Transcription-mediated gene fusion in the human genome (original) (raw)
Abstract
Transcription of a gene usually ends at a regulated termination point, preventing the RNA-polymerase from reading through the next gene. However, sporadic reports suggest that chimeric transcripts, formed by transcription of two consecutive genes into one RNA, can occur in human. The splicing and translation of such RNAs can lead to a new, fused protein, having domains from both original proteins. Here, we systematically identified over 200 cases of intergenic splicing in the human genome (involving 421 genes), and experimentally demonstrated that at least half of these fusions exist in human tissues. We showed that unique splicing patterns dominate the functional and regulatory nature of the resulting transcripts, and found intergenic distance bias in fused compared with nonfused genes. We demonstrate that the hundreds of fused genes we identified are only a subset of the actual number of fused genes in human. We describe a novel evolutionary mechanism where transcription-induced chimerism followed by retroposition results in a new, active fused gene. Finally, we provide evidence that transcription-induced chimerism can be a mechanism contributing to the evolution of protein complexes.
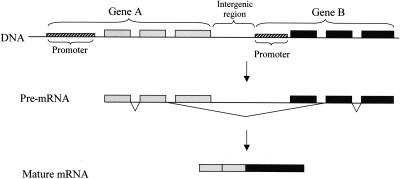
Eukaryotic genes are generally well defined on the genome. Transcription usually begins from a transcription start site, which is guided by the promoter, and ends at a regulated termination point (Zhao et al. 1999; Proudfoot et al. 2002). Consecutive genes are usually separated from each other by intergenic, nonexpressed regions (Lander et al. 2001). In recent years, however, evidence for the existence of mammalian transcripts that span two adjacent, independent genes, have emerged. Typically, such chimeric transcripts begin at the promoter of the upstream gene and end at the termination point of the downstream gene. The intergenic region is spliced out of the transcript as an intron, so that the resulting fused transcripts possess exons from the two different genes (Fig. 1). This phenomenon was coined intergenic splicing or cotranscription and considered extremely rare. In human, only a handful of such transcription-induced chimeras (TICs; see also the accompanying paper by Parra et al. 2006 in this issue) were so far reported (Table 1).
Figure 1.
A model for transcription-induced chimerism. The transcribed region spans both consecutive genes. When the pre-mRNA is spliced, it involves a 5′ splice site at the upstream gene and a 3′ splice site at the downstream gene, thus removing the intergenic region from the mature fused mRNA. The product is a hybrid mRNA containing exons from both genes.
Table 1.
Human transcription-induced chimeras described in the literature
Fused RNAs were shown to be regulated and to have unique expression patterns. For example, the HHLA1-OC90 fusion transcript is restricted to teratocarcinoma cell lines while absent from normal cells (Kowalski et al. 1999). In the case of LY75-CD302 (CD205-DCL1) fusion, the chimera is predominant in Hodgkin and Reed-Strenberg cell lines (Kato et al. 2003). The fusion can change the properties of the participating proteins, or change their localization, such as in the case of Kua-UBE2Vl fusion, where the fused protein is localized to the cytoplasm, while UBE2Vl is a nuclear protein (Thomson et al. 2000). The most characterized human fusion transcript is of two members of the TNF ligand family, TNFSF12 (previously known as TWEAK) and TNFSF13 (APRIL), which represent a type-II transmembrane and a secreted protein, respectively (Pradet-Balade et al. 2002). The fused protein, composed of TNFSF12 cytoplasmic and transmembrane domains fused to the TNFSF13 C-terminal domain, is expressed and translated endogenously in human primary T cells and monocytes. The fused protein is membrane anchored and presents the TNFSF13 receptor-binding domain at the cell surface. It is a biologically active ligand, stimulating cycling in T- and B-lymphoma cell lines (Pradet-Balade et al. 2002).
As no systematic effort was carried out to detect TIC events across the human genome, the extent of this phenomenon and its implications are unknown. In this study, we systematically identified TICs in human and discovered unique features that characterize them. We describe unappreciated roles of TICs in evolution of proteins and protein complexes, and demonstrate novel implications on regulation and function of genes.
Results and Discussion
Computational identification of transcription-induced chimerism events
To characterize the phenomenon of TIC in a genome-wide manner, we first clustered ESTs and cDNAs from GenBank version 136 onto the human genome sequence (build 33) using the LEADS software platform (Sorek et al. 2002; Sorek and Safer 2003) (Table 2; see Methods). The clustering phase resulted in 26,057 clusters of expressed sequences aligned to the genome, containing at least one RNA sequence. We next searched for clusters reporting on fusion between two genes. To avoid cases in which such clusters are caused by natural antisense overlaps, we used the “Antisensor” algorithm (Yelin et al. 2003) to identify and separate such clusters into discrete sense and antisense genes.
