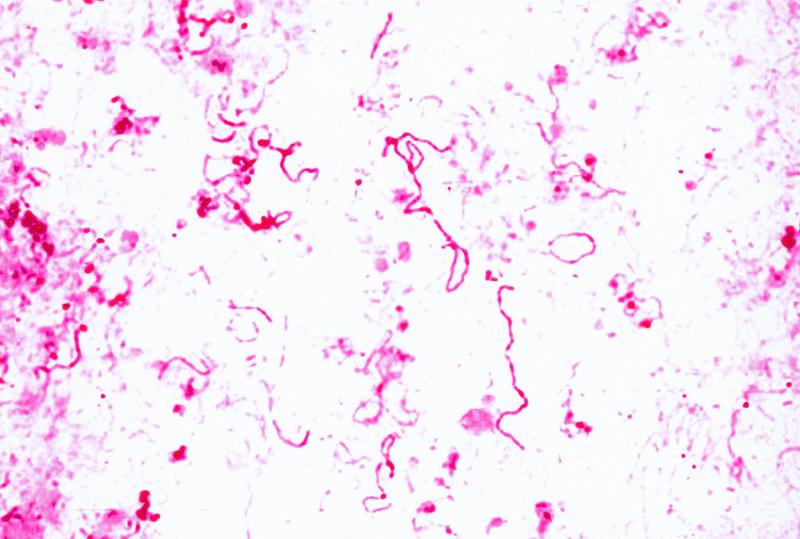
Numerous gram-negative coccobacilli were observed in the amniotic fluid along with numerous neutrophils. Anaerobic culture of the amniotic fluid on blood and chocolate agar resulted in very small gray colonies, <1 mm in diameter, following 72 h of incubation. Gram stain of the colonies revealed gram-negative coccobacilli, including some filamentous forms (Fig. 1). There was no growth on blood agar incubated under aerobic conditions, nor was there anaerobic growth on kanamycin and vancomycin or Mueller-Hinton agars upon subculturing. Viral cultures were negative. Since the bacterium resembled L. sanguinegens, it was inoculated into chopped meat glucose (CMG) broth and incubated under aerobic conditions (4). This medium did not sustain growth, as evidenced by the lack of turbidity of the medium. The organism was extremely fastidious and did not survive beyond the third subculture. There was insufficient growth to perform biochemical or fatty acid analysis.
FIG. 1.
Gram stain of amniotic fluid colonies demonstrating gram-negative pleomorphic bacilli.
The isolate was identified and characterized by PCR amplification of the 16S rRNA gene by using broad-range eubacterial primers FD1 and RD1 (17, 23). The PCR product was directly sequenced as described previously (17). A 1,493-nucleotide consensus sequence was created and edited with DNAsis software (Hitachi Corporation) and compared with the sequences deposited in the GenBank database. The submitted sequence had only 96% homology to L. sanguinegens (GenBank accession no. L37789).
Phylogenetic analysis.
Based on its unique source of isolation, inability to grow on known special media, such as CMG broth, and unique 16S rDNA sequence, it was evident that this bacterium was related to, but different from, L. sanguinegens (4). The phylogenetic relationship of L. amnionii with other species of the bacterial division Fusobacteria, including species of the genus Leptotrichia, was inferred by evolutionary distance, parsimony, and maximum-likelihood analyses. Figure 2 shows a representative evolutionary distance dendrogram. Parsimony and maximum-likelihood analyses gave qualitatively similar results. Bootstrap resampling of data provided strong support for a specific association of L. amnionii with other members of the genus Leptotrichia. L. sanguinegens was identified as the closest neighbor of L. amnionii (bootstrap values of 99 and 100% for distance and parsimony analyses, respectively).
Clinical significance.
Three species of Leptotrichia, L. buccalis, L. trevisanii, and L. sanguinegens (also called Sneathia sanguinegens) (1), have been associated with human infections (Table 1). Fifty-nine percent of the patients were immunosuppressed due to malignancy. Four cases of L. sanguinegens bacteremia were associated with pregnancy, and two neonates were infected (4).
TABLE 1.
