Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome (original) (raw)
Abstract
Transcriptional promoters comprise one of many classes of eukaryotic transcriptional regulatory elements. Identification and characterization of these elements are vital to understanding the complex network of human gene regulation. Using full-length cDNA sequences to identify transcription start sites (TSS), we predicted more than 900 putative human transcriptional promoters in the ENCODE regions, representing a comprehensive sampling of promoters in 1% of the genome. We identified 387 fragments that function as promoters in at least one of 16 cell lines by measuring promoter activity in high-throughput transient transfection reporter assays. These positive functional results demonstrate widespread use of alternative promoters. We show a strong correlation between promoter activity and the corresponding endogenous RNA transcript levels, providing the first experimental quantitative estimate of promoter contribution to gene regulation. Finally, we identified functional regions within a randomly selected subset of 45 promoters using deletion analyses. These experiments showed that, on average, the sequence -300 to -50 bp of the TSS positively contributes to core promoter activity. Interestingly, putative negative elements were identified -1000 to -500 bp upstream of the TSS for 55% of genes tested. These data provide the largest and most comprehensive view of promoter function in the human genome.
The regulation of human gene expression is a critical, highly coordinated, and complex process. Gene regulation plays a crucial role in virtually every biological process from coordinating cell division to responding to extracellular stimuli and directing transcription during development (Pirkkala et al. 2001; Ahituv et al. 2004; Blais and Dynlacht 2004). While knowledge of regulation at the level of individual genes is progressing, global characterization of gene regulation currently represents one of the major challenges and fundamental goals for biomedical research. An initial step in achieving this goal is the comprehensive identification of transcriptional regulatory elements in the human genome. Towards this end, the ENCODE (Encyclopedia of DNA Elements) project began in 2004 as a collective effort of many laboratories to identify the functional elements in 1% of the human genome (The ENCODE Project Consortium 2004). In this paper, we describe our efforts to identify and study the transcriptional promoters in the ENCODE regions.
Promoters are the best-characterized transcriptional regulatory sequences in complex genomes because of their predictable location immediately upstream of transcription start sites (TSS). They are often described as having two separate segments: core and extended promoter regions. The core promoter is generally within 50 bp of the TSS, where the preinitiation complex forms and the general transcription machinery assembles. The extended promoter can contain specific regulatory sequences that control spatial and temporal expression of the downstream gene (for review, see Butler and Kadonaga 2002). Despite a substantial body of literature describing transcriptional promoters, because of the 3′ bias in isolation and synthesis of cDNAs (Kimmel and Berger 1987) and the existence of alternative promoters regulating alternative RNA isoforms (Landry et al. 2003), the identification of the true start sites for all human transcripts is far from complete. Several groups have recently developed large resources of full-length enriched cDNA sequences, including the Database of Transcriptional Start Sites (DBTSS), which contains 11,234 human genes (Suzuki et al. 2002, 2004), as well as the Mammalian Gene Collection (MGC), which contains 12,228 genes (Gerhard et al. 2004). These databases provide sequences enriched for the 5′ ends of genes, but they still contain a significant number of incomplete and artifactual sequences, emphasizing the need for further experimental validation to identify the true TSS and corresponding promoters of all the genes in the human genome. The Eukaryotic Promoter Database is one such resource, but it currently contains only 1871 human promoters (Cavin Perier et al. 1998; Praz et al. 2002), a small fraction of the estimated total.
In previous work we used full-length MGC sequences to predict more than 10,000 distinct human promoters. A random sampling of 150 predicted segments from this data set showed that more than 90% of predicted promoters had significant activity in at least one of four cell lines tested (Trinklein et al. 2003). The full-length cDNA databases have grown substantially since our initial work. By using the most up-to-date sequences generated by the MGC, DBTSS, RefSeq, and other cDNA sequences in GenBank, we predicted all TSS within the ENCODE region, including those of known highly tissue-specific genes. We tested these putative promoter fragments in 16 diverse human cell lines using transient transfection reporter assays. Here, we describe the identification and functional characterization of nearly 400 functional promoters in the ENCODE region, including those driving transcription of 66 genes with functional alternative promoters.
In addition to expanding the catalog of known functional promoters, we addressed several important biological questions regarding promoter function. We calculated the correlation of endogenous transcript levels and promoter activity for a sample of genes. While other transcriptional regulatory elements, such as enhancers, silencers, and insulators, all modulate the function of promoters and affect steady-state RNA levels in vivo, we quantify the contribution of promoters and demonstrate that in many cases promoters play a key role in controlling RNA levels.
We also studied the promoter activities of deletion constructs for a set of 45 promoters, allowing the identification of core promoter elements and other elements within the extended promoter that contribute to regulation of transcription initiation. Finally, we identified significant overlap between functional promoter regions and binding of TBP-associated factor (TAF1, also TAFII250) and RNA Polymerase 2 (POLR2B) and elements conserved among mammalian genomes, each of which were identified in independent experiments done by other ENCODE Consortium members. Together these results reveal an unprecedented view of promoter activity in 1% of the human genome and lend insight into promoter function in the genome as a whole.
Results
921 predicted ENCODE promoters
By aligning 153,645 human cDNAs to the genome and merging transcripts with overlapping exons on the same strand, we predicted 38,412 gene models in the human genome (see Methods). In agreement with previous observations, approximately 13,450 (35%) of these contained only putative single-exon transcripts (Imanishi et al. 2004). From these gene models, we predicted 56,940 potential TSS in the genome, with roughly half of the genes predicted to have multiple promoters. Within the 30 Mb of the ENCODE region, there were 613 gene models, 27% of which were comprised of single-exon transcripts, many of unknown function. We predicted a total of 921 TSS associated with these gene models. These predictions overlap nearly 80% of the 875 known genes (July 2003 freeze of UCSC Genome Browser) and 74% of Ensembl genes (July 2003 freeze of UCSC Genome Browser) (Karolchik et al. 2003). Consistent with our genome-wide estimates, we predicted that 45% of the ENCODE genes had more than one promoter, which is substantially higher than previous estimates (Landry et al. 2003). While there are a number of well-characterized single-exon genes (Hentschel and Birnstiel 1981; Gentles and Karlin 1999), we considered that the large number of putative single-exon transcripts identified in the full-length cDNA libraries might result from genomic poly(A) stretches or other library artifacts. As a result, we tested only a sample of the predicted single-exon promoters. All together, we cloned 642 putative promoters and measured their promoter activities in 16 cell lines. These included 528 putative promoters based on multi-exon transcript and 114 single exon-based predictions and represent 443 of our gene models (Table 1).
Table 1.
