Inhibition of DNA Binding by Differential Sumoylation of Heat Shock Factors (original) (raw)
Abstract
Covalent modification of proteins by the small ubiquitin-related modifier SUMO regulates diverse biological functions. Sumoylation usually requires a consensus tetrapeptide, through which the binding of the SUMO-conjugating enzyme Ubc9 to the target protein is directed. However, additional specificity determinants are in many cases required. To gain insights into SUMO substrate selection, we have utilized the differential sumoylation of highly similar loop structures within the DNA-binding domains of heat shock transcription factor 1 (HSF1) and HSF2. Site-specific mutagenesis in combination with molecular modeling revealed that the sumoylation specificity is determined by several amino acids near the consensus site, which are likely to present the SUMO consensus motif to Ubc9. Importantly, we also demonstrate that sumoylation of the HSF2 loop impedes HSF2 DNA-binding activity, without affecting its oligomerization. Hence, SUMO modification of the HSF2 loop contributes to HSF-specific regulation of DNA binding and broadens the concept of sumoylation in the negative regulation of gene expression.
Covalent modification of target proteins by SUMO (sumoylation) regulates numerous biological functions, such as transcriptional activity and subcellular localization (16). The mammalian SUMO family is composed of four paralogs, SUMO-1, -2, -3, and -4, of which SUMO-2 and SUMO-3 are very similar (11, 16). SUMO conjugation utilizes a multistep enzymatic pathway, in which proteolytically processed SUMO initially forms a thioester bond with Sae1/2 (Aos1/Uba2), the SUMO E1 activating enzyme (8, 17). The SUMO moiety is subsequently transferred to Ubc9, the single SUMO E2 conjugating enzyme, which usually binds the target protein through the consensus tetrapeptide, ΨKXE, where Ψ denotes a hydrophobic residue and K the target lysine, to which SUMO becomes attached (4, 35, 37). In addition, the Ubc9-substrate interaction may be facilitated by SUMO E3 ligases, which increase sumoylation efficiency in a substrate-specific manner, either through accelerating the transfer of SUMO from Ubc9 to the substrate or by merely providing a scaffold (16). Nonetheless, the enzymatic activity required for substrate modification can be carried out by the E1 and E2 enzymes alone (8).
Sumoylated ΨKXE motifs are usually found in unstructured protein regions, in which the consensus site is accessible to the sumoylating machinery. In some cases, additional specificity determinants are necessary for efficient sumoylation. For RanGAP1, residues C terminal to the consensus site make critical contacts with Ubc9, and this region is required for the sumoylation (4, 37). Residues immediately adjoining the consensus site can also significantly affect sumoylation efficiency (35). Additional regulation of sumoylation is achieved through other posttranslational modifications of the target protein. Sumoylation has been shown to be counteracted by phosphorylation (23, 28, 29), but it can also be positively regulated by phosphorylation, as represented by heat shock factor 1 (HSF1). Upon activation, HSF1 is transiently sumoylated on lysine 298, which requires the phosphorylation of serine 303 adjacent to the consensus site (13). Hence, SUMO modification is elaborately regulated, and the SUMO substrate specificity can be determined by regulatory elements outside the consensus site.
The mammalian HSF family comprises three members, HSF1, -2, and -4. All HSFs share structurally conserved domains, of which the most preserved is the N-terminal looped helix-turn-helix DNA-binding domain (DBD) (32). Accordingly, all HSF members bind to similar target sequences, i.e., arrays of inverted pentameric NGAAN repeats that constitute the heat shock elements (HSEs) (3, 44). Nevertheless, different HSF members convey distinct biological functions. Whereas HSF1 activates transcription of several stress-induced genes in response to various proteotoxic stresses, HSF2 appears not to be involved in stress responses but has been implicated in differentiation and development (18, 32, 43). The functional difference between HSF1 and HSF2 might be mediated through a subset of HSF-specific target genes, since they bind to HSEs in slightly different ways (20).
The loop in the conserved HSF DBD has been shown to be important in determining HSF-specific DNA binding (1). Unlike many other looped helix-turn-helix transcription factors, the HSF loop does not make contacts with DNA (24, 42) but is located at the interface between neighboring HSF monomers and may take part in several protein-protein interactions depending on the HSE architecture. Accordingly, deletion of the loop significantly weakens the DNA binding of yeast HSF, possibly by interfering with the formation of the first HSF trimer (7).
Since several functionally different HSF members have evolved in vertebrates and the loop has been shown to play an important role in determining HSF-specific features, it is not surprising that the loop is not well conserved between HSF1 and HSF2. However, the loops of both HSFs contain a SUMO consensus site. The loop of HSF2 is readily modified by SUMO (10), but the loop of HSF1 is not (13, 15), indicating that additional specificity determinants are involved in the differential sumoylation of the HSFs and that HSF2 loop sumoylation contributes to HSF-specific regulation. Here, we have analyzed the molecular basis for the HSF2-specific loop sumoylation. Loop swaps between HSF1 and HSF2 show that the loop alone is able to control sumoylation, also in an E3-independent manner. Site-specific mutagenesis and molecular modeling of the HSF1 and HSF2 DBDs reveal several residues that play critical roles in presenting the SUMO consensus motif of the HSF2 loop to Ubc9. We extend our analysis to include the biological function of HSF2 loop sumoylation and demonstrate that HSF2 sumoylated on K82 has a dramatically impaired capability to bind HSEs. Thus, the negative regulation of HSF2 by SUMO modification is due to inhibition of DNA-binding activity.
MATERIALS AND METHODS
Plasmid constructs.
Plasmids encoding green fluorescent protein (GFP)-SUMO-1, His-SUMO-1, glutathione _S_-transferase (GST)-SUMO-1, GST-Ubc9, GST-Sae1/2, human myc-HSF2, and murine FLAG-HSF2, HSF1, HSF2, HSF1L2, and HSF2L1 have been described previously (1, 2, 13, 19, 31, 41). His-hemagglutinin (HA)-SUMO-2 was a kind gift from Jacob Seeler (Pasteur Institute, Paris, France). Point mutations (QuikChange site-directed mutagenesis kit; Stratagene) were confirmed by sequencing. For construction of GST-Loop1 and GST-Loop2, oligonucleotides containing the loop sequence followed by a translation termination codon were ligated into pGEX-4T-2 (Amersham). pGEX-4T-2 was cleaved with the EcoRI and NotI enzymes. The following forward (F) and reverse (R) oligonucleotides were used: FLoop1, 5′AATTCCCATTGAGCAGGGTGGCCTGGTCAAGCCTGAGAGAGATGACACCGAGTTCTAGGC3′; RLoop1, 5′GGCCGCCTAGAACTCGGTGTCATCTCTCTCAGGCTTGTCCAGGCCACCCTGCTCAATGGG3′; FLoop2, 5′AATTCCCATCGAATCTGGAATTATCAAACAGGAAAGAGATGGCCCTGTTGAATTTTAGGC3′; and RLoop2, 5′GGCCGCCTAAAATTCAACAGGGCCATCTCTTTCCTGTTTGATAATTCCAGATTCGATGGG3′.