Table 2.
Data flow of the computational search for transcription-induced chimeras
| Processa | Resulting data |
|---|---|
| 1 Data download | cDNA data from GenBank version 136 |
| 2 Alignment and clustering | 26,057 clustersb of expressed sequences aligned to the genome |
| 3 Sense/antisense separation | 29,613 clusters on separate strands |
| 4 Computational detection of gene fusion | 322 pairs of fused genes |
| 5 Filtering out alignment artifacts | 281 pairs of fused genes |
| 6 Manual filtration of artifacts | Final data set: 212 pairs of fused genes |
To isolate reliable events of TIC we looked for clusters that contain at least two nonoverlapping cDNA sequences that are annotated as “complete CDS.” To avoid contaminations, we demanded that the sequences connecting these two cDNAs will be canonically spliced, and will share at least one splice site with each of the two separate genes. This demand also screens out cases of naturally overlapping genes (Veeramachaneni et al. 2004). To discard cases of alignment artifacts, in which two consecutive homologous genes were falsely connected, we filtered out connecting sequences having high-scoring alignments in both genes. Our computational search resulted in the identification of 281 putative TICs. Manually inspecting these 281 putative events revealed 55 spurious cases, where the two fused RNAs were apparently parts of one gene (see Methods). These cases, as well as an additional 14 inconclusive events, were discarded (Table 2).
After removing the above-described artifacts, we acquired a reliable data set of 212 TIC events (Supplemental Table S1). The data set contained 421 genes, with four of them participating in more than one fusion (i.e., there was evidence for their fusion both with their upstream and downstream neighboring genes). Of the 212 fusion events, 54 (25%) were supported by more than one expressed sequence, suggesting that in most cases the fusion event is relatively rare or confined to a certain tissue/condition.
To understand the extent to which these events are conserved between species, we searched evidence for our 212 TICs in ESTs of other mammals (see Methods). In 22 cases (10%), we were able to identify an EST from another species supporting the human TIC (Supplemental Table S1). This rate of conservation is similar to the reported 11% conservation of alternative splicing events between human and mouse (Yeo et al. 2005). Overall, 70 (33%) of the TIC events in our set were supported by multiple sequence evidence, i.e., either by more than one human sequence and/or by additional sequences from other species.
As mentioned above, 13 human TICs were reported previously (Table 1). Of these, five were supported by ESTs, with 3/5 supported by one EST only, indicating that even one supporting spliced EST can reliably report on true transcriptional fusion. For the remaining eight reported events, there was no EST showing their existence. We therefore conclude that the 421 genes we detected are only a subset of the actual number of fused genes in human.
To test the possibility that transcription-induced chimerism is cancer induced, we used EST library annotations to extract the histological origin (cancer/normal) of each EST. We then compared the histology distribution of the fusing ESTs with the general distribution in all ESTs. Of the fusing sequences, 51% originate from normal tissues, compared with 46% in the entire EST population. These results indicate that the transcription-induced chimeras present in our data are not the outcome of a cancerous condition.
Unique intergenic splicing patterns and intergenic distance bias in fusion events
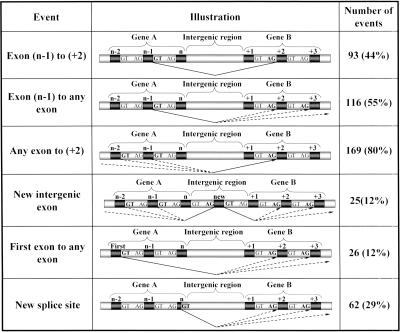
We further analyzed the TIC events to understand the splicing patterns of the fused transcript. The most abundant intergenic splicing type, occurring in 44% of the events (93/212), was between the n-1 exon (one before last) of the upstream gene and the second exon (+2) of the downstream gene (Fig. 2). In this type of fusion, a novel intron is created, spanning the region between the 5′ splice site (donor) of the last intron and the 3′ splice site (acceptor) of the first intron of the upstream and downstream genes, respectively. This abundant fusion-generating splicing pattern usually results in removal of the intergenic region, along with the 3′UTR and 5′UTR of the upstream and downstream genes, respectively. This pattern was also the most abundant in the set of 13 known cases; in nine of them, at least one of the fusion forms was of the (n-1) to (+2) type.
Figure 2.