Promoter activity by class
| Total | Positive | Total | Positive | HiConf | Positive | ||
|---|---|---|---|---|---|---|---|
| Multi-exon | 528 | 351 (66.3%) | Longest | 320 | 75.0% | 247 | 79.4% |
| Alternate | 208 | 53.3% | 159 | 57.9% | |||
| Single-exon | 114 | 36 (31.6%) | Longest | 70 | 35.7% | 27 | 44.4% |
| Alternate | 44 | 25.0% | 20 | 20.0% |
Identification of 387 functional promoters in the ENCODE region
We defined the level of activity of a cloned promoter in our transient transfection assay as a transformed ratio of firefly luciferase (experimental) to Renilla luciferase (transfection control) signal, normalizing for transfection efficiency and allowing comparison between experiments. As described in the Methods, we considered the threshold for positive promoter activity as three standard deviations above the mean ratio of the 102 negative control DNA fragments. We considered a fragment as a functional promoter if it had activity exceeding this threshold. We identified 1-3 outliers per cell type within the 102 negative controls, estimating a false-positive rate for the assay of 1%-3%. Using the thresholds defined for each cell type individually, 387 fragments, representing 303 unique gene models, in the ENCODE region showed promoter activity in at least one of the 16 cell types. We observed a much higher validation rate among promoters predicted by multi-exon gene models (66%) than among those predicted by single exon transcripts (32%) (Table 1). Predicted alternative promoters were less likely to show significant activity than predictions based on longest cDNAs in each gene model. Finally, our high confidence predictions were most likely to be active promoters.
In addition to these classes, we note that the ENCODE region, like the remaining 99% of the human genome contains a prominent class of divergently transcribed genes regulated by putative bidirectional promoters. In agreement with our previously published work (Trinklein et al. 2004), we identified 44 and tested 32 promoters involved in bidirectional gene pairs and found that 31 functioned in at least one of the tested cell types. All of those tested in both orientations functioned bidirectionally.
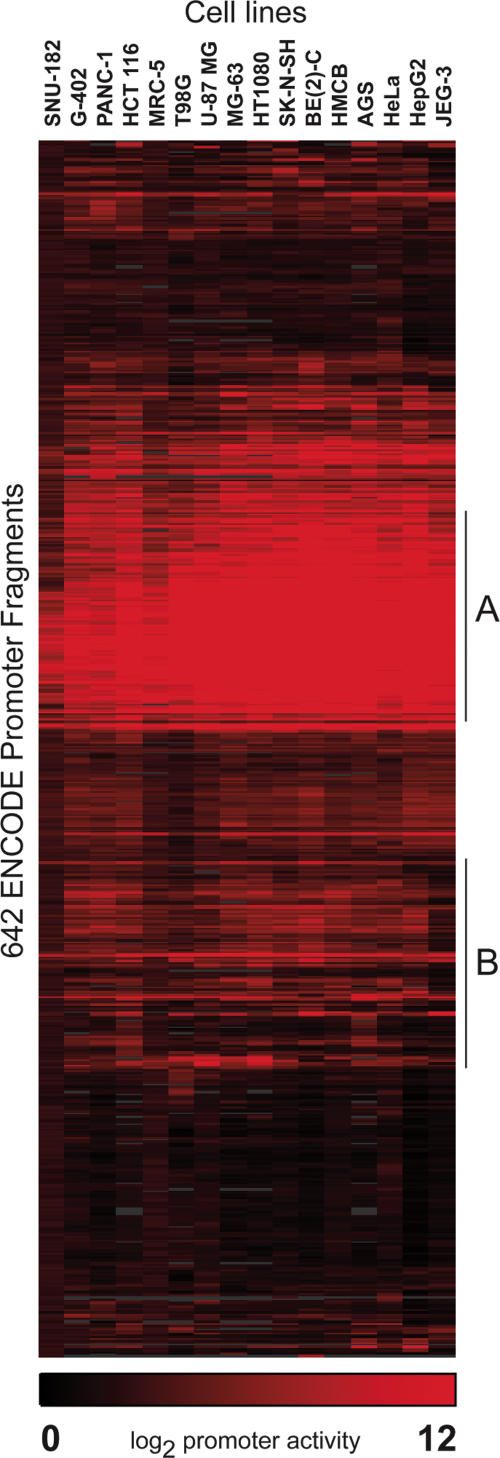
Overall, 60% of the putative promoter fragments we tested were functional in at least one cell type (Fig. 1). Many of these exhibited a high degree of variation in promoter activity between cell types (Fig. 1B), suggesting that regulatory elements within the extended promoter guide cell-type specific expression, even taken out of genomic context. We do not expect the promoter assays to recapitulate perfectly the regulation of the endogenous gene, but we found several instances in which the promoters directed cell-type specific expression similarly in vitro as they do in vivo. For example, the promoter of the hepatocyte growth factor (MET) gene was active in only seven of the 16 cell lines and was most highly active in one of the liver cell lines, HepG2. This is consistent with the expression of MET in a variety of tissues, but predominantly liver and other tissues of mesenchymal origin (Rubin et al. 1993). The osteoclast-associated receptor (OSCAR) promoter was active in only four cell lines, one of which is MG-63, an osteosarcoma cell line. This gene is thought to be expressed exclusively in osteoclasts (Kim et al. 2002). Although our data support the expression of this gene in osteoclasts, we observed promoter activity in additional tissues, suggesting that our assay does not capture all of the regulation controlling the specific expression of this gene. In addition to tissue-specific activity, we identified a prominent cluster of 118 promoters (30% of the total) that had strong, ubiquitous activity in all 16 cell lines (Fig. 1A). Within this cluster, 101 promoter fragments (86%) overlapped CpG Islands, as predicted by the UCSC Genome Browser Database (Karolchik et al. 2003). These data indicate a close association between the presence of CpG dinucleotides and strong, ubiquitous promoter activity. However, 12% (25/202) of the fragments we tested that overlap CpG islands had no promoter activity in any of the 16 cell types. Overlap of CpG islands with the predicted TSS was less common in these 25 cases, but we did not observe a significant difference in CpG content or length between functional and nonfunctional promoters overlapping CpG islands. These data suggest that while the CpG island overlap is an important indicator, it is not sufficient to predict promoter activity.
Figure 1.
Clustergram of 642 putative promoter fragments. The clustergram illustrates the hierarchical clustering of promoter activity among 16 diverse cell lines. Each row indicates the promoter activity of a fragment in each of the cell lines, with red indicating the degree of activity and black indicating no activity. Promoter activity has been normalized and log transformed to reflect comparable values between cell lines. Area A represents a cluster of promoter fragments with strong, ubiquitous activity in all cell lines and area B represents a cluster of promoter fragments that exhibit variable function across the 16 cell types.
Sequence characteristics of promoters
The global sequence content as well as the presence of known DNA motifs within this large data set provide additional insight into promoter function. Because many promoters overlap CpG islands, there is a strong shift in the distribution of GC content in functional promoters. All active promoter fragments have a significantly higher GC content (57%) than putative promoter fragments with no observed activity (48%). The overlap with CpG islands and increased GC content within active promoters is the most striking sequence characteristic distinguishing functional promoters from predicted but nonfunctional promoters in our assay.
