Regulation of Alternative Splicing by the ATP-Dependent DEAD-Box RNA Helicase p72 (original) (raw)
Abstract
Although a number of ATP-dependent RNA helicases are important for constitutive RNA splicing, no helicases have been implicated in alternative RNA splicing. Here, we show that the abundant DEAD-box RNA helicase p72, but not its close relative p68, affects the splicing of alternative exons containing AC-rich exon enhancer elements. The effect of p72 was tested by using mini-genes that undergo different types of alternative splicing. When the concentration of p72 was increased in transient transfections, the inclusion of enhancer-containing CD44 alternative exons v4 and v5 increased using a mini-gene that contained these exons and their flanking introns inserted into a β-globin gene. Other types of alternative splicing were not impacted by altering p72 concentrations. Mutation of the p72 helicase ATP-binding site or deletion of the carboxy-terminal region of the protein reduced the ability of the transfected protein to affect CD44 variable exon splicing. Use of in vitro extracts overexpressing p72 indicated that p72 becomes associated with complexes containing precursor RNA. Helicases have been implicated both in altering RNA-RNA interactions and in remodeling RNA-protein complexes. CD44 exon v4 contains a potential internal secondary structure element that base pairs the 5′ splice site with a region inside the exon located between enhancer elements. Mutations that destroyed this complementarity modestly increased inclusion in the absence of p72 but still responded to increasing p72 concentration like the wild-type exon, suggesting that p72 might have effects on protein-RNA interactions. In agreement with this hypothesis, p72 was not able to restore the inclusion of an exon mutated for its major enhancer element. Our results suggest that RNA helicases may be important alternative splicing regulatory factors.
Pre-mRNA splicing is a complicated process that requires the participation of a number of RNA-binding proteins in RNA recognition and assembly of the active spliceosome. One interesting and abundant class of proteins associated with the splicing process from yeast to humans is the family of ATP-dependent RNA helicases (2, 4, 11, 22, 28, 35, 36, 40, 44). Multiple helicases have been implicated in the correct recognition of RNA during spliceosome formation, rearrangement of the spliceosome, and regeneration of snRNPs between rounds of splicing. The helicases involved in splicing contain a highly conserved set of sequence domains, including an ATP-binding site, a DEAD box, an RNA-binding domain, a domain important for conformational changes, and several other domains still unassigned as to function. In addition, each helicase is characterized by unique N- and C-terminal domains thought to distinguish the functions of individual helicases and provide protein-protein interactions.
A number of the splicing helicases are common between vertebrates and yeast. One common helicase found in vertebrates is p68 (12, 13, 14), recently shown to be part of an in vitro-reconstituted spliceosome (30), although its role in spliceosome assembly or function has not been defined. A close relative of p68 is p72 (22) (Fig. 1). The latter protein has a unique N-terminal domain containing repeats of the sequence RGG, a sequence feature common to hnRNP proteins (17, 21), and a C-terminal domain rich in serine and glycine and terminating with a polyproline region, which is also characteristic of other RNA-binding proteins important for splicing (1, 8, 9, 18, 33, 38, 39, 45). The function of p72 is unknown at present, but p72 and p68 have been recently reported to bind an RNA transcriptional cofactor (steroid receptor RNA activator [SRA]) and functionally coactivate estrogen receptor α (43).
FIG. 1.
Comparison of the human p68 and p72 ATP-dependent DEAD-box RNA helicases. Identical amino acids are shaded in black. The DEAD box is boxed, conserved sequences common to the ATP-dependent helicases are underlined, and RGG sequences unique to p72 are overlined. Alignment was provided by ALIGN and BOXSHADE.
ATP-dependent helicases use the binding of ATP to increase the affinity and specificity of binding of the helicase to RNA (25). Recent experiments have indicated that helicases may also play an important role in modifying protein-RNA interactions (3, 15, 18, 24). This latter property raises the intriguing possibility that helicases may play a role in alternative RNA processing via the ATP-dependent regulation of the binding of exon-specific alternative processing factors (24, 35, 44). Here, we present evidence that the human RNA helicase p72, but not p68, affects alternative processing of certain alternative exons in the human CD44 gene. This effect requires the ATP-binding domain within the helicase but does not require major secondary structure features within the exon. The helicase was also demonstrated to be stably associated with a CD44 precursor RNA containing the affected alternative exon in an in vitro splicing extract. Other alternative processing decisions were not affected by p72. Our results suggest that ATP-dependent helicases may be important proteins for the recognition of alternative exons by splicing machinery.
MATERIALS AND METHODS
Plasmids and transfections.
cDNA clones coding for p72 and mutant K142R cloned into pcDNA3.1/Flag for in vivo expression were kindly provided by S. Kato (University of Tokyo, Tokyo, Japan). All clones contained a Flag tag at the N terminus. Clones giving rise to mutated or truncated p72 proteins were engineered by site-directed mutagenesis and sequence verified. Production of p72 proteins following transfections was monitored by Western blotting with an anti-Flag antibody (anti-Flag M2 monoclonal antibody; Sigma Chemical Co.). A cDNA clone isolated from a human breast cancer cell line (MCF-7) and coding for p68 was obtained from H. Endoh (University of Tokyo). The cDNA was cloned into an N-terminal His- and myc-tagged pcDNA3 vector and used for transfection experiments. The clone was sequenced, and protein expression was checked with a purified monoclonal anti-mouse antibody directed against the His tag (anti-His-horseradish peroxidase antibody, catalog no. R931-25; Invitrogen). The Y-box protein (YB-1) cDNA clone has been described previously (41). The reporter CD44 (41), α-globin (29), adenovirus (29), and calcitonin/calcitonin gene-related peptide (CT/CGRP) (26, 27) mini-genes used in this study have all been described previously.
