The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings (original) (raw)
Abstract
The Drosophila Bithorax Complex encodes three well-characterized homeodomain proteins that direct segment identity, as well as several noncoding RNAs of unknown function. Here, we analyze the iab-4 locus, which produces the microRNAs iab-4-5p and iab-4-3p. iab-4 is analogous to miR-196 in vertebrate Hox clusters. Previous studies demonstrate that miR-196 interacts with the Hoxb8 3′ untranslated region. Evidence is presented that miR-iab-4-5p directly inhibits Ubx activity in vivo. Ectopic expression of mir-iab-4-5p attenuates endogenous Ubx protein accumulation and induces a classical homeotic mutant phenotype: the transformation of halteres into wings. These findings provide the first evidence for a noncoding homeotic gene and raise the possibility that other such genes occur within the Bithorax complex. We also discuss the regulation of mir-iab-4 expression during development.
Keywords: microRNA, iab-4, Ultrabithorax, homeotic gene
Classical genetic studies suggest that the Bithorax Complex (BX-C) contains as many as nine homeotic genes (Lewis 1978). However, only three encode Hox proteins, Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B) (Martin et al. 1995). The bulk of genomic DNA comprising the BX-C is thought to function as _cis_-regulatory DNA that controls the timing and site of Hox expression (Sanchez-Herrero et al. 1985). Nevertheless, it has been known for more than 15 years that intergenic regions of the BX-C are extensively transcribed (Cumberledge et al. 1990; Bae et al. 2002; Drewell et al. 2002). The possible functional activities of the noncoding RNAs have received little attention, despite the fact that these transcripts, including iab-4, are expressed in restricted domains along the anterior-posterior axis, like conventional Hox genes (Cumberledge et al. 1990).
Hox gene clusters contain conserved miRNAs. For example, miR-10 is located within the Drosophila Antennapedia gene complex (ANT-C) between the Hox genes Deformed and Sex combs reduced (Lagos-Quintana et al. 2001). The sequence and location of miR-10 are conserved in vertebrate Hox complexes (Tanzer et al. 2005). Sex combs reduced has been proposed as a direct miR-10 target in insects (Brennecke et al. 2005). A second group of Drosophila miRNAs map to a hairpin located at the distal end of the iab-4 locus (Aravin et al. 2003), which resides between abd-A and Abd-B (Fig. 1A). miRNAs were cloned from both arms of this hairpin and are termed iab-4-5p and iab-4-3p (Aravin et al. 2003). miR-iab-4-5p was recently predicted to regulate Ubx activity (Stark et al. 2003; Grun et al. 2005). Although vertebrates lack an iab-4 ortholog, as defined by sequence identity, a different miRNA, miR-196, resides at an analogous position adjacent to the posterior-most HOX 9-13 paralogs. Tissue culture assays, in vivo cleavage products, and transgenic lacZ “sensors” indicate that miR-196 inhibits Hoxb8 activity (Mansfield et al. 2004; Yekta et al. 2004). Despite these provocative target relationships, no phenotypes have been associated with any Hox miRNA.
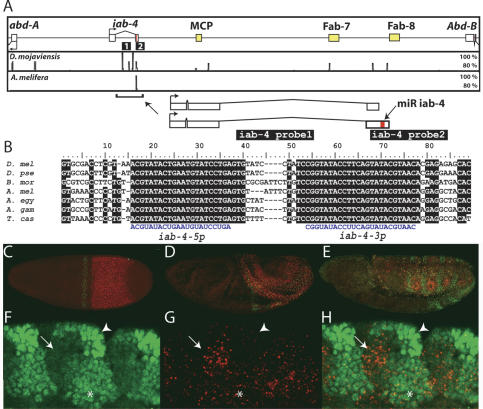
Figure 1.
