Requirement of TRAP/Mediator for Both Activator-Independent and Activator-Dependent Transcription in Conjunction with TFIID-Associated TAFIIs (original) (raw)
Abstract
The multiprotein human TRAP/Mediator complex, which is phylogenetically related to the yeast SRB/Mediator coactivator, facilitates activation through a wide variety of transcriptional activators. However, it remains unclear how TRAP/Mediator functions in the context of other coactivators. Here we have identified a previously uncharacterized integral subunit (TRAP25) of the complex that is apparently metazoan specific. An antibody that is specific for TRAP25 allowed quantitative immunodepletion of essentially all TRAP/Mediator components from HeLa nuclear extract, without detectably affecting levels of RNA polymerase II and corresponding general transcription factors. Surprisingly, the TRAP/Mediator-depleted nuclear extract displayed severely reduced levels of both basal and activator-dependent transcription from DNA templates. Both activities were efficiently restored upon readdition of purified TRAP/Mediator. Moreover, restoration of basal and activator-dependent transcription to extracts that were simultaneously depleted of TRAP/Mediator and TFIID (TBP plus the major TAFIIs) required addition of both TBP and associated TAFIIs, as well as TRAP/Mediator. These observations indicate that TAFIIs and Mediator are jointly required for both basal and activated transcription in the context of a more physiological complement of nuclear proteins. We propose a close mechanistic linkage between these components that most likely operates at the level of combined effects on the general transcription machinery and, in addition, a direct role for Mediator in relaying activation signals to this machinery.
Transcriptional regulation of eukaryotic protein-encoding genes requires the concerted function of distinct classes of factors (reviewed in reference 40). These include (i) RNA polymerase II (Pol II) and its associated general transcription factors (GTFs) (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), which are sufficient for low levels of accurately initiated transcription from DNA templates in vitro (generally referred to as basal transcription), (ii) site-specific DNA-binding regulatory factors, and (iii) transcriptional cofactors, which mediate responsiveness of Pol II and GTFs to various activators and repressors. Cofactors (coactivators or corepressors) have been operationally defined as factors that, while exhibiting no intrinsic site-specific DNA binding, preferentially influence the function of DNA binding regulatory factors without affecting basal (regulatory factor-independent) transcription.
Early biochemical studies employing metazoan systems reconstituted with partially purified factors pointed to TATA box-binding protein (TBP)-associated factors (TAFIIs) in TFIID (7, 16, 37) and positive cofactors (PC1, PC2, PC3, and PC4) within the USA fraction (30) as coactivators required for the function of diverse activators on DNA templates (reviewed in references 4, 40, and 45). Parallel studies with Saccharomyces cerevisiae revealed the existence of a coactivator or Mediator (8) that interacts with Pol II to form a Pol II holoenzyme (19, 44) and that consists of genetically identified SRB and transcriptional regulatory proteins, as well as novel Mediator subunits (reviewed in references 23 and 32). Subsequently, and underscoring the evolutionary conservation of transcriptional mechanisms, several metazoan Mediator-like multiprotein complexes were identified (reviewed in reference 28). These include TRAP (9), SMCC (13), ARC (33), DRIP (38), NAT (43), murine Mediator (18), human Mediator (2), CRSP (41) and the USA-derived and TRAP-related complex PC2 (27). Except for minor differences in composition, the metazoan complexes are rather similar and likely reflect either the same cellular entity (Mediator) or derived or modified forms (28).
The TRAP and SMCC complexes, for which equivalence has been rigorously demonstrated (13, 17), consist of circa 25 polypeptides that include the phylogenetically conserved subunits RGR1, SRB10, MED7, MED6, SRB11, SRB7, SOH1, and NUT2, as well as subunits that are only found in the metazoan complex. The latter include, but are not limited to, the various TRAP polypeptides originally reported as part of the thyroid hormone receptor-associated TRAP complex (28). The identities of several additional subunits that appear to be specifically associated with TRAP/Mediator, especially those clustered in the 20- to 25-kDa range, remain unknown.
The isolation of distinct coactivators has raised the issue of whether they act in a functionally redundant or in a synergistic manner, or indeed whether they reflect alternative activation pathways. In the case of USA-derived positive cofactors, previous studies demonstrated functional synergism between unfractionated USA and TAFIIs (6), between PC4 and TAFIIs (11), and between the Mediator-like PC2 and PC4 and PC3 (27). However, the relative contributions of Mediator components and TFIID-associated TAFIIs to activated transcription have remained unclear. Consistent with earlier biochemical studies in yeast systems, where efficient Mediator-dependent activation was observed in transcription assays that contained TBP in place of intact TFIID (19), genetic analysis revealed that at least a subset of TFIID-specific TAFIIs were dispensable, when individually depleted, for general activator function in yeast cells (reviewed in reference 12). Furthermore, in several cases where a strong TAFII dependence was seen in these genetic studies, the TAFII requirement was found to be dictated by core promoter sequences (42). Consistent with these observations, TBP was found to support efficient function of the model activator Gal4-VP16 both in an immunodepleted HeLa nuclear extract lacking TFIID-specific TAFIIs (34) and in a purified cell-free system from HeLa cells (46), as well as the function of the thyroid hormone receptor (TR)-TRAP complex in a purified cell-free system (10). In contrast, and in agreement with the earlier studies indicating TAFII requirements for optimal function of various activators in partially purified metazoan cell-free systems (reviewed in reference 45), a recent study showed a strict TAFII requirement for the function of a different activator (Sp1) in an assay system reconstituted with purified human Pol II, GTFs, and Mediator-like CRSP (41). Moreover, another recent study showed that yTAFII40, a specific component of yeast TFIID, is essential for most transcription in vivo, which strongly suggests that some TAFIIs may be required as general cofactors and not just as core promoter recognition factors (20).
As part of our ongoing structural and functional characterization of TRAP/Mediator, we report here a novel bona fide subunit of this complex (designated TRAP25). We have exploited the ability of antibodies directed against this subunit to efficiently deplete TRAP/Mediator from nuclear extract to explore broader TRAP/Mediator functions and to revisit issues of coactivator redundancy versus coactivator synergy in the context of a more physiological complement of nuclear proteins. Our results indicate that in this system, which may also contain a number of negative cofactors (reviewed in reference 25), TRAP/Mediator and TFIID-associated TAFIIs act synergistically to support the function of various activators via diverse core promoter sequences. We further find, consistent with a report that appeared while this article was in preparation (31), that TRAP/Mediator is also needed for residual levels of activator-independent (basal) transcription in this system.
MATERIALS AND METHODS
Cloning of TRAP25 cDNA and generation of specific antibody.
