Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex (original) (raw)
Abstract
Pyramidal neurons of the cerebral cortex display marked layer- and subtype-specific differences in their axonal projections and dendritic morphologies. Here we show that transcription factor Zfp312 is selectively expressed by layer V and VI subcortical projection pyramidal neurons and their progenitor cells. Knocking down Zfp312 with small interfering RNAs dramatically reduced the number of subcortical axonal projections from deep-layer pyramidal neurons and altered their dendritic morphology. In contrast, misexpression of Zfp312 in cortically projecting pyramidal neurons of layers II and III induced the expression of Tbr1, a transcription factor enriched in deep-layer neurons, and the formation of ectopic subcortical axonal projections. Thus, our results indicate that transcription factor Zfp312 plays a critical role in layer- and neuronal subtype-specific patterning of cortical axonal projections and dendritic morphologies.
Keywords: development, neocortex, transcription factor
The development of the cerebral cortex requires the correct molecular specification of neuronal identity and the proper formation of neuronal connections. The majority of cortical neurons are pyramidal neurons, which extend long axonal projections both within and beyond the cortex (1-3). Pyramidal neurons display marked layer- and subtype-specific differences in their axonal projections and dendritic morphologies (1-6). The axons of pyramidal neurons in layers II and III form synaptic connections solely with other cortical neurons. In contrast, the majority of layer V and VI pyramidal neurons project axons to subcortical targets, comprising the collective output of the cortex. Layer VI pyramidal neurons project to the thalamus, whereas other subcortical regions, including the brainstem and spinal cord, receive cortical projections mainly from layer V pyramidal neurons.
Cortical progenitor cells give rise to pyramidal neurons in an inside-first, outside-last sequential manner (3, 6-8). Deep-layer neurons originate from early progenitors in the ventricular zone (VZ), whereas upper-layer neurons arise from late progenitors. Although laminar position is normally correlated with the type of axonal projection, it is not laminar position but the timing of neuron generation that determines the axonal target (3, 6-8). Cortical progenitors at the earliest stage of neurogenesis are multipotent, exhibiting the ability to generate multiple types of pyramidal neurons (8). Later in neurogenesis, the developmental potential of these progenitors becomes progressively restricted to the generation of only upper-layer neurons (8). It has been proposed that genetic programs control this restriction process. However, active molecular determinants for the generation and differentiation of deep-layer neurons remain unknown.
Layer- and neuronal subtype-specific molecular markers, many of which are transcription factors, have been identified in the cerebral cortex (3, 9-12). To explore the molecular development of deep-layer pyramidal neurons, we started with genome-wide expression analysis and identified Zinc finger protein 312 (Zfp312; also known as Fezl and Fez1) as a transcription factor selectively expressed by layer V and VI subcortical projection pyramidal neurons and their progenitors. Through a series of molecular manipulations in mouse embryos, we demonstrate that Zfp312 is necessary for the normal development and sufficient for the ectopic formation of subcortical axonal projections of pyramidal neurons.
Methods
Animals. Experiments were carried out with CD1 mice in accordance with a protocol approved by the Committee on Animal Research at Yale University. The morning of a detectable vaginal plug and the first neonatal day were considered to be embryonic day 0.5 (E0.5) and postnatal day 0 (P0), respectively.
Affymetrix Microarrays and RT-PCR Analysis. Total RNA was isolated from freshly dissected brain tissue by using the TRIzol reagent and cDNA was synthesized by using SuperScript (Invitrogen). Messenger RNA expression profiles were determined by the Affymetrix GeneChip Mouse Expression Set 430 (for detailed procedure and results, see Supporting Text, which is published as supporting information on the PNAS web site). For quantitative real-time RT-PCR, predesigned primer and probe sets were obtained from Applied Biosystems or generated by using PrimerExpress. Thermocycling was carried out by using the Applied Biosystems 7900 system and monitored by TaqMan 5′ exonuclease assay or SYBR Green I dye detection. Gapdh levels were used for normalization.
In Situ Hybridization (ISH). Adult mice were perfused with 4% paraformaldehyde (PFA). Embryos were fixed by immersion in PFA for 24 h. Brains were cryoprotected in graded sucrose solutions and frozen. ISH was performed on cryosections (40 μm) or whole mounts by using digoxigenin (DIG)-labeled riboprobe corresponding to nucleotides 1241-1734 of mouse Zfp312 (GenBank accession no. NM_080433). Hybridization was performed overnight at 60°C, and the signal was detected with an alkaline phosphatase-conjugated anti-DIG antibody and NBT/BCIP (Roche).
DNA Cloning and Constructs. Detailed information on the generation of the Zfp312-GFP, CLEG-Zfp312, Zfp312-siRNAs, and the appropriate control constructs is published in Supporting Text and Table 1, which are published as supporting information on the PNAS web site.
N2a Cells and Immunoblot Analysis. N2a cells were cultured and transfected by using FuGENE 6 (Roche Diagnostics) as described (13). Two days after transfection, cells were either examined directly under a fluorescent microscope or sorted by FACS for GFP-positive cells for Western blotting (13). In Utero Electroporation. For in utero gene transfer by electroporation, 1-2 μl of DNA solution (4 μg/μl) was injected into the lateral ventricle and electroporated (five 50-ms pulses of 40 V with 950-ms intervals) as described (14).