We determined the presence of previously characterized promoter-specific motifs in our functionally characterized promoters by doing a simple pattern match for the consensus sequences within our functional promoters. We identified 61 functional promoters (16% of total) containing a TATA-box (TATA(T/A)(T/A)) and 72 functional promoters (19% of total) containing a CAAT (CCAAT) box. However, in agreement with previous work, we did not find any significant correlation between the presence of these motifs and promoter activity (Trinklein et al. 2003). This suggests that while these motifs may be functionally important, there is no universally required element within promoters necessary for promoter activity.
Using a set of constrained elements identified for all ENCODE targets based on comparisons of human genomic sequence to orthologous sequence from 6-9 mammalian species for each target (Cooper et al. 2005), we characterized the extent of constraint in the 500-bp functional promoters that we identified. We found that 12.5% of bases within functional promoters are constrained, whereas 10% of bases within nonfunctional promoters were constrained. Both of these are well above the total of 4.3% constrained bases in 30 Mb of the ENCODE regions as defined by these methods. Interestingly, the vast majority of constraint above random is observed within ±50 bp from the transcription start site (Supplemental Fig. 1). The peak of conservation we observe at position +1 relative to the TSS is very encouraging as it speaks to the accuracy of our TSS predictions. These data also suggest that the basal elements are more likely to be evolutionarily constrained. However, the extended promoter contains more constraint than expected by chance, showing evidence for a reduced but still significant density of functional and constrained elements in this region.
More than 20% of genes have functional alternative promoters
We predicted multiple promoters, each regulating a unique RNA isoform, for 45% of multi-exon genes in the ENCODE regions and have functional data supporting multiple active promoters for approximately 22% of the gene models that we tested in the transient assay. Most of these (54/66) had two functional promoters, but the UDP glycosyltransferase 1 gene (UGT1A10) shows evidence for seven functional promoters. Despite requiring full-length clones for alternative promoter prediction, only half of alternative promoter predictions were validated. This may be explained by highly tissue-specific alternative promoters or by annotated full-length cDNAs that are not truly full length. Interestingly, in some cases, use of these alternative promoters results in predicted altered protein products. Of the 66 gene models with more than one functional promoter, 42 alternative isoforms have similarity to each other, and only six have identical amino acid sequences. The remaining 18 result in protein products with no significant similarity to each other. Our method of defining gene models can be affected by chimeric transcripts or misaligned cDNAs. In these cases, two potentially unrelated transcripts can be included in the same gene model, and these transcripts define alternative promoters of the same gene model with different open reading frames (ORFs). Six of the 18 cases mentioned above involve short single-exon transcripts that overlap one or more exons in a longer multi-exon gene, and it is not surprising that these transcripts have different predicted ORFs. On manual inspection, we observed that in 10 of the remaining 12 cases, transcripts derived from alternative promoters have a similar exon structure with the exception of the 5′ exons. These transcripts use an alternative start codon that results in a completely different ORF. These proteins may have important biological functions of their own, or the existence of an alternate promoter and downstream transcript may act as a regulatory mechanism for the functional protein. Work from other groups has provided examples in which a secondary, unrelated protein, sharing coding exons with a primary transcript, plays a role in the regulation of the primary transcript (Yang et al. 1998). In some cases, these transcripts may act as regulatory RNAs, creating no protein at all, or they may be completely unrelated genes, sharing exonic sequences.
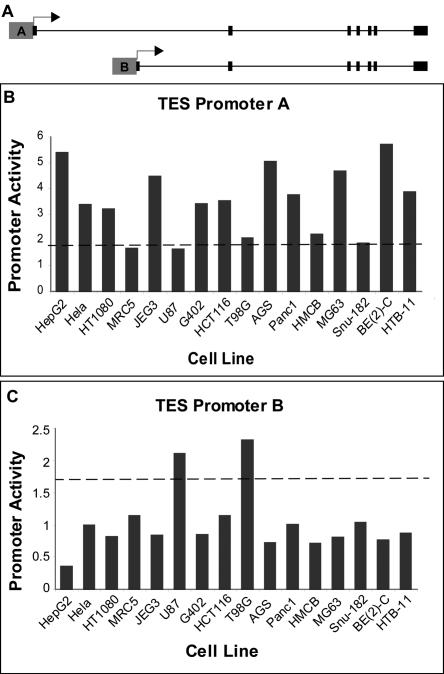
In addition to changing the amino acid sequence of the protein, alternative promoters provide distinct regulation for alternate isoforms of the same gene. Our results indicate that 60% of alternative promoter pairs have significantly different expression patterns among the 16 cell lines we tested. For example, the testin (TES) gene has evidence for two promoters. The TES gene is ubiquitously expressed and has three isoforms and two putative promoters (Tatarelli et al. 2000). We found one promoter active in two of the brain cell lines (Fig. 2B) and a second promoter active in twelve remaining cell lines (Fig. 2C). In this case, the protein product is unaffected by the alternative promoter, but these promoters may be used to provide differential regulation of this gene in various tissues. Looking closely at the data from Tatarelli et al. (2000), we see that expression in the brain is much lower than in other tissues, and this may be explained by the use of an alternative promoter. This is just one example of alternative promoters functioning to differentially regulate transcription of alternate RNA isoforms.
Figure 2.
Two promoters differentially regulate testin gene. (A) Gene structure of testin (TES) gene. (B,C) Promoter activity for promoters of the TES gene in 16 tested cell types represented as a transformed firefly luciferase/Renilla luciferase ratio. (B) Promoter A shows activity in 12 of the 16 tissues but little activity in two brain cell lines, U87 and T98G. (C) Promoter B has significant activity only in U87 and T98G, both brain cell lines.
Functional regions within extended promoter fragments
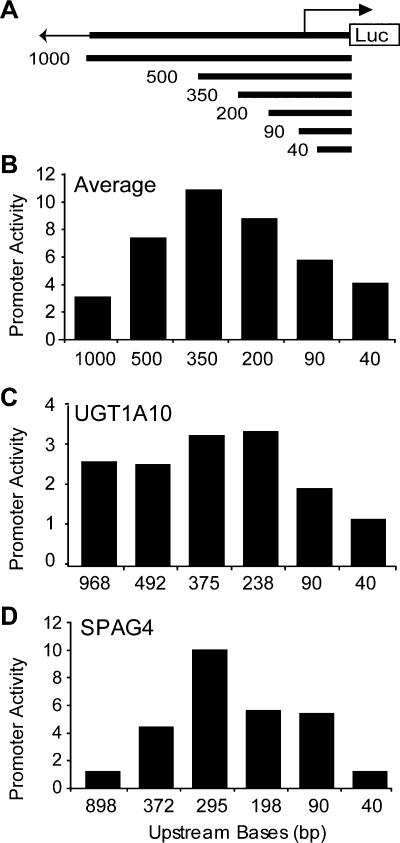
To understand further the functional elements within the extended promoter region, we generated reporter constructs with a series of nested deletions for 45 of the promoters that were active in the transient assay. The deletion fragments (described in Methods) range in size from 40 bp to 1000 bp and were cloned upstream of the luciferase gene as diagrammed in Figure 3A. These fragments were assayed for promoter activity as before and the average activity for each deletion construct illustrates a number of interesting points (Fig. 3B). First, promoter activity decreases with deletion of sequences between 350 bp to 40 bp upstream of the TSS, indicating the presence of positive elements between -350 and -40 bp relative to the TSS in many of these promoters. We found that in 17 of 25 cases, the presence of 40 bp upstream of the predicted transcription start site was sufficient for basal activity that was significantly above background, but only five of these core promoter fragments had activity that was at least 90% of the 500-bp extended promoter fragment.