Cell culture and transient transfections.
Cos7 cells were maintained at 37°C in a humidified 5% CO2 atmosphere in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 2 mM l-glutamine, penicillin, and streptomycin. Confluent cell plates were trypsinized and 5 × 106 cells resuspended in 400 μl OptiMEM (Gibco-BRL) together with approximately 20 μg DNA. Cells were electroporated (975 μF, 240 V) in BTX cuvettes using a Bio-Rad Gene Pulser and allowed to recover for 24 h prior to further treatments. For harvesting, cells were suspended in cold medium and washed with cold phosphate-buffered saline.
Coimmunoprecipitation and immunoblotting.
For analysis of endogenously sumoylated HSF2, cells were lysed by boiling in sodium dodecyl sulfate (SDS) as described previously (33). Lysates were precleared with nonspecific immunoglobulin G-Sepharose (Amersham) and HSF2 immunoprecipitated with anti-FLAG (αFLAG) M2 (Sigma) with 100 μl of a 50% slurry of protein G-Sepharose (Amersham) for 2 h at 4°C. The beads were washed six times with 1% Triton X-100 in phosphate-buffered saline. For analysis of HSF2 trimer formation, immunoprecipitation was done essentially as described by Hietakangas et al. (13), except that FLAG-HSF2 was immunoprecipitated with αFLAG M2. Immunoprecipitated proteins were run on 7% SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane (Protran nitrocellulose; Schleicher & Schuell), and immunoblotted using antibodies against SUMO-1, SUMO-2/3 (Zymed Laboratories, Inc.), HSF2 (39), and FLAG and Myc epitopes (Sigma). For the input control, membranes were blotted with αHsc70 antibodies (SPA-815; StressGen). Immunocomplexes were visualized using enhanced chemiluminescence (ECL; Amersham).
Purification of recombinant proteins.
The recombinant His-tagged HSF chimeras were purified as described by Ahn et al. (1), except bacteria were grown at 30°C. For purification of GST-tagged proteins, the Escherichia coli strain BL21 was transformed using the manufacturer's protocol (Stratagene). Bacteria were grown to an optical density at 600 nm of 0.5 to 0.7, induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and held at 30°C for 3 h. The harvested bacteria were resuspended in cold lysis buffer (100 mM NaCl, 50 mM Na2HPO4, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 1 μg/μl leupeptin, 1 μg/μl aprotinin), lysed with lysozyme, and sonicated. Lysates were incubated with _S_-hexylglutathione beads (Sigma) for 2 h at 4°C and washed extensively with lysis buffer containing 0.5% Triton X-100. GST-tagged proteins were eluted in 50 mM Tris-HCl (pH 8.0)-50 mM NaCl-0.5 mM EDTA-20% glycerol-50 μM ZnCl2-0.5 mM phenylmethylsulfonyl fluoride-5 mM dithiothreitol containing 0.3 mM glutathione and dialyzed against in vitro sumoylation buffer.
In vitro sumoylation and GST pull-down assays.
For in vitro sumoylations, 150 ng E1, 800 ng E2, and 1 μg SUMO-1 were incubated with recombinant or reticulocyte lysate-translated proteins at 30°C for 60 to 120 min in 20 mM HEPES (pH 7.4)-110 mM potassium acetate-2 mM magnesium acetate-0.5 mM EGTA. In vitro translation was performed using the TNT-coupled transcription-translation system (Promega) in the presence of [35S]methionine (Perkin-Elmer). Sumoylation reactions were stopped by addition of denaturing buffer and analyzed by Western blotting using antibodies against HSF2, SUMO-1, or GST (Upstate Biotechnology). For GST pull-down assays, 10 μg of recombinant GST or GST-Ubc9 was bound to 15 μl of a 50% slurry of _S_-hexylglutathione beads in 250 μl in vitro sumoylation buffer at 4°C for 2 h. After removal of unbound GST proteins, in vitro-translated proteins were added, together with 200 μl sumoylation buffer containing 0.1% Triton X-100. After binding at room temperature (RT) for 30 min, the beads were washed with reaction buffer containing 0.1% Triton X-100. Bound proteins were released by boiling in denaturing buffer and analyzed by Western blotting.
Modeling of HSF1, HFS2, and the complex with Ubc9.
Models for the DBDs of HSF1 (residues 16 to 105) and HSF2 (residues 8 to 97) were made using the 2.0-Å resolution X-ray structure of the Kluyreromyces HSF DBD (Protein Data Bank code 1FBU) (12) as the template structure and using the program Modeler version 7 (26, 36). The sequences of the DBDs of HSF1 and HSF2 share 47.2% and 47.8% sequence identity, respectively, with the K. lactis sequence, and there are no gaps in the alignment of HSF2 with 1FBU and only a single one-residue gap and a single one-residue insertion between HSF1 and 1FBU.
The modeling of the complexes of the DBDs of HSF1 and HSF2 with Ubc9 was based on the following data and assumptions. The structure of Ubc9 is defined by the 2.5-Å resolution X-ray structure of the complex between human Ubc9 and the C-terminal domain of mouse RanGAP1 (Protein Data Bank code 1KPS) (4). In this X-ray structure, the active site of Ubc9 is bound to the SUMO consensus motif of RanGAP1, and the highly conserved motif was used to pinpoint the localized interactions that would take place with other domains, including the DBD of HSF2, that have the SUMO consensus motif. To properly position the consensus motifs of the DBDs with respect to Ubc9, the SUMO consensus motifs of the modeled DBDs were superimposed with the SUMO consensus motif from RanGAP1 bound to Ubc9. The unbound HSF loop containing the SUMO consensus motif is highly mobile, as evidenced, e.g., by the nuclear magnetic resonance structure solutions of the Drosophila HSF (PDB code 1HKT) (42), where the loop position varies dramatically by up to 12 Å. The geometry for residues L525 to E528 of RanGAP1 in the structure of the complex with Ubc9 was applied to residues V75 to E78 of HSF1 and I74 to E77 of HSF2, using the program Sybyl (Tripos Corp.). As a result, the flexible loops from the models of the DBDs of HSF1 and HSF2 took on the local conformation of the sumoylation tetrapeptide of RanGAP1 as seen in the known RanGAP1-Ubc9 structure, and the resulting modeled complexes had good surface-to-surface complementarity (data not shown).
EMSA and oligonucleotide pull-down assay.