Human expressed sequence tag (EST) clone AI806623 was obtained from Research Genetics and completely sequenced. This EST clone contained the complete open reading frame for TRAP25. The full-length TRAP25 cDNA was inserted into 6His-pET11d vector by PCR. Recombinant TRAP25 expressed in Escherichia coli was insoluble. The protein was solubilized in denaturing buffer (6 M guanidine HCl, 20 mM Tris [pH 7.9], and 500 mM NaCl) and purified by nickel-nitrilotriacetic acid-agarose column chromatography. The purified protein was separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) and visualized by using an ice-chilled 0.25 M KCl solution. The protein band was cut out for antibody production (Covance Research Products Inc.).
TRAP/Mediator purification.
Affinity purification of TRAP/Mediator from nuclear extract was performed essentially as previously described (13). Briefly, 5-ml aliquots of nuclear extracts prepared from cell lines expressing FLAG-tagged CDK8 (13), FLAG-tagged NUT2 (27), or FLAG-tagged TR (9) were adjusted to 300 mM KCl and 0.5% NP-40 and incubated with 100 μl of M2-agarose beads (IBI/Kodak) at 4°C for 6 h with rotation. After four 5-ml washes with BC300-0.1% NP-40, proteins were eluted from beads by incubation at 4°C for 30 min with 100 μl of BC100-0.05% NP-40 plus 0.2 mg of FLAG peptide/ml.
Highly purified TRAP/Mediator was obtained from nuclear extract prepared from cell lines expressing FLAG-tagged NUT2 (27). Briefly, after precipitation with 33% ammonium sulfate, the resuspended protein was loaded on a phosphocellulose P11 column at BC400. Protein fractions eluting between 0.5 and 0.85 M KCl were collected and dialyzed to BC300. Finally, affinity purification of TRAP/Mediator on M2 agarose was performed as described above.
In vitro transcription.
Transcription reactions were carried out in a final volume of 25 μl and contained 50 ng of each supercoiled plasmid DNA template, 2 or 5 μl of HeLa nuclear extract (at ∼10 mg/ml), 20 mM HEPES (pH 8.2), 11 to 16% glycerol, 4 mM MgCl2, 60 mM KCl, 8 mM dithiothreitol, 0.5 mM ATP and CTP, 5 μM UTP, 0.1 mM 3′-_O_-methyl-GTP, 16 μCi (0.6 MBq) of [α-32P]UTP, 0.4 mg of bovine serum albumin per ml, and 20 U of RNasin (Promega). Reaction mixtures were incubated at 30°C for 1 h, at which time the UTP concentration was increased to 25 μM and 15 U of RNase T1 was added. After 30 min of incubation at 30°C, the reactions were extracted with phenol-chloroform in the presence of 150 μl of stop solution (0.4 mM sodium acetate [pH 5.2], 13 mM EDTA, 0.33% SDS, and 0.67 mg of yeast tRNA per ml). The aqueous layer was precipitated by ethanol and analyzed by gel electrophoresis followed by autoradiography. Correctly initiated transcripts were quantitated by PhosphorImager analysis with a STORM 840 system (Molecular Dynamics).
Immunodepletion from nuclear extract.
Since recombinant His-tagged TRAP25 was totally insoluble in E. coli, the His-tagged TRAP25 was purified with a nickel-nitrilotriacetic acid-agarose column under denaturing conditions (6 M guanidine HCl, 0.05 M NaCl, 20 mM HEPES [pH 7.6], 100 mM EDTA). The purified His-tagged TRAP25 was cross-linked under the above conditions with CNBr-activated Sepharose 4B (Pharmacia) that had been pretreated with 1 mM HCl. After mixing for 2 h at room temperature, the cross-linking reaction was blocked by adding Tris buffer (pH 8.0) to a final concentration of 0.1 M. The resin was washed three times with 0.1 M sodium acetate (pH 4.0)-0.5 M NaCl and three times with equilibration buffer (0.1 M Tris [pH 8.0], 0.5 M NaCl) before use.
For immunodepletion of TRAP/Mediator, anti-TRAP25 antisera were purified by passage through a column containing His-tagged TRAP25 protein, which was covalently cross-linked to CNBr-activated Sepharose 4B (Pharmacia). Bound anti-TRAP25 antibodies were eluted with 200 mM glycine (pH 2.5). The antibodies were further purified and concentrated by binding to protein A-Sepharose (Pharmacia) and then cross-linked to protein A with dimethylpimelidate (Sigma). One milliliter of HeLa nuclear extract in BC100 was passed four times over a 1-ml cross-linked anti-TRAP25-protein A-Sepharose column. The flowthrough fractions were used for Western blotting and in vitro transcription.
TBP and TAFII depletions were carried out in BC500, and depleted extracts were dialyzed in BC100 essentially as described (34).
MS.
Protein sequencing using mass spectrometry (MS) and tandem MS (MS/MS) was carried out as described previously (35), with the exception that 18O-labeled H2O was omitted. Briefly, the Coomassie blue-stained protein band was in-gel digested with trypsin and the recovered peptides were analyzed using an electrospray ion trap mass spectrometer (LCQ; Finnigan MAT, San Jose, Calif.) coupled on-line with a capillary high-pressure liquid chromatograph (LC) (C18 Magic MS column, Magic 2002; Michrom BioResources, Auburn, Calif.). The flow was split with a Magic precolumn capillary splitter assembly (Michrom BioResources). The LC/MS was programmed to run in a data-dependent fashion, in which the mass spectrometer switched to the MS/MS mode to acquire collision-induced dissociation spectra, once an ion signal was detected to exceed a preset value in the MS mode during the entire LC run. Data derived from the collision-induced dissociation spectrum were used to search a compiled protein database that was composed of the nonredundant protein database and a six-reading-frame translated EST database to identify the protein.
RESULTS
Molecular cloning of a full-length cDNA encoding TRAP25.
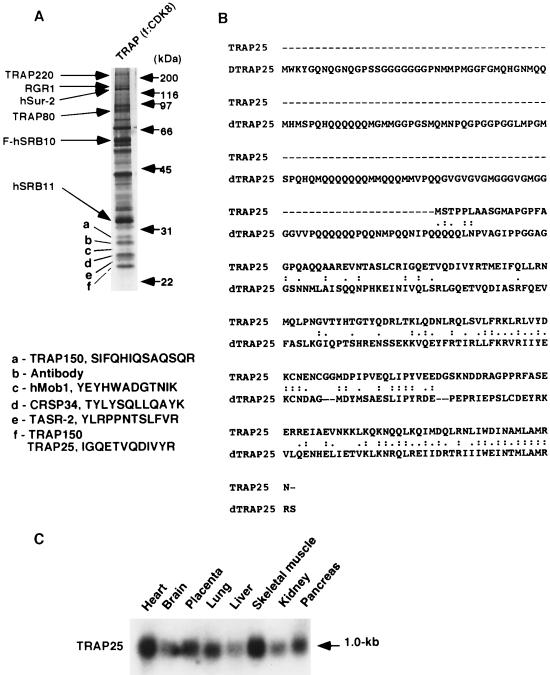
Our earlier analysis (27) of the polypeptide composition of the TRAP/Mediator complex revealed, in addition to the subunits whose sequences were previously identified (13, 17), two or three polypeptides in the 20- to 25-kDa range that coeluted with the complex regardless of whether it was purified from a FLAG-NUT2- or a FLAG-SRB10/CDK8-expressing cell line (13, 27). These bands were also consistently observed in PC2 preparations that had been chromatographed over several columns (27), strongly suggesting that they represent authentic TRAP/Mediator subunits. To determine the identities of these bands, we electrophoretically resolved a TRAP/Mediator preparation that was affinity purified from the FLAG-SRB10/CDK8-expressing cell line (13) (Fig. 1A). The bands (a to f) corresponding to these polypeptides were excised and subjected to mass-spectrometric sequence analysis.