For transfections, we used HeLa cells on 10-cm-diameter plates in the presence of Lipofectamine (Gibco/BRL) according to the manufacturer's instructions. Total cell RNA was isolated 48 h posttransfection with TRIzol (Gibco/BRL) following the manufacturer's instructions. Splicing patterns were evaluated by reverse transcription (RT)-PCR using low-cycle PCR (15 to 20 cycles) determined previously to provide accurate estimations of relative levels of spliced product RNAs. CD44-spliced RNA product analysis was performed with 5′-end-radiolabeled primers specific for β-globin sequences in the flanking exons (5′ AGACACCATGCATGGTGCACC and 3′ CCTGATCAGCGAGCTCTAG). These primers amplified no RNA from untransfected HeLa cells. Amplification conditions were 30 s at 94°C, 30 s at 62°C and 1 min at 72°C. Product DNA was denatured and displayed on 5 to 6% urea gels. Spliced adenovirus and α-globin product RNAs were assayed with pcDNA3 5′ and pcDNA3 3′ oligonucleotide primers, which are complementary to pcDNA3 vector sequences that flank the inserted α-globin and adenovirus sequences, as previously described (29). Detection of alternatively spliced and polyadenylated RNAs from the CT/CGRP gene entailed the use of mixed primers specific for alternative exons 4 and 5 coupled with a primer specific for the common upstream exon as described previously (26). RNA products from all amplifications were quantified in a phosphorimager and expressed as the percentage of total product RNA in each species. Plasmid pBR322 digested with _Hpa_II was used in all displayed gels for product size determination.
In vitro analysis.
The in vitro transcription templates used to generate RNAs were produced by cloning downstream of the T7 or SP6 promoter. Capped 32P-labeled in vitro RNA transcripts were prepared from linearized templates as described previously (26). An in vitro version of exon v4 contained sequences from −38 to +56 of the exon. The control RNA was a region of the human α-globin second intron containing G-rich elements. Nuclear extracts were prepared from transfected cells to provide an extract enriched in p72 as previously described (26). After 48 h of transfection, cells were harvested and nuclear splicing extract was prepared by standard techniques. Uniformly labeled substrates were produced following previously described protocols (34). Immunoprecipitation of substrate RNA associated with p72 was carried out as follows. Anti-Flag M2-agarose affinity gel (Sigma)-conjugated beads were washed five times with NET solution (50 mM Tris [pH 7.5], 150 mM NaCl, 0.05% NP-40). Labeled, in vitro-transcribed RNA was incubated with mock- or p72-transfected HeLa splicing nuclear extract for 10 min. The reactions were mixed with the conjugated beads and 400 μl of NET solution and incubated for 2 h at 4°C with shaking. The beads were washed five times with NET solution and digested with proteinase K for 20 min at 55°C. Released RNA was displayed on a 5% urea denaturing gel.
RESULTS
Increasing the concentration of the p72 RNA helicase activates inclusion of CD44 alternative exons v4 and v5.
The observed cofractionation of human p68 with an in vitro-reconstituted spliceosome (30) suggested that this helicase and its relatives might be the first class of helicases to examine for possible effects on alternative processing. Our attention was drawn to p72 because of suggestions that it might be associated with RNAs that bind YB-1 (43; D. Auboeuf and B. W. O'Malley, unpublished observation). It was recently demonstrated that YB-1 binds an alternatively spliced RNA containing an AC-rich splicing enhancer sequence (41). YB-1 also binds a transcriptional coactivator RNA (SRA) which contains AC-rich sequences (Auboeuf and O'Malley, unpublished). The helicase p72 has recently been reported to bind SRA (43). Therefore, it seemed possible that p72 could bind and affect the alternative splicing of AC-rich exons.
The AC-rich exon that we studied is the fourth alternative exon from the human CD44 gene. The human CD44 gene undergoes extensive alternative RNA processing (10, 37). At least 10 alternative exons (termed variable exons and numbered v1 through v10) exist in a block within the central portion of the gene (Fig. 2A, diagram 1). To study CD44 alternative splicing, we used a mini-gene containing the region of the CD44 gene encompassing alternative exons v4 and v5 and the natural intron sequences around and between them inserted into the human β-globin gene (Fig. 2A, diagram 2). This mini-gene produces RNA in which 20 to 30% of the product includes both exons v4 and v5 (Fig. 2B, lane 1). Inclusion of exon v4 is dependent upon an AC-rich exon enhancer within exon v4 (Fig. 2A, diagram 3) that binds YB-1. Increasing the concentration of YB-1 in a cotransfection experiment increases the inclusion of exon v4 in product RNA from 25 to 85% (41).
FIG. 2.
