Microarray Analysis of the Mycobacterium tuberculosis Transcriptional Response to the Acidic Conditions Found in Phagosomes (original) (raw)
Abstract
We used microarrays and real-time reverse transcription-PCR to analyze the global transcriptional response of Mycobacterium tuberculosis to low pH in vitro, which may mimic an environmental signal encountered by phagocytosed mycobacteria. Eighty-one genes were differentially expressed >1.5-fold, including many involved in fatty acid metabolism. The most highly induced genes showed homology with nonribosomal peptide synthetases/polyketide synthases.
Among the first steps in human infection with Mycobacterium tuberculosis is phagocytosis of the bacteria by macrophages in the lung. The bacilli can survive and multiply in this normally hostile environment because the phagosomes do not fully mature into phagolysosomes (4, 8, 48, 52). Phagosomes containing mycobacteria begin to acidify rapidly after phagocytosis. If the bacilli are viable, the pH drops below 6 and then may rise over several hours to approximately 6.5. Dead mycobacteria, however, do not block phagosomal acidification, and the pH drops rapidly to 5.5 or lower (21, 37, 49). These data suggest active inhibition of phagosomal acidification by the bacilli. Acidification itself may be a signal used by mycobacteria to induce the expression of genes needed to alter phagosomal maturation.
Whole-genome microarrays have been used previously to determine gene expression patterns of tubercle bacilli in response to several different environmental conditions (34, 42, 54). One key to the effective use of microarrays for expression studies is to have a well-controlled experimental protocol in which RNA can be rapidly extracted from uniform populations of cells. In our investigation of changes in gene expression following phagocytosis, we chose to use an in vitro acid shock instead of isolating phagocytosed mycobacteria to avoid complications due to difficulties in rapidly isolating mycobacterial RNA from a mixture of mycobacteria and macrophages and in synchronizing phagocytosis and obtaining a uniform population of mycobacteria.
Microarray analysis of transcription after acid shock.
M. tuberculosis H37Rv (TMC102) bacteria were grown in Middlebrook 7H9 broth (Difco, Detroit, Mich.) supplemented with 10% (vol/vol) albumin-dextrose-catalase (Difco) and 0.05% (vol/vol) Tween 80 (Sigma, St. Louis, Mo.) at 37°C to an _A_600 of ≅0.5 on a rotating platform (50 rpm). Bacteria were harvested by centrifugation (5 min, 6,000 × g) at room temperature, resuspended in fresh prewarmed 7H9-T (pH 6.9) medium, and allowed to recover for 3 h at 37°C with shaking. Cells were harvested by centrifugation (1 min, 25,000 × g, 37°C), resuspended in either prewarmed 7H9-T (pH 6.9) or acidic 7H9-T (pH 5.5) medium, and incubated at 37°C with shaking. At 15 and 30 min, samples of each culture were removed and RNA was extracted by a modification of the method of DesJardin (10). The overnight acid shock was performed essentially as described above, except that the cells were incubated for 18 h in the normal or acid broth and the starting cell density was _A_600 ≅ 0.28 to ensure that the final cell density would be near what was used for the short-term acid shocks. The absence of residual DNA was confirmed by the lack of a product after 25 cycles of a PCR with primers specific for the hspX/acr gene. RNA integrity was monitored by agarose gel electrophoresis, and concentration was measured by spectrophotometry. For the various analyses, two replicate RNA preparations were used for the 15-min time points and one each was used for the 30-min and 18-h time points.
M. tuberculosis microarrays, provided by S. Johnston (University of Texas-Southwestern, Dallas, Tex.), consisted of the M. tuberculosis 70-mer oligonucleotide genome set (Operon Technologies, Alameda, Calif.), representing all open reading frames (ORFs) annotated in the H37Rv genome sequencing project (9). Oligonucleotides were robotically spotted in duplicate onto poly-l-lysine-coated slides and blocked by the succinic anhydride-sodium borate-1-methyl-2-pyrrolidinone method as previously described (16).
Fluorescently labeled cDNAs were produced by reverse transcription (RT) of total M. tuberculosis RNA (normal or acid shocked) with Superscript II (Life Technologies) in the presence of Cy3-dCTP or Cy5-dCTP (Amersham Pharmacia, Piscataway, N.J.) by using “arbitrary” decamers to prime cDNA synthesis. Arbitrary decamers were produced by pooling three oligonucleotide syntheses in which an A, G, C, or T residue was randomly incorporated at each position, except that a G or C residue was randomly incorporated at every third position and the first biased G or C substitution fell in either the first, second, or third position of the oligonucleotide in one of the three syntheses. This design takes into account the high overall G+C content of M. tuberculosis and the strong G/C bias in the third codon position of some ORFs (3, 9, 43). An equal mass of each RNA sample was labeled with each dye. For 20-μl RT reaction mixtures, 3 μg of RNA and 2 μg of arbitrary decamers were resuspended in 10 μl of nuclease-free H2O, heated for 10 min at 70°C, and snap-cooled on ice. Ice-cold RT reaction mix (9 μl) containing 200 U of Superscript II reverse transcriptase was added to give final concentrations of 1× first-strand buffer (Life Technologies); 0.01 M dithiothreitol; 250 μM (each) dATP, dGTP, and dTTP; and 62.5 μM dCTP. One microliter of Cy3- or Cy5-dCTP (1 nmol/μl) was added, and the reaction mixtures were incubated for 10 min at room temperature to allow primer annealing and then for 2 h at 42°C. Reactions were stopped by addition of 1 μl of 20 mM EDTA, and the remaining RNA was degraded by base-catalyzed hydrolysis as previously described (22). After the addition of 4 μg of sheared salmon sperm carrier DNA, fluorescent probes were purified with Centrisep columns (Princeton Separations, Adelphia, N.J.).