Immunohistochemistry (IHC). The custom anti-mouse Zfp312 antibody was raised in rabbits against the peptide SVGPTATPSAKDLARTVQS (Alpha Diagnostic). Primary antibodies used were αZfp312 (1:100), αGFP (1:3,000; A11122, Molecular Probes), αTbr1 (1:1,000; ref. 3), and αPou3f3 (1:2,000; sc6028, Santa Cruz Biotechnology). IHC was performed as described (15).
Axonal Tracing. The retrograde tracer BDA-3k (Dextran-3000 MW conjugated to tetramethylrhodamine; Molecular Probes) was injected through a Hamilton syringe into designated regions of adult mice in a Kopf stereotaxic instrument. After recovery and survival of 14 days, brains were fixed by perfusion and cryosections were processed for Zfp312 ISH as described above. The expression of Zfp312 was examined for colocalization with BDA-3k under a fluorescent microscope.
Somato-Dendritic Analysis. The cell body size, dendritic arborization, and spines of diaminobenzidine-immunolabeled GFP+ neurons in the primary motor cortex were quantified by 3D reconstruction using the Neurolucida system (Microbrightfield). To determine laminar distribution, sections through the primary somatosensory cortex were divided into 10 horizontal bins from superficial to deep, and the percentage of GFP+ neurons in each bin was calculated.
Results
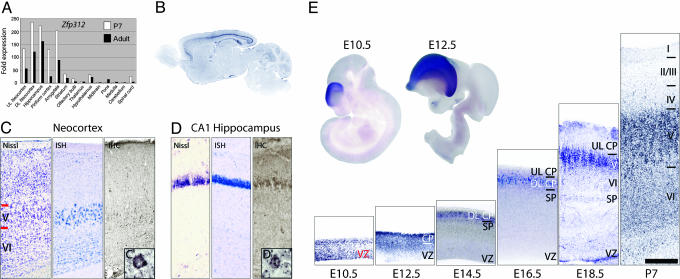
Zfp312 Expression in Subcortically Projecting Neurons and Their Progenitors. To identify transcription factors that may regulate the identity and connectivity of subcortically projecting neurons, we searched for transcripts selectively expressed in deep layers of the developing neocortex. We used the Affymetrix GeneChip Mouse Expression Set 430 to compare global gene expression in neocortical layers I-IV, layers V and VI, hippocampus, and striatum microdissected from P7 mice. Of the 16,930 genes and ESTs present in the neocortex and hippocampus, 56 with neocortical expression enriched in the deep layers (Fig. 7, which is published as supporting information on the PNAS web site) and were chosen for validation and secondary screening using quantitative real-time RT-PCR and ISH. One of the candidate transcripts, Zfp312, was highly enriched in neocortical layer V, and to a lesser extent in layer VI, the CA fields of the hippocampus, the piriform cortex, and the amygdala of the P7 and adult mouse brain (Fig. 1 A and B). Recently, Zfp312 has also been identified by other groups as a marker of deep-layer neocortical neurons (11, 12, 16, 17, 27, ‡). The mouse Zfp312 gene is a homolog of the Xenopus and zebrafish Forebrain-specific embryonic zinc finger gene (18-21), which contains six C2H2-type zinc-finger motifs, suggesting that it functions as a transcription factor. Consistent with this, Zfp312 localizes to nuclei of pyramidal neurons in layers V and VI and hippocampus (Fig. 1 C and D) as well as the nuclei of N2a cells when misexpressed (Fig. 3_A_). The zebrafish homolog of Zfp312 is required for the development and function of forebrain monoaminergic neurons (21). Mice lacking Zfp312 exhibit hyperactive behavior and have a reduced number of subplate neurons as well as diminished connections between the cortex and thalamus (16). The restricted expression pattern of this transcription factor and its involvement in brain development make it a promising candidate for controlling the identity and connectivity of layer V and VI pyramidal neurons.
Fig. 1.
Zfp312 expression in early VZ progenitors and deep-layer pyramidal neurons. (A) Quantitative real-time RT-PCR analysis of Zfp312 expression. (B) Sagittal section of adult brain hybridized with Zfp312 riboprobe. (C and D) Nissl staining and Zfp312 ISH and immunohistochemistry in neocortex and CA1 hippocampus. (Insets) Nuclear localization of Zfp312 in neocortical layer V (_C_′) and hippocampal (_D_′) pyramidal neurons. (E) Zfp312 ISH in whole-mount embryos and sagittal sections of embryonic neocortical wall and P7 neocortex. Zfp312 is expressed by early ventricular zone (VZ) progenitors (E10.5 and E12.5). From E14.5, Zfp312 is down-regulated by VZ progenitors, and _Zfp312_-expressing neurons are localized solely in the subplate (SP), deep-layer cortical plate (DL CP), and layers V and VI and absent in the upper-layer CP (UL CP) and layers II to IV. (Scale bar, 275 μm in C, 140 μm in D, and 200 μm in E.)