Increasing the in vivo concentration of p72 RNA helicase increases inclusion of CD44 variable exons v4 and v5. (A) CD44 alternative splicing. In diagram 1, the exon structure of the human CD44 gene is depicted. Constitutive exons are shaded black, and alternative exons (termed variable exons) are shaded gray. In diagram 2, the employed CD44 mini-gene in which CD44 variable exons v4 and v5 and their surrounding intron sequences have been inserted into a β-globin mini-gene driven from the CMV promoter is depicted. Globin exons are shaded white, and CD44 exons are shaded gray. In diagram 3, the sequence of CD44 exon v4 is shown. Exon sequences are capitalized; intron sequences are lowercase. The AC-rich exon enhancer (ACE) is underlined. (B) Transfection of p72 or p68. The mini-gene shown in panel A was cotransfected into HeLa cells with 0, 1, 2, or 4 μg of an expression plasmid coding for full-length human p72 (lanes 1 through 4) or human p68 (lanes 5 through 8). RNA splicing patterns were characterized by RT-PCR amplification of total cell RNA with primers specific for the exons flanking the CD44 variable exons. Bands corresponding to the inclusion of no CD44 exons (the skip product), one CD44 variable exon (exons v4 and v5 are of almost identical length), or both CD44 variable exons are indicated by the shading in of the boxes between the gels. The identity of each band was confirmed by sequencing of PCR products. Numbers below the gels and lane numbers indicate the percentage of product RNA containing both exons v4 and v5 as determined by scanning of gels in a phosphorimager. wt, wild type.
In an initial set of experiments, the CD44 mini-gene depicted in Fig. 2A was cotransfected into HeLa cells with increasing concentrations of an expression plasmid coding for p72 (Fig. 2B, lanes 1 through 4). Expression of higher levels of p72 increased the inclusion of both exons v4 and v5 from 27 to 71% (Fig. 2B). The inclusion of both CD44 exons was increased, resulting in greater production of a spliced product RNA that contained both exons v4 and v5. This effect was similar to that previously observed when concentrations of the RNA-binding protein YB-1 were increased (41). To see if there was some specificity for a particular helicase in the activation, a similar experiment was performed using the closely related human p68 instead of p72. In this case, however, no activation of splicing of the CD44 alternative exons was observed (Fig. 2B, lanes 5 through 8) despite the production of equal amounts of p68- and p72-transfected proteins (data not shown), suggesting differences in the ability of the two closely related helicases to activate variable exon recognition in early spliceosome assembly.
An RNA helicase could theoretically affect multiple steps in RNA biogenesis. To control for any potential effects on RNA stability, we compared the ability of p72 to affect alternative splicing to its ability to change the level of expression of mini-genes containing cDNAs corresponding to the two observed products of splicing from the CD44 mini-gene (data not shown). Cotransfecting the p72 expression plasmid with cDNA mini-genes containing either both or neither CD44 variable exons produced uniform RNA signals regardless of the level of expression of p72, suggesting that increasing the levels of p72 neither stabilized nor destabilized one of the two products from the alternative processing decision. This result suggests that the effect of p72 is on the level of splicing of the CD44 mini-gene RNA.
Activation of CD44 variable splicing requires multiple domains within the p72 helicase protein.
ATP-dependent RNA helicases have multiple conserved domains required for various aspects of activity (2). To gain further insight into the action of p72 on CD44 alternative splicing, we created and assayed mutant versions of p72. Multiple mutations were created and are diagrammed in Fig. 3A. Each of the mutant versions contained a Flag tag to facilitate the detection of protein following transfection. Each of the expression plasmids containing mutated p72 sequences, including the extensive deletions, directed the synthesis of protein as demonstrated by the appearance of a protein of the correct molecular weight following transfection, as detected by the anti-Flag antibody (Fig. 3C and data not shown). Furthermore, fractionation of transfected cells into nuclear and cytoplasmic fractions indicated that transfected proteins were present in the nuclear fraction (data not shown), presumably because the N-terminal domain thought to be responsible for nuclear transport was retained in all constructs.
FIG. 3.
Maximum stimulation of CD44 alternative splicing required multiple domains within p72. Deletion and point mutants of p72 were prepared and assayed by cotransfection with the CD44 mini-gene. (A) Diagram of the constructed mutations. Domains of p72 are indicated by shaded boxes. Point mutations are indicated by the residue number of the mutation and the alteration. p300 was a translational stop mutation introduced at amino acid 300, and p437 was a truncation mutant at amino acid 437 lacking the SG-rich C-terminal tail of the protein. All mutants contained an N-terminal Flag tag equivalent to that present in the wild-type protein. (B and D) Quantification of the ability of each mutant protein to affect the inclusion of CD44 variable exons v4 and v5. The percentage of RNA resulting from the inclusion of variable exons v4 and v5 was calculated from phosphorimager tracing of a gel of RT-PCR amplification of RNAs from cotransfections of the CD44 mini-gene and various forms of p72 as described in the legend to Fig. 2. Standard deviations from multiple experiments are indicated. The last bar indicates the level of the inclusion in the absence of cotransfected p72. wt, wild type. (C) Western blot of total cell protein following transfection with the indicated p72 genes. Detection was with the anti-Flag antibody.