Competitive microarray hybridizations were performed in duplicate with both dye arrangements (i.e., Cy3-labeled acid plus Cy5-labeled normal cDNAs and Cy5-labeled acid plus Cy3-labeled normal cDNAs) in 55 μl of hybridization buffer (2.6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.2% sodium dodecyl sulfate, 0.7 mg of sheared salmon sperm DNA/ml). The mixed cDNAs were denatured by being heated to 95°C for 2 min, loaded onto microarrays under Lifterslip coverslips (Erie Scientific, Portsmouth, N.H.), and hybridized for approximately 13 h at 67°C. Arrays were washed for 3 min each in 1× SSC-0.03% sodium dodecyl sulfate, 0.2× SSC, and 0.05× SSC at room temperature and then dried by centrifugation (5 min, 1,000 × g).
Fluorescence intensity data from each array were collected with a GenePix 4000 scanner with GenePix Pro 3.0 software (Axon, Union City, Calif.). Data were normalized based on total intensity of good-quality spots above background for each channel, and ratios of acid-shocked to normal cDNA were calculated based on normalized data. In this assay, the ratio of signal from the acid-treated sample to that of the normal sample for a given ORF should represent the relative abundance of the transcripts of that ORF under the two conditions.
The fluorescence ratio of the acid-treated to the normal sample was approximately 1 for the vast majority of the 4,295 ORFs represented on the arrays (see additional information at http://www.cdc.gov/ncidod/dastlr/TB/). Only 23 ORFs displayed ratios of >1.5-fold in comparisons of acid to normal conditions. Most of these were induced <2-fold, but one cluster, Rv3083 to Rv3089, showed considerably higher levels of induction (Table 1). The proximity and orientation of these ORFs to each other, as well as the high level of induction with respect to the rest of the genome, strongly suggested that these genes form an operon. Based on arrangement and expression levels, three other gene clusters which may be operons (Rv1130-Rv1131, Rv2428-Rv2429, and Rv3616c to Rv3614c) were also identified from the microarray data. Many of the 23 induced genes encode products with proposed or known functions, including eight proteins thought to be involved in intermediary metabolism and respiration; two thought to be involved in lipid metabolism; two thought to be involved in virulence, detoxification, or adaptation; one thought to be involved in cell wall and cell processes; and one regulatory protein (http://genolist.pasteur.fr/TubercuList). The remaining nine had no recognizable features or annotations by which to predict functions.
TABLE 1.
M. tuberculosis genes induced >1.5-fold by 15-min acid shocka
| ORF no. | Gene name | Avg induction ratiob | Gene product |
|---|---|---|---|
| Rv3083 | 10.3 ± 5.24 | Probable monooxygenase | |
| Rv3084 | lipR | 4.7 ± 4.96 | Probable acetyl hydrolase |
| Rv3085 | 4.1 ± 2.49 | Short-chain ADH | |
| Rv3086 | adhD | 8.3 ± 3.22 | Zinc-containing ADH |
| Rv3087 | 9.6 ± 2.10 | Conserved hypothetical protein | |
| Rv3088 | 3.4 ± 1.31 | Conserved hypothetical protein | |
| Rv3089 | fadD13 | 7.3 ± 3.10 | Acyl-CoA synthase |
| Rv0467 | icl | 2.2 ± 0.28 | Isocitrate lyase |
| Rv0468 | fadB2 | 1.5 ± 0.64 | 3-Hydroxyacyl-CoA dehydrogenase |
| Rv3614c | 1.8 ± 0.16 | Conserved hypothetical protein | |
| Rv3615c | 1.9 ± 0.14 | Conserved hypothetical protein | |
| Rv3616c | 1.8 ± 0.41 | Conserved hypothetical protein | |
| Rv1130 | 1.8 ± 0.40 | Conserved hypothetical protein | |
| Rv1131 | gltA1 | 1.7 ± 0.21 | Citrate synthase 3 |
| Rv2428 | ahpC | 1.8 ± 0.31 | Alkyl hydroperoxide reductase |
| Rv2429 | ahpD | 1.8 ± 0.26 | Member of AhpC/TSA family |
| Rv1245c | 1.8 ± 0.79c | Putative dehydrogenase | |
| Rv2007c | fdxA | 1.8 ± 0.32 | Ferredoxin |
| Rv1404 | 1.7 ± 0.04 | Transcriptional regulator (MarR family) | |
| MT0196 | 1.6 ± 0.54c | Hypothetical protein | |
| Rv1169c | PE | 1.6 ± 0.28 | PE family protein |
| Rv0582 | 1.5 ± 0.87c | Hypothetical protein | |
| Rv2450c | (rpfE) | 1.5 ± 0.24 | Conserved hypothetical protein (resuscitation-promoting factor) |
Fifty-eight genes appeared to be repressed >1.5-fold after acid treatment (Table 2), including the five-gene kas/FAS II operon (Rv2243 to Rv2247) (45). The acid-repressed genes are predicted to be involved in cell wall and cell processes (eight genes), intermediary metabolism (eight genes), lipid metabolism (seven genes), regulation (two genes), phage or insertion sequences (two genes), PE/PPE family proteins (two genes), and information pathways (one gene). The remaining 28 ORFs encode hypothetical (13 genes), conserved hypothetical (8 genes), and unknown (7 genes) proteins (http://genolist.pasteur.fr/TubercuList).