Buffer C extracts (12 μg) from transfected cells were incubated with a 32P-labeled oligonucleotide representing the proximal HSE of the human hsp70 promoter. The protein-DNA complexes were analyzed on a native 4% polyacrylamide gel (27). For electrophoretic mobility shift assay (EMSA) with in vitro-translated HSF2, wild-type and K82R HSF2 were translated as described above, but nonlabeled methionine was used. After sumoylation in vitro for 2 h, equal aliquots of the reaction mixtures were used for EMSA. The input samples were incubated for the same duration and at the same temperature as the binding reactions to enable direct comparison between the amount of HSF2 sumoylation and DNA-binding activity The HSE-containing oligonucleotides 5′-biotin-AACGAGAATCTTCGAGAATGGCT-3′ and 5′-AGCCATTCTCGAAGATTCTCGTT-3′ and the corresponding scrambled control oligonucleotides 5′-biotin-AACGACGGTCGCTCCGCCTGGCT-3′ and 5′-AGCCAGGCGGAGCGACCGTCGTT-3′ were from Oligomer (Helsinki, Finland). Buffer C extracts containing 40 mM _N_-ethylmaleimide were incubated with 0.5 μM annealed oligonucleotide in binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2 mM EDTA, 10% glycerol). Salmon sperm DNA was added (0.5 μg/μl), and proteins were allowed to bind the nucleotide for 30 min at RT. The samples were precleared with CL-4B Sepharose (Sigma) for 30 min at 4°C, and the remaining DNA was precipitated with 15 μl of a 50% slurry of UltraLink streptavidin gel (Pierce) for 1 h at 4°C. Following centrifugation, flowthrough samples were taken from lysates incubated with the scrambled oligonucleotide. Bound fractions were washed twice with binding buffer and three times with binding buffer containing 0.2% Triton X-100. DNA-bound proteins were eluted with denaturing buffer, followed by SDS-polyacrylamide gel electrophoresis and Western blotting using a polyclonal αHSF2 antibody.
RESULTS
HSF2 is modified by endogenous SUMO-2/3.
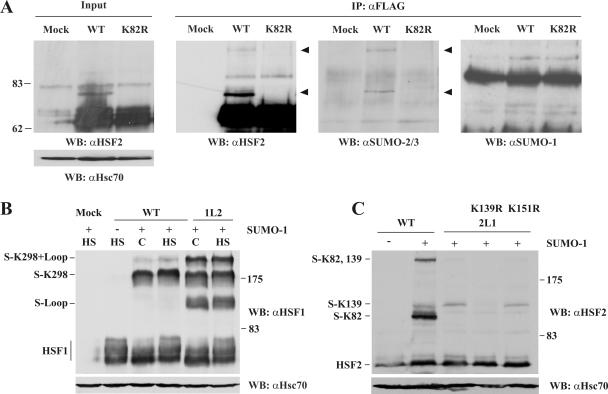
To analyze endogenous HSF2 sumoylation, we transiently transfected Cos7 cells with HSF2-FLAG expression vectors. After denaturing cell lysis, HSF2 was immunoprecipitated with an αFLAG antibody, followed by Western blotting with antibodies against SUMO-1, SUMO-2/3, and HSF2. Consistent with previous studies (10), the major sumoylated form of HSF2 was abrogated by the K82R mutation (Fig. 1A). Interestingly, the slowly migrating forms of HSF2 did not show immunoreactivity towards an αSUMO-1 antibody, but instead showed immunoreactivity towards an antibody that recognizes both SUMO-2 and -3 isoforms. Moreover, a second SUMO-2/3-reactive form was detected in the immunoprecipitates (Fig. 1A). As HSF2 has previously been suggested to be modified on a secondary site (10), we mutated two other consensus-site lysines, at positions 139 and 151, to arginines and subjected the mutants to sumoylation assays. The K139R mutation inhibited the accumulation of two slower-migrating forms of HSF2, whereas the sumoylation pattern was unaffected by K151R (Fig. 1C and data not shown). Thus, HSF2 is modified by SUMO-2/3 on lysines K82 and K139. However, under the conditions used in this study, the majority of HSF2 sumoylation occurred on K82 in the DBD.
FIG. 1.
HSF2 is modified by SUMO-2/3. A. Cos7 cells were transfected with FLAG-HSF2 (wild type [WT]) or FLAG-HSF2 K82R (K82R). HSF2 was immunoprecipitated (IP) using an αFLAG antibody, and samples were analyzed by Western blotting (WB) with antibodies against HSF2, SUMO-2/3, or SUMO-1. Arrowheads denote the HSF2-sumoylated species recognized by αHSF2 and αSUMO-2/3 antibodies. Hsc70 was used as a loading control. B. The loop of HSF2 is sumoylated when inserted into HSF1. Cos7 cells were transfected with plasmids encoding HSF1 (WT) or HSF1Loop2 (1L2) together with GFP-SUMO-1 (+) or empty plasmid (−). Cells were heat shocked at 42°C for 15 min (HS) or maintained at 37°C (C). Cell extracts were subsequently analyzed by Western blotting with αHSF1 and αHsc70 antibodies. The sumoylated species are indicated on the left. The decrease in mobility of HSF1 in heat-shocked samples is due to hyperphosphorylation (39). Notably, coexpression of HSF1 and SUMO-1 in Cos7 cells led to constitutive HSF1 sumoylation, in contrast to the heat-inducible sumoylation previously observed in other cell types (13, 15). C. The loop of HSF1 is not sumoylated when inserted into HSF2. Cos7 cells were transfected with HSF2 (WT), HSF2Loop1 (2L1), or 2L1 mutants together with GFP-SUMO-1 (+) or empty plasmid (−). Cell extracts were analyzed by Western blotting with αHSF2 and αHsc70 antibodies. Notably, SUMO coexpression led to increased total levels of HSF2, which is likely due to the inhibitory effect of sumoylation on HSF2 DNA binding (see Fig. 4).
The SUMO consensus sites of the HSF1 and HSF2 loops are differentially sumoylated.
Although the exposed loops of both HSF1 and HSF2 DBD contain a SUMO consensus site, the DBD of HSF1 is not modified by SUMO (13, 15). To determine the basis for this difference, we used chimeric constructs in which the loop of HSF1 has been inserted into HSF2 (HSF2Loop1) and vice versa (HSF1Loop2) (1). As seen in Fig. 1B, wild-type HSF1 is modified by SUMO on a single lysine residue previously identified as K298, located in the regulatory domain (13, 15). When the loop of HSF1 was replaced with that of HSF2, three sumoylated HSF1 species appeared (Fig. 1B). These bands correspond to a loop-sumoylated form, sumoylation of K298, and a double sumoylation, since a K298R mutation abolished the two upper bands (data not shown). Conversely, the HSF1 loop effectively inhibited DBD sumoylation when inserted into HSF2 (Fig. 1C), indicating that the sumoylation specificity is provided by the HSF loop itself.
Ubc9 determines the differential sumoylation of the HSF1 and HSF2 loops.
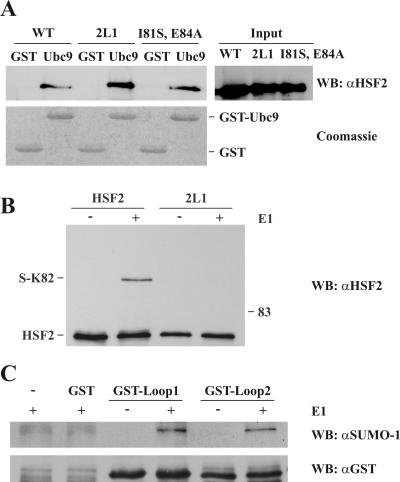
Since the HSF2 DBD with the HSF1 loop was not sumoylated, we sought to determine the mechanism for the differential sumoylation of HSF1 and HSF2 loops. A plausible explanation for the difference seen in Fig. 1C (compare lanes 2 and 3) is that the HSF1 loop contains an element impairing the interaction with Ubc9. To investigate this possibility, we performed pull-down assays using GST or GST-Ubc9 as bait for HSF2 and HSF2Loop1 translated in rabbit reticulocyte lysates (Fig. 2A). Surprisingly, both HSF2 and HSF2Loop1 bound Ubc9 with similar efficiency. The binding was not dependent on the consensus site surrounding K82, since mutation of the ΨKXE motif did not decrease Ubc9 binding. Moreover, Ubc9 binding was unaffected by a simultaneous disruption of the K82 and K139 consensus sites (data not shown). These results suggest that the stable interaction between Ubc9 and HSF2 is mediated by regions outside the K82 and K139 SUMO consensus sites of HSF2.