Maximal activation of CD44 splicing by p72 was dependent upon the use of an expression vector coding for wild-type protein. The most deleterious mutants were those that altered the ATP-binding domain of the protein. Use of a mutant version of p72 that contained a point mutation within the ATP-binding domain previously shown to inactivate ATP binding (K142R) (2) prevented activation (Fig. 3B), suggesting that activation requires the binding and/or cleavage of ATP. A second mutation within the ATP domain (T143A [Fig. 3A and D]) was slightly less inhibitory but still significantly reduced the ability of the protein to activate exon inclusion above the level seen in cells not transfected with any isoform of p72 (Fig. 3D, far right). Two mutations, S277L and W276G, altered a SAT sequence beginning at amino acid 227. In other helicases, the SAT sequence is thought to reside in a domain necessary for a conformational switch necessary for RNA binding (2, 25, 28). These mutations eliminated approximately 50% of the ability of the helicase to activate inclusion but did not completely eliminate it (Fig. 3A and D). Similar activity was observed for a mutation that altered the DEAD box to DEAH (mutant D249H [Fig. 3A and D]). Two other mutations that produced truncated p72 proteins were created. One of these, p300, terminated the protein at amino acid 300, upstream of the conserved domains IV, V, and VI common to this class of helicases. A second mutant p72, p437, terminating at amino acid 437, contained all of the conserved domains but lacked the C-terminal region unique to individual helicases and postulated to be involved in protein-protein interactions. The deletion removing both the C-terminal domain and domains IV, V, and VI was severely inhibited for activation, noticeably more so than the mutant lacking only the C-terminal domain. This result suggests the potential importance of these domains in activation, which could reflect either the putative RNA-binding activities of these domains or contact with the ATP of domain VI. Unfortunately, we were not able to test this more directly by deleting just the domain thought to be important in RNA binding in other helicases (domain IV) because such a deletion did not result in stable protein production.
Potential for the action of p72 through RNA secondary structure.
It seemed possible that the role of p72 in CD44 exon recognition was related to relieving an inhibitory secondary structure adopted by one or both of the activated exons. We concentrated on exon v4 because of previous studies indicating the pivotal role of this exon and its sequences in the recognition of both exons v4 and v5 (41). Although the internal regions of the exon have minimal potential for secondary structure due to the prevalence of AC-rich sequences, one obvious potentially negative secondary structure exists. The 5′ splice site of exon v4 is immediately followed by G-rich sequences that could effectively base pair to a region of the exon located between the second and third AC-rich regions of the exon, thereby sequestering the 5′ splice site in a stem loop (Fig. 4A). This postulated base pairing is very similar to the inhibitory base pairing that operates in the Tau exon and represses inclusion of an alternative exon in this gene (16). To test this hypothesis, several mutations were made within the exon or within the G-rich element downstream of the 5′ splice site (Fig. 4A). These exons were tested for the ability to direct in vivo inclusion of exons v4 and v5 in the presence or absence of cotransfected p72 (Fig. 4B).
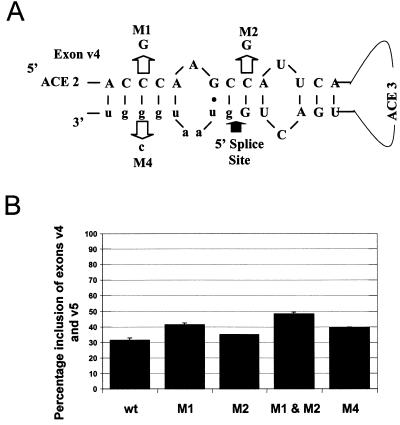
FIG. 4.
A stem loop created by base pairing of exon sequences to a region of the intron immediately downstream of the exon v4 5′ splice site may function to minimize exon inclusion. (A) Diagram of a potential stem loop at the 3′ end of exon 4. Base pairing and introduced mutations that should disrupt the stem are indicated. Exon sequences are in capitals, and intron sequences are in lowercase. Two of the AC-rich elements (ACE 2 and ACE 3) are indicated, as is the 5′ splice site. (B) Levels of inclusion of exons v4 and v5 of the mutants in the absence of cotransfecting p72. Transfections and quantification are as described in the legend to Fig. 3.
The postulated stem loop has two 4- to 5-base regions of complementarity in the stem. The 5-base region was targeted by mutation in either the exon or intron portion of the complementary region. Both mutations increased inclusion levels from 30 to 40% in the absence of cotransfecting p72. Mutation of the 4-base complementarity site increased inclusion less effectively. Mutating both regions increased inclusion levels to almost 50%, suggesting that inclusion levels are partially suppressed by secondary structure in the absence of additional helicase. All mutants responded maximally to increased p72 just like the wild-type exon (data not shown). These results suggest that secondary structure may play some role in the regulation of CD44 alternative exons. The inability of the mutants to cause constitutive inclusion of the exons as occurs with similar mutations in the Tau gene (16), however, suggests either other secondary structure features not obvious from either visual or computer analysis of the exon or a role of p72 in other aspects of exon recognition.
Activation of CD44 alternative splicing requires the exon v4 AC-rich enhancer.