TABLE 2.
M. tuberculosis genes repressed >1.5-fold by 15-min acid shocka
| ORF no. | Gene name | Avg repression ratiob | Gene product |
|---|---|---|---|
| MT1812 | 2.4 ± 0.17 | Hypothetical protein | |
| Rv2563 | 2.4 ± 0.06 | Possible membrane protein | |
| Rv2243 | fabD | 2.1 ± 0.10 | Malonyl CoA-[ACPc] transacylase |
| Rv2244 | acpM | 1.8 ± 0.41 | ACP (meromycolate extension) |
| Rv2245 | kasA | 2.2 ± 0.24 | β-Ketoacyl-ACP synthase (meromycolate extension) |
| Rv2246 | kasB | 2.1 ± 0.32 | β-Ketoacyl-ACP synthase (meromycolate extension) |
| Rv2247 | accD6 | 1.7 ± 0.26 | Acetyl/propionyl-CoA carboxylase β subunit |
| Rv3415c | 2.1 ± 0.16 | Conserved hypothetical protein | |
| MT1968 | 2.1 ± 0.04 | PPE family protein | |
| Rv1387 | 2.0 ± 0.23 | PPE family protein | |
| Rv2551c | 2.0 ± 0.03 | Hypothetical protein | |
| Rv1585c | 1.9 ± 0.10 | φRV1 phage-related protein | |
| Rv1315 | murA | 1.9 ± 0.12 | UDP-_N_-acetylglucosamine-1-carboxyvinyltransferase |
| Rv3249c | 1.9 ± 0.19 | Transcriptional regulator (TetR/AcrR family) | |
| Rv3250c | rubB | 1.8 ± 0.35 | Rubredoxin B |
| Rv3251c | rubA | 1.5 ± 0.32 | Rubredoxin A |
| Rv3145 | nuoA | 1.8 ± 0.42 | NADH dehydrogenase chain A |
| MT2803.1 | 1.8 ± 0.13 | Hypothetical protein | |
| Rv0475 | 1.7 ± 0.29 | Possible exported protein | |
| Rv2846c | efpA | 1.7 ± 0.65 | Putative efflux protein |
| MT2367.1 | 1.7 ± 0.03 | Hypothetical protein | |
| MT3454 | 1.7 ± 0.09 | Hypothetical protein | |
| MT1118.2 | 1.7 ± 0.03 | Hypothetical protein | |
| Rv2281 | pitB | 1.7 ± 0.55 | Phosphate permease |
| MT0506 | 1.7 ± 0.25 | Hypothetical protein | |
| Rv0965c | 1.7 ± 0.16 | Conserved hypothetical protein | |
| Rv3163c | 1.6 ± 0.11 | Possible membrane protein | |
| Rv3469c | mhpE | 1.6 ± 0.15 | Probable 4-hydroxy-2-oxovalerate aldolase |
| MT2365 | 1.6 ± 0.47 | Hypothetical protein | |
| Rv2545 | 1.6 ± 0.24 | Conserved hypothetical protein | |
| Rv0972c | fadE12 | 1.6 ± 0.32 | Acyl-CoA dehydrogenase |
| Rv3295 | 1.6 ± 0.28 | Transcriptional regulator (TetR/AcrR family) | |
| MT0868 | 1.6 ± 0.05 | Hypothetical protein | |
| MT3024 | 1.6 ± 0.83 | Hypothetical protein | |
| Rv3565 | aspB | 1.6 ± 0.00 | Aspartate aminotransferase |
| Rv3643 | 1.6 ± 0.10 | Hypothetical protein | |
| Rv2689c | 1.6 ± 0.75 | Weak similarity to RNA methyltransferases | |
| MT1401 | 1.6 ± 0.12 | Hypothetical protein | |
| MT3436 | 1.6 ± 0.44 | Hypothetical protein | |
| Rv1897c | 1.6 ± 0.65 | Conserved hypothetical protein | |
| Rv2803c | 1.6 ± 0.09 | Hypothetical protein | |
| Rv3445c | 1.6 ± 0.05 | Hypothetical protein | |
| Rv3151 | nuoG | 1.6 ± 0.46 | NADH dehydrogenase chain G |
| Rv3907c | pcnA | 1.6 ± 0.52 | Polynucleotide polymerase |
| Rv3361c | 1.6 ± 0.36 | Conserved hypothetical protein | |
| Rv3655c | 1.6 ± 0.37 | Hypothetical protein | |
| Rv1645c | 1.5 ± 0.33 | Conserved hypothetical protein | |
| Rv3481c | 1.5 ± 0.30 | Possible membrane protein | |
| Rv3679 | 1.5 ± 0.50 | Possible anion transporter | |
| Rv2305 | 1.5 ± 0.45 | Hypothetical protein | |
| MT2405 | 1.5 ± 0.57 | Hypothetical protein | |
| Rv3541c | 1.5 ± 0.09 | Hypothetical protein | |
| MT2038 | 1.5 ± 0.54 | Conserved hypothetical protein | |
| Rv2893 | 1.5 ± 0.32 | Similar to alkanal monooxygenase alpha chain | |
| MT0740.1 | 1.5 ± 0.44 | Hypothetical protein | |
| Rv2599 | 1.5 ± 0.45 | Conserved hypothetical protein | |
| Rv2791c | 1.5 ± 0.44 | Transposase | |
| Rv3409c | choD | 1.5 ± 0.27 | Cholesterol oxidase |
To confirm the validity of the 1.5-fold cutoff level, the SAM (significance analysis of microarrays) application (50) was utilized to identify statistically significant differentially expressed genes. Nearly all (20 of 23 induced, 58 of 58 repressed) genes listed in Tables 1 and 2 were shown to be significant at stringent levels of analysis (median number falsely called significant genes = 4.12, median false discovery rate = 2.86%).