Comparison of clinical features of Leptotrichia species isolated until 2001
| Organisms | Source | Patient age (yr) | Sexa | Clinical condition | Reference |
|---|---|---|---|---|---|
| L. buccalis | Blood | 15 | M | Lymphocytic lymphoma | 22 |
| L. buccalis | Blood | 7 | F | Acute leukemia | 22 |
| L. buccalis | Blood | 19 | M | Acute leukemia | 22 |
| L. buccalis | Blood | 73 | F | Ovarian carcinoma | 22 |
| L. buccalis | Blood | 46 | M | Lymphocytic leukemia, cavitary pneumonia | 11 |
| L. buccalis | Blood | 67 | F | Lymphatic leukemia | 7 |
| L. buccalis | Blood | 14 | M | Osteogenic sarcoma | 14 |
| L. buccalis | Blood | 9 | F | Medullary aplasia | 14 |
| L. buccalis (4 cases) | Blood | 43 ± 3 | NKb | Bone marrow transplantation | 9 |
| L. sanguinegens | Blood | 100 | F | Pneumonia | 4 |
| L. sanguinegens (4 cases) | Blood | NK | Pregnancy/postpartum fever | 4 | |
| L. sanguinegens | Blood | <1 (35 wk) | F | Suspected sepsis | 4 |
| L. sanguinegens | Blood | <1 (41.5 wk) | M | Suspected sepsis | 4 |
| Leptotrichia sp. | Blood | 50 | M | Myelogenous leukemia | 13 |
| L. trevisanii | Blood | 46 | M | Myelogenous leukemia | 21 |
| L. amnionii | Amniotic fluid | 27 | F | Pregnancy/intrauterine fetal demise | This report |
L. buccalis has been well characterized and is part of the normal oral flora. It has been isolated from babies <1 year of age, and 40% of babies seem to be carriers (19). L. trevisanii is a recently identified bacterium that was recovered from a patient with myeloid leukemia. L. sanguinegens has been proposed as an agent of postpartum and neonatal bacteremia. At this time, we suspect that L. amnionii is indigenous to the urogenital tract and is an opportunist in the appropriate clinical situations. Like other _Leptotrichia-_related clinical cases, this bacterium was isolated from a clinical condition that is physiologically somewhat analogous to having the same stress and immunosuppression as an underlying malignancy. Hanff et al. described the presence of a strong odor from two neonatal cases of infection possibly due to L. sanguinegens (4), as was detected from the amniotic fluid in our case. It appears that L. amnionii is not a blood-loving microbe like L. sanguinegens, because it failed to grow on blood agar. Additional clinical isolates will help establish its true ecological niche and pathogenic potential.
Description of Leptotrichia amnionii sp. nov.
The name “_L. amnionii_” (am′ 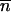 .on.
.on.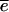 .
.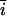 . L. gen. n., amnionii) is derived from the word “amnion.” The organism is characterized by pleomorphic coccobacillus, long, nonmotile, fusiform cells. Some cells are joined end to end in a filamentous form. L. amnionii grows anaerobically on blood agar after 3 days of incubation and is closely related to L. sanguinegens based on its 16S rDNA sequences.
. L. gen. n., amnionii) is derived from the word “amnion.” The organism is characterized by pleomorphic coccobacillus, long, nonmotile, fusiform cells. Some cells are joined end to end in a filamentous form. L. amnionii grows anaerobically on blood agar after 3 days of incubation and is closely related to L. sanguinegens based on its 16S rDNA sequences.
Acknowledgments
We thank Karen Park, Carol Murray, and Teresa Aspeslet for technical assistance, Alice Stargardt for help in preparing the manuscript, and Norman Pace for helpful discussions.
This work was supported in part by grants from the Marshfield Medical Research Foundation.