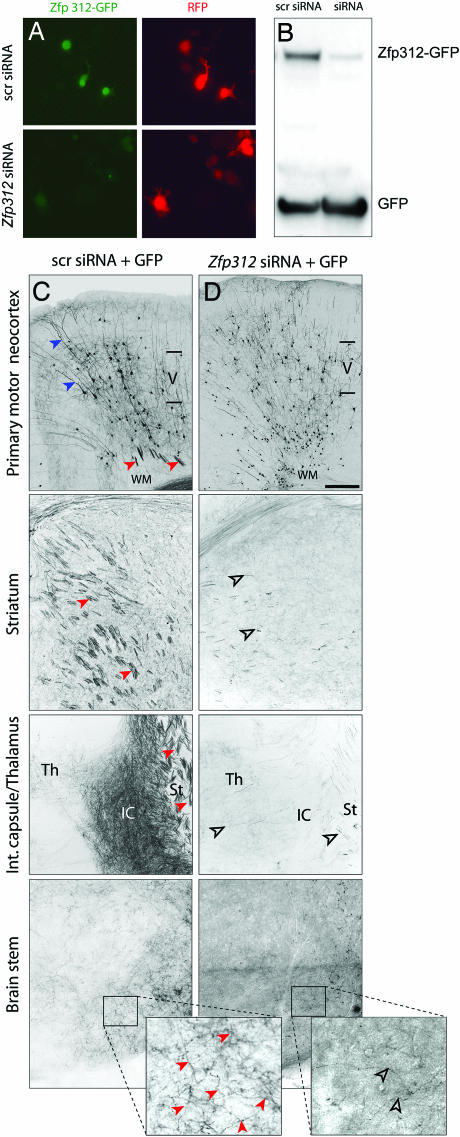
Fig. 3.
Zfp312 is required for subcortical projections and axonal fasciculation. (A and B) Zfp312 siRNAs dramatically decreased the expression and nuclear localization of Zfp312-GFP in N2a cells. N2a cells were transfected with Zfp312-GFP in the presence of 10-fold excess of pCRLH (A) or pLVTH (B) expressing either scr siRNAs or Zfp312 siRNAs. GFP from pLVTH was used as an internal control (B). pLVTH expressing either scr siRNA (C) or Zfp312 siRNAs (D) was delivered by in utero electroporation to cortical VZ progenitors at E12.5 and analyzed at P14. GFP+ apical dendrites (blue arrowheads) and axons (red arrowheads) were present in the control cortex. In control brains, numerous GFP+ axons were present in the white matter (WM), striatum (st) internal capsule (IC), and thalamus (Th) and formed axonal fascicles (red arrowheads). GFP+ subcortical axons were dramatically reduced in brains expressing Zfp312 siRNAs and do not form fasciculated bundles (open arrowheads). (Scale bar, 300 μm in C and D.)
To explore the possible role of Zfp312 in the development of deep-layer projection neurons, we examined its expression using ISH in embryonic and postnatal mouse brains (Fig. 1_E_). Consistent with recent reports (16-18, 22, 27, ‡), Zfp312 was expressed by early cortical VZ progenitor cells, which generate neurons destined for the subplate zone and deep layers of the neocortex. At E12.5, Zfp312 transcripts were highly enriched in the postmigratory pyramidal neurons forming the cortical plate situated beneath the pial surface. During late embryonic and early postnatal development, Zfp312 expression disappeared from cortical progenitors and was restricted to the subplate and the prospective layer V and VI pyramidal neurons.
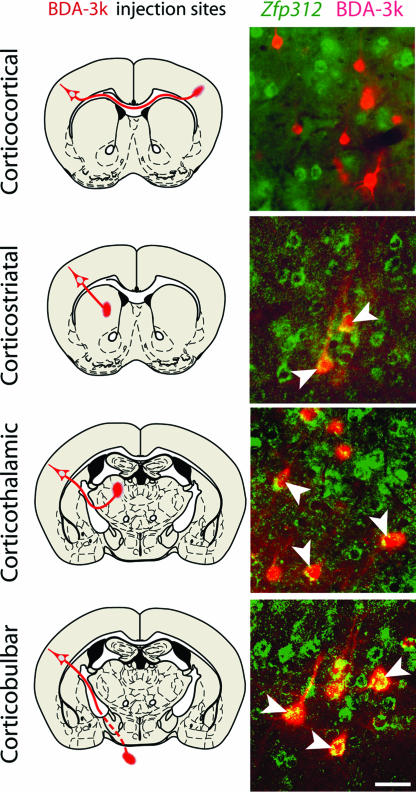
Cortical layers V and VI contain pyramidal neurons that extend either subcortical or cortical axons (1-3, 5, 10). To determine the projection targets of _Zfp312_-expressing neurons, we combined BDA-3k retrograde axonal tracing with ISH for Zfp312 (n = 12 animals). Zfp312 transcripts colocalized with a vast majority of retrogradely labeled deep-layer neurons with axonal projections to the striatum (84 ± 2%), thalamus (96 ± 2%), and brainstem (98 ± 2%), but not with the neurons that have callosal projections to the contralateral neocortex (0%) (Fig. 2). These results demonstrate that Zfp312 is a developmentally regulated transcription factor selectively expressed by the subcortically projecting deep-layer pyramidal neurons and their VZ progenitor cells.
Fig. 2.