Confirmation and extension of microarray data with real-time RT-PCR.
Total RNA (1 μg) from either normal or acid-shocked M. tuberculosis was converted to cDNA with Superscript II reverse transcriptase as recommended by the manufacturer with 250 ng of arbitrary decamer primers. The resulting cDNA was used as a template for semiquantitative hybridization probe real-time PCR (TaqMan-style) assays (23). Briefly, equal amounts of acid-shocked and normal cDNAs were added to PCR mixtures containing 0.5 U of Platinum Taq polymerase, 1× Platinum Taq polymerase buffer (Life Technologies), 6 mM MgCl2, 1.25 mM (each) deoxynucleoside triphosphates, 0.2 μM (each) primers, and 0.2 μM probe. Primers and probes were designed with Primer Express software (Applied Biosystems, Foster City, Calif.), and hybridization probes were synthesized with 5′-FAM reporter dyes (Applied Biosystems) and 3′ QSY-7 dark quenchers (Molecular Probes, Eugene, Oreg.) by the CDC Scientific Resources Program (see additional information at http://www.cdc.gov/ncidod/dastlr/TB/). Duplicate reaction mixtures were heated to 95°C for 2 min and then cycled 40 times at 95°C for 15 s and 60°C for 45 s in an ABI 7700 real-time PCR instrument (Applied Biosystems). Change in fluorescence threshold crossing (Δ_C[infi]t_) values were used to compare acid-shocked and normal template levels for each gene relative to an internal control.
Genes were chosen for analysis by real-time RT-PCR based upon the level and reproducibility of changes seen in the microarray experiments or because of suspected involvement in altering phagosome maturation or intracellular survival. The 17 selected genes included three putative operons (Rv3083 to Rv3089, Rv1130 to Rv1132, and Rv2243 to Rv2247) and Rv0467 and Rv3249c (Fig. 1 to 4). Rv1481 was included as an internal control. It encodes a hypothetical integral membrane protein and was previously shown to be stably expressed under both normal and 15-min acid shock conditions (M. A. Fisher, unpublished data).
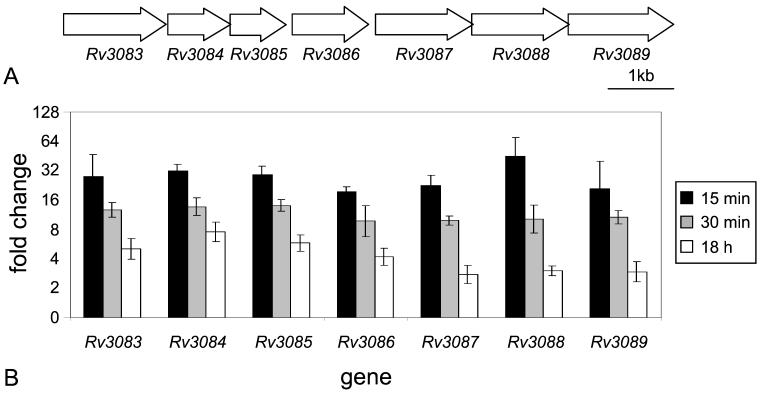
FIG. 1.
Genomic organization and real-time RT-PCR results of Rv3083 to Rv3089 (fadD13). (A) Schematic representation of the arrangement of ORFs Rv3083 to Rv3089 in the M. tuberculosis H37Rv genome. (B) Real-time semiquantitative RT-PCR was performed on RNA isolated from M. tuberculosis cultures incubated in either acidic or normal medium for 15 min, 30 min, and 18 h. Data are presented as the mean change of expression ± standard deviation for each gene between acid and normal conditions.
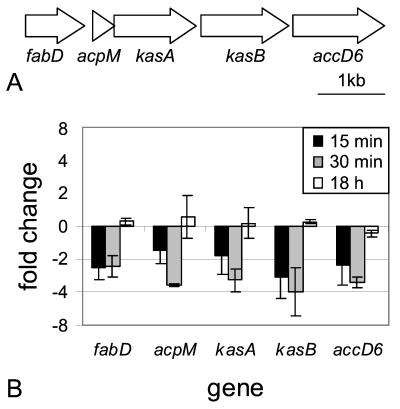
FIG. 4.
Genomic organization and real-time RT-PCR results of Rv2243 (fabD) to Rv2247 (accD6). (A) Schematic representation of the arrangement of ORFs Rv2243 to Rv2247 in the M. tuberculosis H37Rv genome. (B) Real-time semiquantitative RT-PCR was performed on RNA isolated from M. tuberculosis cultures incubated in either acidic or normal medium for 15 min, 30 min, and 18 h. Data are presented as the mean change of expression ± standard deviation for each gene between acid and normal conditions.