FIG. 1.
Isolation and cDNA cloning of TRAP25. (A) Analysis of affinity-purified (M2-agarose) TRAP/Mediator from an f:CDK8 cell line. Samples were analyzed by SDS-11.5% PAGE and stained with silver. Bands (a to f) in the 25-kDa range were excised for mass-spectrometric sequence analysis. Peptide sequences thus obtained and corresponding proteins are indicated. (B) Amino acid sequence of TRAP25. Amino acid sequences that were identified by MS are underlined. An amino acid sequence comparison of human (TRAP25) and Drosophila TRAP25 (dTRAP25) homologue is also shown. The alignment was performed with MacVector software, and identities (double dots) and similarities (single dots) are indicated. (C) Northern analysis of a multiple-tissue mRNA blot hybridized with a TRAP25 cDNA probe showing expression of TRAP25 in human tissues.
Bands a and f appear, on the basis of derived sequences, to be proteolytic degradation products of the previously reported TRAP150α. Band d contained sequences from a polypeptide that was reported both in PC2 (referred to as p37 in reference 27) and in CRSP (referred to as CRSP34 in reference 41). Band b corresponded to immunoglobulin light chain derived from the affinity resin. Additional sequences not previously found in Mediator preparations were obtained from bands c, e, and f. The sequence YEYHWADGTNIK in band c identified ESTs (AI393039 and AI557507) that encode the human ortholog of yeast Mob1, which has been linked to the CCR4-NOT complex (21). However, several lines of evidence (data not shown) suggest that Mob1 is not an authentic, stoichiometric component of TRAP/Mediator. The sequence YLRPPNTSLFVR in band e identified ESTs (AF047448 and AF067730) that encode TASR-2 (translocated in liposarcoma-associated serine-arginine protein 2). TASR-2 may be involved in RNA splicing via the Pol II C-terminal domain (15, 47). TASR-2 was not further characterized but, like Mob1, is unlikely to be an authentic TRAP/Mediator subunit. However, it remains possible that these proteins are representative of a variety of cellular components (other than the basic transcription apparatus) with which TRAP/Mediator normally interacts.
Band f yielded the additional sequence IGQETVQDIVYR, which identified an EST (AI806623) that encodes a hitherto-uncharacterized protein (termed TRAP25 based on evidence presented below) with a predicted molecular weight of 20 kDa (Fig. 1B). A Drosophila TRAP25 ortholog (CG17183 gene product), which exhibited localized identity of 35% and similarity of 53% with human TRAP25 (Fig. 1B), was found in the database. S. cerevisiae and Caenorhabditis elegans do not appear to possess an equivalent protein, potentially implicating this subunit in signaling processes restricted to higher metazoans. A Northern blot analysis (Fig. 1C) indicated ubiquitous expression of a 1-kb TRAP25 mRNA, although elevated levels were seen in heart, placenta, and skeletal muscle. This pattern is qualitatively similar to that previously reported for other TRAP/Mediator subunits (17).
TRAP25 is a bona fide component of TRAP/Mediator.
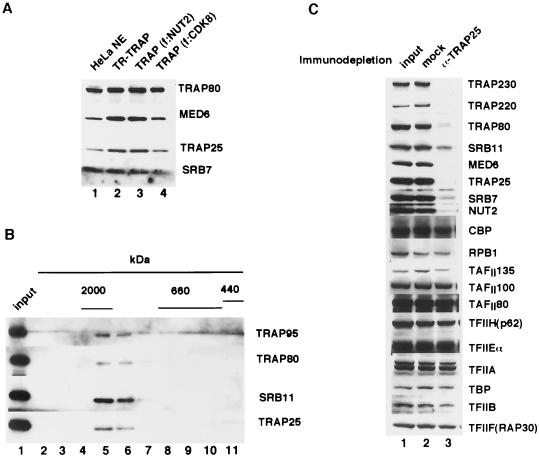
Toward establishing that TRAP25 is a bona fide subunit of TRAP/Mediator, we first expressed this protein in bacteria and raised antisera against the purified recombinant protein. Immunoblot analysis (Fig. 2A) with the TRAP25-specific antiserum revealed the presence of TRAP25 in all preparations of TRAP/Mediator regardless of how they were obtained, either through association with FLAG-TR or through intrinsic FLAG-tagged subunits (NUT2 or SRB10/CDK8). Furthermore, the amount of TRAP25 relative to other authentic subunits (TRAP80, MED6, and SRB7) was the same for the purified complexes and for unfractionated nuclear extract, strongly suggesting that TRAP25 is a stoichiometric component of TRAP/Mediator. To further demonstrate that TRAP25 is stably associated with TRAP/Mediator complex, TRAP/Mediator purified from epitope-tagged SRB10/CDK8 cells was subjected to gel filtration chromatography on Superose 6 (Fig. 2B). Consistent with previous reports (13), immunoblot analysis of the resulting fractions with antibodies against representative subunits revealed that the majority of the TRAP/Mediator eluted as a 1.8-MDa complex. Importantly, the bulk of the complex coeluted with TRAP25 (Fig. 2B, lanes 5 and 6). This result provides additional strong evidence for the presence of TRAP25 in the TRAP/Mediator complex.
FIG. 2.
TRAP25 is a bona fide component of TRAP/Mediator. (A) Complexes from the indicated cell lines (f:TR, lane 2) were analyzed by immunoblotting with anti-TRAP25 antibodies. Representative TRAP/Mediator subunits were also probed, as indicated. Total HeLa cell nuclear extract was also included (lane 1) as a control. (B) TRAP/Mediator purified from the f:CDK8 cell line was chromatographed (in 300 mM KCl) over a Superose 6 gel filtration column. Fractions were immunoblotted with the indicated antibodies. The elution range of molecular mass markers is indicated at the top. (C) An anti-TRAP25 antibody depletes TRAP/Mediator from HeLa nuclear extract. HeLa nuclear extract (input) was passed over a control column containing immobilized preimmune antibodies (mock) or over a column containing anti-TRAP25. Unbound extract was immunoblotted with the indicated antibodies.