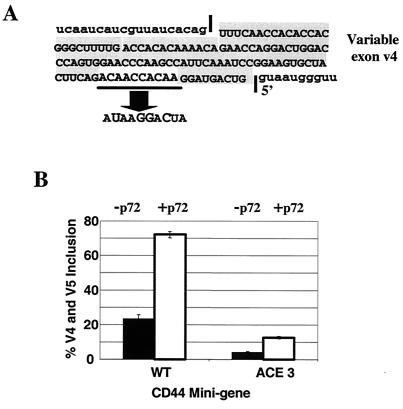
We have previously detected an AC-rich enhancer element within exon v4 required for inclusion and which binds YB-1 (41). A mutant version of the CD44 mini-gene in which the AC-rich enhancer was altered from CAACCACAA to UAAGGACUA (Fig. 5A) did not respond to increasing concentrations of p72 for the production of an RNA containing both v4 and v5 (Fig. 5B) to the extent that wild-type RNA did. Therefore, increasing the concentration of p72 could not compensate for the absence of the enhancer sequence. There was a positive effect on the ability of exon v5 to be included by using the mutant RNA (data not shown). V5 is also known to contain an enhancer sequence different from the enhancer sequence in v4 (19). The observation that p72 cannot overcome the mutation in exon v4 that binds enhancer-binding proteins but continues to enhance the inclusion of enhancer-containing v5 suggests that p72 can activate enhancer-mediated exon inclusion for CD44 v4 and v5. A low level of p72-mediated stimulation of the mutant was observed. Because the AC-rich element enhancer mutation ACE 3 should have no effect on secondary structure features, it seems likely that this residual enhancement results from a relieving of the secondary structure features within the RNA.
FIG. 5.
The effect of p72 is dependent upon a wild-type exon enhancer within CD44 variable exon v4. (A) The sequence of CD44 exon v4 including the AC-rich enhancer (ACE) and a mutant version of the enhancer (ACE 3). The wild-type and ACE 3 mutant mini-genes were cotransfected with wild-type p72, and RNA splicing phenotypes were assayed by RT-PCR as described in the legend to Fig. 2. The mutant produces RNA including exon v5 but not exon v4 (41). (B) Quantification of the ability of p72 to produce RNA that includes exons v4 and v5. Quantification was performed as described in the legend to Fig. 3.
Increasing the concentration of p72 has no impact on the splicing of other alternatively spliced RNAs.
Because helicases have been reported to be involved in multiple steps of RNA splicing, it was possible that the effect of increasing the concentration of p72 was a general effect on splicing and not a gene-specific effect. To begin to address this possibility, we assayed the ability of p72 to alter the alternative splicing of precursor RNA that typically changes its splicing properties when cotransfected with well-known splicing regulatory proteins such as SR-rich proteins or hnRNP proteins. We used four different test mini-genes to assay the ability of p72 to generically affect alternative splicing. These genes represented a spectrum of alternative splicing choices and responses to different factors. All four genes were driven by the same promoter as the CD44 mini-gene to eliminate problems that might be associated with differential transcription.
The first utilized mini-gene contained the alternatively processed region of the human CT/CGRP gene. This mini-gene directs the processing of two RNAs, one truncating at the poly(A) site of alternative exon 4, the other skipping exon 4 to terminate with exons 5 and 6. When HeLa cells are transfected with this mini-gene, approximately half of the product RNA comes from each pathway. The use of exon 4 can be stimulated by increasing the concentrations of the SR-rich proteins SRp20 or ASF/SF2 or the hnRNP protein PTB (26). Altering the concentration of p72 had no impact on the processing of this RNA (Fig. 6A), nor did altering the concentration of YB-1, the other protein able to affect CD44 splicing. CT/CGRP alternative splicing is regulated by an intron enhancer located downstream of exon 4 that resembles a pseudo mini-exon and binds U1 snRNPs, SRp20, and PTB. Both SRp20 and PTB stimulate the binding of U1 snRNPs to the enhancer. The inability of p72 to affect this processing choice suggests that the helicase does not influence the binding of U1 snRNPs to 5′ splice sites.
FIG. 6.
Increasing the concentration of p72 had minimal impact on the alternative splicing of other pre-mRNAs capable of splicing choices. Wild-type p72 was cotransfected with mini-genes containing alternative exons 4 through 6 from the human CT/CGRP genes, a constructed adenovirus gene with a first exon containing two 5′ splice sites, and a construct containing a weakened β-globin internal exon. (A) The utilized CT/CGRP gene has exons 4 through 6 of the human CT/CGRP gene and their flanking introns fused to exon 1 from the adenovirus major late transcription unit. Alternative splicing of the mini-gene results in two polyadenylated (An) products: one ending in exon 4 and one resulting from the skipping of exon 4 and inclusion of exons 5 and 6. Alternative products were detected by simultaneous RT-PCR with oligonucleotides (a, b, and c) specific for exons 4 and 5 (26). The effects of p72 were compared to those of cotransfection with the protein YB-1, which has an effect on CD44 alternative splicing (41). Product bands resulting from the usage or skipping of exon 4 are indicated. The percentage of product RNA resulting from the inclusion of exon 4 as determined in the phosphorimager is shown below the gel. (B) The diagrammed mini-gene consisting of sequences from the second exon of the major late adenovirus transcription unit was cotransfected with p72. RNA products resulting from the use of the proximal and distal 5′ splice site are indicated. (C) The diagrammed mini-gene was a derived β-globin gene containing an internally deleted internal exon previously shown to be impaired for inclusion in HeLa cells. RNA products resulting from inclusion or skipping of the middle exon are indicated.
This interpretation was bolstered by experiments with two other commonly employed alternative splicing substrates. Both of these substrates contained a first exon with duplicated 5′ splice sites. We used two such substrates, one containing a first exon and splice sites from adenovirus and a second containing sequences from human α-globin. The adenovirus substrate changes usage of the 5′ splice site in the presence of increasing concentrations of SR-rich proteins. The α-globin substrate does not respond to SR-rich proteins (29). Neither substrate responded to increasing concentrations of either p72 (Fig. 6B and data not shown) or YB-1 (data not shown), again suggesting that p72 does not affect the binding of either U1 snRNPs or typical SR-rich proteins to 5′ splice sites.