The most highly induced genes after acid shock as determined by microarray analysis comprise the putative Rv3083-to-Rv3089 operon (Table 1; Fig. 1). Real-time RT-PCR data showed that all seven genes are induced at similar levels, approximately 17- to 33-fold, after a 15-min acid shock. These genes continue to show induction levels of 10- to 14-fold after 30 min of acid treatment and 3- to 8-fold after 18 h of acid treatment (Fig. 1B).
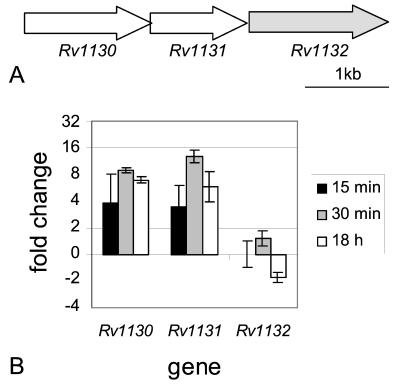
Another putative operon, Rv1130-Rv1131 (Table 1; Fig. 2A), showed only a twofold induction in one 15-min acid-treated sample but an approximately eightfold induction in a second 15-min sample. These genes were induced 9- to 13-fold in the 30-min and 6- to 7-fold in the 18-h acid shock samples (Fig. 2B). A third gene in this cluster, Rv1132, showed no induction after 15 min, very slight induction after 30 min, and slight repression after 18 h of acid treatment (Fig. 2B).
FIG. 2.
Genomic organization and real-time RT-PCR results of Rv1130 to Rv1132. (A) Schematic representation of the arrangement of ORFs Rv1130 to Rv1132 in the M. tuberculosis H37Rv genome. Rv1132 is shaded to indicate that, although it appears to be in the proper arrangement to be coexpressed with Rv1130-gltA1, it is not regulated in a similar manner. (B) Real-time semiquantitative RT-PCR was performed on RNA isolated from M. tuberculosis cultures incubated in either acidic or normal medium for 15 min, 30 min, and 18 h. Data are presented as the mean change of expression ± standard deviation for each gene between acid and normal conditions.
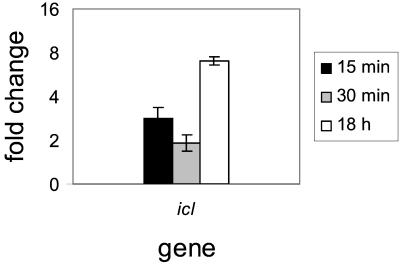
The gene encoding isocitrate lyase 1 (Icl, Rv0467, Fig. 3) was also examined by real-time RT-PCR. Again, this confirmed the induction level (2.5-fold) seen by microarray after 15-min acid shock and also demonstrated slightly less induction at 30 min but significantly higher induction levels (sevenfold) after 18 h of acid treatment (Fig. 3).
FIG. 3.
Real-time RT-PCR results for Rv0467 (icl). Real-time semiquantitative RT-PCR was performed on RNA isolated from M. tuberculosis cultures incubated in either acidic or normal medium for 15 min, 30 min, and 18 h. Data are presented as the mean change of expression ± standard deviation for each gene between acid and normal conditions.
Five of the genes that appeared to be repressed in the microarray data compose what has been referred to as the FAS II or Kas operon (45) (Fig. 4A). Real-time PCR analysis of these genes revealed that they were, in fact, repressed approximately 1.5- to 4-fold after 15 and 30 min of acid treatment, but after overnight acid exposure, expression returned to levels equivalent to those of normal broth controls (Fig. 4B).
Another repressed gene according to microarray experiments was the putative transcriptional regulator Rv3249c. By real-time RT-PCR on 15-min, 30-min, and 18-h acid shock samples, however, little if any repression was detected relative to normal broth controls, emphasizing the need to confirm microarray data with an independent assay (data not shown).
Acidification alters expression of genes involved in lipid metabolism.
Isocitrate lyase is a key enzyme in the glyoxylate pathway which is used to metabolize fatty acids through an acetyl coenzyme A (CoA) intermediate to glucose. The icl genes from both M. tuberculosis and Mycobacterium avium were shown previously to be induced in macrophages (19, 25), and recent evidence demonstrated the requirement of this gene for survival in macrophages and for persistence in mouse lungs (35). The fact that icl is induced by acid (Fig. 3) suggests that M. tuberculosis may sense low pH as a signal that the bacteria are entering the phagosome and should begin to express genes, such as those involved in fatty acid metabolism, needed for long-term survival in that environment. Indeed, four other genes possibly involved in metabolism of fatty acids, fadB2 (Table 1), gltA1 (Fig. 2B), Rv1130 (Fig. 2B), and Rv1169c (Table 1), are also induced by short-term acid exposure. Rv0468 (FadB2) is involved in β-oxidation of fatty acids. It is adjacent to icl and is regulated in a similar manner, but at reduced levels based on this and previous microarray data (34, 42). Rv1131 (GltA1, citrate synthase 3) catalyzes the formation of citrate from acetyl-CoA and oxaloacetate, the end product of the glyoxylate pathway. Rv1130, a conserved hypothetical protein, is homologous to the Salmonella enterica serovar Typhimurium PrpD protein required for propionate metabolism (26-28) and to MmgE, which is encoded by an ORF immediately downstream of citrate synthase 3 in an operon with genes thought to be involved in β-oxidation of fatty acids in Bacillus subtilis (6). Rv1169c displays homology to triacylglycerol lipases.