Structure, conservation, and expression of the iab-4 miRNA. (A) A 120-kb interval of the D. melanogaster Bithorax Complex that includes the homeobox genes abd-A and Abd-B, the noncoding RNA gene iab-4, and three transcribed boundary elements (MCP, Fab-7, and Fab-8). Shown are VISTA plots depicting highly conserved sequences in the distantly related Drosophilid Drosophila mojaviensis (regions of >80% identity in a window of 50 nt) and the honeybee A. mellifera (regions of >80% identity in a window of 150 nt). Note that all of these conserved regions are associated with transcribed noncoding elements. Depicted at higher magnification are two iab-4 splice forms, an ∼2.1-kb transcript containing the iab-4 miRNA hairpin and a shorter transcript that terminates just 5′ of the miRNAs. The two iab-4 probes used in this study (iab-4 probe1 and iab-4 probe2) are shown as black bars below the schematics in A. (B) Perfect conservation of the iab-4-5p and iab-4-3p miRNAs in diverse arthropods: D. melanogaster (D. mel) D. pseudoobscura (D. pse), Bombyx mori (B. mor), A. mellifera (A. mel), A. aegypti (A. egy), A. gambiae (A. gam), and T. castaneum (T. cas). (_C_-E) Time course of iab-4 (red) and Ubx (green) nascent transcription: stage 5 (C), stage 8 (D), stage 11 (E). Nascent iab-4 transcription was detected with an intronic probe marked as probe 1 in A. (_F_-H) Complementary expression of iab-4 RNA (red) and Ubx protein (green) in a stage 12 embryo. Shown is a maximum projection through a sagittal section revealing the ectoderm and underlying mesoderm (approximately five cell layers). Domains of highest Ubx protein levels do not express iab-4 (arrowheads), whereas cells with highest levels of iab-4 do not accumulate Ubx (arrows). Other regions show low levels of coexpression (asterisk).
miRNAs are short, 21-24-nt RNAs that attenuate protein synthesis by binding complementary sites in target mRNAs (Lai 2003; Bartel 2004). An unexpectedly modest amount of base-pairing appears to underlie target recognition. Experimental and computational studies have converged on the principle of “seed-pairing,” whereby ∼7 continuous Watson-Crick base pairs at the 5′-end of the miRNA mediate target recognition (Lai 2002; Brennecke et al. 2005; Lewis et al. 2005). The limited sequence requirement for miRNA-mRNA interactions has fueled current proposals that a third or more of all mRNAs may be regulated by miRNAs (Lewis et al. 2005; Xie et al. 2005). As tallies of miRNA loci continue to grow (with current estimates for humans ranging from 800 to 1000) (Bentwich et al. 2005; Berezikov et al. 2005), the network of possible miRNA:target interactions will expand.
Only a small number of miRNA:target interactions have been studied in vivo. Here we present evidence that iab-4 microRNAs selectively attenuate Ubx activity in vivo. The Ubx 3′ untranslated region (3′ UTR) contains predicted target sites for miR-iab-4-5p and expression of a _GFP-Ubx-3_′ UTR “sensor” transgene is repressed by ectopic expression of a mir-iab-4 minigene. This minigene also reduces Ubx protein levels in haltere imaginal discs, thereby inducing a classical homeotic transformation of halteres into wings. Taken together, these results suggest that the iab-4 transcription unit encodes an authentic homeotic regulatory gene. We suggest that additional noncoding RNAs correspond to “missing” homeotic genes in the Bithorax complex, and discuss novel mechanisms of iab-4 regulation during development.
Results and Discussion
Complementary patterns of iab-4 and Ubx expression
Among the sequenced Drosophilids, there are only a few highly conserved sequences in the 120-kb region separating abd-A and Abd-B in the BX-C (Fig. 1A). Three are located in regions that flank known insulator elements (Karch et al. 1994; Zhou et al. 1996; Barges et al. 2000). A fourth conserved region is located within the 3′ region of the iab-4 transcription unit (Cumberledge et al. 1990; summarized in Fig. 1A). The iab-4 locus contains regulatory DNAs that control abd-A expression (Karch et al. 1990). It also produces two spliced, polyadenylated transcripts that differ by the presence or absence of the highly conserved 3′ sequences (Fig. 1A). This region contains a single ∼100-nucleotide (nt) pre-miRNA hairpin structure that encodes two stable miRNAs: iab-4-5p and iab-4-3p (Aravin et al. 2003).
The pre-mir-iab-4 hairpin is conserved both in sequence and genomic location in the mosquitoes Anopheles gambiae and Aedes aegypti, the honeybee Apis mellifera, and the flour beetle Tribolium castaneum (Fig. 1B), species that last shared a common ancestor roughly 400 million years ago. In fact, it is by far the best conserved sequence in the abd-A/Abd-B interval among Drosophilid and non-Drosophilid genomes (Fig. 1A). It is notable that the sequences corresponding to the mature iab-4-5p and iab-4-3p miRNAs (each forming one arm of the hairpin) are perfectly conserved among this broad spectrum of insects.