In order to prove definitively that TRAP25 is an integral component of TRAP/Mediator, we used our TRAP25 antiserum to show that depletion of TRAP25 from nuclear extract simultaneously resulted in depletion of other TRAP/Mediator subunits (Fig. 2C). As expected, immunoblot analysis of extract that had been passed through an anti-TRAP25 antibody column revealed that TRAP25 was quantitatively removed by this treatment (compare lanes 1 and 2 with lane 3). Furthermore, as predicted, the majority of TRAP/Mediator subunits were also efficiently depleted. Exceptions included SRB11/cyclin C, which was only partially depleted; this likely reflects its presence not only in TRAP/Mediator but also in a kinase-cyclin pair with SRB10/CDK8 that can exist in an unassociated form (13, 27). We therefore conclude that TRAP25 is a bona fide TRAP/Mediator subunit.
We also examined if the TRAP25 antibody depleted Pol II and its associated GTFs. Immunoblotting analysis showed negligible reductions in the amounts of Pol II, TFIIA, TFIIB, TFIID, TFIIH, TFIIE, and TFIIF in the supernatant (Fig. 2C), despite the fact that this depletion experiment was performed under mild salt conditions (125 mM KCl). Therefore, a majority of TRAP/Mediator appears not to be tightly associated with Pol II and other GTFs, consistent with previous results (13, 43). However, as minor differences would not be evident from the present analysis, it remains possible that a subpopulation of TRAP/Mediator complexes is associated with Pol II (S. Malik and R. G. Roeder, unpublished results).
Function of TRAP/Mediator in nuclear extract.
Our previous functional analyses of TRAP/Mediator (13, 17, 27) employed an in vitro assay system that was reconstituted from highly purified Pol II, GTFs, and the USA-derived positive cofactor, PC4. The Mediator-depleted extract described above now provided an opportunity to assess the function of TRAP/Mediator in a less purified system that presumably contains a more normal complement of both positively and negatively acting nuclear factors.
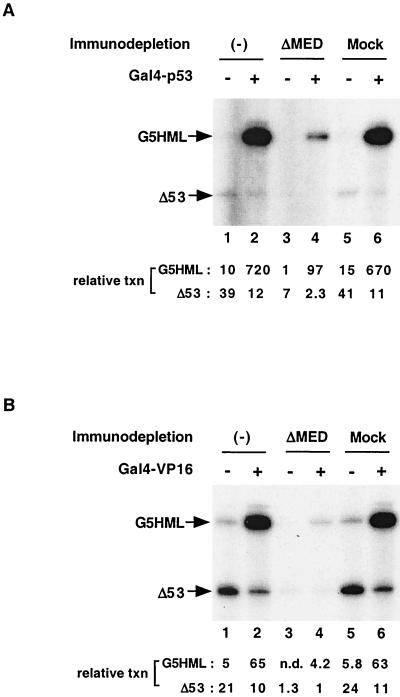
We thus tested the Mediator-depleted extract for its ability to support activation by GAL4-p53 (Fig. 3A) and GAL4-VP16 (Fig. 3B), which consist of the DNA binding domain of the yeast activator GAL4 fused, respectively, to the activation domains of the tumor suppressor p53 and the viral activator VP16. Standard transcription assays contained two templates. The G5HML plasmid has five copies of a GAL4 binding site upstream of a hybrid core promoter and permits us to assay GAL4-fused activation domains. The MLΔ53 plasmid contains only the core promoter sequences (−53 to +10) from the adenovirus major late promoter and allows us to monitor basal (activator-independent) transcription. The G5HML is thus a weaker core promoter and yields lower levels (circa fourfold) of basal activity relative to the MLΔ53 promoter (Fig. 3, lanes 1) (46). Under our conditions, GAL4-p53 (Fig. 3A, lanes 1 and 2) activated transcription 72-fold and GAL-VP16 (Fig. 3B, lanes 1 and 2) activated transcription 13-fold in untreated extracts. By contrast, in the Mediator-depleted extract (ΔMED) (Fig. 3) the absolute levels of activated transcription were dramatically reduced for both GAL4-p53 (ca. 7.5-fold) (Fig. 3A, lane 2 versus lane 4) and GAL4-VP16 (ca. 15-fold) (Fig. 3B, lane 2 versus lane 4). In a control extract, which had been passed over a column containing preimmune antiserum, the levels of activated transcription were essentially unaltered (Fig. 3, lanes 2 versus lanes 6). We conclude that depletion of Mediator from nuclear extract severely compromises its ability to support high levels of activated transcription.
FIG. 3.
Depletion of TRAP/Mediator from nuclear extract reduces basal and activated transcription. (A) Transcription activation by Gal4-p53 is suppressed in the anti-TRAP25 antibody-depleted nuclear extract. In vitro transcription reaction mixtures contained 50 μg of each nuclear extract (lanes 1 and 2, untreated; lanes 3 and 4, anti-TRAP25 antibody-depleted [ΔMED]; lanes 5 and 6, mock depleted) and 50 ng each of pMLΔ53 (Δ53) and pG5HML (G5HML) templates. Relative transcription levels, determined by phosphorimager analysis, are indicated. (B) In vitro transcription reactions were performed as for panel A except that Gal4-VP16 was used as an activator.
We also noted that the level of basal transcription, as monitored from both the MLΔ53 template and the G5HML template in the absence of added activators, was reduced 6- to 18-fold following depletion of Mediator (compare lanes 1, 3, and 5 in Fig. 3A and, especially, 3B). Thus, despite the low absolute levels, the level of activation appears to remain unchanged in the presence and absence of Mediator. While this residual activation might possibly be attributed to an alternative mechanism (involving a distinct coactivator activity), the underlying reason is currently unclear. It may arise as well from trace amounts of TRAP/Mediator that are not detected by immunoblot analysis but whose effects would be evident in the more sensitive transcription assay. The slight reduction in transcription from the MLΔ53 template upon activator addition (e.g., lane 1 versus lane 2) is normal and has previously been attributed to competition between the two templates for limiting factors.