Some of the acid-repressed genes also impact metabolic pathways, including fatty acid metabolism. For example, decreased expression of the Kas/FAS II operon was seen by microarray (Table 2) and real-time RT-PCR (Fig. 4B) after acid shock. This operon encodes five proteins (FabD, AcpM, KasA, KasB, and AccD6) which are required for the extension of mycolic acids in mycobacteria (32). It is not surprising that the biosynthetic enzymes of one of the most abundant cell wall molecules in M. tuberculosis are modulated by the external environment, that this energy-intensive biosynthetic pathway is transiently down-regulated in response to an environmental insult, and that this pathway returns to normal levels as the bacteria adapt to growing at the lower pH.
Acidification induces a putative nonribosomal peptide synthetase/polyketide synthase (NRPS/PKS).
The genes in the putative Rv3083-to-Rv3089 operon were induced at considerably higher levels than any others in these experiments (Table 1; Fig. 1B). BLAST searches revealed that this cluster of genes is present in all strains of M. tuberculosis subjected to large-scale sequencing efforts (H37Rv, CDC1551, and 210) as well as in Mycobacterium bovis, and homologs of all genes except Rv3086 appear on a single contig of the unfinished M. avium genome (http://www.tigr.org), although the arrangement differs. Rv3083 to Rv3089 are annotated as encoding two conserved hypothetical proteins (Rv3087 and Rv3088) or as having unknown, but probable, functions (Table 1). Further in silico investigation, however, revealed that these gene products display many of the features of NRPSs.
Most NRPSs, termed type I by analogy to type I PKSs, are very large proteins, encoding all functions required for peptide synthesis on a single polypeptide (31, 41). Each module typically contains an adenylation domain and two thiolation domains separated by a condensation domain, and terminal modules often contain a thioesterase domain. The process of nonribosomal peptide synthesis is illustrated at http://www.cdc.gov/ncidod/dastlr/TB/. Many of the typical NRPS domains are functional when expressed as individual polypeptides (12, 14, 20, 30, 38). Furthermore, Du et al. have discovered genes in the bleomycin biosynthetic cluster which encode proteins homologous to individual domains of NRPSs and suggest the existence of type II NRPSs, analogous to type II PKSs, in which the domains are encoded on individual polypeptides (7, 13, 14).
Conserved domain (RPS-BLAST) and SMART utilities (2, 39, 40) reveal that the Rv3083-to-Rv3089 gene products display homologies to many NRPS domains. For example, Rv3089 (FadD13) contains an AMP-binding domain (Pfam00501), as do most adenylation domains of NRPSs. Rv3087 contains a partial condensation domain (Pfam00668) like those responsible for peptide bond formation in NRPSs. Rv3088 is 25% identical and 41% similar to Rv3087, and both products contain the HHXXXDG catalytic site of condensation domains (46). Rv3084 (LipR) contains a partial thioesterase domain (Pfam00975), including the conserved GXSXG motif containing the catalytic serine residue (17, 36). Rv3083 was annotated as a monooxygenase, as it contains a partial flavin-binding monooxygenase-like domain (Pfam00743). A recent report demonstrated the existence of a monooxygenase domain in MtaG, the final NRPS unit of the myxothiazol biosynthetic cluster (44). The monooxygenase domain of this protein is thought to be involved in the formation of the unusual amide terminus of myxothiazol, a mitochondrial electron transport chain inhibitor.
The only domain present in typical NRPSs but not represented in this putative operon is the thiolation domain, which contains a phosphopantetheine moiety that allows interaction of amino acid thioesters with the condensation domain. Several proteins in M. tuberculosis, including PKSs, CoA ligases, and peptide synthetases end with phosphopantetheine binding domains. One of these proteins might provide the necessary thiolation domain activity in this presumptive type II NRPS. Alternatively, activated amino acids could be supplied on soluble intermediates. Indeed, soluble molecules resembling the typical thiolation domain-linked aminoacyl- or acyl-phosphopantetheine thioesters are substrates for various domains (condensation, thioesterase, and ketoreductase) in NRPSs and the mechanistically related PKSs (15, 18, 24, 47). Additionally, Belshaw et al. demonstrated that free aminoacyl-CoA thioesters, which can be synthesized by aminoacyl-tRNA synthases (29), are sufficiently stable to act as soluble intermediates in nonribosomal peptide synthesis (5).
The two remaining genes in this cluster, Rv3085 and Rv3086, may also have activities related to NRPSs. Rv3085 contains a short-chain alcohol dehydrogenase (ADH) domain (Pfam00106), and PSI-BLAST reveals homology to polyketide reductases (e.g., PgaD from Streptomyces sp. PGA64; 27% identical, 42% similar). Rv3086 (AdhD) appears to be a member of the zinc-containing ADH (ADH-Zn) family. ADH-Zn domains (Pfam00107) are also found in M. tuberculosis PKSs including _pks1_and ppsC as well as other PKSs such as those in the mixed NRPS-PKS biosynthetic cluster for rapamycin in Streptomyces hygroscopicus (41). In these gene products, the ADH-Zn domains are often followed by partial short-chain ADH domains, as found in Rv3085. Based on these homologies, Rv3085 and/or Rv3086 could be involved in modification of the incoming substrate, product, or both, of a type II NRPS or could be acting analogously to the respective ADH domains in a PKS (41).