We used one last substrate that contained a weak constitutive exon derived from the second exon of the human β-globin gene, which appears to be stochastically included in RNA on the basis of the lack of strength of its splice sites, further compromised by the shortness of the exon (5, 6). Inclusion of this exon was not enhanced by altering the concentration of either p72 or p68 (Fig. 6C); in fact, increasing concentrations of both proteins slightly depressed inclusion. Other studies have indicated that the binding of U2 snRNPs to the 3′ splice site of this exon is rate determining for recognition (5, 6). The inability of p72 to influence the inclusion of this exon suggests that p72 cannot alter the binding of U2 snRNPs to generic 3′ splice sites in the absence of other factors. Taken together, the results shown in Fig. 6 suggest that p72 may be restricted to altering only selected RNA-processing events through interactions with exon-specific factors or structures.
p72 binds to complexes containing CD44 exon RNA sequences in an in vitro splicing assay.
The effects of p72 on CD44 splicing suggest that p72 might be present in splicing complexes. To see if p72 associates with splicing precursor RNAs in vitro, we asked ourselves if antibodies to p72 could immunoprecipitate a precursor RNA containing CD44 exon v4 sequences from an in vitro splicing reaction. To avoid potential problems from the cross-reactivity of antibodies against p72 caused by the high degree of conservation of helicase domains, we used an extract from cells overexpressing a Flag-tagged version of p72 and an anti-Flag antibody (Fig. 7A). This Flag-tagged antibody detects p72 in the supplemented extract but no protein species in a normal extract (data not shown).
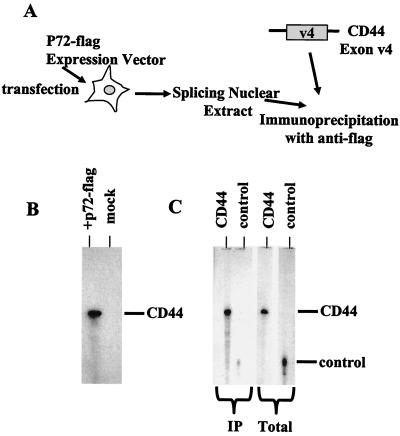
FIG. 7.
p72 associates with precursor RNA in in vitro splicing extracts. (A) HeLa cells were transfected with an expression vector for p72-Flag. Standard splicing extract was prepared from the nuclei of these cells. Radiolabeled substrate RNAs containing a portion of the CD44 exon v4, which was comprised of AC-rich sequences or a control, were added to the extract and incubated under splicing conditions for 10 min. Radiolabeled RNA was immunoprecipitated with the anti-Flag antibody. (B) Immunoprecipitation of CD44 RNA sequences from extract made from cells expressing tagged p72 versus mock-transfected cells is shown. Equal amounts of an RNA containing CD44 exon v4 sequences were added to extracts made from p72- or mock-transfected HeLa cells. After a 10-min incubation under splicing conditions, reactions were immunoprecipitated with anti-Flag antibodies and immunoprecipitated RNAs were displayed on a denaturing gel. (C) Comparison of the ability of p72 to associate with CD44 exon 4 or control RNA sequences is shown. Equal amounts of CD44 or a control RNA (Total lanes) were incubated with the extract expressing p72-Flag and immunoprecipitated (IP lanes) with anti-Flag antibodies after 10 min of incubation. Immunoprecipitated RNAs are indicated.
We used the Flag antibodies to detect RNA-protein complexes formed on RNA containing CD44 v4 sequences and compared the level of these complexes to the level formed by using a control RNA lacking any AC-rich sequences. After a 10-min incubation in splicing extract under splicing conditions, the anti-Flag antibody immunoprecipitated an RNA containing CD44 v4 sequences but was unable to immunoprecipitate equal amounts of an RNA containing the control sequences (Fig. 7B and C). These results suggest that p72 is associated in complexes formed with CD44 precursor RNA sequences and suggest a direct involvement of p72 in exon recognition during splicing.
DISCUSSION
ATP-dependent DEAD-box helicases are prominent components of the pre-mRNA splicing machinery (4, 11, 31, 40, 44). The best-characterized members of the family play roles in rearranging the spliceosome or its components and require the RNA helicase activities of the proteins to alter RNA-RNA interactions and provide fidelity in these interactions. The functions of other members of the family are less clear, although recent experiments suggest that helicases may play a significant role in mediating RNA-protein interactions during assembly and remodeling of the spliceosome (3, 15, 18, 24). p68 is an abundant, reasonably ubiquitously expressed member of the vertebrate helicase family and has been observed as a component of an in vitro-assembled spliceosome (13, 14, 30). p72 was isolated as a very similar protein (22), with approximately 90% homology in the core helicase domains to p68. In the non-core helicase domains, the two proteins are dissimilar. Both the N- and C-terminal regions of p72 contain motifs common to proteins most commonly identified as regulators of mammalian alternative splicing. In particular, the N terminus contains multiple copies of RGG, a sequence thought to bind RNA and present in a number of hnRNP proteins (17, 21). The C terminus is rich in SG repeats and ends with a polyproline sequence, also common to RNA-binding proteins (8, 20, 32, 38, 39, 45). These characteristics suggest that p72 could have a distinct function from p68 and raise interest in the possibility that helicases could play a role in alternative RNA splicing.