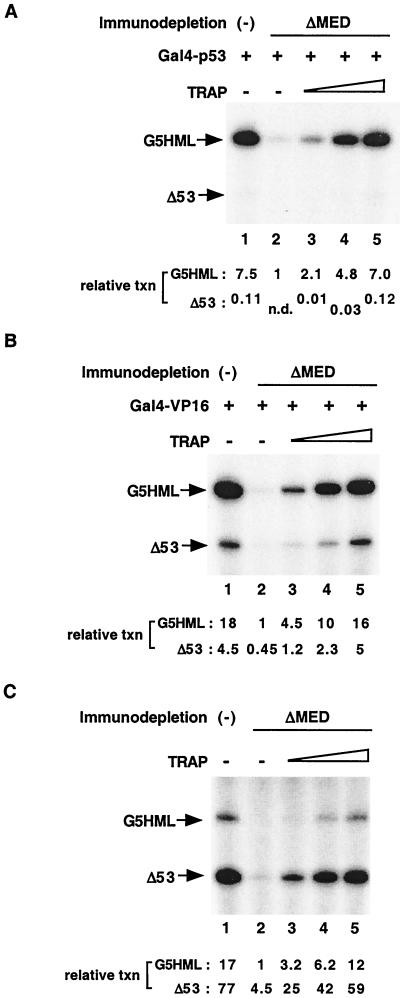
We next asked if purified TRAP/Mediator could restore the ability of Mediator-depleted extract to support activation by GAL4-p53 and GAL4-VP16. For this purpose, TRAP/Mediator that had been affinity-purified from nuclear extract from our epitope-tagged NUT2 cell-line, and which additionally had been fractionated over phosphocellulose P11 (see Materials and Methods), was added back to transcription reactions performed with the depleted extract (Fig. 4A and B). In this experiment, TRAP/Mediator was added over a concentration range that, as determined by immunoblot analysis (data not shown), either was just under (Fig. 4A and B, lanes 3 and 4) or actually approximated (lane 5) the concentration of Mediator in standard nuclear extracts.
FIG. 4.
Restoration of activated and basal transcription by exogenously added purified TRAP/Mediator. (A) Increasing amounts of purified TRAP/Mediator (lanes 3 to 5) were added to anti-TRAP25 antibody-depleted nuclear extract (ΔMED) in in vitro transcription reactions. Reaction mixtures were as for Fig. 3 except that they contained 20 μg of each nuclear extract. As determined by immunoblotting, the amount of TRAP/Mediator added to the reactions corresponded to approximately 20% (lane 3), 70% (lane 4), or 100% of the TRAP/Mediator concentration in untreated nuclear extract. GAL4-p53 was used as the activator in all reactions. Lane 1 shows the product of a control reaction with untreated extract. (B and C) In vitro transcription reactions were performed as for panel A except that GAL4-VP16 was used as the activator (B) or no activator was added (C).
Upon addition of increasing amounts of TRAP/Mediator to Mediator-depleted extract, in which activation by GAL-p53 (Fig. 4A, lane 2) and GAL-VP16 (Fig. 4B, lane 2) was virtually undetectable, activated transcription was enhanced in an essentially linear dose-dependent fashion (lanes 3 to 5). Indeed, for both GAL4-p53 and GAL4-VP16, full recovery of activity was observed when the normal concentration of Mediator in nuclear extracts was reached (compare lanes 5 and 1 in Fig. 4A and B). These results confirm that diminution of coactivator activity following depletion of Mediator is due solely to its absence and not to depletion of associated factors.
We also observed that basal transcription from the MLΔ53 template was fully restored upon complementation of the depleted extract with TRAP/Mediator. This is especially evident in Fig. 4B (compare lane 5 and lane 1). Nonetheless, to ensure that these Mediator effects on basal transcription are entirely independent of activators, we repeated the depletion-and-complementation experiment in the absence of any exogenous activators (Fig. 4C). Once again, basal transcription activity from both G5HML and MLΔ53 templates was both severely reduced in Mediator-depleted extract (Fig. 4C, lane 2) and fully restored by complementation with purified TRAP/Mediator (lane 5 versus lane 1). Together, these results demonstrate that, in nuclear extract, Mediator plays a dominant role in regulating both activator-dependent and activator-independent transcription.
Joint requirement of TRAP/Mediator and TAFIIs in TFIID for activated transcription.
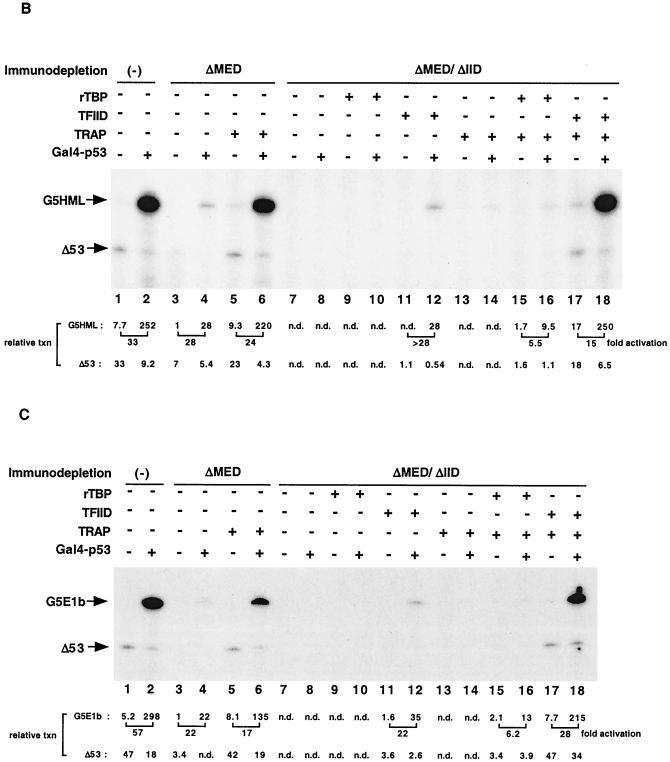
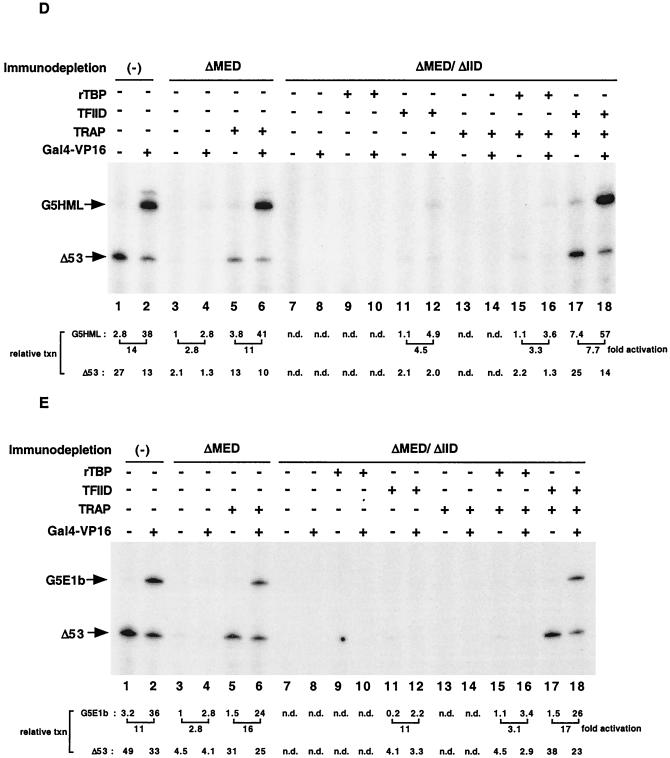
We also made use of our ability to deplete Mediator from nuclear extracts to address the outstanding question of whether the coactivator activities of Mediator and TAFIIs within TFIID are functionally redundant or synergistic. We therefore prepared HeLa nuclear extract lacking both TFIID and Mediator by sequential passages over columns containing immobilized antibodies against TBP and TAFII100 (to deplete TFIID) (34) and against TRAP25 (to deplete Mediator). As judged by immunoblot analysis of representative TFIID subunits (TBP, TAFII135, and TAFII100), and consistent with previous results (34), the extract (ΔMED/ΔIID) was efficiently depleted of TFIID (Fig. 5A, lane 2 versus lane 1). Similarly, the extract was simultaneously depleted of Mediator. Importantly, none of the other GTFs tested showed any significant decrease. Our strategy was to supplement these extracts with Mediator and either TBP or intact TFIID, either singly or in combination. Thus, together with an extract (ΔMED) that was selectively depleted of only Mediator (see above) (Fig. 5A, lane 3), this provided a system for assessing the relative contribution of TFIID-associated TAFIIs and TRAP/Mediator in transcription. These assays were performed both with the above-described G5HML template (Fig. 5B and D) and with a template (G5E1b) in which the GAL4 sites are located upstream of the adenovirus E1b core promoter, which contains a relatively weak TATA box (Fig. 5C and E).