Zfp312 is exclusively expressed by subcortically projecting neurons. (Left) Schematic depictions of stereotaxical injection sites of retrograde tracer BDA-3k and expected projection routes (red). (Right) Representative images of neurons in the adult primary motor neocortex retrogradely labeled with BDA-3k (red) and colabeled by Zfp312 ISH (green). Zfp312+ neurons project to the striatum, thalamus, and brainstem (arrowheads). (Scale bar, 50 μm.)
Zfp312 Is Required for the Formation of Subcortical Axonal Projections. The specific expression of Zfp312 in early VZ progenitor cells and the postmitotic deep-layer pyramidal neurons suggests that Zfp312 may play a role in their generation, migration, or differentiation. To examine these possibilities, we used small interfering RNA (siRNA) to knock down Zfp312 activity. Two different siRNA sequences, each targeting a specific region of the mouse Zfp312 transcript, and corresponding control scrambled (scr) sequences were cloned into pLVTH or pCRLH, siRNA vectors that coexpress GFP or RFP, respectively (Fig. 8, which is published as supporting information on the PNAS web site). The ability of siRNAs to knock down Zfp312 expression was confirmed in N2a cells coexpressing Zfp312-GFP and Zfp312 siRNAs or scr siRNAs. Compared with control scr siRNAs, Zfp312 siRNAs dramatically reduced mRNA levels (data not shown), GFP fluorescence (Fig. 3_A_), and protein levels (Fig. 3_B_) of the Zfp312-GFP construct, confirming that siRNA induced robust knockdown of Zfp312 expression.
To knock down Zfp312 expression in vivo, we performed in utero electroporation to codeliver two Zfp312 siRNAs (1:1) into neocortical VZ progenitors at E12.5, when layer V and VI neurons are generated (3, 6, 23, 24). Electroporated brains were analyzed at P14 by using diaminobenzidine immunohistochemistry to enhance the detection of GFP. Cells expressing siRNAs were reliably identified by the coexpression of GFP, which can be used to reveal complete dendritic arborizations and axonal projections. TUNEL analysis of DNA fragmentation revealed no difference in cell death between GFP+ neurons expressing Zfp312 siRNAs and scr siRNAs at P0 (n = 1; Fig. 9, which is published as supporting information on the PNAS web site), indicating that Zfp312 inactivation did not affect neuronal survival. GFP+ pyramidal neurons expressing scr siRNAs or Zfp312 siRNA were found in all cortical layers (n = 2) (Figs. 3 C and D, and 5_A_), because the electroporation transfected early VZ progenitor cells, which eventually gave rise to pyramidal neurons of all layers. This finding indicates that the expression of Zfp312 siRNAs did not interfere with the generation and migration of pyramidal neurons. Layer V and VI pyramidal neurons expressing scr siRNA exhibited typical pyramidal morphology and sent descending fasciculated bundles of GFP+ axons to the white matter, striatum, and internal capsule (Fig. 3_C_, red arrowheads). In striking contrast, neurons expressing Zfp312 siRNAs sent a dramatically reduced number of GFP+ axonal projections to subcortical structures (Fig. 3_D_). Furthermore, the remaining subcortical GFP+ axons lacked the typical tightly bundled organization (Fig. 3_D_, open arrowheads). Taken together, these results indicate that Zfp312 is not required for the generation, migration, or survival of neurons in layers V and VI. However, Zfp312 is necessary for the formation and fasciculation of subcortical axonal projections.
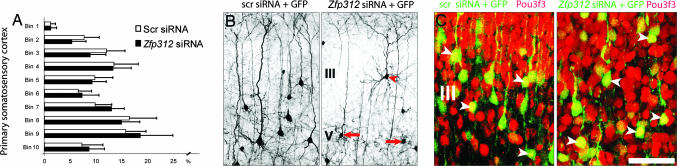
Fig. 5.
Zfp312 siRNAs do not alter the generation, migration, or development of upper-layer pyramidal neurons in the primary somatosensory cortex electroporated at E12.5 with pLVTH. (A) Quantification of laminar distribution of GFP+ neurons at P14 showing no differences between neurons expressing scr siRNA and Zfp312 siRNA (P > 0.05 for each bin). Error bars indicate SD. (B) Zfp312 siRNAs alter morphology of layer V (arrows) but not layer III pyramidal neurons (arrowhead) at P14. (C) Zfp312 siRNAs do not affect the expression of upper-layer marker Pou3f3 (red) at P0 (arrowheads). (Scale bar, 200 μm in B and 400 μm in C.)