Double-label RNA FISH and antibody staining was used to determine the relative expression patterns of iab-4 RNA and Ubx RNA/protein accumulation during embryonic development (Fig. 1C-E). The iab-4 primary transcript is strongly expressed in the presumptive abdomen, mainly in the progenitors of the second (A2) through seventh (A7) segments (see Cumberledge et al. 1990). There is also a weak transient stripe of expression in anterior regions. Ubx RNA is distributed in a strong stripe in parasegment 6, but only low levels are seen in regions of the presumptive abdomen containing high levels of iab-4 transcript. No Ubx protein is detected at this early stage, possibly due to the time required to transcribe the entire ∼80-kb locus (Shermoen and O'Farrell 1991).
During the rapid phase of germband elongation, Ubx protein becomes detectable in the abdomen (Fig. 1D). At the conclusion of germband elongation, the Ubx and iab-4 patterns are largely complementary in the dorsal ectoderm (Fig. 1E). During segmentation of the germband, the Ubx protein shows complex modulation in the dorsal ectoderm (Fig. 1F-H). There are three apparent levels of Ubx protein distribution in these regions: high, low, and none (Fig. 1F). We note an inverse correlation between the levels of Ubx protein and the sites of iab-4 expression (Fig. 1G,H). The strongest expression of iab-4 occurs in regions having the lowest accumulation of Ubx protein (Fig. 1F-H, arrows), whereas intermediate and low levels coincide with sites of diminished Ubx accumulation (Fig. 1F-H, asterisks); there is little or no iab-4 expression in those cells containing the highest levels of Ubx protein (Fig. 1F-H, arrowheads). These observations are consistent with the possibility that Ubx protein synthesis might be modulated by one or both iab-4 miRNAs. Direct support for this possibility stems from the analysis of a GFP-Ubx transgene containing the 3′ UTR sequence from Ubx. This transgene displays slightly diminished expression in abdominal regions containing high levels of iab-4 transcripts (data not shown; see below).
The Ubx 3′ UTR is directly targeted by miR-iab-4-5p
Computational studies have identified the Ubx 3′ UTR as a likely target of regulation by iab-4-5p (Stark et al. 2003; Grun et al. 2005). Of the seven potential sites identified by Stark and colleagues (Fig. 2A), five exhibit conserved and canonical seed pairing of six or more nucleotides (Fig. 2B). Of these, sites #3 and #6 are perfectly conserved among sequenced Drosophilids and have seeds of at least 7 nt, a length sufficient for efficient in vivo recognition by miRNAs (Brennecke et al. 2005; Lai et al. 2005); site #7 also has a 7-mer seed match that is conserved in some species (see below).
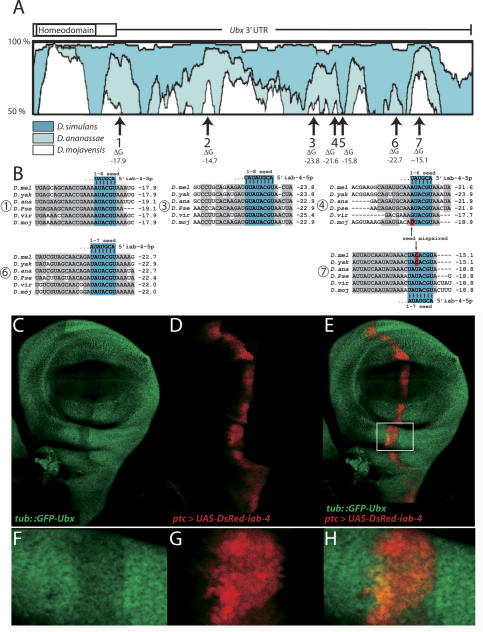
Figure 2.
Identification and validation of iab-4-5p target sites in the Ubx 3′ UTR. (A) Conserved regions of the Ubx 3′ UTR revealed by VISTA plots between D. melanogaster and other Drosophilids (dark blue, Drosophila simulans; light blue, D. ananassae; white, D. mojaviensis). All but one of seven putative iab-4-5p target sites (Stark et al. 2003) correspond to islands of conserved sequence (numbered arrows); free energies of predicted iab-4-5p:target duplexes are noted below. (B) Alignments and free energies of likely iab-4-5p target sites from six divergent Drosophilids: D. melanogaster (D. mel), D. yakuba (D. yak), D. ananassae (D. ana), D. pseudoobscura (D. pse), D. virilis (D. vir), and D. mojaviensis (D. moj). Conserved Ubx 3′ UTR sequences are shaded gray; nucleotides that pair with the iab-4-5p seed are shaded blue. Note apparent compensatory mutations between strong/weak sites in different species (in red, arrows). For example, site #4 is likely active in D. melanogaster but inactive in D. mojaviensis, while site #7 probably inactive in D. melanogaster but active in D. mojaviensis. (_C_-H) Evidence for direct recognition of the Ubx 3′ UTR by iab-4-5p. Shown is expression of a _tub-GFP-Ubx 3_′ UTR sensor transgene (green) in a background where a UAS-DsRed-iab-4 transgene has been activated using ptc-Gal4 (red). The Ubx sensor is specifically down-regulated in _mir-iab-4_-misexpressing cells marked by expression of DsRed. (_F_-H) Close-ups of the boxed region in E.