FIG. 5.
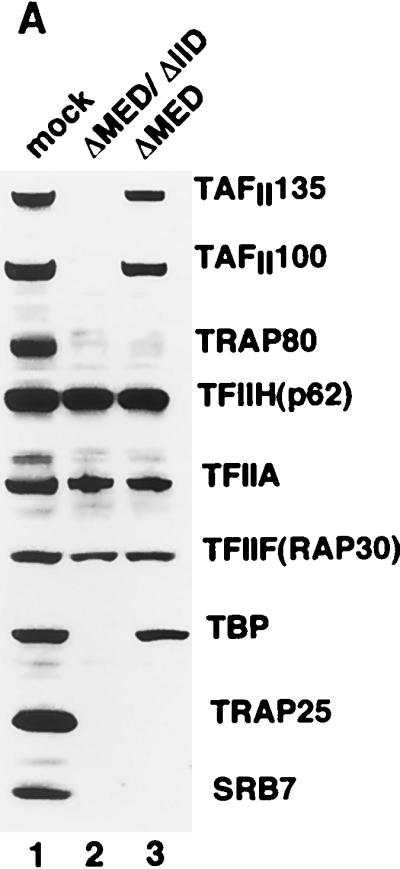
Synergistic effect of TRAP/Mediator and TFIID-associated TAFIIs on transcription activation in vitro. (A) Immunoblot analysis of TFIID- and TRAP/Mediator-depleted HeLa nuclear extract (Material and Methods) with the indicated antibodies. Lane 1, mock-depleted extract; lane 2, nuclear extract depleted with anti-TBP, anti-TAFII100, and anti-TRAP25 antibodies (ΔMED/ΔIID); lane 3, anti-TRAP25-depleted nuclear extract (ΔMED). Each lane was loaded with 20 μg of each nuclear extract and analyzed on a 4-to-20% gradient SDS-PAGE gel, followed by immunoblotting. (B and C) In vitro transcription reaction mixtures contained the indicated nuclear extract plus 25 ng each of pMLΔ53 and either pG5HML (B) or pG5E1b (C) in a 12.5-μl reaction volume. GAL4-p53 (50 ng), recombinant TBP (rTBP, 3 ng), affinity-purified TFIID containing an equivalent molar amount of TBP (as determined by immunoblotting), and an amount of TRAP/Mediator corresponding to that in standard nuclear extract were added where indicated. (D and E) In vitro transcription reactions were performed as for panels B and C, respectively, except that Gal4-VP16 (50 ng) was the activator.
As expected, ΔMED/ΔIID extract failed to support both basal and activated transcription by GAL-p53 and GAL-VP16 (Fig. 5B to E, lanes 7 and 8 versus lanes 1 and 2). Furthermore, whereas the activated transcription in ΔMED extracts could be readily recovered upon supplementation with purified TRAP/Mediator in this experiment (Fig. 5B to E, lanes 6 versus lanes 2 and 4), consistent with earlier data (Fig. 4A and B), this was not sufficient to restore activity to the ΔMED/ΔIID extract (Fig. 5B to E, lanes 14 versus lanes 2 and 8). We also added affinity-purified TFIID to the ΔMED/ΔIID extract in amounts that were empirically determined to support baseline levels (in the absence of exogenous Mediator) of transcription equivalent to those seen with the ΔMED extract (Fig. 5B to E, compare lanes 12 with lanes 4). This resulted in the addition of an amount of TFIID whose TBP content was only about a fifth of that in standard extract, which is to be expected in view of the fact that TBP is shared among several nuclear multiprotein complexes. Under these conditions, addition of an equivalent molar amount of TBP had negligible effects (Fig. 5B to E, compare lanes 9 and 10 with lanes 7 and 8). When the ΔMED/ΔIID extract was supplemented with both TFIID and TRAP/Mediator, the resulting transcription levels for both GAL-p53 (Fig. 5B and C) and GAL-VP16 (Fig. 5D and E) were virtually indistinguishable from those observed with control extract (lanes 18 versus lanes 2).
In sharp contrast, when the ΔMED/ΔIID extract was supplemented with TBP and TRAP/Mediator, the absolute transcription levels remained low (less than 5% of that with intact TFIID) (Fig. 5B and E, lanes 16 versus lanes 18). Similarly, although TBP could support low-level (circa three- to sixfold activation) activated transcription (Fig. 5B to E, lanes 15 and 16 versus lanes 17 and 18) in the presence of TRAP/Mediator, the results clearly show that TAFIIs contribute an additional potential that allows a further 5-fold enhancement (8- to 28-fold activation in the presence of TFIID and TRAP/Mediator). At the same time, because basal levels are also elevated significantly as a result of the combined effects of TFIID and TRAP/Mediator (Fig. 5B to E, lanes 17 versus lanes 7) but much less so with TBP and TRAP/Mediator (lanes 15 versus lanes 7), the final extent of transcription is significantly higher in the former situation. We note, however, that the TBP- and TFIID-nucleated basal transcription in this system is critically dependent on the amount of exogenously added factor. Thus, the levels of basal transcription reconstituted with f:TFIID and TRAP/Mediator were similar to those with rTBP and TRAP/Mediator when a fivefold excess of exogenous f:TFIID and rTBP was added to the reaction mixtures (data not shown).
As indicated above, these assays were performed with templates containing either combined TATA and initiator elements (G5HML) or a relatively weak TATA (G5E1b) element within the core promoter. The fact that the two templates showed qualitatively similar results indicates that, at least with respect to the limited number of core promoters tested, the observed synergy between TFIID-specific TAFIIs and Mediator is not a function of core promoter sequences.