Here, we document that increasing the concentration of p72 increases the in vivo inclusion of two human CD44 alternative exons, v4 and v5. Both of these exons have regulatory sequences within the exon. Exon v4 has at least one enhancer sequence with an AC-rich motif that was recently demonstrated to bind the single-stranded RNA-binding protein YB-1 (41). Exon v5 contains an enhancer that responds to SR-rich proteins (19). The inclusion of both exons responded to increased concentrations of p72. Other alternative splicing decisions, even those in the same vector backbone, did not respond to alterations of p72 concentration, suggesting that the observed effect was not due to a general positive effect on splicing activity or splicing factors. Similarly, increasing the concentration of p72 was not able to overcome the negative effect of mutation of the AC-rich enhancer within exon v4, again suggesting no general effect of p72 on splicing. Nor did p72 seem to have an effect on the stability or transport of the spliced product RNAs. Finally, the effect of p72 on CD44 splicing was promoter independent (data not shown), suggesting that the step in RNA biogenesis affected by p72 was RNA splicing.
How might helicases affect alternative exon recognition? Two models seem possible: (i) alteration of RNA-protein interactions or (ii) alteration of RNA-RNA interactions. With respect to the former, it is possible that p72 serves a role in removing factors that bind to silencer elements. CD44 exon v5 has been reported to bind hnRNP A1 to a silencer region within the exon. Although no silencer has yet been identified for CD44 exon v4, certain regions of the exon have not yet been probed for the possible presence of silencing elements. The absolute requirement for the ATP-binding domain within p72 for activation is consistent with a role for the helicase in remodeling RNA-protein interactions.
We also obtained evidence suggesting a potential role for RNA secondary structure in regulating CD44 exon v4 recognition. The exon has two obvious potential secondary structures that could sequester the 5′ splice site with a stem loop. The first of these is a loose complementation between the AC-rich exon enhancer sequences and the 5′ splice site of exon v4, and the second is a potential stem loop in which sequences between two of the AC-rich enhancers and G-rich sequences downstream of the 5′ splice site could base pair. We do not think that the former is related to exon recognition because single mutations in the AC-rich blocks were either inhibitory or neutral and introduction of multiple mutations did not result in cumulative activation. The second potentially interesting base pairing, however, may be related to exon v4 recognition. Disruption of the stem of this structure by mutation did cause a modest increase in exon v4 inclusion. It did not, however, result in full inclusion of the exon. Nor did it cause a nonresponse to p72 expression levels. Like p72, the other protein shown to be involved in exon v4 recognition, YB-1, has been reported to affect RNA structure. The Escherichia coli relative of YB-1, the bacterial cold shock protein A (CspA), destabilizes RNA secondary structures (42). Therefore, it is possible that both p72 and YB-1 participate to alter CD44 exon structure and thereby influence splicing. Use of in vitro extracts demonstrated that p72 was present in a complex containing precursor RNA, indicating that the protein is present in the spliceosome. We were unable, however, to demonstrate a direct interaction between the helicase and RNA by UV cross-linking assays; therefore, it is possible that the helicase enters the complex through protein-protein interactions.
Our experiments are the second observation of association of p72 and YB-1 with an RNA (43; Auboeuf and O'Malley, unpublished). The transcriptional coactivator SRA (23) binds both p72 and YB-1. In this instance, p72 and p68 were reported to act via the estrogen receptor α-activation function 1 domain to promote transcriptional coactivation (43). Interestingly, both YB-1 and p72 have been postulated to be both transcriptional regulators and splicing regulators (7, 42). It will be interesting to determine if the role of both proteins in splicing is to provide a bridge that facilitates the communication between alternative splicing and transcription.
Acknowledgments
Special thanks go to members of the Cooper and Berget laboratories for helpful suggestions and critical comments. We thank the Cell Culture Center (Endotronics Inc.) for large-volume HeLa cell growth.
This work was supported by grants from the U.S. Army (DAMD17-96-1-6084) and the NIH (GM38526 and GM58019) to S.M.B. and NICHD HD-08818 to B.W.O.