In current target-finding approaches, greater confidence is usually ascribed to those miRNA-binding sites that are conserved in the greatest number of analyzed species. Curiously, the putative iab-4-5p target sites with the lowest free energy are not necessarily the best conserved. Instead, there appear to be compensatory changes among different iab-4-5p-binding sites in individual Ubx 3′ UTRs. For example, site #4 exhibits canonical 6-mer seed pairing in four species of Drosophila, but contains a G:U base pair in Drosophila virilis and a seed mismatch in Drosophila mojavenesis and is likely nonfunctional in these two species. Conversely, site #7 is mispaired in D. melanogaster and Drosophila yakuba, but is conserved as a strong 7-mer seed-paired site in D. mojavenesis, Drosophila pseudoobscura, Drosophila ananassae, and D. virilis. These observations suggest that individual target sites may be evolutionarily labile, and in vivo regulation depends on the net complement of both high- and low-affinity sites contained in the target mRNA. These compensatory changes in strong and weak target sites are reminiscent of the evolution of individual Bicoid-binding sites in the eve stripe 2 enhancers present in divergent Drosophilids (Ludwig et al. 1998).
Direct evidence for iab-4:Ubx miRNA interactions was obtained using a tub::_GFP-Ubx 3_′ UTR transgene (the “Ubx sensor”). This construct directs ubiquitous expression of the GFP coding sequence fused to the Ubx 3′ UTR, and wing imaginal discs bearing the Ubx sensor display relatively uniform expression of GFP. Ectopic expression of UAS-DsRed under the control of ptc-Gal4, which directs expression along the anterior-posterior border of the disc, has little or no effect on the distribution of GFP staining (Brennecke et al. 2005).
We next assayed the expression of the Ubx sensor in the presence of ectopic iab-4 miRNAs. For this purpose, a transgene was created that contains DsRed and 400 base pairs (bp) from iab-4 encompassing the entire 100-bp 3′ hairpin sequence (UAS-DsRed-iab-4). As shown previously, transgenes of this type direct the expression of biologically active miRNAs in cells that are labeled by expression of DsRed (Stark et al. 2003). When driven by ptc-Gal4 in wing imaginal discs, Ubx sensor levels were specifically diminished in those cells expressing the iab-4 transgene (Fig. 2C-E). Detailed analysis of the DsRed-iab-4 and GFP-Ubx expression profiles suggests that repression of the Ubx sensor by ectopic iab-4 miRNA is dose-sensitive (Fig. 2F-H). These data constitute in vivo evidence that iab-4 miRNAs specifically recognize target sequences in the Ubx 3′ UTR and thereby attenuate Ubx protein synthesis.
Ectopic iab-4 inhibits endogenous Ubx and induces a Ubx-like homeotic transformation
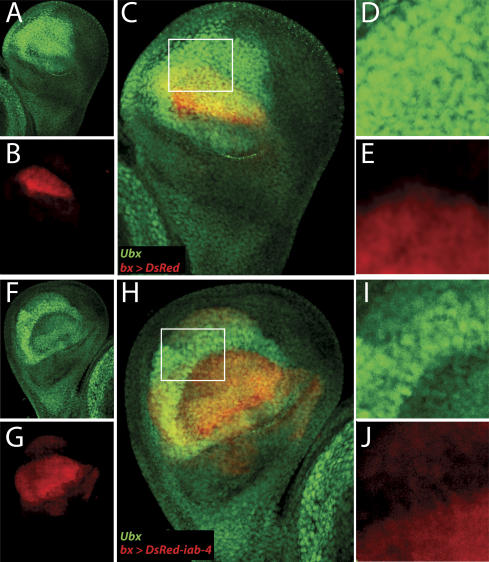
Ubx protein is broadly distributed throughout the haltere imaginal disc, where it imposes haltere identity by repressing the expression of many genes that otherwise direct wing development (e.g., Weatherbee et al. 1998). This repression is very sensitive to Ubx levels, and consequently, even partial loss of Ubx function can transform halteres into wings (i.e., Fig. 4B, below). Haltere discs were examined for the accumulation of Ubx protein in the absence or presence of ectopic iab-4 miRNAs. Ubx is detected at high levels in most of the cells of the presumptive pouch. Expression of DsRed alone using bx-Gal4, which is active in the presumptive dorsal region of the pouch, did not affect Ubx accumulation (Fig. 3A-E). In contrast, haltere discs expressing UAS-DsRed-iab-4 under the control of bx-Gal4 displayed strongly reduced levels of Ubx protein (Fig. 3F-J). Thus, as seen for the Ubx sensor in wing discs, ectopic iab-4 miRNA inhibits accumulation of endogenous Ubx protein.