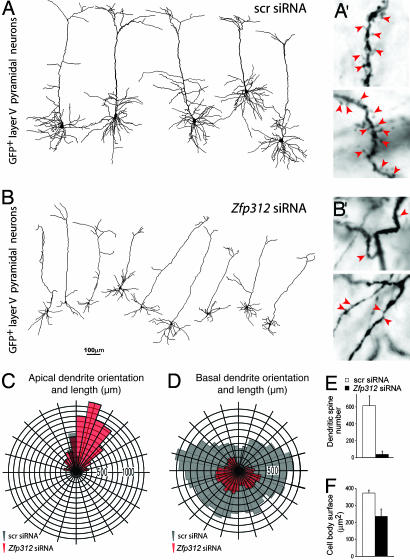
Zfp312 Is Required for the Dendritic Development of Deep-Layer Pyramidal Neurons. To assess whether somato-dendritic development was affected by Zfp312 silencing, we performed 3D reconstruction and analysis of large GFP+ pyramidal neurons in layer V of the primary motor cortex from serial sections of P14 brains using the Neurolucida system (n = 7 scr siRNAs; n = 10 Zfp312 siRNAs). As expected, GFP+ neurons expressing scr siRNAs displayed normal apical dendrites extending toward the pial surface and multiple basal dendrites extending laterally from the base of the pyramidal cell body (Figs. 3_C_ and 4_A_). In contrast, GFP+ deeplayer pyramidal neurons expressing Zfp312 siRNAs had significantly smaller cell bodies (Fig. 4_F_) and showed a decrease in the complexity of basal dendrites (Fig. 4 B and D). Most strikingly, the total number of dendritic spines (Fig. 4 _B_′ and E) and the radial length of basal dendrites were severely reduced (Fig. 4_D_). In addition, we found that the radial orientation, but not the length, of apical dendrites (main shaft) was disrupted (Fig. 4_C_). Additional dendritic quantifications are presented in Fig. 10, which is published as supporting information on the PNAS web site. Thus, in addition to its role in the formation of subcortical projections, Zfp312 controls the development of dendritic arborization and spines of large layer V pyramidal neurons.
Fig. 4.
Zfp312 regulates pyramidal morphology, dendritic patterning, and spine number. (A and B) Neurolucida 3D reconstructions of layer V neurons in the primary motor cortex electroporated at E12.5 with pLVTH expressing either control (scr) siRNAs or Zfp312 siRNAs and analyzed at P14. Zfp312 siRNAs significantly reduced basal dendritic tree complexity (D; P = 0.001), spine number (arrowheads in _A_′ and _B_′, quantification in E; P < 0.001), and soma size (F; P < 0.001) and affected the vertical orientation of apical dendrites (C; P = 0.027).
Upper-Layer Pyramidal Neurons Develop Normally with Zfp312 siRNA. We next examined whether the knockdown of Zfp312 in early progenitor cells and deep-layer neurons would affect the migration and development of later-born upper-layer pyramidal neurons that normally project to other cortical areas. Analysis at P14 of the laminar distribution of GFP+ neurons expressing Zfp312 siRNAs showed no apparent disruption of neuronal migration (Figs. 3_D_ and 5_A_). These neurons also developed normal pyramidal-shape morphology and dendritic arborization (Fig. 5_B_). In addition, numerous GFP+ axons projected via the corpus callosum to the contralateral cortical hemisphere, suggesting that these neurons form normal axonal projections. Pyramidal neurons in layers II and III expressing Zfp312 siRNAs were immunolabeled for transcription factor Pou3f3 (Fig. 5_C_) (also known as Brn1), a marker of upper-layer neurons (9). These results indicate that Zfp312 inactivation in early VZ progenitors and deep-layer pyramidal neurons does not affect the generation, migration, or development of upper-layer cortically projecting neurons.
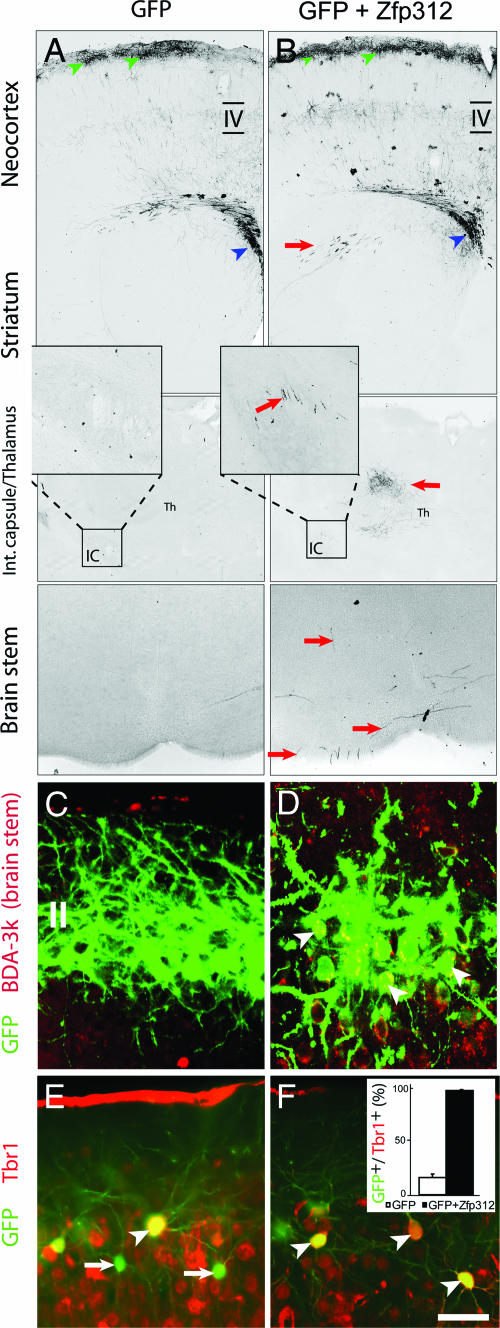
Misexpressing Zfp312 in Upper-Layer Callosal Neurons Induces Ectopic Subcortical Projections. We next investigated whether the misexpression of Zfp312 in layer II and III cortically projecting neurons, which normally do not express Zfp312, would induce the formation of ectopic subcortical axonal projections. The coding region of mouse Zfp312 was cloned into pCLEG, a retroviral vector containing GFP (Fig. 8). Control pCLEG or pCLEG_-Zfp312_ plasmids were delivered into cortical VZ progenitor cells by in utero electroporation at E17, when the last of the callosal pyramidal neurons destined for the upper part of layer III and layer II are generated (3, 6, 23, 24). Previous studies have shown that the production of corticospinal pyramidal neurons continues until E15.5 (23). Our own analysis of pyramidal neurons electroporated in utero with control pCLEG confirmed that the last subcortically projecting cortical neurons are generated at E15.5 (Fig. 11, which is published as supporting information on the PNAS web site). Therefore, undertaking electroporation at E17 ensures that Zfp312 is introduced only into late cortical progenitors and cortically projecting layer II and III neurons normally not expressing