Figure 3.
Reporter activity of promoter deletion constructs. (A) Diagram of promoter deletion constructs. (B) Average promoter activity observed for each of the 6 constructs of decreasing upstream sequence (1000 bp, 500 bp, 350 bp, 200 bp, 90 bp, 40 bp). The average represents normalized activity of constructs in 45 promoters and seven cell lines (HT1080, HeLa, HCT116, G-402, AGS, T98G, and JEG3). The promoter activity, assayed in triplicate and represented as normalized firefly luciferase/Renilla luciferase ratio, provides a transfection-normalized value to compare activity within and between cell lines. (C) Average activities of promoter fragments for the UDP glycosyltransferase gene (UGT1A10) across seven cell types. (D) Average activities of sperm-associated antigen 4 (SPAG4) promoter fragments across seven cell types. The 898-bp fragment of the SPAG4 promoter shows considerably less activity than the 372-bp fragment.
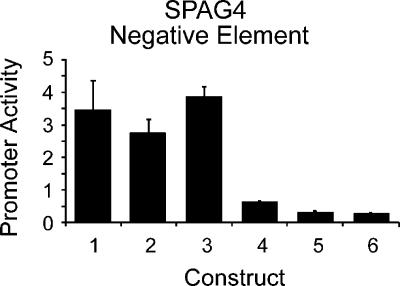
We also observed that, on average, the 500-bp and 1000-bp promoter fragments showed decreased activity compared with the corresponding 350-bp fragment. Overall, we see a reduction in activity of the larger fragments, but we observed a range of behaviors for individual promoters (Fig. 3C,D). Like the sperm-associated antigen 4 (SPAG4) promoter (Fig. 3D), many (12/22) of the 1000-bp and 500-bp fragments showed significantly less activity than the 350-bp fragment of the same promoter in all seven tested cell types. These results suggest the presence of negative regulatory elements in the region -350 to -1000 bp upstream of the TSS for many of these genes. We examined the sequences of these fragments and could not identify any simple sequence elements such as stop codons or long repetitive stretches beyond what is expected by chance, nor could we identify any significant secondary structure to explain these results (data not shown). We conducted experiments to demonstrate that the change in activity we observed was not a result of increased plasmid size by cloning the 500-bp promoter in duplicate or cloning 500 bp of random sequence upstream of the 500-bp promoter (Fig. 4, cf. construct 1 with constructs 2 and 3).
Figure 4.
Negative regulatory element in SPAG4 promoter. Average promoter activity across two cell types, HT1080 and HCT116, of six constructs: Construct 1, SPAG4 372-bp fragment; Construct 2, SPAG4 372-bp promoter cloned in tandem duplicate to control for size; Construct 3, 500 bp of random sequence cloned upstream of the SPAG4 372-bp promoter; Construct 4, SPAG4 898-bp fragment; Construct 5, SPAG4 -898 to -372 fragment cloned upstream of heterologous promoter A; Construct 6, SPAG4 -898→372 fragment upstream of heterologous promoter B. Error bars indicate one standard deviation from the mean of four replicates of each construct.
To test further the hypothesis that these fragments contain a negative regulatory element, we cloned the -1000- to -500-bp fragments of five promoters upstream of two 40-bp heterologous promoters that are otherwise highly active in these cell types (Fig. 4, constructs 5 and 6). These results strongly support the presence of a negative element in this region of the SPAG4 promoter. Of the five fragments we examined, we found evidence that three of these contain negative regulatory elements (see Supplemental data). The others may act as position-specific or gene-specific negative elements.
Endogenous transcript levels correlate with promoter activity
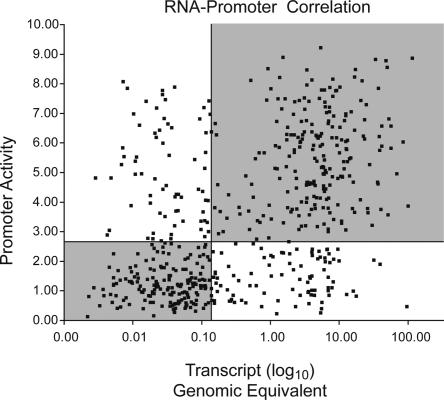
Given the variety of transcriptional regulatory elements known to exist outside of the promoter regions of genes as well as post-transcriptional regulatory mechanisms, we wished to quantify the extent to which the activity of promoter fragments correlates to the steady state endogenous transcript levels in the same cell types. We used quantitative RT-PCR to assay the absolute endogenous transcript levels for 35 genes whose promoter activity we measured in reporter assays in 14 cell types. In addition, we collected more comprehensive data for 96 additional genes in one cell type. We observed a correlation of r = 0.53 between endogenous RNA levels and the promoter activity predicted by its TSS (Fig. 5). To assess the significance of this correlation, we calculated the correlation coefficient of randomized data 1000 times. The average correlation coefficient of these randomized data sets was 0.026 with a standard deviation of 0.04, indicating that the observed correlation is highly significant compared with random (P < 10–12). This correlation indicates that the extended promoter fragments contain many of the elements important for regulating the transcription of these genes in vivo.
Figure 5.
Scatterplot of endogenous RNA transcript levels versus promoter activity. RNA levels, expressed as absolute genomic equivalents, are plotted on the _x_-axis and the normalized promoter activity is shown on the _y_-axis. We calculated the correlation coefficient, r = 0.53. (R2 = 0.28). Quadrants' boundaries are set by the median RNA transcript level (0.17 genomic equivalents) and median promoter activity (2.69 firefly luciferase/Renilla luciferase ratio).
The RNA data also allows us to assess false-positive and false-negative rates, which indicate how well promoter activity predicts in vivo RNA transcript levels. Across 14 cell types and 35 genes, we find 58/273 (21%) active promoter fragments have no detectable RNA transcript and 72/217 (33%) inactive promoters have detectable RNA transcript. There are a variety of biological explanations for these apparent discrepancies. Promoters that function in our assay but do not seem to function in vivo can be explained by a promoter taken out of context, removed from epigenetic signals or relevant regulatory sequences or by an RNA with low abundance and high turnover. These data also confirm our expectation that for a fraction of expressed genes, we have incorrectly predicted the promoter. Nonetheless, the degree of correlation we observed indicates we have captured much of the regulatory sequence relevant to gene expression.