FIG. 2.
Ubc9 determines the differential sumoylation of the HSF1 and HSF2 loops. A. HSF2 and HSF2Loop1 interact equally well with Ubc9. In vitro-translated HSF2 proteins were incubated with recombinant GST or GST-Ubc9 as indicated. The ΨKXE-motif was disrupted by the I81S and E48A mutations. Bound fractions were released with denaturing buffer and subjected to Western blotting (WB) with an αHSF2 antibody. Equal amounts of GST and GST-Ubc9 were determined by Coomassie blue staining. WT, wild type. B. HSF2Loop1 is poorly sumoylated in vitro. Recombinant HSF2 and HSF2Loop1 were sumoylated in vitro using recombinant GST-Sae1/2 (E1), GST-Ubc9 (E2), and GST-SUMO-1. For controls, reaction buffer (−) was added instead of GST-Sae1/2 (+). Reactions were stopped by addition of denaturing buffer and analyzed by Western blotting using an αHSF2 antibody. C. The structural context determines the differential sumoylation of the HSF1 and HSF2 loops. GST-tagged peptides containing the loop of HSF1 or HSF2 were sumoylated in vitro as for panel B. Reactions were stopped by addition of denaturing buffer and analyzed by Western blotting using a SUMO-1-specific antibody. Equal input levels were determined by reblotting the membrane with an αGST antibody.
The similar binding of Ubc9 to HSF2 and HSF2Loop1 prompted us to investigate whether Ubc9 was able to determine the difference in sumoylation of HSF2 and HSF2Loop1 in the absence of E3 ligases. Bacterially expressed HSF2 and HSF2Loop1 were subjected to in vitro sumoylation, using only recombinant Sae1/2 (E1), Ubc9 (E2), and SUMO proteins. Replacing the HSF2 loop with that of HSF1 abolished the DBD sumoylation in the absence of an E3 ligase (Fig. 2B), implying that the specificity of loop sumoylation was mediated by the loop-Ubc9 interaction. The results were not due to differential sumoylation efficiency of recombinant or in vitro-translated proteins, since reticulocyte lysate-translated HSF2Loop1 was as poorly sumoylated as the recombinant protein (data not shown). Therefore, Ubc9 is capable of distinguishing between the loops of HSF2 and HSF1. Although stable Ubc9 binding to HSF2 likely occurs through a region outside the loop, the loop can provide Ubc9 with a local recognition site required for efficient sumoylation.
To investigate whether the primary amino acid sequences of the loops determine the differential regulation, we fused the 16-amino-acid loop sequences of HSF1 (I84 to F99) and HSF2 (I76 to F91) to a GST tag. Bacterially expressed GST-Loop1 and GST-Loop2 were subjected to in vitro sumoylation as described above. The short peptides representing the HSF1 and HSF2 loops were sumoylated similarly (Fig. 2C), suggesting that the primary amino acid sequence of the loops is not per se responsible for the differential sumoylation but is instead involved in the regulation within the structural context of the loop.
Sumoylation of the HSF2 loop is regulated by residues outside the consensus site.
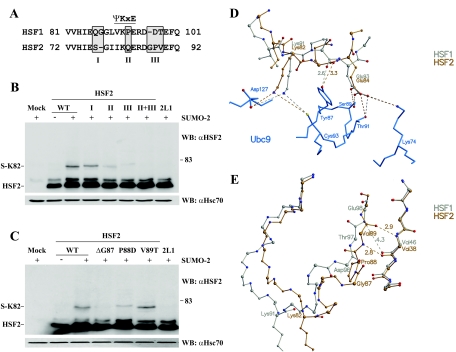
To elucidate the amino acids of the loop that are involved in regulating HSF2 sumoylation, we performed site-directed mutagenesis, changing a single residue or clusters of amino acids of HSF2 to the corresponding amino acids of HSF1 (Fig. 3A). As seen in Fig. 3B, mutations at the N-terminal part of the loop (mutation I) had little or no effect on the sumoylation efficiency. Replacement of glutamine with a proline at the consensus site (mutation II) decreased sumoylation, but HSF2 sumoylation was most prominently impaired by the mutation of three residues C terminal to the consensus site (mutation III) (Fig. 3B). The effect of the triple mutation was enhanced by the addition of Q83P (mutations II and III), abrogating HSF2 sumoylation to the same extent as the HSF1 loop. Moreover, introduction of the amino acids of HSF2 regions II and III into HSF1 conferred the same sumoylation pattern as observed for HSF1Loop2 (data not shown).
FIG. 3.
SUMO modification of the HSF2 loop is regulated by residues neighboring the consensus site. A. Schematic presentation of the nonconserved regions between the HSF1 and HSF2 loops. Mutations I, II, and III were introduced at the shaded sites, in which the amino acids of HSF2 were replaced with the corresponding amino acids of HSF1. B. Mutation of the GPV motif (mutation III) reduces HSF2 loop sumoylation. Extracts from cells transfected with wild-type HSF2 (WT) or HSF2 mutants containing HSF1-specific residues (I, II, and III), together with His-HA-SUMO-2 (+) or empty plasmid (−), were analyzed by Western blotting (WB) using an αHSF2 antibody. Equal protein input was determined by Hsc70 levels. C. Both the deletion of G87 and a P88D mutation reduce the sumoylation of the HSF2 loop. The experiment was done as for panel B. D. Close-up depiction of the interface between the ΨKXE motif from HSF1 (silver) or HSF2 (gold) and Ubc9 (blue). In comparison with HSF2, the presence of proline in HSF1 would lead to a much stronger hydrogen bond with Y87 of Ubc9 (the distance is smaller, 2.6 versus 3.3 Å, and the angle of bonding is ideal, nearly 180°). Such tight binding might prevent Y87 from altering its position as part of a catalytic mechanism and slow the release of sumoylated product. The effect of proline on its own would not be expected to completely prevent catalysis in the case of HSF1, since proline would not affect the relative positioning of the lysine and glutamate of the SUMO consensus motif. E. The region C terminal to the motif in HSF1 relative to HSF2 would limit the ability of the loop to place the sumoylation motif in an appropriate binding position. Here, the HSF model structures were superimposed over the elements of regular secondary structure of the DBDs but not over the loop regions, in order to reveal any relative displacements of the sumoylation motif. As a consequence of the unique structural properties of G87 and P88, in HSF2 two separate strong hydrogen bonds would be formed between main-chain atoms of V89 and V38, connecting the loop region to the final β-strand of the DBD sheet, stabilizing the structure, and facilitating better coordination of the VKQE tetrapeptide within the active site of Ubc9. In comparison, these strong hydrogen bonds would be replaced in HSF1 by a single, long and weak interaction between the main-chain nitrogen of E98 and oxygen atom of V46. Thus, in HSF1, the C-terminal end of the loop would be more detached from the β-sheet and more flexible than in HSF2, hampering Ubc9 from recognizing and docking VKPE of the loop to the active site of Ubc9. Due to the sequence differences C terminal to the sumoylation motif, in the modeled structures the nonsumoylated lysine of HSF1 is displaced by >4 Å with respect to the sumoylated lysine of HSF2.