Zfp312. Brains of electroporated animals were examined at P14 for the presence of ectopic GFP+ axons in the internal capsule and subcortical targets. In control brains (n = 16), GFP+ neurons were present at the uppermost part of the neocortex corresponding to layers II and III (Fig. 6_A_) and GFP+ axons were present only in the white matter around the striatum, corpus callosum, and cortex. Pyramidal neurons misexpressing Zfp312 migrated to layers II and III (Fig. 6_B_). Remarkably, the upper-layer neurons misexpressing Zfp312 (n = 36 animals) showed numerous GFP+ axons in the striatum, internal capsule, thalamus, and brainstem, in addition to their normal callosal projections (Fig. 6_B_). The induction of ectopic subcortical projections by Zfp312 was confirmed by BDA-3k retrograde axonal tracing (n = 6 animals) from the pons (Fig. 6 C and D). Collectively, these results indicate that Zfp312 is necessary for the formation and sufficient for the induction and direction of ectopic, subcortical projections.
Fig. 6.
Zfp312 misexpression in layer II/III callosal neurons induces Tbr1 and ectopic subcortical axonal projections. Neocortical VZ progenitors were electroporation in utero at E17 with pCLEG or pCLEG-Zfp312 and analyzed by GFP immunohistochemistry at P14. (A) Control GFP+ neurons in the uppermost layer (green arrowheads) send axons through the white matter and corpus callosum (blue arrowheads). _Zfp312_-misexpressing but not control GFP+ neurons have ectopic subcortical axons (A and B; red arrows) and are retrogradely labeled by BDA-3k injected into the pons (C and D; arrowheads). (E and F)A small number of neurons electroporated with control pCLEG expressed Tbr1 (E), whereas the vast majority of neurons misexpressing Zfp312 coexpressed Tbr1 (F) (arrowheads). (Scale bar, 500 μm in A and B,50 μm in C and D, and 90 μm in E and F.)
Upper-Layer Neurons Misexpressing Zfp312 Express Tbr1. Because Zfp312 is a transcription factor, we investigated whether its misexpression would alter the molecular identity of upper-layer pyramidal neurons. P14 brains electroporated with either control pCLEG or pCLEG expressing Zfp312 at E17 were immunostained with antibodies against T-box brain gene 1 (Tbr1), a transcription factor with enriched expression in subplate and layer VI neurons (refs. 3 and 24; see also Fig. 7_B_). Functionally, Tbr1 is necessary for the development of subplate neurons and deep-layer pyramidal neurons and their corticofugal connections (24). In control GFP+ neurons, 15 ± 2% of upper-layer neurons were immunolabeled for Tbr1 (Fig. 6_E_). In contrast, virtually all _Zfp312_-misexpressing upper-layer neurons (98 ± 2%) were immunolabeled for Tbr1 (Fig. 6_F_). Based on this finding, we conclude that Zfp312 misexpression can respecify upper-layer cortically projecting neurons to adopt certain molecular properties of deep layer subcortically projecting pyramidal neurons.
Discussion
A Dual Role of Zfp312 in the Specification of Early Progenitors and the Postmitotic Development of Subcortically Projecting Neurons. We show here that the mouse transcription factor Zfp312 is required for the development of axonal projections and dendritic morphology of subcortically but not cortically projecting pyramidal neurons. Zfp312 is a highly specific marker for deep-layer pyramidal neurons in all areas of the neocortex, and this specificity begins with the VZ progenitor cells that generate these neurons. The absence of Zfp312 transcripts in late cortical progenitors suggests that Zfp312 expression is actively shut off when upper-layer neurons are generated, which is consistent with the possibility that Zfp312 acts as a specific molecular determinant of early VZ progenitor cells.
Our study demonstrates that the silencing of Zfp312 in early cortical progenitor cells and deep-layer neurons does not affect their generation, migration, or survival. However, it does disrupt the formation of axonal projections and dendritic arborizations, which occur postmitotically. Additionally, Zfp312 is required for the development of the soma and dendritic spines of large layer V pyramidal neurons. Taken together, our data show that, in addition to a possible role in the specification of early VZ progenitor cells, Zfp312 also has multiple functions in the postmitotic development of their progenies, the deep layer pyramidal neurons.