In addition to these genes, we measured the correlation between transcript levels and promoter activity for 11 genes with alternative promoters. In many cases, genes with two promoters and unique RNA isoforms showed activity consistent with one another (see Supplemental Fig. 2). Of the 11 genes with alternative promoters that we tested, seven had promoter activity patterns that matched the trends seen in the corresponding transcript levels. These data provide further evidence that promoters and alternative promoters contribute significantly to the control of RNA levels within a cell and that we are able to recapitulate aspects of this regulation with the transient transfection assay.
Functional promoters co-occur with TAF1, POLR2B binding
Other researchers in the ENCODE Consortium have generated data useful to understanding the activity of the promoters we have identified. Specifically, chromatin IP-microarray experiments examining the occupancy of two promoter-binding proteins, TAF1 and POLR2B, have been produced by our collaborators (Kim et al. 2005) and confirmed in a reporter assay in our own laboratory. These experiments measure ChIP-enriched targets by genomic tiling microarray hybridization. Using a stringent cutoff for identification of binding (P < 10–4 for TAF experiments and P < 10–6 for POLR2B experiments), we compared functional promoter fragments with regions bound by these two transcription factors and made the following observations (Table 2). Of the 258 functional promoters we identified in the two cell types common to our experiments (HCT116 and HeLa), approximately half overlapped either TAF1 or POLR2B sites identified by chromatin IP. Conversely, of the 177 TAF1 binding sites and 203 POLR2B binding sites we tested in the reporter assay, over 80% showed significant activity. Finally, of promoters bound to both POLR2B and TAF1, 85% had significant promoter activity.
Table 2.
Locations of promoter-binding factors, TAF1 and POLR2B overlap functional promoters
| Factor binding sitesa | Promoter predictions overlapping sitesb | Tested promoters overlapping sitesc | Functional promoters overlapping sitesd | |
|---|---|---|---|---|
| TAF1 | 426 | 248 | 177 | 143 (81%) |
| POLR2B | 553 | 288 | 203 | 162 (80%) |
Discussion
Comparison to previous functional promoter studies
The experiments presented here represent the comprehensive functional testing of DNA fragments likely to be transcriptional promoters in a selected 1% of the human genome. Overall, 60% of the predicted promoters showed significant activity in at least one cell type in the transient transfection reporter assay. The fraction of active promoters is substantially lower than the 90% positives established in a previous, smaller study we described in 2003 (Trinklein et al. 2003). One likely explanation for the discrepancy is that the promoters predicted in our previous work relied exclusively on full-length cDNA sequences from an early version of the MGC. This early collection was likely biased towards highly expressed genes, and consequently, the promoters we initially predicted were upstream of ubiquitously and highly expressed genes. In addition, the ENCODE targets contain many genes known to be highly tissue specific, including the genes of the HOXA cluster and the beta (HBB) and alpha (HBA1/2) globin gene clusters. The promoters of these genes are less likely to be active in a limited panel of cell lines, where factors necessary for transcription initiation may be absent.
Because of the distinct goal of identifying all functional promoters in this region, the method used to predict promoters in the ENCODE region was also considerably different than that used in our previous study, which aimed to verify predictions based exclusively on the MGC full-length cDNA collection. By using alignments of all the cDNAs in GenBank, we included promoter predictions based on weak evidence (either there was no full-length clone to validate the prediction or only a single cDNA supported the existence of a TSS). This strategy introduced false predictions but allowed a more complete identification of promoters within the ENCODE region. In support of this, our data for bidirectional promoters is directly comparable with previous work and shows a similar high validation.
As with the earlier experiment (Trinklein et al. 2003), false-negative results arise because of the artificial nature of the transient reporter assay. By cloning the promoter fragment in a plasmid, we require the cloned fragment to function independently, and we may not be able to detect the activity of promoters that require elements outside the 500 bp that we tested. Although we must take care in analyzing negative results, using a large number of random fragments as a baseline for no activity ensures that positive results are more definitive. With a false-positive rate of 2%, we are confident that the vast majority of positive promoter activity identified by our assay represents biologically relevant promoter activity. The data we present here represents one of the largest functional promoter data sets and provides a valuable resource for a large number of researchers studying these regions.
A significant fraction of transcripts of unknown function have functional promoters
Several recent studies have shown that a significantly larger fraction of the genome is transcribed than previously thought (Kapranov et al. 2002; Bertone et al. 2004). It remains to be seen whether these “transcripts of unknown function” (TUFs) have an important biological activity and, if so, how their expression is regulated. About half of the single-exon gene models and a much smaller fraction of multi-exon gene models that we predicted for this study fit the category of TUFs, lacking a known function or an ORF of longer than 100 amino acids. We must cautiously interpret negative results, but the considerable difference in validation between the single-exon-based prediction and multi-exon-based predictions suggests a biological difference between the two classes. This difference suggests that either a larger fraction of TUFs are cDNA library or alignment artifacts or that their promoters are less likely to function in the experiments we have designed. Nevertheless, our data indicate that one third of the sequences upstream of these single-exon transcripts are functional promoters, and the presence of an ORF of at least 100 amino acids is not predictive of promoter function in this class of transcripts. In accordance with the low abundance of some of the TUFs, two thirds of active TUF promoters function in at least one but no more than 10 of the 16 cell types tested, while less than half of the multi-exon predicted promoters meet these criteria, suggesting that TUFs may be more likely to be expressed in a specific time or place. While these data support the hypothesis that some TUFs are regulated and biologically important, the possibility exists that these transcripts are in regions of the genome that have leaky transcriptional activity and the reason for their existence is the presence of a spurious upstream promoter-like sequence. Ongoing experiments within the ENCODE Consortium to characterize the regulatory elements of novel transcribed regions will prove helpful in determining which of the TUFs are functionally relevant and specifically regulated.
Core promoters and upstream regulatory elements
Our observations that 68% of 40-bp core promoter fragments maintain basal promoter activity and that these fragments contain much of the constraint observed in promoters emphasize the importance of the core promoter. However, the deletion analyses we report also demonstrate that additional regulatory sequences are present throughout the extended promoter. Successive removal of sequences in the -350- to -40-bp region of the promoters significantly reduces promoter activity in the transient transfection assay, indicating that these regions contain positive regulatory elements. In contrast, the region upstream of -350 tends to contain elements that negatively affect transcription initiation. This trend was particularly striking within a few of the -1000- to -500-bp regions.