REFERENCES
- 1.Arning, S., P. Gruter, G. Bilbe, and A. Kramer. 1996. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA 2**:**794-810. [PMC free article] [PubMed] [Google Scholar]
- 2.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12**:**123-133. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J. Y., L. Stands, J. P. Staley, R. R. Jackups, L. J. Latus, and T. H. Chang. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7**:**227-232. [DOI] [PubMed] [Google Scholar]
- 4.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24**:**192-199. [DOI] [PubMed] [Google Scholar]
- 5.Dominski, Z., and R. Kole. 1992. Cooperation of pre-RNA sequence elements in splice site selection. Mol. Cell. Biol. 12**:**2108-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominski, Z., and R. Kole. 1994. Identification of exon sequences involved in splice site selection. J. Biol. Chem. 269**:**23590-23596. [PubMed] [Google Scholar]
- 7.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol. Cell. Biol. 19**:**5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Ge, H., P. Zuo, and J. L. Manley. 1991. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 26**:**373-382. [DOI] [PubMed] [Google Scholar]
- 9.Ghisolfi, L., G. Joseph, F. Amalric, and M. Erard. 1992. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 267**:**2955-2959. [PubMed] [Google Scholar]
- 10.Günthert, U. 1993. CD44: a multitude of isoforms with diverse functions. Curr. Top. Microbiol. Immunol. 184**:**47-63. [DOI] [PubMed] [Google Scholar]
- 11.Hamm, J., and A. I. Lamond. 1998. Spliceosome assembly: the unwinding role of DEAD-box proteins. Curr. Biol. 8**:**R532-R534. [DOI] [PubMed] [Google Scholar]
- 12.Hirling, H., M. Scheffner, T. Restele, and H. Stahl. 1989. RNA helicase activity associated with the human p68 protein. Nature 339**:**562-564. [DOI] [PubMed] [Google Scholar]
- 13.Iggo, R. D., and D. P. Lane. 1989. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 8**:**1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iggo, R. D., D. J. Jamieson, S. A. MacNeill, J. Southgate, J. McPheat, and D. P. Lane. 1991. p68 RNA helicase: identification of a nucleolar forma and cloning of related genes containing a conserved intron in yeasts. Mol. Cell. Biol. 11**:**1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291**:**121-124. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, Z., J. Cote, J. W. Kwon, A. M. Goate, and J. Y. Wu. 2000. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 20**:**4036-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 11**:**2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kistler, A. L., and C. Guthrie. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomeal ATPase. Genes Dev. 15**:**42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.König, H., H. Ponta, and P. Herrlich. 1998. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 17**:**2904-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins U1 70K and Drosophila splicing regulators. Cell 66**:**383-394. [DOI] [PubMed] [Google Scholar]
- 21.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11**:**363-371. [DOI] [PubMed] [Google Scholar]
- 22.Lamm, G. M., S. M. Nicol, F. V. Fuller-Pace, and A. I. Lamond. 1996. p72: a human nuclear DEAD box protein highly related to p68. Nucleic Acids Res. 24**:**3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanz, R. B., N. J. McKenna, S. A. Onate, U. Albrecht, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97**:**17-27. [DOI] [PubMed] [Google Scholar]
- 24.Linder, P., N. K. Tanner, and J. Banroques. 2001. From RNA helicases to RNPases. Trends Biochem. Sci. 26**:**339-340. [DOI] [PubMed] [Google Scholar]
- 25.Lorsch, J. R., and D. Herschlag. 1998. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry 37**:**2194-2206. [DOI] [PubMed] [Google Scholar]
- 26.Lou, H., R. F. Gagel, and S. M. Berget. 1996. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 10**:**208-219. [DOI] [PubMed] [Google Scholar]
- 27.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19**:**1978-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luking, A., U. Stahl, and U. Schmidt. 1998. The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33**:**259-296. [DOI] [PubMed] [Google Scholar]
- 29.McCullough, A. J., and S. M. Berget. 1997. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 17**:**4562-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20**:**46-50. [DOI] [PubMed] [Google Scholar]
- 31.Ohno, M., and Y. Shimura. 1996. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 10**:**997-1007. [DOI] [PubMed] [Google Scholar]
- 32.Patton, J. G., E. B. Porro, J. Galceran, P. Tempst, and B. Nadal-Ginard. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7**:**393-406. [DOI] [PubMed] [Google Scholar]
- 33.Pinol-Roma, S., M. S. Swanson, J. G. Gall, and G. Dreyfuss. 1989. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell Biol. 109**:**2575-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramchatesingh, J., A. M. Zahler, K. M. Neugebauer, M. B. Roth, and T. A. Cooper. 1995. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol. Cell. Biol. 15**:**4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwer, B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8**:**113-116. [DOI] [PubMed] [Google Scholar]
- 36.Schwer, B., and T. Meszaros. 2000. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 23**:**6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Screaton, G. R., M. V. Bell, D. G. Jackson, F. B. Cornelis, U. Gerth, and J. I. Bell. 1992. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. USA 89**:**12160-12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sillekens, P. T., R. P. Beijer, W. J. Habets, and W. J. Venrooij. 1988. Human U1 snRNP-specific C protein: complete cDNA and protein sequence and identification of a multigene family in mammals. Nucleic Acids Res. 16**:**8307-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sillekens, P. T., W. J. Habets, R. P. Beijer, and W. J. Venrooij. 1987. cDNA cloning of the human U1 snRNA-associated A protein: extensive homology between U1 and U2 snRNP proteins. EMBO J. 6**:**3841-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92**:**315-326. [DOI] [PubMed] [Google Scholar]
- 41.Stickeler, E., S. D. Fraser, A. Honig, A. L. Chen, S. M. Berget, and T. A. Cooper. 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 20**:**3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tafuri, S. T., and A. P. Wolffe. 1990. Xenopus Y-box-binding transcription factors: molecular cloning, functional analysis and developmental regulation. Proc. Natl. Acad. Sci. USA 87**:**9028-9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, M., J. Yanagisawa, H. Kitagawa, K. Takeyama, S. Ogawa, Y. Arao, M. Suzawa, Y. Kobayashi, T. Yano, H. Yoshikawa, Y. Masuhiro, and S. Kato. 2001. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor α coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 20**:**1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Will, C. L., and R. Luhrmann. 2001. Molecular biology: RNP remodeling with DexH/D boxes. Science 291**:**1916-1917. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, M., P. D. Zamore, M. Carmo-Fonseca, A. I. Lamond, and M. R. Green. 1992. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. USA 89**:**8769-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]