Putative Mechanisms by Which Zfp312 Functions. Mechanistically, Zfp312 may be involved in the specification of neuronal identity via the regulation of other transcription factors. Our results showed that Zfp312 misexpression induced the expression of transcription factor Tbr1, which is required for the development of corticofugal projections (24). Alternatively, Zfp312 may directly regulate the expression of guidance molecules that control the response of neuronal processes to external cues. The peak of Zfp312 expression occurred in postmitotic neurons during the formation and refinement of axonal projections and synapses, which is consistent with the latter possibility.
A previous study showed that, in a genetic knockout of Fezl (Zfp312), thalamocortical axons were reduced in numbers and exhibited aberrant projections (16). Because Zfp312 expression is absent from the dorsal thalamus, defects in thalamocortical projections are likely non-cell-autonomous. According to the hand-shake hypothesis, thalamocortical axons grow in the direction opposite to, but associate with, corticofugal axons from the deep layers of the cortex (25). Therefore, the aberrant thalamocortical projections in Zfp312 knockouts may occur via the disruption of corticothalamic projections, which we observed in our study. We did not find a reduction of the subplate as described in Zfp312 mutant mice (16), possibly because of the timing of our experiments on or after E12.5, when most subplate neurons had been generated (3, 6, 23, 24). No other cortical abnormalities were reported (16), suggesting that our experimental approach may uncover additional roles for Zfp312 in cortical development. Our RNA interference strategy offers possible advantages compared with genetic knockouts, by allowing for gene inactivation within a specific developmental period and in a mosaic to facilitate the analysis of cell autonomous effects. Importantly, possible compensatory mechanisms are minimized. Analysis of the mouse genome revealed that Zfp312 has a highly similar homolog BC049157, or Zfp312-like, which is also expressed by pyramidal neurons (Fig. 12, which is published as supporting information on the PNAS web site). The zinc-finger binding domains of Zfp312 and Zfp312-like are 95.7% identical, indicating that these two proteins very likely bind the same DNA sequences and may functionally compensate for each other.
Implications for the Evolution and Development of the Neocortex. The six-layered structure of the neocortex is unique to mammals. Evolutionary differences in Zfp312 may underlie phylogenetic differences in the development of subcortical projections (2, 25, 26). Thus, we investigated whether Zfp312 might have undergone adaptive changes during the evolution of the cerebral cortex. In silico amino acid sequence analyses of Zfp312 orthologs in six mammalian and four nonmammalian species revealed that Zfp312 is remarkably well-conserved in mammals (92.2% identity) (Figs. 12 and 13, which are published as supporting information on the PNAS web site). Between mammalian and nonmammalian species, the conservation is very high within the zinc finger region but lower in other regions. Additionally, several regions in the protein are completely conserved within mammals but are absent from non-mammals. These include a polyglycine repeat region, which may function as a flexible hinge to facilitate protein interactions. These observations indicate that significant amino acid changes occurred during the divergence of the mammalian lineage, rapidly arose to fixation, most likely due to strong positive selection, and have remained virtually unchanged since. This finding is consistent with the possibility that Zfp312 played a role in the evolution of long-range subcortical axonal projections of the neocortex.
Note. After the completion of these experiments, Molyneaux et al. (27) reported that Fezl (Zfp312) is required for the birth and specification of corticospinal motor neurons.
Supplementary Material
Supporting Information
Acknowledgments
We thank Susan Liu-Chen and Mariamma Pappy for help with experiments and members of the Sestan laboratory for constructive discussions. We also thank Gord Fishell (New York University) and Nicholas Gaiano (Johns Hopkins University) for pCLE, Robert Hevner (University of Washington) for αTbr1, Jun-ichi Miyazaki (Osaka University Medical School) for pCAGG-mRFP, Shrikant Mane for help with microarrays, Pasko Rakic for use of confocal microscope, Didier Trono (Swiss Federal Institute of Technology) for pLVTH, and David McCor-mick and Leonard Kaczmarek for helpful comments on this manuscript. This study was supported by National Institutes of Health Grants NS054273 and HD045481, the Kavli Institute for Neuroscience, the Tourette Syndrome Association, the Whitehall Foundation (N.S.), and the Canadian Institutes of Health Research (K.Y.K.).
Author contributions: J.-G.C., M.-R.R., K.Y.K., and N.Š. designed research; J.-G.C., M.-R.R., and K.Y.K. performed research; J.-G.C., M.-R.R., and K.Y.K. contributed new reagents/analytic tools; J.-G.C., M.-R.R., K.Y.K., and N.Š. analyzed data; and J.-G.C., M.-R.R., K.Y.K., and N.Š. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: VZ, ventricular zone; E_n_, embryonic day n;P_n_, postnatal day n; ISH, in situ hybridization.
Footnotes
‡
Chen B., Schaevitz L. R. & McConnell, S. K. (2005) Soc. Neurosci. Abstr. 29, 140.5 (abstr.).