These experiments can lead to interesting hypotheses about gene regulation. For example, our experiments demonstrate a negative element within the SPAG4 promoter meeting the criteria for classically defined silencers (Ogbourne and Antalis 1998). The SPAG4 gene is expressed exclusively in spermatid cells during tail elongation (Tarnasky et al. 1998) and an element located between -372 and -898 from the TSS could act to control tissue-specific expression of this gene by inhibiting expression in other cell types. While tissue-specific expression initiated by a tissue-specific positive element is common, precedence for tissue-specific regulation by a negative element has also been previously established in neurons, where gene expression is controlled by the neuron-restrictive silencer element and the factor that binds it (Schoenherr and Anderson 1995; Schoenherr et al. 1996). The fragments containing negative elements that we have identified provide a detailed resource for researches interested in the regulation of these genes.
Regulatory contribution of promoters to endogenous transcript levels
One of the fundamental questions in the field of gene expression is the relative contribution of the extended promoter region to the regulation of transcription. Long-range regulatory elements, such as enhancers, silencers, and insulators, have been identified and shown to play an important role in spatial and temporal regulation of gene expression, particularly during development (Howard and Davidson 2004). However, the extent of this type of regulation remains to be seen. Furthermore, epigenetic alterations, such as DNA methylation and covalent histone modification, also contribute to gene expression by altering chromatin conformation (Lunyak et al. 2004). Post-transcriptional mechanisms affecting mRNA processing and stability also play a role in regulating steady-state mRNA levels (Meyer et al. 2004; Wilusz and Wilusz 2004). With all of these contributing factors, there is little experimental evidence to allow a quantitative estimate of the contribution of promoters to human gene expression on a large scale. Our studies of promoter activity in the ENCODE region gave us the unique opportunity to measure the correlation of promoter function with mRNA transcript levels.
The steady-state mRNA levels we measured are affected by a variety of transcriptional and post-transcriptional factors, all of which would be expected to reduce the correlation between promoter function and mRNA levels. Nevertheless, we observed a remarkably high correlation between promoter activity and the levels of endogenous mRNA in each cell type, indicating that extended promoters play a significant role in regulating transcript levels. Based on the calculated correlation coefficient of 0.53 (R), 28% (R2) of the variation observed in transcript levels can be attributed to differences in promoter activity. This is likely an underestimate of overall promoter contribution because of the inherent experimental noise in the promoter activity measurements and mRNA quantification. Most genes likely require a combination of regulatory inputs. The continuous distribution of correlations between promoter function and mRNA levels among genes supports this hypothesis. Experimental noise certainly contributes to this continuous distribution; however, the wide distribution supports the notion that some genes are regulated entirely by their promoter, while other genes rely on other elements to control expression. Genes that show strong correlation between promoter and RNA levels could be studied further by mutational analysis to locate the specific regions of the promoter that confer the observed regulation.
Integrating data to reveal promoter function
The integration of multiple data sets generated by the ENCODE Consortium serves to validate the different experimental approaches. The locations of active promoters and TAF1 and POLR2B binding sites throughout the ENCODE regions overlapped significantly. Of the sites bound by both TAF1 and POLR2B, and that were tested in our reporter assays, 85% were active promoters. The strong overlap between the positive results of the two experiments serves to validate both approaches as they independently identify many of the same functional promoters. The minority of fragments that were bound by both factors but were not functionally active in the reporter assays could represent sites where the preinitiation complex was assembled but paused and not transcriptionally active (Krumm et al. 1992, 1995). Additional work measuring the levels of the endogenous transcripts of these genes could confirm which sites represent paused complexes rather than false-positive chromatin IP results or false-negative reporter data.
Most surprisingly, we found many examples of active promoters measured in our assay that did not bind either TAF1 or POLR2B binding. Although this is partly due to the stringent threshold we set for TAF1 and POLR2B binding, one biological explanation is that long-range negative elements acting on these promoters in vivo prevent TAF1 and POLR2B from binding and when taken out of their genomic context and separated from negative elements, these fragments act as promoters in the transient-reporter system. This may reflect true biological activity relevant in certain cell types or under certain conditions.
Furthermore, we identified seven genes with active promoters that do not bind either TAF1 or RNAP II but have detectable transcripts in the cell lines tested. The possibility exists that factor binding at these promoters is more difficult to detect because the DNA-protein interactions are harder to capture by chromatin immunoprecipitation for a variety of reasons. Alternatively, some of these promoters may not be bound by TAF1 and do not require TAF1 to initiate transcription. In support of this hypothesis, previous work shows that a temperature-sensitive TAF1 allele in mammalian cells does not have a global defect in RNAP II transcription demonstrating that not all transcription requires TAF1 (Wang and Tjian 1994; Suzuki-Yagawa et al. 1997). As more promoters are identified and characterized, it is becoming clear that only a small fraction of promoters contain a TATA-box and other elements previously thought to be features of the general promoter. Indeed, as more promoters are functionally characterized, the concepts of the “general transcription machinery” and “basal promoter elements” will be continuously refined.
The data we present represents a functional study of 1% of all human promoters. Our data, in combination with other data generated for the ENCODE region, provide new opportunities to identify regulatory elements and better understand the transcriptional regulatory code of human cells. In addition to providing biological insight, the combination of these experimental data sets with complete sequence conservation and motif data may eventually facilitate more accurate promoter prediction throughout the genome.
Methods
Predicting human promoters based on full-length cDNA sequences
We predicted the locations of promoters for genes in the ENCODE region as previously described with some modifications (Trinklein et al. 2003, 2004). We downloaded all human cDNA alignments from the July 2003 freeze with at least 95% identity, available from UCSC Genome Browser (Karolchik et al. 2003), which totaled 153,642 alignments. These cDNAs represented all available cDNAs in GenBank at that time. Using the alignments of these cDNAs to the genome, we defined gene models by merging all alignments with at least 1 bp of exon overlap on the same strand. For each gene model, one TSS was defined as the 5′-most base of the gene model; however, single-exon transcripts were not permitted to extend 5′ ends of multi-exon genes. Alternative TSS were based only on annotated full-length clones whose 5′ ends were at least 500 bp downstream from the previously defined TSS. Throughout the paper, we define alternative promoters as distinct sequences resulting in transcription of alternate RNA isoforms.
Cloning and plasmid preparation
We used Primer3 software to design primers by inputting 600 bp of upstream sequence and 100 bp downstream of the predicted TSS (Rozen and Skaletsky 2000). Each primer pair was required to flank the TSS. To the 5′ end of each primer, we added 16-bp tails to facilitate cloning by the Infusion Cloning System (BD Biosciences, Clontech cat. no. 639605) (left primer tail: 5′-CCGAGCTCTTACGCGT-3′; right primer tail: 5′-CTTAGATCGCAGATCT-3′). We amplified the fragments using the touchdown PCR protocol previously described (Trinklein et al. 2004) and Titanium Taq Enzyme (BD Biosciences, Clontech, cat. no 639210). To clone our PCR amplified fragments using the Infusion Cloning System, we combined 2 μL of purified PCR product and 100 ng of linearized pGL3-Basic vector (Promega). We added this mixture to the infusion reagent and incubated at 42°C for 30 min. After incubation, the mixture was diluted and transformed into competent cells (Clontech cat. No. 636758). We screened clones for insert by PCR and positive clones were prepared as previously described. We quantified DNA with a 96-well spectrophotometer (Molecular Devices, Spectramax 190) and standardized concentrations to 50 ng/μL for transfections.