To examine further which residues of the GPV tripeptide were crucial for sumoylation, each residue was mutated to the corresponding HSF1 residue. Both the deletion of glycine 87 (ΔG87) and replacement of proline 88 with aspartic acid (P88D) caused the protein to be sumoylated inefficiently, whereas the V89T mutation had no effect (Fig. 3C). Taking all of the mutagenesis data into consideration, we conclude that the differential sumoylation of HSF1 and HSF2 DBDs is regulated mainly outside the consensus site, through HSF-specific residues within the loop.
To evaluate the possible consequences of the sequence differences between HSF1 and HSF2 on sumoylation, we modeled the DBDs of HSF1 and HSF2 in complex with Ubc9. The presence of proline in HSF1 or in the HSF2 Q83P mutant, which decreased the sumoylation of the mutant HSF2 (Fig. 3B), alters the backbone of the SUMO target motif, since proline will be in the cis conformation (Fig. 3D). Consequently, the main-chain oxygen atoms on either side of the proline would be oriented differently from the corresponding oxygen atoms in native HSF2. The main-chain oxygen atom of proline would turn towards the active site of Ubc9 (Fig. 3D), leading to a shorter interaction distance with the hydroxyl group of Y87 of Ubc9, and the angle is nearly ideal for a hydrogen bond. Thus, if VKPE of HSF1 was positioned within the Ubc9 active site as depicted in the model, the proline would lock the tyrosine into place (Fig. 3D), possibly reducing flexibility at the active site, which may interfere with sumoylation. The presence of proline, however, would not affect the relative positions of the lysine and glutamate side chains, and these effects would be in line with the experimentally observed modest sumoylation of the HSF2 Q83P mutant (Fig. 3B).
As shown in Fig. 3A, in the variable region C terminal to the ΨKXE motif (III), HSF2 contains a tripeptide, G87-P88-V89, and HSF1 contains a dipeptide, D96-T97. In the modeled structures, this region lies adjacent to a β-strand in the DBD (Fig. 3D) and does not interact directly with Ubc9. Instead, this region would participate in interactions that affect the flexibility of the SUMO target loop. In HSF2, the replacement of proline 88 and deletion of glycine 87 led to drastic reductions in sumoylation compared to wild-type HSF2 (Fig. 3B and C). Thus, the length of the C-terminal region is critical for HSF2 sumoylation as well as the combination of the flexible glycine and rigid proline. The combination of glycine and proline in HSF2 would lead to a main-chain conformation not achievable by the corresponding region in HSF1 or by other amino acids. This is shown in Fig. 3E, where the main chain of HSF2 with the GPV tripeptide is rotated by almost 180° with respect to the DT dipeptide in HSF1. As a result, the β-strand and the C terminus of the sumoylated loop would be more tightly packed on the β-sheet in HSF2. In contrast, the shorter C-terminal end of the loop in HSF1 would be more detached from the β-sheet, making it more flexible. If we superimpose HSF1 onto HSF2 only over the major secondary structures to show the effect of the structural differences along the C-terminal ends of the HSF loops, then nonsumoylated K91 in HSF1 would be displaced in position by as much as 4 Å from the sumoylated K82 in HSF2 (Fig. 3E). Our model suggests a plausible mechanism by which the C-terminal sequence in the HSF1 loop impedes the recognition and docking of the SUMO consensus tetrapeptide to the active site of Ubc9.
Sumoylation of the HSF2 loop inhibits the DNA-binding activity of HSF2.
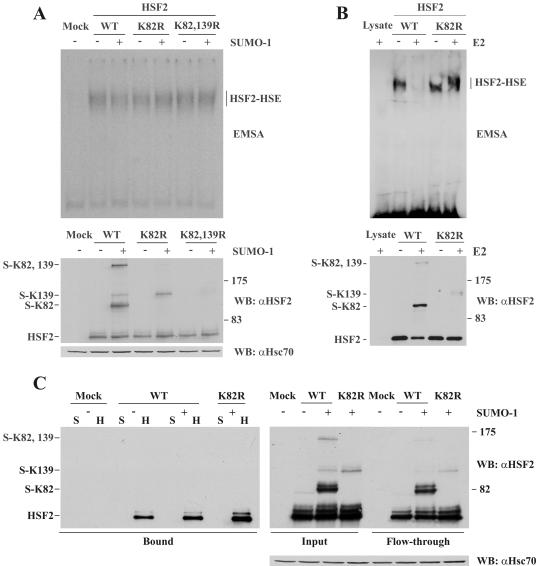
The interactions between adjacent HSF monomers across the tail-to-tail interface involve hydrogen bonding between “turn” and “loop” residues (24). In mammalian HSF1 and HSF2, the loop residues engaged in these interactions align to the SUMO consensus site (data not shown), which indicates that conjugating the SUMO moiety to the HSF2 loop would prevent or modulate the DNA binding of HSF2. Surprisingly, the opposite effect has been presented, i.e., that SUMO-1 modification of HSF2 is required for its DNA-binding activity (10, 14). Using EMSA, we examined the binding of wild-type and nonsumoylatable HSF2 to an oligonucleotide representing the proximal HSE of the hsp70 promoter. HSE-binding activity was detected in samples containing exogenous HSF2, and this activity was unaffected by coexpression of SUMO-1 (Fig. 4A). In contrast to earlier studies (10, 14), K82R had no effect on HSF2 DNA-binding activity (Fig. 4A). Notably, coexpression of SUMO-1 led to a significant increase in the total levels (nonsumoylated plus sumoylated forms) of HSF2, while the levels of nonsumoylated HSF2 remained constant. Since the HSE-binding activity was equal in all samples, the sumoylated HSF2 species might not contribute to the total HSE-binding activity.
FIG. 4.