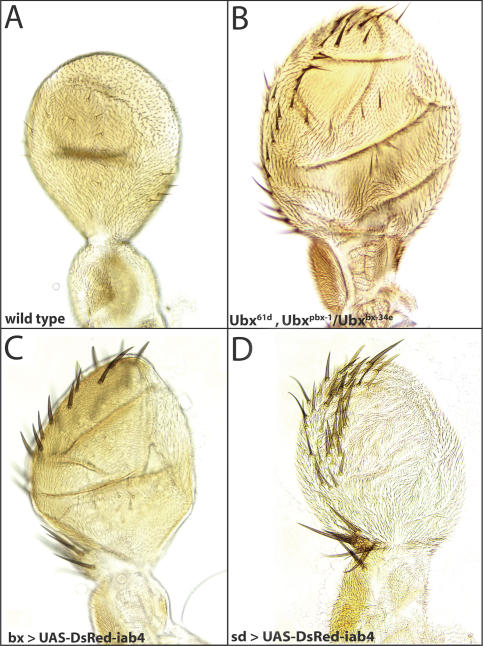
Figure 4.
Directed expression of mir-iab-4 induces a homeotic transformation. (A) Wild-type haltere, which contains small lightly pigmented sensilla but lacks the triple row of sensory bristles seen in wings. (B) Haltere-to-wing transformation in a mild Ubx loss-of-function mutant background. Misexpression of iab-4 miRNA hairpin in bx-Gal4/Y; UAS-DsRed-mir-iab-4 (C) and sd-Gal4, UAS-DsRed-mir-iab-4 (D) animals induces a similar haltere-to-wing transformation.
Figure 3.
Expression of mir-iab-4 in the haltere disc reduces endogenous Ubx protein accumulation. (_A_-E) Normal expression of Ubx protein (green) in bx-Gal4/Y; UAS-DsRed/+ haltere discs. (A) Ubx is detected throughout the pouch region of the haltere disc. (B) DsRed (red) marks bx-Gal4 activity in the dorsal compartment of the presumptive haltere pouch. (C) Merge. (D,E) Close-ups of the region boxed in C. (_F_-H) bx-Gal4/Y; UAS-DsRed-mir-iab-4 haltere disc exhibits reduced accumulation of Ubx protein (green) in the presence of ectopic mir-iab-4 (red). (I,J) Close-ups of the region boxed in H.
The effect of iab-4 miRNA misexpression on adult haltere development was examined. The wild-type haltere contains small lightly pigmented sensilla but lacks the triple row of sensory bristles at the leading margin seen in wings (Fig. 4A). In contrast, halteres that developed from discs expressing UAS-DsRed-iab-4 under the control of bx-Gal4 or scalloped-Gal4 are flattened and elongated in the proximal-distal axis, and exhibit an extensive row of sensory bristles at the leading margin (Fig. 4C,D). All of these phenotypes are strongly indicative of a classic haltere-to-wing homeotic transformation (Roch and Akam 2000).