Intergenic splicing patterns. Exons (dark boxes) are numbered from 1 to n. Percentage of events is calculated out of the 212 events detected in our computational search. Thin triangles mark the intergenic splicing pattern. “GT” and “AG” stand for the 5′ and 3′ splice sites, respectively. Splice sites used for the intergenic splicing are in bold. Patterns are not mutually exclusive, and hence, the percentages sum up to more than 100%.
Figure 2 lists the distribution of the fusion-generating splicing patterns we observed. The most abundant donor site in the upstream gene is located in the last intron (55%) and the most abundant acceptor site in the downstream gene is located at the first intron (80%). This indicates a strong tendency to separately drop the last and first exons of the upstream and downstream genes, respectively, and exactly explains the 44% of cases indicated above, which drop both first and last exons. In 12% of the events, a novel exon, residing between the two fused genes, appears in the fusion transcript (and not in any of the nonconnecting sequences). The fact that such novel exons originate from the intergenic region rules out the possibility that the fusion is mediated by _trans_-splicing, because in the _trans_-splicing option, the intergenic region would not have been transcribed.
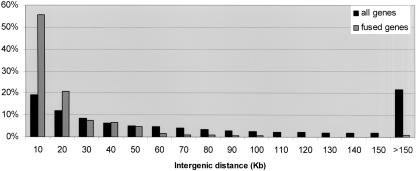
To test whether there is a preference for specific intergenic distance between fused genes, we calculated the intergenic distance distribution of the 212 fusion events, and compared it with the distance distribution of 12,395 human adjacent genes (see Methods). As shown in Figure 3, fused genes tend to reside closer on the genome than the entire human gene pair population; the median distance between fused genes was 8.5 kb, compared with a median of 48 kb for the entire gene population. This indicates that the mechanism involved in TIC generation strongly prefers shorter distances between the genes. However, our data show that gene fusions can also occur over large gene distances: In 5% of the fusion events, the distance between fused genes exceeded 50 kb.
Figure 3.
Intergenic distances distribution. Compared are a data set of “all genes,” representing distances between 12,395 consecutive RefSeq pairs residing on the same strand in the human genome, and “fused genes,” representing distances between the 212 genes in the current analysis. _x_-axis, intergenic distance in kilobase, divided to bins of 10 kb (i.e., the “20” bin corresponds to distances between 10,001 and 20,000 bp). _y_-axis, percent of gene pairs out of each data set. Notably, fused genes tend to reside much closer on the genome than the entire population of gene pairs.
RT–PCR experimental validation
To experimentally test our predicted TIC events, we selected 10% of the data (20 events) for RT–PCR screening using RNAs from a panel of 19 different tissues and cell lines (See Methods). For nine of the events, fusion was detected in at least one of the tissues tested. Six of these events were found to be ubiquitously expressed, while the remaining three were tissue specific (see Table 3). Figure 4 presents examples of validated fusions. Our success rate (9/20) suggests that roughly half of the events we identified are bona fide fusion events in human. However, some of the events we could not detect might be expressed in a tissue/condition other than the ones we tested, so the number of events that actually exist in human RNAs might be higher.
Table 3.
Fusion events tested by RT–PCR
| Gene 1a | Gene 2b | Number of sequences supporting fusionc | Upstream gene (WT) detectedd | Fusion RNA detectede |
|---|---|---|---|---|
| NME1 | NME2 | 30 | + | + |
| NME2 | WDR50 | 1 | + | – |
| RBM14 | RBM4 | 20 | + | – |
| BCL2L2 | PABPN1 | 1 | + | + (Testis) |
| TMPRSS3 | TFF1 | 1 | + | – |
| CTBS | GNG5 | 15 | + | + |
| MAG | CD22 | 1 | + | – |
| PIR | FIGF | 1 | – | + (Brain) |
| SERPINA5 | SERPINA3 | 1 | + | – |
| RPL17 | LOC497661 | 1 | + | + |
| NM_003947 | NM_007064 | 2 | + | + |
| HOXC10 | HOXC5 | 1 | + | – |
| SPN | QPRT | 1 | + | + (Farage) |
| NM_033542 | NM_018478 | 9 | + | + |
| GIMAP1 | GIMAP5 | 1 | – | – |
| HIF1A | SNAPC1 | 1 | + | – |
| C1QA | C1QG | 1 | – | – |
| MIA | RAB4B | 1 | + | + |
| S100A3 | S100A4 | 1 | – | – |
| MUC1 | KRTCAP2 | 1 | + | – |
| PIP5K1A | PSMD4 | 0 | + | + Retrogene |
| + TIC (HepG2) |
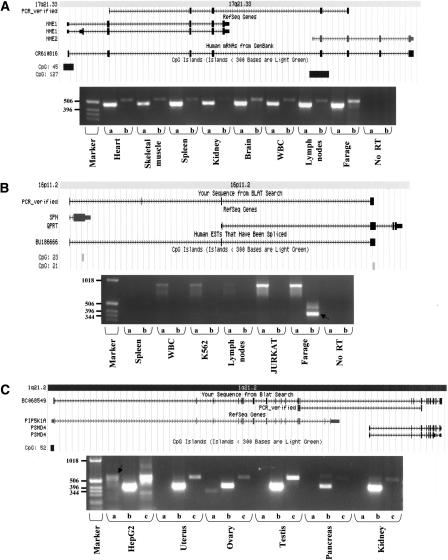
Figure 4.