Sumoylation of K82 inhibits the DNA binding of HSF2. A. EMSA performed on extracts from Cos7 cells transfected with the indicated plasmids (upper panel). The HSF2-HSE complex is indicated on the right. Input HSF2 levels were analyzed by Western blotting (WB) with an αHSF2 antibody (middle panel), and the αHsc70 antibody was used as a loading control (lower panel). WT, wild type. B. EMSA performed on reticulocyte lysate-translated WT or K82R HSF2 after sumoylation in vitro in the presence (+) or absence (−) of Ubc9 (upper panel). Aliquots of the sumoylation reactions were incubated at RT for 20 min and analyzed by Western blotting with an αHSF2 antibody (lower panel). C. Pull-down assay using an oligonucleotide containing three inverted NGAAN repeats (H) or a scrambled control (S). Extracts from Cos7 cells transfected with the indicated plasmids were incubated with an (NGAAN)3 oligonucleotide. After the binding reaction, a flowthrough sample was taken and unbound proteins were removed by washing. The bound fraction was released by addition of denaturing buffer and analyzed by Western blotting using an αHSF2 antibody (left panel). Equal stoichiometry of sumoylated and nonsumoylated HSF2 species in the beginning (input) and end (flowthrough) of the binding reaction is shown in the right panel. Hsc70 is shown for equal loading.
Since the total cellular HSF2 levels were modulated by exogenous expression of SUMO in Cos7 cells (Fig. 4A), we investigated the effect of sumoylation on HSF2 DNA-binding activity in vitro. Equal amounts of HSF2 and K82R proteins translated in reticulocyte lysates were subjected to sumoylation in vitro and subsequently incubated with the HSE-containing probe. In accordance with previous studies (25, 38), we found that HSF2 produced in reticulocyte lysates possessed constitutive DNA-binding activity (Fig. 4B). However, after incubation of HSF2 with the sumoylation machinery, a dramatic decrease in HSF2 DNA-binding ability was observed (Fig. 4B). The SUMO-mediated inhibition was specifically due to the modification of HSF2, since the DNA binding of the K82R mutant was unaltered in the presence of the sumoylation machinery.
To further analyze the negative effect of sumoylation on the DNA-binding activity of HSF2 in cells, we used biotin-tagged HSE-containing oligonucleotides for precipitation of HSF2 forms with the highest affinity for HSEs. Since HSF2 has been shown to prefer short HSEs (21), we designed an HSE containing three perfect NGAAN pentamers (see Materials and Methods). To exclude the possibility that the large GFP moiety fused to SUMO would interfere with the DNA binding of sumoylated HSF2, His-tagged SUMO-1 was overexpressed together with HSF2 or HSF2 K82R, and cell extracts were incubated with the HSE-containing oligonucleotide or a scrambled control. Following binding, a flowthrough sample from the scrambled control was analyzed to verify that the sumoylation of HSF2 had not been reduced during the binding reaction. As shown in Fig. 4C, despite an estimated 1:1 stoichiometry between nonsumoylated and sumoylated HSF2 in the input and flowthrough samples (right panel), we were unable to detect sumoylated HSF2 in the HSE-bound fraction (left panel). Identical results were obtained when the pull-down assay was repeated with a biotin-tagged version of the oligonucleotide representing the proximal HSE from the hsp70 promoter or with GFP-SUMO-1 or His-HA-SUMO-2 with either oligonucleotide (data not shown). These results indicate that HSF2 species sumoylated on K82 are excluded from DNA binding.
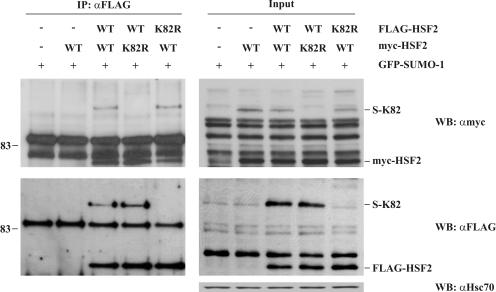
Since HSFs bind to DNA as trimers (20), the SUMO-mediated inhibition of HSF2 DNA-binding activity could be due to interference with HSF2 trimerization or to decreased affinity of the oligomeric HSF2 for HSEs. To examine whether the SUMO moiety would modulate the physical interaction between HSF2 monomers, Cos7 cells were transfected with GFP-SUMO-1 together with the wild-type or K82R HSF2, tagged with either FLAG or Myc epitopes. After immunoprecipitation of FLAG-HSF2, bound Myc-HSF2 was analyzed with Western blotting with antibodies against the Myc epitope. As seen in Fig. 5, the sumoylation of HSF2 on K82 did not interfere with HSF2 oligomerization, since the sumoylated fraction of Myc-HSF2 was pulled down as efficiently as the nonsumoylated fraction. Therefore, we conclude that the inhibitory role of HSF2 DBD sumoylation is not likely to be mediated by HSF2 trimer formation.
FIG. 5.
Sumoylation on K82 does not interfere with HSF2 trimer formation. Cos7 cells were transfected with GFP-SUMO-1 together with empty plasmid or the indicated variants of Myc-HSF2 and FLAG-HSF2. FLAG-HSF2 was immunoprecipitated (IP) with antibodies against the FLAG epitope, and bound Myc-HSF2 was released by addition of denaturing buffer and detected by Western blotting (WB) using antibodies against the Myc epitope. Immunoprecipitated material is shown in the left panel and the input is shown in the right panel. Sumoylated and nonsumoylated Myc-HSF2 bind to FLAG-HSF2 with similar efficiency. Hsc70 was used as a loading control. WT, wild type.
DISCUSSION
Rigidification of the consensus site is important for efficient sumoylation of HSF2.
Despite the increasing number of identified sumoylated proteins, the determinants for SUMO substrate specificity are largely unknown. Since the majority of protein sumoylation occurs on ΨKXE motifs, additional requirements for Ubc9 conjugation and SUMO target specificity may easily be overlooked. The differentially sumoylated DBDs of HSF1 and HSF2 present an interesting example of the high specificity of SUMO conjugation. The accessible loop of both HSFs contains the SUMO consensus site, but only that of HSF2 is modified by SUMO. The ability of the HSF2 loop to be sumoylated is maintained also in the chimeric HSF1Loop2 protein, showing that the loop per se is a major contributor to SUMO substrate specificity. This contribution is nevertheless independent of E3 ligases, since discernible differences in the sumoylation of the loop chimeras were obtained in vitro using purified E1 and E2 enzymes only. Mutation of nonconserved amino acids within the HSF loops revealed that the DBD sumoylation was determined by the combined action of several amino acids. Molecular modeling of the HSF DBDs suggested that the functions of the critical residues were mechanistically different.
The transfer of the SUMO moiety from the Ubc9-SUMO thioester to an exposed consensus site necessitates a correct positioning of the target lysine to the Ubc9 active site and a subsequent conjugation of SUMO to the substrate. Although the HSF-specific residues at the C terminus of the loop do not participate in direct interactions with Ubc9, they define the structural boundaries of the loop and, consequently, binding of Ubc9 to the consensus site. The importance of the GP motif at the C terminus of the HSF2 loop suggests that although SUMO conjugation sites are generally found at unstructured regions, a certain rigidity of the area surrounding the consensus sequence is required for proper formation of the multiple contacts with the Ubc9 active site. This agrees with the earlier observation that unstructured peptides containing the SUMO consensus motif are only weakly modified (Fig. 2C) (22). Our functional analyses also revealed that a proline within the consensus site can interfere with sumoylation. The modeled Ubc9-HSF1 complex suggests that the proline in the consensus site could restrain Y87 of Ubc9 through hydrogen bonding, and this may explain why proline is rarely found within sumoylated consensus sites. Importantly, Y87 has been shown to contact the consensus motif through multiple interactions, and a Y87A mutation disrupts the sumoylation of several substrates (4). Strengthening of these interactions by proline at the consensus site, while having little effect on the positioning of the SUMO target lysine, may lead to increased rigidity within the Ubc9 active site and thereby to reduced catalytic activity.