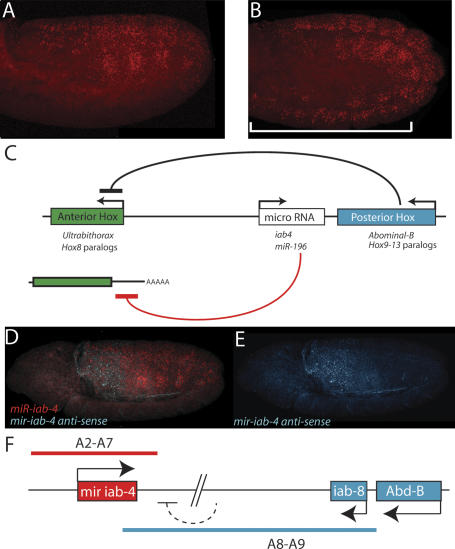
The demonstration that miR-iab-4 represses the anterior Hox gene Ubx might be relevant to the phenomenon of “posterior prevalence” (Duboule and Dolle 1989). Polycomb mutant embryos have previously been observed to derepress Hox gene expression, resulting in broad misexpression of all Hox genes (e.g., Pirrotta 1997). Ultimately, ectopic expression of posterior Hox genes (e.g., Abd-B or Hox9-13) leads to the transcriptional repression of anterior Hox genes (e.g., Ubx or Hox8 paralogs) (Choi et al. 2000). We observe that Polycomb mutant embryos also derepress iab-4 expression throughout the embryo (Fig. 5, cf. B and A). Therefore, misexpression of iab-4 miRNAs may contribute to the repression of Ubx function observed in the Polycomb mutant background. Thus, posterior prevalence may arise from the dual utilization of protein-based/transcriptional mechanisms and miRNA-based/post-transcriptional mechanisms (Fig. 5C).
Figure 5.
Regulation and function of iab-4. (A,B) Expression of iab-4 in wild-type (A) and Pc1 (B) stage 12 embryos. The domain of iab-4 transcription is expanded anteriorly in the Pc mutant, which correlates with suppression of Ubx activity in this mutant background. (C) Similar regulatory and functional arrangement of the BX-C and the vertebrate HOX clusters. Posteriorly expressed Hox proteins repress the transcription of anterior Hox genes; posteriorly expressed microRNAs (iab-4-5p and iab-4-3p in insects, and miR-196 in vertebrates) repress anterior Hox genes at a post-transcriptional level (either by transcript cleavage or inhibition of productive translation). (D,E) Double RNA FISH staining to localize iab-4 transcripts (red) and a noncoding RNA produced from the opposite strand (blue). The opposite transcript is not detected with iab-4 probes until the onset of germband elongation, presumably due to the time it takes for Pol II to progress from the iab-8 promoter located ∼75 kb away. In germband-elongated embryos the two transcripts are expressed in complementary patterns. The probes used for hybridization reside within 2 kb of one another within the iab-4 transcription unit and are described in Figure 1. (F) Summary of the strand-exclusion model. A large transcript derived from the Abd-B/iab-8 domain might preclude transcription from the iab-4 promoter on the opposite strand.
Regulation and function of iab-4 miRNAs
The detailed analysis of intergenic transcripts in the abd-A/Abd-B interval suggests a “strand exclusion” model for iab-4 regulation (summarized in Fig. 5F). The iab-4 locus is unusual in that both strands are transcribed (Bae et al. 2002). However, each strand displays a distinct pattern of expression. The strand producing the iab-4 pri-miRNA is broadly expressed in the A2-A7 region of the germband, while the other strand is expressed in A8 and A9 (e.g., Fig. 5D,E). Double RNA FISH assays suggest that the miR strand is initially expressed in A2-A8, but expression is lost in A8 as transcription of the other strand progresses from the iab-8 domain. Perhaps transcription from one strand diminishes transcription from the other (Fig. 5F). Although the detailed mechanism may be different, these observations are evocative of the mutually exclusive expression of Xist and Tsix RNAs on mammalian X chromosomes (e.g., Rougeulle and Avner 2004). Additional target predictions for miR-iab-4 imply that exclusion of iab-4 expression from A8 might be important for stable accumulation of other potential iab-4 target mRNAs, such as Abd-B (Enright et al. 2003; Stark et al. 2003).
Traditionally, recessive loss-of-function mutations are used to assess the in vivo activities of patterning genes. The principal argument for iab-4:Ubx interactions in development rests with the analysis of dominant gain-of-function phenotypes arising from the misexpression of miR-iab-4 in the haltere imaginal disks (see Figs. 3, 4). The specificity of the resulting haltere-to-wing homeotic transformation correlates with reduced levels of Ubx protein accumulation specifically in the regions where miR-iab-4 products are misexpressed. It is likely that there is redundancy in the transcriptional repression of Ubx by Abd-A and Abd-B products, and the inhibition of Ubx protein synthesis by iab-4 miRNAs. Indeed, noncoding genes in the BX-C were mainly identified by dominant mutations such as chromosomal rearrangements (e.g., Lewis 1978). Therefore, misexpression assays may prove effective in analyzing the function of other Hox noncoding genes.