REFERENCES
- 1.Collins, M. D., L. Hoyles, E. Tornqvist, R. von Essen, and E. Falsen. 2001. Characterization of some strains from human clinical sources which resemble “_Leptotrichia sanguinegens_”: description of Sneathia sanguinegens sp. nov., gen. nov. Syst. Appl. Microbiol. 24**:**358-361. [DOI] [PubMed] [Google Scholar]
- 2.Duperval, R., S. Beland, and J. A. Marcoux. 1984. Infective endocarditis due to Leptotrichia buccalis: a case report. Can. Med. Assoc. J. 130**:**422-424. [PMC free article] [PubMed] [Google Scholar]
- 3.Gao, S. J., and P. S. Moore. 1996. Molecular approaches to the identification of unculturable infectious agents. Emerg. Infect. Dis. 2**:**159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanff, P. A., J. A. Rosol-Donoghue, C. A. Spiegel, K. H. Wilson, and L. H. Moore. 1995. Leptotrichia sanguinegens sp. nov., a new agent of postpartum and neonatal bacteremia. Clin. Infect. Dis. 20(Suppl. 2)**:**S237-S239. [DOI] [PubMed] [Google Scholar]
- 5.Holt, J. G., N. R. Krieg, P. H. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed., p. 297. Williams & Wilkins, Baltimore, Md.
- 6.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180**:**366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohler, J. L., D. Raoult, H. Gallais, M. Pons, Y. Peloux, and P. Casanova. 1982. Septicemia due to Leptotrichia buccalis in an immunosuppressed patient. Sem. Hop. 58**:**1767-1768. (In French.) [PubMed]
- 8.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 9.Lark, R. L., S. A. McNeil, K. VanderHyde, Z. Noorani, J. Uberti, and C. Chenoweth. 2001. Risk factors for anaerobic bloodstream infections in bone marrow transplant recipients. Clin. Infect. Dis. 33**:**338-343. [DOI] [PubMed] [Google Scholar]
- 10.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29**:**173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgenstein, A. A., D. M. Citron, B. Orisek, and S. M. Finegold. 1980. Serious infection with Leptotrichia buccalis. Report of a case and review of the literature. Am. J. Med. 69**:**782-785. [DOI] [PubMed] [Google Scholar]
- 12.Nakou, M., F. H. Mikx, P. J. Oosterwaal, and J. C. Kruijsen. 1987. Early microbial colonization of permucosal implants in edentulous patients. J. Dent. Res. 66**:**1654-1657. [DOI] [PubMed] [Google Scholar]
- 13.Patel, J. B., J. Clarridge, M. S. Schuster, M. Waddington, J. Osborne, and I. Nachamkin. 1999. Bacteremia caused by a novel isolate resembling Leptotrichia species in a neutropenic patient. J. Clin. Microbiol. 37**:**2064-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reig, M., F. Baquero, M. Garcia-Campello, and E. Loza. 1985. Leptotrichia buccalis bacteremia in neutropenic children. J. Clin. Microbiol. 22**:**320-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327**:**293-301. [DOI] [PubMed] [Google Scholar]
- 16.Relman, D. A. 1999. The search for unrecognized pathogens. Science 284**:**1308-1310. [DOI] [PubMed] [Google Scholar]
- 17.Shukla, S. K., D. N. Vevea, D. N. Frank, N. R. Pace, and K. D. Reed. 2001. Isolation and characterization of a black-pigmented Corynebacterium sp. from a woman with spontaneous abortion. J. Clin. Microbiol. 39**:**1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Struckmann, B. Nonhoff, M. Lenke, A. Vilbig, T. Ludwig, A. Bode, K. H. Schleifer, and W. Ludwig. 1996. ARB: a software environment for sequence data. [Online.] ARB Project, Technical University of Munich, Munich, Germany. http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps.
- 19.Sutter, V. L. 1984. Anaerobes as normal oral flora. Rev. Infect. Dis. 6(Suppl. 1)**:**S62-S66. [DOI] [PubMed] [Google Scholar]
- 20.Swofford, D. L. 1999. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 21.Tee, W., P. Midolo, P. H. Janssen, T. Kerr, and M. L. Dyall-Smith. 2001. Bacteremia due to Leptotrichia trevisanii sp. nov. Eur. J. Clin. Microbiol. Infect. Dis. 20**:**765-769. [DOI] [PubMed] [Google Scholar]
- 22.Weinberger, M., T. Wu, M. Rubin, V. J. Gill, and P. A. Pizzo. 1991. Leptotrichia buccalis bacteremia in patients with cancer: report of four cases and review. Rev. Infect. Dis. 13**:**201-206. [DOI] [PubMed] [Google Scholar]
- 23.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173**:**697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]