In toto, the genes of the Rv3083-to-Rv3089 putative operon display significant homologies to most of the key NRPS domains and, hence, might form a multienzyme type II NRPS like that hypothesized by Du and Shen (14). Interestingly, many bioactive molecules such as the immunosuppressant cyclosporine (53), the antitumor drug bleomycin (13), and the vancomycin and beta-lactam group antibiotics (1, 51) are built from peptides produced by NRPSs. Furthermore, two M. tuberculosis proteins which are thought to be NRPSs, Nrp and MbtB, have been implicated in facilitating the survival and growth of mycobacteria in macrophages (11, 33). Taken together, these findings suggest that a bioactive product made by this hypothetical multicomponent enzyme could play a role in the phagosomal acidification-inhibiting activity of mycobacteria or in another facet of intracellular survival. Further studies, such as those investigating the behavior of strains lacking these genes and biochemical characterization of the putative gene products, will be required to elucidate the precise roles of these gene products in the inhibition of phagosomal acidification or intracellular survival of M. tuberculosis.
Acknowledgments
We thank Brian Holloway, Karen McKaustland, and Robin Scarborough of the NCID Scientific Resources Program for oligonucleotide synthesis and expert advice on real-time PCR and microarray processing. We thank Stephen Johnston and Adel Talaat of the University of Texas-Southwestern, Center for Biomedical Inventions, for providing microarrays and technical advice on setting up microarray studies.
REFERENCES
- 1.Aharonowitz, Y., J. Bergmeyer, J. M. Cantoral, G. Cohen, A. L. Demain, U. Fink, J. Kinghorn, H. Kleinkauf, A. MacCabe, and H. Palissa. 1993. Delta-(l-alpha-aminoadipyl)-l-cysteinyl-d-valine synthetase, the multienzyme integrating the four primary reactions in beta-lactam biosynthesis, as a model peptide synthetase. Bio/Technology 11**:**807-810. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, G. E., and P. M. Sharp. 1996. Codon usage in the Mycobacterium tuberculosis complex. Microbiology 142**:**915-925. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, J. A., and P. D. Hart. 1971. Response of cultured macrophages to Mycobacterium tuberculosis with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 134**:**713-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belshaw, P. J., C. T. Walsh, and T. Stachelhaus. 1999. Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284**:**486-489. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, E. M., B. W. Beall, and C. P. Moran, Jr. 1996. A σE-dependent operon subject to catabolite repression during sporulation in Bacillus subtilis. J. Bacteriol. 178**:**4778-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carreras, C. W., and C. Khosla. 1998. Purification and in vitro reconstitution of the essential protein components of an aromatic polyketide synthase. Biochemistry 37**:**2084-2088. [DOI] [PubMed] [Google Scholar]
- 8.Clemens, D. L., and M. A. Horwitz. 1995. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J. Exp. Med. 181**:**257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393**:**537-544. [DOI] [PubMed] [Google Scholar]
- 10.DesJardin, L. E. 1999. Isolation of Mycobacterium tuberculosis RNA from sputum. Methods Mol. Med. 48**:**133-139. [DOI] [PubMed] [Google Scholar]
- 11.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97**:**1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dieckmann, R., Y. O. Lee, H. van Liempt, H. von Dohren, and H. Kleinkauf. 1995. Expression of an active adenylate-forming domain of peptide synthetases corresponding to acyl-CoA-synthetases. FEBS Lett. 357**:**212-216. [DOI] [PubMed] [Google Scholar]
- 13.Du, L., C. Sanchez, M. Chen, D. J. Edwards, and B. Shen. 2000. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC 15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 7**:**623-642. [DOI] [PubMed] [Google Scholar]
- 14.Du, L., and B. U. Shen. 1999. Identification and characterization of a type II peptidyl carrier protein from the bleomycin producer Streptomyces verticillus ATCC 15003. Chem. Biol. 6**:**507-517. [DOI] [PubMed] [Google Scholar]
- 15.Ehmann, D. E., J. W. Trauger, T. Stachelhaus, and C. T. Walsh. 2000. Aminoacyl-SNACs as small-molecule substrates for the condensation domains of nonribosomal peptide synthetases. Chem. Biol. 7**:**765-772. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303**:**179-205. [DOI] [PubMed] [Google Scholar]
- 17.Feller, G., M. Thiry, and C. Gerday. 1991. Nucleotide sequence of the lipase gene lip2 from the antarctic psychrotroph Moraxella TA144 and site-specific mutagenesis of the conserved serine and histidine residues. DNA Cell Biol. 10**:**381-388. [DOI] [PubMed] [Google Scholar]
- 18.Gokhale, R. S., D. Hunziker, D. E. Cane, and C. Khosla. 1999. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem. Biol. 6**:**117-125. [DOI] [PubMed] [Google Scholar]
- 19.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96**:**11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guenzi, E., G. Galli, I. Grgurina, E. Pace, P. Ferranti, and G. Grandi. 1998. Coordinate transcription and physical linkage of domains in surfactin synthetase are not essential for proper assembly and activity of the multienzyme complex. J. Biol. Chem. 273**:**14403-14410. [DOI] [PubMed] [Google Scholar]
- 21.Hackam, D. J., O. D. Rotstein, W. Zhang, S. Gruenheid, P. Gros, and S. Grinstein. 1998. Host resistance to intracellular infection: mutation of natural resistance-associated macrophage protein 1 (Nramp1) impairs phagosomal acidification. J. Exp. Med. 188**:**351-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29**:**548-550, 552-554, 556. [DOI] [PubMed]