This study provides evidence that iab-4 encodes a novel homeotic regulatory activity, which functions, at least in part, by producing miRNAs inhibiting Ubx. iab-4 miRNAs may regulate additional target mRNAs. For example, computational analyses identify homothorax as another potential target of interest (Grun et al. 2005). Homothorax works in parallel with the Hox cofactor Extradenticle and various Hox proteins to control the patterning of legs and antennae (for review, see Mann and Morata 2000). It is also possible that downstream transcriptional targets of Ubx (“realizators”) might be modulated by iab-4 miRNAs. Additional noncoding RNAs in the BX-C, such as cbx, pbx and bxd (e.g., Lipshitz et al. 1987), might also possess homeotic regulatory activities and account for the remaining genes identified by Lewis (1978).
Materials and methods
Immunohistochemistry and in situ hybridization
Embryo fixation, probe labeling, and nascent transcript RNA FISH were performed according to Kosman et al. (2004) with the following modifications. The sequence of the iab-4 probes were previously described (Bae et al. 2002). RNA probes labeled with FITC-, DIG-, and Biotin-conjugated UTP were detected as follows: DIG, sheep α DIG (Roche); Biotin, mouse α Biotin (Roche); FITC, rabbit α FITC (Molecular Probes). The secondary detection was as follows: donkey α sheep Alexa Fluor 555 (Molecular Probes), donkey α mouse Alexa Fluor 488 (Molecular Probes), and chicken α rabbit Alexa Fluor 647 (Molecular Probes). Ubx protein was detected using mouse α-Ubx (FP3.83) (White and Wilcox 1984). For detection of GFP and Ubx, we fixed discs in 4% paraformaldehyde and probed them with rabbit α-GFP (1:1000; Molecular Probes) or mouse α-Ubx FP3.83 (1:10).
Drosophila strains
The UAS-miR-iab4 construct was generated by cloning a 400-bp fragment centered around the mir-iab-4 hairpin downstream of pUAST-DSred2 (Stark et al. 2003). The Ubx sensor was cloned by inserting genomic DNA including the Ubx 3′ UTR and 400 bp of downstream genomic DNA into tub-GFP (Brennecke et al. 2003). These constructs were injected into w1118 embryos, and multiple transgenic lines were established. Other Drosophila lines were obtained from the Bloomington Stock Center, including scalloped-GAL4, bxMS1096-GAL4, and various Ubx and Pc alleles.
Acknowledgments
We acknowledge Nipam Patel for antibodies, David Bilder for fly stocks, and Gerald Rubin for his support during this project. We also thank Robert Zinzen and Kimberly Mace for discussions and helpful comments on the manuscript, and Oliver Zill who provided important help during the initial phases of this work. This study was funded by grants from the NIH (GM34431 to M.L. and GM72395 to M.R.), and grants from the Leukemia and Lymphoma Society (LLS #3096-05) and the Burroughs Wellcome Foundation (CABS 1004721) to E.C.L.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1372505.
Corresponding authors.
References
- Aravin A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5**:** 337-350. [DOI] [PubMed] [Google Scholar]
- Bae E., Calhoun, V.C., Levine, M., Lewis, E.B., and Drewell, R.A. 2002. Characterization of the intergenic RNA profile at Abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 99**:** 16847-16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S., Mihaly, J., Galloni, M., Hagstrom, K., Muller, M., Shanower, G., Schedl, P., Gyurkovics, H., and Karch, F. 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127**:** 779-790. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116**:** 281-297. [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37**:** 766-770. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H., and Cuppen, E. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120**:** 21-24. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113**:** 25-36. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark, A., Russell, R.B., and Cohen, S.M. 2005. Principles of microRNA-target recognition. PLoS Biol. 3**:** e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.H., Oh, C.T., Kim, S.H., Kim, Y.T., and Jeon, S.H. 2000. Effects of Polycomb group mutations on the expression of Ultrabithorax in the Drosophila visceral mesoderm. Mol. Cells 10**:** 156-161. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Zaratzian, A., and Sakonju, S. 1990. Characterization of two RNAs transcribed from the _cis_-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 87**:** 3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell R.A., Bae, E., Burr, J., and Lewis, E.B. 2002. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 99**:** 16853-16858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. and Dolle, P. 1989. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8**:** 1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. 2003. MicroRNA targets in Drosophila. Genome Biol. 5**:** R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Wang, Y.L., Langenberger, D., Gunsalus, K.C., and Rajewsky, N. 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1**:** e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F., Bender, W., and Weiffenbach, B. 1990. abdA expression in Drosophila embryos. Genes & Dev. 4**:** 1573-1587. [DOI] [PubMed] [Google Scholar]