DISCUSSION
The main conclusions of this work are (i) that TRAP25 is a novel, integral subunit of human Mediator, (ii) that Mediator is required for both basal (activator-independent) and activated transcription in the context of nuclear extract, and (iii) that Mediator and TAFIIs within TFIID function synergistically in both basal and activated transcription in this system.
Role of metazoan Mediator in basal transcription.
Initial characterization of the yeast Mediator complex had pointed to three distinct associated biochemical activities. It was found to play a critical role as a coactivator (the function most closely identified with the Mediator) and to stimulate basal transcription activity in reaction mixtures reconstituted with Pol II and GTFs, leading to the hypothesis that the two activities may be interdependent (19). Functions in both basal and activated transcription were also evident from analyses of extracts from yeast with a deletion mutation in the SRB5 subunit of Mediator (44). In vitro studies also showed that Mediator can stimulate the Pol II carboxy-terminal repeat domain kinase activity of TFIIH (19). Although our earlier functional analysis of the various human Mediator complexes (13, 17) clearly revealed their coactivator potential for a wide range of activators (including the thyroid hormone receptor in conjunction with the retinoid X receptor, GAL4-p53, and GAL4-VP16), the effects on activator-independent transcription were not as marked. Our previous reconstituted transcription system relied on an additional coactivator, PC4, that, in addition to promoting activated transcription, strongly suppresses basal transcription (11, 26). Similarly, in experiments in which ARC (33) and DRIP (38) coactivator effects were seen only in the context of chromatin templates, basal transcription may have been suppressed (either by histones or by negative cofactors in the crude chromatin assembly extracts) to such an extent that it was refractory to stimulation by Mediator alone. By contrast, the unfractionated nuclear extract-based assays employed here and by Mittler et al. (31) have helped uncover a prominent basal function of human Mediator. By thus extending our initial characterization, these findings now suggest that, consistent with the phylogenetic conservation of some subunits, the fundamental mechanisms by which the Mediator functions may also be conserved from yeast to humans.
It is likely that basal effects of Mediator are readily observed in an unfractionated nuclear extract because transcription in such a system is subject to a much broader range of both positively and negatively acting cofactors than exist in the purified reconstituted systems. Thus, it may be that net transcription, both basal and activated, in less purified systems entails a major antirepression component and that one function of Mediator is to help overcome restrictions imposed by negatively acting cofactors. These cofactors are presumably distinct from histones and PC4 but could include cofactors such as NC2 (reviewed in reference 25). It perhaps is relevant that in pure systems, basal transcription (in the absence of PC4) is relatively high, providing another possible reason why basal effects of Mediator have been undetected (26).
Alternative, but not mutually exclusive, mechanisms by which Mediator could promote basal transcription involve direct interactions with the basal transcription machinery. Indeed, in yeast, genetic interactions of SOH1 with Pol II and TFIIB have been described (reviewed in reference 5). However, it should be noted that while SOH1 is an integral subunit of the human Mediator (13; Malik and Roeder, unpublished), it has not been documented as a yeast Mediator subunit. Furthermore, in vitro recruitment experiments have revealed that the yeast holoenzyme (consisting minimally of Pol II and Mediator) is dependent on both TBP and TFIIB for incorporation into the preinitiation complex formation (39). It is conceivable, therefore, that the Mediator could directly stabilize the preinitiation complex. A more dynamic effect of the Mediator on basal transcription also might be exerted via its ability to modulate TFIIH activities (19). Although it has been proposed that the SRB10/CDK8 kinase subunit of the human Mediator down-regulates TFIIH activity by phosphorylating its cyclin H subunit (1), the yeast Mediator preparation that was shown to stimulate TFIIH carboxy-terminal repeat domain kinase activity lacked the SRB10 kinase (19).
Synergistic transcriptional activation by Mediator and TAFIIs.
Our results also have revealed that, in unfractionated systems, Mediator and TAFIIs (in TFIID) functionally synergize to give high levels of activated transcription that result, albeit only in part, from favorable TAFII- and Mediator-dependent basal effects discussed above. Although at apparent variance (but see below) with earlier results (34) indicating that the TBP subunit of TFIID could support activated transcription in a similar unfractionated system in the absence of associated TAFIIs, the present experiments have clearly served to define experimental conditions in which functional synergy between Mediator and TFIID-specific TAFIIs can be readily observed. Our use of near-physiological concentrations of exogenously added factors in depleted extracts may account in part for the present results. However, it also remains possible that our particular protocol (depletion of both TFIID and Mediator and subsequent complementation with highly purified preparations of Mediator and TFIID) results in conditions that favor multiple rounds of transcription and thus render the system TAFII dependent (34). The combined depletion of TFIID and Mediator may also result in depletion of components that facilitate TAFII-independent transcription and that are not present in the highly purified TFIID and Mediator preparations. Nonetheless, the synergistic function of TFIID and Mediator is in accord with the coactivator properties ascribed to each (9, 13, 45) and with recently published data showing synergistic effects of TFIID and Mediator in highly purified reconstituted systems (14, 27, 41).
While the role of TFIID-specific TAFIIs as conventional coactivators remains somewhat controversial, a consensus is emerging that they may play major roles in core promoter selectivity both in yeast and in metazoans (reviewed in reference 12). This is especially evident from studies showing an absolute requirement for metazoan TAFIIs in basal transcription from promoters that contain divergent core promoter elements, including initiators and downstream promoter elements (DPEs), but often no TATA element (3, 22, 29). TAFIIs are also required for functional synergy between TATA and initiators or DPEs, indicating promoter-selective functions on composite core promoters (29). Preliminary indications are that, mechanistically, this may involve direct interactions with core promoter sequences, especially in the case of promoters made up of divergent (non-TATA) promoter elements (3). The recently published results of Park et al. (36), who examined the relative contribution of TAFIIs and Mediator in activation of several Drosophila promoters, are also in agreement with the hypothesis that TFIID-associated TAFIIs are primarily involved in core promoter effects. These authors observed that a TATA-box-containing promoter, Adh, could be activated by GAL4-VP16 (through upstream GAL4 sites) with only TBP (in the absence of TAFIIs) and Mediator. However, activation through the engrailed promoter, which lacks a TATA box but contains a DPE, required intact TFIID and Mediator.