- 23.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6**:**986-994. [DOI] [PubMed] [Google Scholar]
- 24.Holzbaur, I. E., R. C. Harris, M. Bycroft, J. Cortes, C. Bisang, J. Staunton, B. A. Rudd, and P. F. Leadlay. 1999. Molecular basis of Celmer's rules: the role of two ketoreductase domains in the control of chirality by the erythromycin modular polyketide synthase. Chem. Biol. 6**:**189-195. [DOI] [PubMed] [Google Scholar]
- 25.Honer Zu Bentrup, K., A. Miczak, D. L. Swenson, and D. G. Russell. 1999. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181**:**7161-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horswill, A. R., and J. C. Escalante-Semerena. 2001. In vitro conversion of propionate to pyruvate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry 40**:**4703-4713. [DOI] [PubMed] [Google Scholar]
- 27.Horswill, A. R., and J. C. Escalante-Semerena. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179**:**928-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horswill, A. R., and J. C. Escalante-Semerena. 1999. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 181**:**5615-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakubowski, H. 1998. Aminoacylation of coenzyme A and pantetheine by aminoacyl-tRNA synthetases: possible link between noncoded and coded peptide synthesis. Biochemistry 37**:**5147-5153. [DOI] [PubMed] [Google Scholar]
- 30.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39**:**15513-15521. [DOI] [PubMed] [Google Scholar]
- 31.Keating, T. A., and C. T. Walsh. 1999. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3**:**598-606. [DOI] [PubMed] [Google Scholar]
- 32.Kremer, L., A. R. Baulard, and G. S. Besra. 2000. Genetics of mycolic acid biosynthesis, p. 173-190. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 33.Lagier, B., V. Pelicic, D. Lecossier, G. Prod'hom, J. Rauzier, C. Guilhot, B. Gicquel, and A. J. Hance. 1998. Identification of genetic loci implicated in the survival of Mycobacterium smegmatis in human mononuclear phagocytes. Mol. Microbiol. 29**:**465-475. [DOI] [PubMed] [Google Scholar]
- 34.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41**:**423-437. [DOI] [PubMed] [Google Scholar]
- 35.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406**:**735-738. [DOI] [PubMed] [Google Scholar]
- 36.Naggert, J., A. Witkowski, J. Mikkelsen, and S. Smith. 1988. Molecular cloning and sequencing of a cDNA encoding the thioesterase domain of the rat fatty acid synthetase. J. Biol. Chem. 263**:**1146-1150. [PubMed] [Google Scholar]
- 37.Oh, Y. K., and R. M. Straubinger. 1996. Intracellular fate of Mycobacterium avium: use of dual-label spectrofluorometry to investigate the influence of bacterial viability and opsonization on phagosomal pH and phagosome-lysosome interaction. Infect. Immun. 64**:**319-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5**:**631-645. [DOI] [PubMed] [Google Scholar]
- 39.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28**:**231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95**:**5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarzer, D., and M. A. Marahiel. 2001. Multimodular biocatalysts for natural product assembly. Naturwissenschaften 88**:**93-101. [DOI] [PubMed] [Google Scholar]
- 42.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98**:**7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shinnick, T. M. 1987. The 65-kilodalton antigen of Mycobacterium tuberculosis. J. Bacteriol. 169**:**1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silakowski, B., H. U. Schairer, H. Ehret, B. Kunze, S. Weinig, G. Nordsiek, P. Brandt, H. Blocker, G. Hofle, S. Beyer, and R. Muller. 1999. New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 274**:**37391-37399. [DOI] [PubMed] [Google Scholar]
- 45.Slayden, R. A., R. E. Lee, and C. E. Barry III. 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38**:**514-525. [DOI] [PubMed] [Google Scholar]
- 46.Stachelhaus, T., H. D. Mootz, V. Bergendahl, and M. A. Marahiel. 1998. Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J. Biol. Chem. 273**:**22773-22781. [DOI] [PubMed] [Google Scholar]
- 47.Stindl, A., and U. Keller. 1993. The initiation of peptide formation in the biosynthesis of actinomycin. J. Biol. Chem. 268**:**10612-10620. [PubMed] [Google Scholar]
- 48.Sturgill-Koszycki, S., U. E. Schaible, and D. G. Russell. 1996. _Mycobacterium_-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 15**:**6960-6968. [PMC free article] [PubMed] [Google Scholar]
- 49.Sturgill-Koszycki, S., P. H. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. K. Fok, R. D. Allen, S. L. Gluck, J. Heuser, and D. G. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263**:**678-681. [DOI] [PubMed] [Google Scholar]
- 50.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98**:**5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Wageningen, A. M., P. N. Kirkpatrick, D. H. Williams, B. R. Harris, J. K. Kershaw, N. J. Lennard, M. Jones, S. J. Jones, and P. J. Solenberg. 1998. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5**:**155-162. [DOI] [PubMed] [Google Scholar]
- 52.Via, L. E., D. Deretic, R. J. Ulmer, N. S. Hibler, L. A. Huber, and V. Deretic. 1997. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272**:**13326-13331. [DOI] [PubMed] [Google Scholar]
- 53.Weber, G., K. Schorgendorfer, E. Schneider-Scherzer, and E. Leitner. 1994. The peptide synthetase catalyzing cyclosporine production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr. Genet. 26**:**120-125. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96**:**12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]