- Karch F., Galloni, M., Sipos, L., Gausz, J., Gyurkovics, H., and Schedl, P. 1994. Mcp and Fab-7: Molecular analysis of putative boundaries of _cis_-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22**:** 3138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D., Mizutani, C.M., Lemons, D., Cox, W.G., McGinnis, W., and Bier, E. 2004. Multiplex detection of RNA expression in Drosophila embryos. Science 305**:** 846. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294**:** 853-858. [DOI] [PubMed] [Google Scholar]
- Lai E.C. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30**:** 363-364. [DOI] [PubMed] [Google Scholar]
- ____. 2003. microRNAs: Runts of the genome assert themselves. Curr. Biol. 13**:** R925-R936. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Tam, B., and Rubin, G.M. 2005. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes & Dev. 19**:** 1067-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B. 1978. A gene complex controlling segmentation in Drosophila. Nature 276**:** 565-570. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120**:** 15-20. [DOI] [PubMed] [Google Scholar]
- Lipshitz H.D., Peattie, D.A., and Hogness, D.S. 1987. Novel transcripts from the ultrabithorax domain of the bithorax complex. Genes & Dev. 1**:** 307-322. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Patel, N.H., and Kreitman, M. 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: Rules governing conservation and change. Development 125**:** 949-958. [DOI] [PubMed] [Google Scholar]
- Mann R.S. and Morata, G. 2000. The developmental and molecular biology of genes that subdivide the body of Drosophila. Ann. Rev. Cell Dev. Biol. 16**:** 243-271. [DOI] [PubMed] [Google Scholar]
- Mansfield J.H., Harfe, B.D., Nissen, R., Obenauer, J., Srineel, J., Chaudhuri, A., Farzan-Kashani, R., Zuker, M., Pasquinelli, A.E., Ruvkun, G., et al. 2004. MicroRNA-responsive `sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 36**:** 1033-1034. [DOI] [PubMed] [Google Scholar]
- Martin C.H., Mayeda, C.A., Davis, C.A., Ericsson, C.L., Knafels, J.D., Mathog, D.R., Celniker, S.E., Lewis, E.B., and Palazzolo, M.J. 1995. Complete sequence of the bithorax complex of Drosophila. Proc. Natl. Acad. Sci. 92**:** 8398-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. 1997. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 13**,** 314-318. [DOI] [PubMed] [Google Scholar]
- Roch F. and Akam, M. 2000. Ultrabithorax and the control of cell morphology in Drosophila halteres. Development 127**:** 97-107. [DOI] [PubMed] [Google Scholar]
- Rougeulle C. and Avner, P. 2004. The role of antisense transcription in the regulation of X-inactivation. Curr. Top. Dev. Biol. 63**:** 61-89. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E., Vernos, I., Marco, R., and Morata, G. 1985. Genetic organization of Drosophila bithorax complex. Nature 313**:** 108-113. [DOI] [PubMed] [Google Scholar]
- Shermoen A.W. and O'Farrell, P.H. 1991. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67**:** 303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke, J., Russell, R.B., and Cohen, S.M. 2003. Identification of Drosophila MicroRNA targets. PLoS Biol. 1**:** E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer A., Amemiya, C.T., Kim, C.B., and Stadler, P.F. 2005. Evolution of microRNAs located within Hox gene clusters. J. Exp. Zoolog. B Mol. Dev. Evol. 304**:** 75-85. [DOI] [PubMed] [Google Scholar]
- Weatherbee S.D., Halder, G., Kim, J., Hudson, A., and Carroll, S. 1998. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes & Dev. 12**:** 1474-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.A. and Wilcox, M. 1984. Protein products of the bithorax complex in Drosophila. Cell 39**:** 163-171. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu, J., Kulbokas, E.J., Golub, T.R., Mootha, V., Lindblad-Toh, K., Lander, E.S., and Kellis, M. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434**:** 338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih, I.H., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304**:** 594-596. [DOI] [PubMed] [Google Scholar]
- Zhou J., Barolo, S., Szymanski, P., and Levine, M. 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes & Dev. 10**:** 3195-3201. [DOI] [PubMed] [Google Scholar]