Negative control fragment selection
We chose a total of 102 fragments similar in length to the experimental fragments to assay as negative controls. Twenty-four fragments were picked from coding exons that were at least 5 kb from a predicted TSS. We chose the remaining 78 size-matched fragments randomly from the ENCODE regions. Because they were randomly chosen fragments, the GC content was similar to the ENCODE-wide average of approximately 43%. We designed primers and followed all downstream protocols identically to those performed for putative promoter fragments.
Cell culture, transient transfections, and reporter gene activity assays
We obtained each of the 16 cell lines [AGS, Be(2)-C, G-402, HCT116, HepG2, HeLa, HMCB, HT1080, JEG-3, MG-63, MRC-5, Panc-1, SK-N-SH, SNU-182, T98G, and U-87 MG] from American Type Culture Collection (ATCC) and grew them in the media suggested by ATCC (see Supplemental Methods for more information).
We performed transfections of cultured human cell lines as previously described (Trinklein et al. 2004). We seeded 5,000-10,000 cells per well in 96-well plates (see Supplemental Methods). Twenty-four hours after seeding, we cotransfected 50 ng of experimental firefly luciferase plasmid with 10 ng of Renilla luciferase control plasmid (pRL-TK, Promega cat. no. E2241) in duplicate using 0.3 μL of FuGene (Roche) transfection reagent per well. Cells were lysed 24-48 hr post-transfection, depending on cell type. We measured firefly luciferase and Renilla luciferase activity using the PE Wallac Luminometer and the Dual Luciferase Kit (Promega, cat. no. E1960). We followed the protocol suggested by the manufacturer with the exceptions of injecting 60 μL each of the firefly luciferase and Renilla luciferase substrate reagents and reading for 5 sec.
Data analysis and verification
We reported all data as a transformed ratio of firefly luciferase to Renilla luciferase. We determined the mean ratio of the 102 negative controls and eliminated outliers by Dixon's test (Dixon 1950). By this test, 0-3 outliers were identified in each cell line. Only two outliers appeared in multiple cell types. We assessed the activity of putative promoters by defining a threshold three standard deviations above the mean ratio of the negatives. We normalized for comparison between cell types by dividing each ratio by the mean ratio of the negative controls for that cell type adding one and taking the log2 of each ratio [Activity = log2((firefly luciferase/Renilla luciferase)/AvgNeg+1)]. To verify our data, we prepared 48 promoters independently to assess reproducibility. Each sample began with a new transformation, bacterial culture, DNA extraction, quantification, and transfection. We assayed promoter activity in four cell lines and found a correlation of 0.93 between transformed firefly luciferase/Renilla luciferase ratios of the two independent samples.
Sequence analysis and comparative studies
For motif discovery, we divided promoters into clusters, based on the clustering displayed in Figure 1, and used MEME (Bailey and Elkan 1994) to search for motifs over represented within each cluster. High GC content confounded the search and no significant motifs were identified. We also used Bioprospector (Liu et al. 2001) to identify motifs which differentiated between functional and nonfunctional promoters but did not recover any significant motifs.
Constrained elements were identified for all ENCODE target regions based on analyses performed by other members of the ENCODE consortium (G.M. Cooper and A. Sidow, unpubl.). We used constrained element annotations generated for the October 2004 ENCODE sequence data freeze (The ENCODE Project Consortium 2004), using Genomic Evolutionary Rate Profiling (GERP) (described in detail in Cooper et al. 2005) analyses of multiple sequence alignments built using MLAGAN alignment software (Brudno et al. 2003). These constrained elements collectively cover 4.3% of all human ENCODE bases, and all elements are statistically significant at 95% confidence (Cooper et al. 2005) (see Supplemental materials). More information, along with updated constrained element annotations and scores, will be available through the ENCODE portal of the UCSC genome browser (http://genome.ucsc.edu/ENCODE).
Promoter deletions series
For each of 45 promoters, we designed additional amplicons and constructed plasmids with promoter inserts averaging 1000, 330, 210, 90, and 40 upstream bases, in addition to the 500-bp fragments already cloned. (Primer sequences are available as Supplemental materials.) We subcloned each of the smaller fragments from the original promoter and amplified the 1000-bp fragments from genomic DNA. We cloned these fragments using restriction enzymes and ligation as described previously (Trinklein et al. 2003, 2004). After cloning, the constructs were transfected and assayed as described above in seven cell lines: HT1080, HCT116, AGS, T98G, U87 MG, HeLa, and JEG-3.
RNA preparation and cDNA synthesis
We isolated RNA using Qiagen RNA/DNA Mini Kit (cat. no. 14123) from duplicate samples of 14 cell types (AGS, G-402, HCT116, HeLa, HepG2, HMCB, HT1080, JEG-3, MG-63, MRC-5, Panc-1, SNU-182, T98G, and U-87 MG). We grew each cell line in monolayer and lysed 4 × 106 cells in 0.5 mL of lysis buffer. We resuspended RNA pellets in 100 μL of RNase-free water. We then reverse transcribed the RNA samples by using a mix of random hexamers, poly-T first strand synthesis primers, and Superscript reverse transcriptase (Invitrogen).
Quantitative RT-PCR
We designed amplicons to the cDNA sequence of each gene and performed real-time PCR to quantitate the absolute amount of cDNA for each gene (amplicon size range between 60-100 bp). Each reaction contained 3.5 mM MgCl2, 0.125 mM dNTPs, 0.5 μM forward primer, 0.5 μM reverse primer, 0.5X Sybr Green (Molecular Probes), 1U Stoffel fragment (Applied Biosystems), and template DNA in a final volume of 20 μL. For each amplicon we measured a standard curve of 50 ng, 5 ng, 500 pg, and 50 pg total genomic DNA in addition to our replicate cDNA samples. We measured product accumulation for 40 cycles on the Bio-Rad Icycler and calculated the threshold cycle for each dilution of the standard curve. We then performed a linear regression to fit the threshold cycle from our cDNA sample to this standard curve to measure the absolute number of genomic equivalents of that gene in the pool of cDNA from each of the 14 cell lines. We measured the levels of β-actin and GAPDH in each cDNA preparation to normalize for any variation in absolute quantities of cDNA in each prep. We also measured 3 genomic controls to estimate the background levels of contaminating genomic DNA or other background signal. For false-positive and false-negative calculations, RNA transcript was considered detectable at 10-fold over the genomic background controls.