Selected examples of experimentally verified transcription induced chimeras. Shown are snapshots taken from the UCSC genome browser (http://genome.ucsc.edu/), presenting the alignment of expressed sequences to the genome, as well as the location of CpG islands (Gardiner-Garden and Frommer 1987). A total of 70% of the downstream genes involved in TICs possess CpG islands in their 5′ regions, indicating that they are also regulated as single genes. Boxes represent exons, with thinner boxes representing the untranslated regions (UTRs). Arrowed thin lines represent introns. “PCR_verified” represents the chimeric sequence validated by RT–PCR. Beneath are RT–PCR results showing the fusion events (see Methods). (A) Fusion transcript between NME1 and NME2 creating a predicted fused protein. This transcript is supported by 30 ESTs (one is shown in the figure), and was found to be ubiquitously expressed in human tissues. The same fusion event was also detected in mouse ESTs. In the gel image—lanes a indicate the NME1 wild-type (WT) transcript and lanes b indicate the fused (TIC) NME1-NME2 transcript. (B) Fusion transcript between sialophorin (SPN) and quinolinate phosphoribosyltransferase (QPRT) demonstrates the donation of SPN regulatory sequence (5′UTR) to the QPRT transcript. The fusion was experimentally detected in RNAs from the Farage cell line (B-lymphoma). In the gel image—lanes a indicate the wild-type SPN and lanes b indicate the fused (TIC) SPN-QPRT transcript (validated product marked by black arrow). (C) Fusion transcript between phosphatidylinositol-4-phosphate 5-kinase (PIP5K1A) and proteasome 26S subunit non-ATPase 4 (PSMD4) on chromosome 1q21. The fusion event was discovered by identification of retroposed, chimeric processed expressed pseudogene residing on chromosome 10q23. No EST supports this fusion, but it was verified by RT–PCR. The RNA BC068549, expressed from the processed pseudogene on chromosome 10, is shown aligning to both genes. In the gel image—lanes a indicate the fused (TIC) PIP5K1A-PSMD4 transcript on chromosome 1 (validated product marked by black arrow); lanes b indicate the PIP5K1A (WT), and lanes c indicate the active transcription of the retroposed gene from chromosome 10, which was found to be ubiquitously expressed. Primers were designed from regions that are diverged between the fusion transcript and the retrogene, so that each product will be uniquely amplified (Supplemental Table S2). Products were verified by direct sequencing.
Functions of fusion products
What are the possible functions of transcription-induced chimeras? To understand this, we examined the fusion patterns with respect to the resulting ORF. In 53 events in our set (25%), a fusion protein containing coding sequences of both genes (without a premature stop codon) was created. This kind of fusion might generate a bifunctional protein having properties from both original proteins, as happens in the known cases of TWE-PRIL (Pradet-Balade et al. 2002) or Kua-UEV1 (Thomson et al. 2000). An example of this type of fusion, between NME1 and NME2, is presented in Figure 4A.
Another functional impact could be at the transcriptional regulation level. This will occur when the fusion involves only the first exon of the upstream gene, so that the upstream gene mainly contributes its 5′UTR to the fused transcript. Indeed, 26 (12%) of our events correspond to this type of fusion. This will potentially cause the downstream gene to be regulated as the upstream one, both transcriptionally (promoter) and translationally (5′UTR; see Fig. 4B). Multiple variable first exons were described in many human genes, functioning in alternating gene regulation (Zhang et al. 2004). Similarly, the stability of the upstream gene could be influenced if the downstream gene contributes only the 3′ UTR (eight events).
TIC can also be intended to suppress the expression of the upstream gene by the Nonsense Mediated Decay (NMD) mechanism (Hillman et al. 2004). This would occur when the fusion causes a frame-shift that results in a premature stop codon. Indeed, 120 (56%) TIC events in our set are expected to undergo NMD. Frame shift can also result in alternative C terminus of the upstream gene, if the resulting stop codon occurs in the last (or one before last) exon, as in the known ANKHD1-EIF4EBP3 (MASK-BP3) case (Poulin et al. 2003).