Sumoylation inhibits the DNA-binding activity of HSF2.
The sumoylation of the HSF2 DBD is a likely mechanism for HSF-specific regulation. The variability in architecture of different HSEs probably generates differently positioned HSF2 trimers with various binding strengths. Between HSFs bound in a head-to-head orientation, the loop faces outwards and may facilitate interactions with another HSF monomer or other transcription factors. In the tail-to-tail orientation, the loop is less exposed, making contacts with the DBD turn of an adjacent monomer (24). Two of three contacts from the loop are through the polypeptide backbone, suggesting that the nature of these amino acids might not be a strong determinant for DNA binding. Hence, one function of the loop might be to stabilize the tail-to-tail bound HSF complex. Interestingly, yeast and Drosophila HSF seem to bind tail-to-tail HSEs more strongly than head-to-head HSEs (5, 45). Moreover, the deletion of the loop results in a decreased DNA-binding affinity of yeast HSF (7). In support of these previous observations, we found that the SUMO moiety covalently attached to the HSF2 loop severely reduced HSF2 binding to HSEs. First, abrogation of DBD sumoylation by the K82R mutation did not affect DNA binding of HSF2. Second, the DNA-binding activity of wild-type HSF2, but not K82R HSF2, was severely reduced after in vitro sumoylation. Third, employing the high stoichiometry of HSF2 sumoylation in overexpressing cells, the large cellular pool of SUMO-modified HSF2 bound HSEs significantly more weakly than nonsumoylated HSF2. Interestingly, sumoylation also diminishes the DNA-binding activity of other transcription factors, such as SATB2 and TFIID (6, 9), highlighting the role of SUMO in the negative regulation of gene expression.
Our results contradict those of Goodson and coworkers, who reported that sumoylation of HSF2 on K82 would convert HSF2 into a DNA-binding form, as judged by EMSA of in vitro-translated HSF2 that was sumoylated using a combination of purified proteins and HeLa nuclear extracts (10). In contrast, we found that in vitro-translated HSF2 possessed constitutive DNA-binding activity, as has been previously reported (25, 38). Importantly, HSF2 produced in E. coli, where sumoylation does not occur, is also constitutively active for DNA binding (1, 20, 21). We cannot exclude the possibility that the different results are due to the use of HSEs with dissimilar architectures. However, this hypothesis is unlikely, since we could not observe SUMO-dependent HSF2 DNA binding to either an (NGAAN)3 or an (NGAAN)5 HSE. Instead, the amount of HSF2 DNA binding always corresponded to the amount of nonsumoylated HSF2. In different studies, however, the amount of sumoylated HSF2 seems to be surprisingly variable. Previous reports suggest that in extracts from Xenopus and HeLa cells overexpressing HSF2, the whole HSF2 pool is modified by SUMO-1 and migrates at ∼90 kDa (14, 46). In contrast, our results demonstrate that albeit readily modified by SUMO-2/3 upon overexpression, the majority of HSF2 in all tested cell types migrates at ∼70 kDa, corresponding to nonsumoylated HSF2. Accordingly, high levels of the ∼70-kDa form of HSF2 correlate with constitutive HSE-binding activity (30, 34, 39, 40). Based on our data, we conclude that SUMO modification inhibits, rather than activates, the DNA binding of HSF2. This observation may be of great value in elucidating the physiological role of HSF2. HSF2 has recently been proposed to maintain chromatin accessibility of the hsp70 promoter during mitosis, i.e., hsp70 bookmarking (46). In the light of our results, the role of sumoylation in HSF2-mediated bookmarking needs to be further examined. HSF2 has also been assigned critical functions in spermatogenesis and neuronal development (18, 43). However, the lack of known HSF2-specific target genes has posed constraints on the clarification of the in vivo function(s) of HSF2, and therefore, it will be intriguing to further investigate the regulatory role of sumoylation in the expression of HSF2-specific target genes.
Acknowledgments
We thank Ronald Hay, Jorma J. Palvimo, and Jacob Seeler for generously providing plasmids. Jorma J. Palvimo is further acknowledged for valuable discussions and suggestions. We are thankful to Helena Saarento for excellent technical assistance. We acknowledge Johanna K. Ahlskog, Henri Blomster, Eva Henriksson, Aura Kaunisto, Minna Poukkula, and Anton Sandqvist for valuable suggestions and comments on the manuscript.
This work was supported by the Academy of Finland (L.S., K.D., and M.S.J.), the Sigrid Jusélius Foundation (L.S., and M.S.J.), the Finnish Cancer Organizations (L.S.), the Finnish Life Insurance Companies (L.S.), the Turku Graduate School of Biomedical Sciences (V.H.), the Paulo Foundation (V.H.), the Finnish Cultural Foundation (V.H.), and the U.S. National Institutes of Health (grant GM59911, D.J.T.).