In view of the above results and given that this study, in contrast to an earlier report (34), utilized relatively weak core promoters (the adenovirus E1b promoter and the synthetic HML promoter that contains the TATA box from the human immunodeficiency virus promoter and the initiator element from adenovirus ML promoter), it is likely that the TAFII dependence observed here has its basis in how the transcription apparatus senses the promoter sequences. Together with the newly uncovered basal transcription effect of the Mediator reported here and by Mittler et al. (31), it appears that the abilities of human TFIID and Mediator complexes, originally thought only to mediate activator signals to the basal apparatus, to directly modulate the basal machinery and to process activation signals are inextricably conjoined. This implies that one mechanism by which these coactivators function is to “present” the basal machinery in a form that is now responsive to activators. For TFIID (including TBP and TAFIIs) this may take the form of appropriately positioning Pol II and GTFs (i.e., in promoter recognition); for Mediator, this could entail any of the mechanisms discussed above. Interestingly, by this view, and on the basis of the results presented here, Mediator would appear to be functionally indistinguishable from any of the GTFs. However, because it is likely the key component that contacts activators (2, 9, 17, 33, 38) and because the level of activation in the presence of Mediator is significantly higher than what would be expected if it were simply contributing as a basal factor, Mediator is also clearly functioning as a coactivator in the conventional sense. Therefore, the activation process must additionally involve a Mediator-dependent signal transduction event. This notion is also supported by studies indicating that mutations in yeast Mediator subunits may alter function without noticeably affecting interactions of Mediator with activator and Pol II (24). Clearly, future studies on a wide variety of naturally occurring combinations of core promoters and upstream activating sequences, together with detailed mutagenesis of the Mediator components, will be required to further substantiate and elaborate these concepts.
Acknowledgments
We thank Y. K. Kang for a purified TRAP (Flag-NUT2) preparation.
This work was supported by NIH grant CA42567 to R.G.R.
REFERENCES
- 1.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407**:**102-106. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, T. G., M. E. D. Martin, E. Lees, R. P. Ricciard, and A. P. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399**:**276-279. [DOI] [PubMed] [Google Scholar]
- 3.Burke, T. W., and J. T. Kadonaga. 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10**:**711-724. [DOI] [PubMed] [Google Scholar]
- 4.Burley, S. K., and R. G. Roeder. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65**:**769-799. [DOI] [PubMed] [Google Scholar]
- 5.Chang, M., and J. A. Jaehning. 1997. A multiplicity of mediators: alternative forms of transcription cofactors communicate with transcriptional regulators. Nucleic Acids Res. 25**:**4861-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang, C. M., H. Ge, Z. Wang, A. Hoffmann, and R. G. Roeder. 1993. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 12**:**2749-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dynlacht, B. D., T. Hoey, and R. Tjian. 1991. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66**:**563-576. [DOI] [PubMed] [Google Scholar]
- 8.Flanagan, P. M., R. J. Kelleher III, M. H. Sayre, H. Tschochner, and R. D. Kornberg. 1991. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350**:**436-438. [DOI] [PubMed] [Google Scholar]
- 9.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93**:**8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fondell, J. D., M. Guermah, S. Malik, and R. G. Roeder. 1999. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl. Acad. Sci. USA 96**:**1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge, H., and R. G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78**:**513-523. [DOI] [PubMed] [Google Scholar]
- 12.Green, M. R. 2000. TBP-associated factors (TAFs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25**:**59-63. [DOI] [PubMed] [Google Scholar]
- 13.Gu, W., S. Malik, M. Ito, C. X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3**:**97-108. [DOI] [PubMed] [Google Scholar]
- 14.Guermah, M., Y. Tao, and R. G. Roeder. 2001. Positive and negative TAFII functions that suggest a dynamic TFIID structure and elicit synergy with TRAPs in activator-induced transcription. Mol. Cell. Biol. 21**:**6882-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14**:**1415-1429. [PubMed] [Google Scholar]
- 16.Hoffmann, A., E. Sinn, T. Yamamoto, J. Wang, A. Roy, M. Horikoshi, and R. G. Roeder. 1990. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature 346**:**387-390. [DOI] [PubMed] [Google Scholar]
- 17.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3**:**1-20. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95**:**8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77**:**599-608. [DOI] [PubMed] [Google Scholar]
- 20.Komarnitsky, P. B., B. Michel, and S. Buratowski. 1999. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 13**:**2484-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komarnitsky, S. I., Y. C. Chiang, F. C. Luca, J. Chen, J. H. Toyn, M. Winey, L. H. Johnson, and C. L. Denis. 1998. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 18**:**2100-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutach, A. K., and J. T. Kadonaga. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20**:**4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34**:**77-137. [DOI] [PubMed] [Google Scholar]
- 24.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y. J. Kim. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19**:**2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maldonado, E., M. Hampsey, and D. Reinberg. 1999. Repression: targeting the heart of the matter. Cell 99**:**455-458. [DOI] [PubMed] [Google Scholar]
- 26.Malik, S., M. Guermah, and R. G. Roeder. 1998. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc. Natl. Acad. Sci. USA 95**:**2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5**:**753-760. [DOI] [PubMed] [Google Scholar]
- 28.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. 25**:**277-283. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, E., C. M. Chiang, H. Ge, and R. G. Roeder. 1994. TAFs in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 13**:**3115-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meisterernst, M., A. L. Roy, H. M. Lieu, and R. G. Roeder. 1991. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell 66**:**981-993. [DOI] [PubMed] [Google Scholar]
- 31.Mittler, G., E. Kremmer, H. T. M. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2**:**808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69**:**729-749. [DOI] [PubMed] [Google Scholar]
- 33.Näär, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398**:**828-832. [DOI] [PubMed] [Google Scholar]
- 34.Oelgeschläger, T., Y. Tao, Y. K. Kang, and R. G. Roeder. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFs. Mol. Cell 1**:**925-931. [DOI] [PubMed] [Google Scholar]
- 35.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94**:**35-44. [DOI] [PubMed] [Google Scholar]
- 36.Park, J. M., B. S. Gim, J. M. Kim, J. H. Yoon, H. S. Kim, J. G. Kang, and Y. J. Kim. 2001. Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol. Cell. Biol. 21**:**2312-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pugh, B. F., and R. Tjian. 1990. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell 29**:**1187-1197. [DOI] [PubMed] [Google Scholar]
- 38.Rachez, C., H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398**:**824-828. [DOI] [PubMed] [Google Scholar]
- 39.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13**:**49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roeder, R. G. 1998. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp. Quant. Biol. 63**:**201-218. [DOI] [PubMed] [Google Scholar]
- 41.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397**:**446-450. [DOI] [PubMed] [Google Scholar]
- 42.Shen, W. C., and M. R. Green. 1997. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90**:**615-624. [DOI] [PubMed] [Google Scholar]
- 43.Sun, X., Y. Zhang, H. Cho, R. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2**:**213-222. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, C. M., A. J. Koleske, D. M. Chao, and R. A. Young. 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73**:**1361-1375. [DOI] [PubMed] [Google Scholar]
- 45.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21**:**338-341. [PubMed] [Google Scholar]
- 46.Wu, S. Y., E. Kershnar, and C. M. Chiang. 1998. TAFII-independent activation mediated by human TBP in the presence of positive cofactor PC4. EMBO J. 17**:**4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, L., L. J. Embree, S. Tsai, and D. D. Hickstein. 1998. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 273**:**27761-27764. [DOI] [PubMed] [Google Scholar]