Acknowledgments
We thank the ENCODE Consortium members for providing unpublished data and valuable discussion. Specifically, we thank Bing Ren, Tae Hoon Kim, Leah Barrera, Arend Sidow, and Gregory Cooper. We also thank Daryl Thomas, Kate Rosenbloom, and the rest of the UCSC team for creating the database and Webbrowser resources to enable these analyses. Finally, we thank members of the Myers Lab for helpful discussion and encouragement, and specifically Robert Otillar for his discussion on gene models. S.J.C. is funded by the Stanford Genome Training Program (Training Grant NIH 5 T32 HG00044). This work was supported by NIH Grant 1 U01 HG 03162-01 from the National Human Genome Research Institute.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4222606
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Ahituv, N., Rubin, E.M., and Nobrega, M.A. 2004. Exploiting human-fish genome comparisons for deciphering gene regulation. Hum. Mol. Genet. 13 Spec No 2: R261-R266. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2 28-36. [PubMed] [Google Scholar]
- Bertone, P., Stolc, V., Royce, T.E., Rozowsky, J.S., Urban, A.E., Zhu, X., Rinn, J.L., Tongprasit, W., Samanta, M., Weissman, S., et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306 2242-2246. [DOI] [PubMed] [Google Scholar]
- Blais, A. and Dynlacht, B.D. 2004. Hitting their targets: An emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 14 527-532. [DOI] [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., and Batzoglou, S. 2003. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J.E. and Kadonaga, J.T. 2002. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Dev. 16 2583-2592. [DOI] [PubMed] [Google Scholar]
- Cavin Perier, R., Junier, T., and Bucher, P. 1998. The Eukaryotic Promoter Database EPD. Nucleic Acids Res. 26 353-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, G.M., Stone, E.A., Asimenos, G., Green, E.D., Batzoglou, S., and Sidow, A. 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15 901-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, W. 1950. Analysis of extreme values. Ann. Math. Stat. 21 488-506. [Google Scholar]
- The ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306 636-640. [DOI] [PubMed] [Google Scholar]
- Gentles, A.J. and Karlin, S. 1999. Why are human G-protein-coupled receptors predominantly intronless? Trends Genet. 15 47-49. [DOI] [PubMed] [Google Scholar]
- Gerhard, D.S., Wagner, L., Feingold, E.A., Shenmen, C.M., Grouse, L.H., Schuler, G., Klein, S.L., Old, S., Rasooly, R., Good, P., et al. 2004. The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC). Genome Res. 14 2121-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel, C.C. and Birnstiel, M.L. 1981. The organization and expression of histone gene families. Cell 25 301-313. [DOI] [PubMed] [Google Scholar]
- Howard, M.L. and Davidson, E.H. 2004. _cis_-regulatory control circuits in development. Dev. Biol. 271 109-118. [DOI] [PubMed] [Google Scholar]
- Imanishi, T., Itoh, T., Suzuki, Y., O'Donovan, C., Fukuchi, S., Koyanagi, K.O., Barrero, R.A., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. 2004. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2 856-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296 916-919. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N., Takami, M., Rho, J., Josien, R., and Choi, Y. 2002. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J. Exp. Med. 195 201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.H., Barrera, L.O., Qu, C., Van Calcar, S., Trinklein, N.D., Cooper, S.J., Luna, R.M., Glass, C.K., Rosenfeld, M.G., Myers, R.M., et al. 2005. Direct isolation and identification of promoters in the human genome. Genome Res. 15 830-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, A.R. and Berger, S.L. 1987. Preparation of cDNA and the generation of cDNA libraries: Overview. Methods Enzymol. 152 307-316. [DOI] [PubMed] [Google Scholar]
- Krumm, A., Meulia, T., Brunvand, M., and Groudine, M. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes & Dev. 6 2201-2213. [DOI] [PubMed] [Google Scholar]
- Krumm, A., Hickey, L.B., and Groudine, M. 1995. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes & Dev. 9 559-572. [DOI] [PubMed] [Google Scholar]
- Landry, J.R., Mager, D.L., and Wilhelm, B.T. 2003. Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 19 640-648. [DOI] [PubMed] [Google Scholar]
- Liu, X., Brutlag, D.L., and Liu, J.S. 2001. BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 127-138. [PubMed]
- Lunyak, V.V., Prefontaine, G.G., and Rosenfeld, M.G. 2004. REST and peace for the neuronal-specific transcriptional program. Ann. N.Y. Acad. Sci. 1014 110-120. [DOI] [PubMed] [Google Scholar]
- Meyer, S., Temme, C., and Wahle, E. 2004. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39 197-216. [DOI] [PubMed] [Google Scholar]
- Ogbourne, S. and Antalis, T.M. 1998. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem. J. 331 (Pt 1): 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkkala, L., Nykanen, P., and Sistonen, L. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15 1118-1131. [DOI] [PubMed] [Google Scholar]
- Praz, V., Perier, R., Bonnard, C., and Bucher, P. 2002. The Eukaryotic Promoter Database, EPD: New entry types and links to gene expression data. Nucleic Acids Res. 30 322-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365-386. [DOI] [PubMed] [Google Scholar]
- Rubin, J.S., Bottaro, D.P., and Aaronson, S.A. 1993. Hepatocyte growth factor/scatter factor and its receptor, the c-met proto-oncogene product. Biochim. Biophys. Acta 1155 357-371. [DOI] [PubMed] [Google Scholar]
- Schoenherr, C.J. and Anderson, D.J. 1995. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 267 1360-1363. [DOI] [PubMed] [Google Scholar]
- Schoenherr, C.J., Paquette, A.J., and Anderson, D.J. 1996. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. 93 9881-9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Yamashita, R., Nakai, K., and Sugano, S. 2002. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 30 328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Yamashita, R., Sugano, S., and Nakai, K. 2004. DBTSS, DataBase of Transcriptional Start Sites: Progress report 2004. Nucleic Acids Res. 32 D78-D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Yagawa, Y., Guermah, M., and Roeder, R.G. 1997. The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol. Cell. Biol. 17 3284-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnasky, H., Gill, D., Murthy, S., Shao, X., Demetrick, D.J., and van der Hoorn, F.A. 1998. A novel testis-specific gene, SPAG4, whose product interacts specifically with outer dense fiber protein ODF27, maps to human chromosome 20q11.2. Cytogenet. Cell. Genet. 81 65-67. [DOI] [PubMed] [Google Scholar]
- Tatarelli, C., Linnenbach, A., Mimori, K., and Croce, C.M. 2000. Characterization of the human TESTIN gene localized in the FRA7G region at 7q31.2. Genomics 68 1-12. [DOI] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.F., Hartman, S.J., Schroeder, D.I., Otillar, R.P., and Myers, R.M. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14 62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E.H. and Tjian, R. 1994. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science 263 811-814. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J. and Wilusz, J. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20 491-497. [DOI] [PubMed] [Google Scholar]
- Yang, A., Kaghad, M., Wang, Y., Gillett, E., Fleming, M.D., Dotsch, V., Andrews, N.C., Caput, D., and McKeon, F. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2 305-316. [DOI] [PubMed] [Google Scholar]
Web site references
- http://genome.ucsc.edu/ENCODE; ENCODE.