REFERENCES
- 1.Ahn, S. G., P. C. Liu, K. Klyachko, R. I. Morimoto, and D. J. Thiele. 2001. The loop domain of heat shock transcription factor 1 dictates DNA-binding specificity and responses to heat stress. Genes Dev. 15**:**2134-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alastalo, T.-P., M. Hellesuo, A. Sandqvist, V. Hietakangas, M. Kallio, and L. Sistonen. 2003. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J. Cell Sci. 116**:**3557-3570. [DOI] [PubMed] [Google Scholar]
- 3.Amin, J., J. Ananthan, and R. Voellmy. 1988. Key features of heat shock regulatory elements. Mol. Cell. Biol. 8**:**3761-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. J. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108**:**345-356. [DOI] [PubMed] [Google Scholar]
- 5.Bonner, J. J., C. Ballou, and D. L. Fackenthal. 1994. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol. Cell. Biol. 14**:**501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer-Guittaut, M., K. Birsoy, C. Potel, G. Elliott, E. Jaffray, J. M. Desterro, R. T. Hay, and T. Oelgeschlager. 2005. SUMO-1 modification of human transcription factor (TF) IID complex subunits: inhibition of TFIID promoter-binding activity through SUMO-1 modification of hsTAF5. J. Biol. Chem. 280**:**9937-9945. [DOI] [PubMed] [Google Scholar]
- 7.Cicero, M. P., S. T. Hubl, C. J. Harrison, O. Littlefield, J. A. Hardy, and H. C. M. Nelson. 2001. The wing in yeast heat shock transcription factor (HSF) DNA-binding domain is required for full activity. Nucleic Acids Res. 29**:**1715-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desterro, J. M. P., M. S. Rodriguez, G. D. Kemp, and R. T. Hay. 1999. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274**:**10618-10624. [DOI] [PubMed] [Google Scholar]
- 9.Dobreva, G., J. Dambacher, and R. Grosschedl. 2003. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin μ gene expression. Genes Dev. 17**:**3048-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodson, M. L., Y. Hong, R. Rogers, M. J. Matunis, O-K. Park-Sarge, and K. D. Sarge. 2001. SUMO-1 modification regulates the DNA-binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 276**:**18513-18518. [DOI] [PubMed] [Google Scholar]
- 11.Guo, D., M. Li, Y. Zhang, P. Yang, S. Eckenrode, D. Hopkins, W. Zheng, S. Purohit, R. H. Podolsky, A. Muir, J. Wang, Z. Dong, T. Brusko, M. Atkinson, P. Pozzilli, A. Zeidler, L. J. Raffel, C. O. Jacob, Y. Park, M. Serrano-Rios, M. T. Larrad, Z. Zhang, H. J. Garchon, J. F. Bach, J. I. Rotter, J. X. She, and C. Y. Wang. 2004. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36**:**837-841. [DOI] [PubMed] [Google Scholar]
- 12.Hardy, J. A., and H. C. M. Nelson. 2000. Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor. Protein Sci. 9**:**2128-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hietakangas, V., J. K. Ahlskog, A. M. Jakobsson, M. Hellesuo, N. M. Sahlberg, C. I. Holmberg, A. Mikhailov, J. J. Palvimo, L. Pirkkala, and L. Sistonen. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23**:**2953-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilgarth, R. S., L. A. Murphy, C. M. O'Connor, J. A. Clark, O-K. Park-Sarge, and K. D. Sarge. 2004. Identification of Xenopus heat shock transcription factor-2: conserved role of sumoylation in regulating deoxyribonucleic acid-binding activity of heat shock transcription factor-2 proteins. Cell Stress Chaperones 9**:**214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, Y., R. Rogers, M. J. Matunis, C. N. Mayhew, M. L. Goodson, O.-K. Park-Sarge, and K. D. Sarge. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276**:**40263-40267. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73**:**355-382. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16**:**5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallio, M., Y. Chang, M. Manuel, T.-P. Alastalo, M. Rallu, Y. Gitton, L. Pirkkala, M-T. Loones, L. Paslaru, S. Larney, S. Hiard, M. Morange, L. Sistonen, and V. Mezger. 2002. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 21**:**2591-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotaja, N., U. Karvonen, O. A. Jänne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 14**:**5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroeger, P. E., K. D. Sarge, and R. I. Morimoto. 1993. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol. Cell. Biol. 13**:**3370-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroeger, P. E., and R. I. Morimoto. 1994. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol. Cell. Biol. 14**:**7592-7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, D., M. H. Tatham, B. Yu, S. Kim, R. T. Hay, and Y. Chen. 2002. Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 277**:**21740-21748. [DOI] [PubMed] [Google Scholar]
- 23.Lin, J.-Y., T. Ohshima, and K. Shimotohno. 2004. Association of Ubc9, an E2 ligase for SUMO conjugation, with p53 is regulated by phosphorylation of p53. FEBS Lett. 573**:**15-18. [DOI] [PubMed] [Google Scholar]
- 24.Littlefield, O., and H. C. M. Nelson. 1999. A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nat. Struct. Biol. 6**:**464-470. [DOI] [PubMed] [Google Scholar]
- 25.Manuel, M., M. Rallu, M-T. Loones, V. Zimarino, V. Mezger, and M. Morange. 2002. Determination of the consensus binding sequence for the purified embryonic heat shock factor 2. Eur. J. Biochem. 269**:**2527-2537. [DOI] [PubMed] [Google Scholar]
- 26.Marti-Renom, M. A., A. Stuart, A. Fiser, R. Sánchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29**:**291-325. [DOI] [PubMed] [Google Scholar]
- 27.Mosser, D. D., N. G. Theodorakis, and R. I. Morimoto. 1988. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 8**:**84736-84744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17**:**61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller, S., M. Berger, F. Lehembre, J.-S. Seeler, Y. Haupt, and A. Dejean. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275**:**13321-13329. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, S. P., J. J. Gorzowski, K. D. Sarge, and B. Phillips. 1994. Characterization of constitutive DNA-binding activity in mouse carcinoma cells. Mol. Cell. Biol. 14**:**5309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirkkala, L., T-P. Alastalo, X. Zuo, I. J. Benjamin, and L. Sistonen. 2000. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol. Cell. Biol. 20**:**2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirkkala, L., P. Nykänen, and L. Sistonen. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15**:**1118-1131. [DOI] [PubMed] [Google Scholar]
- 33.Poukkula, M., A. Kaunisto, V. Hietakangas, K. Denessiouk, T. Katajamäki, M. S. Johnson, L. Sistonen, and J. E. Eriksson. 2005. Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J. Biol. Chem. 280**:**27345-27355. [DOI] [PubMed] [Google Scholar]
- 34.Rallu, M., M.-T. Loones, Y. Lallemand, R. Morimoto, M. Morange, and V. Mezger. 1997. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc. Natl. Acad. Sci. USA 94**:**2392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276**:**12654-12659. [DOI] [PubMed] [Google Scholar]
- 36.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234**:**779-815. [DOI] [PubMed] [Google Scholar]
- 37.Sampson, D. A., M. Wang, and M. J. Matunis. 2001. The small ubiquitin-related modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276**:**21664-21669. [DOI] [PubMed] [Google Scholar]
- 38.Sarge, K. D., V. Zimarino, K. Holm, C. Wu, and R. I. Morimoto. 1991. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding activity. Genes Dev. 5**:**1902-1911. [DOI] [PubMed] [Google Scholar]
- 39.Sarge, K. D., S. P. Murphy, and R. I. Morimoto. 1993. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13**:**1392-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarge, K. D., O.-K. Park-Sarge, J. D. Kirby, K. E. Mayo, and R. I. Morimoto. 1994. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol. Reprod. 50**:**1334-1343. [DOI] [PubMed] [Google Scholar]
- 41.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. P. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276**:**35368-35374. [DOI] [PubMed] [Google Scholar]
- 42.Vuister, G. W., S. J. Kim, A. Orosz, J. Marquardt, C. Wu, and A. Bax. 1994. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat. Struct. Biol. 1**:**605-614. [PubMed] [Google Scholar]
- 43.Wang, G., J. Zhang, D. Moskophidis, and N. F. Mivechi. 2003. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 36**:**48-61. [DOI] [PubMed] [Google Scholar]
- 44.Xiao, H., and J. T. Lis. 1988. Germline transformation used to define key features of heat-shock response elements. Science 239**:**1139-1142. [DOI] [PubMed] [Google Scholar]
- 45.Xiao, H., O. Perisic, and J. T. Lis. 1991. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell 64**:**585-593. [DOI] [PubMed] [Google Scholar]
- 46.Xing, H., D. C. Wilkerson, C. N. Mayhew, E. J. Lubert, H. S. Skaggs, M. L. Goodson, Y. Hong, O.-K. Park-Sarge, and K. D. Sarge. 2005. Mechanism of hsp70i gene bookmarking. Science 307**:**421-423. [DOI] [PubMed] [Google Scholar]