References
- 1.DeFelipe, J. & Farinas, I. (1992) Prog. Neurobiol. 39**,** 563-607. [DOI] [PubMed] [Google Scholar]
- 2.O'Leary, D. D. & Koester, S. E. (1993) Neuron 10**,** 991-1006. [DOI] [PubMed] [Google Scholar]
- 3.Hevner, R. F., Daza, R. A., Rubenstein, J. L., Stunnenberg, H., Olavarria, J. F. & Englund, C. (2003) Dev. Neurosci. 25**,** 139-151. [DOI] [PubMed] [Google Scholar]
- 4.Whitford, K. L., Dijkhuizen, P., Polleux, F. & Ghosh, A. (2002) Annu. Rev. Neurosci. 25**,** 127-149. [DOI] [PubMed] [Google Scholar]
- 5.Koester, S. E. & O'Leary, D. D. (1993) J. Neurosci. 12**,** 1382-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caviness, V. S., Jr. (1982) Brain Res. 256**,** 293-302. [DOI] [PubMed] [Google Scholar]
- 7.Jensen, K. F. & Killackey, H. P. (1984) Proc. Natl. Acad. Sci. USA 81**,** 964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frantz, G. D. & McConnell, S. K. (1996) Neuron 17**,** 55-61. [DOI] [PubMed] [Google Scholar]
- 9.McEvilly, R. J., de Diaz, M. O., Schonemann, M. D., Hooshmand, F. & Rosenfeld, M. G. (2002) Science 295**,** 1528-1532. [DOI] [PubMed] [Google Scholar]
- 10.Weimann, J. M., Zhang, Y. A., Levin, M. E., Devine, W. P., Brulet, P. & McConnell, S. K. (1999) Neuron 24**,** 819-831. [DOI] [PubMed] [Google Scholar]
- 11.Gray, P. A., Fu, H., Luo, P., Zhao, Q., Yu, J., Ferrari, A., Tenzen, T., Yuk, D. I., Tsung, E. F., Cai, Z., et al. (2004) Science 306**,** 2255-2257. [DOI] [PubMed] [Google Scholar]
- 12.Arlotta, P., Molyneaux, B. J., Chen, J., Inoue, J., Kominami, R. & Macklis, J. D. (2005) Neuron 45**,** 207-221. [DOI] [PubMed] [Google Scholar]
- 13.Roncarati, R., Sestan, N., Scheinfeld, M. H., Berechid, B. E., Lopez, P. A., Meucci, O., McGlade, J. C., Rakic, P. & D'Adamio, L. (2002) Proc. Natl. Acad. Sci. USA 99**,** 7102-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito, T. & Nakatsuji, N. (2001) Dev. Biol. 240**,** 237-246. [DOI] [PubMed] [Google Scholar]
- 15.Sestan, N., Artavanis-Tsakonas, S. & Rakic, P. (1999) Science 286**,** 741-746. [DOI] [PubMed] [Google Scholar]
- 16.Hirata, T., Suda, Y., Nakao, K., Narimatsu, M., Hirano, T. & Hibi, M. (2004) Dev. Dyn. 230**,** 546-556. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, K., Terashima, T., Nishikawa, T. & Takumi, T. (2004) Eur. J. Neurosci. 20**,** 2909-2916. [DOI] [PubMed] [Google Scholar]
- 18.Matsuo-Takasaki, M., Lim, J. H., Beanan, M. J., Sato, S. M. & Sargent, T. D. (2000) Mech. Dev. 93**,** 201-204. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto, H., Yabe, T., Hirata, T., Shimizu, T., Bae, Y., Yamanaka, Y., Hirano, T. & Hibi, M. (2000) Mech. Dev. 97**,** 191-195. [DOI] [PubMed] [Google Scholar]
- 20.Yang, Z., Liu, N. & Lin, S. (2001) Dev. Biol. 231**,** 138-148. [DOI] [PubMed] [Google Scholar]
- 21.Levkowitz, G., Zeller, J., Sirotkin, H. I., French, D., Schilbach, S., Hashimoto, H., Hibi, M., Talbot, W. S. & Rosenthal, A. (2003) Nat. Neurosci. 6**,** 28-33. [DOI] [PubMed] [Google Scholar]
- 22.Sansom, S. N., Hebert, J. M., Thammongkol, U., Smith, J., Nisbet, G., Surani, M. A., McConnell, S. K. & Livesey, F. J. (2005) Development (Cambridge, U.K.) 132**,** 3947-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polleux, F., Dehay, C. & Kennedy, H. (1998) J. Neurosci. 18**,** 9910-9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hevner, R. F., Shi, L., Justice, N., Hsueh, Y., Sheng, M., Smiga, S., Bulfone, A., Goffinet, A. M., Campagnoni, A. T. & Rubenstein, J. L. (2001) Neuron 29**,** 353-366. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Bendito, G. & Molnar, Z. (2003) Nat. Rev. Neurosci. 4**,** 276-289. [DOI] [PubMed] [Google Scholar]
- 26.Rouiller, E. M. & Welker, E. (2000) Brain Res. Bull. 53**,** 727-741. [DOI] [PubMed] [Google Scholar]
- 27.Molyneaux, B. J., Arlotta, P., Hirata, T., Hibi, M. & Macklis, J. D. (2005) Neuron 47**,** 817-831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information