Finally, some of the events seen in our database could represent transcriptional “leakage”, where the transcriptional machinery accidentally ignores the termination of the upstream gene and transcribes through the downstream one. Such a leakage can be a rare, nonregulated stochastic event that does not contribute to the fitness of the organism. Indeed, only 33% of the cases in our database were supported by multiple-sequence evidence, demonstrating the low-occurrence frequency of the majority of our events. In addition, only 10% of TICs were found to be conserved between species. It could also be argued that the low frequency of protein fusion events (25% of the total) is indicative of the stochastic nature of the TIC phenomenon; however, a similar frequency of fusion proteins (23%) was also detected in the subset of events that are conserved between mammals, indicating that nonfusion-protein events can be under selective pressure as well and are hence possibly functional. Overall, although we cannot determine the actual fraction of TICs that is functional, our results suggest that at least a subset of these events have a biological role.
Currently, regulation of TIC is generally uncharacterized. Models for transcription termination indicate that both _cis_-acting sequence elements, as well as _trans_-acting termination factors that belong both to the transcriptional and the splicing machineries, act together to generate an accurate 3′ end (Zhao et al. 1999; Proudfoot et al. 2002). Regulated transcriptional read-through was described in viruses and suggested also for TIC (Hardy and Wertz 1998; Magrangeas et al. 1998). Presumably, both _cis_-acting sequences, such as weak polyadenylation signals, and _trans_-acting suppressors/regulators of the termination machinery, could regulate the transcriptional read-through involved in TIC.
What is the proportion of TIC events in the genome? Our data suggest that ∼2% of all human genes might be involved in such fusion. However, as ESTs are merely a sample of the transcriptome (Sorek et al. 2004), they do not represent all possible transcripts. Indeed, only five of the 13 known cases (40%) are represented in GenBank dbEST, indicating that many more genes might be fused than actually detected. In addition, our strict filtering process removed many events that might actually be real (see Methods). Indeed, an EST-independent search for TICs in the Encode regions suggests that ∼5% of all human genes are involved in TIC (Parra et al. 2006).
Evolution of protein complexes
It has been shown that gene fusion events across genomes can be used for predicting functional associations of proteins, including physical interactions and complex formation (Enright and Ouzounis 2001). This relies on the observation that two proteins that function in the same complex in one organism are frequently fused into a single “Rosetta Stone” protein in another organism (Marcotte et al. 1999). The TIC phenomenon might be a major process supporting this evolutionary mechanism. For example, the nucleoside diphosphate kinase (NDK) complex is a hexamer composed of the “A” (encoded by NME1) and “B” (NME2) polypeptides (Gilles et al. 1991). We detected and experimentally validated a ubiquitously expressed chimeric transcript fusing these two subunits, which codes for a natural NME1-NME2 fusion protein, in agreement with the above hypothesis (Fig. 4A). In the evolutionary future of our species, this fusion might be fixed as a single gene. Additional such cases are described in the accompanying paper by Parra et al (2006).
Intriguingly, we were able to identify a processed pseudogene indicating fusion of the genes PIP5K1A and PSD4. Although no EST supported this fusion event, we verified experimentally the existence of a PIP5K1A-PSD4 fusion transcript in human RNA (Fig. 4C). PIP5K1A and PSD4 consecutively reside on chromosome 1, while the processed pseudogene is found on chromosome 10 as a single exon chain. Moreover, we detected a GenBank cDNA (BC068549) indicating active transcription from the pseudogene itself, suggesting that it is actually an active retrogene. Indeed, we were able to verify experimentally through RT–PCR and sequencing that this retrogene is being transcribed in human tissues (Fig. 4C). This retrogene underwent considerable evolution relative to the original fused transcript (79 mismatches between the sequence of BC068549 and the PIP5K1A-PSD4 locus). Still, translation of BC068549 results in an uninterrupted PIP5K1A-PSD4 fusion protein, suggesting that the new retrogene is under selective pressure and is hence functional.
This unique pseudogene example sets transcription-induced chimerism followed by retro-position as a novel molecular evolutionary mechanism enabling the creation of new, fused “Rosetta Stone” sequences. This mechanism is expected to affect mainly Eukaryotes, where the splicing machinery can efficiently remove the intergenic region. Presumably, additional fused genes were created through this mechanism during the evolutionary history of metazoa.
Conclusions
We have demonstrated that transcription-induced chimerism is much more widespread in the human transcriptome than initially appreciated, forming yet an additional layer of protein diversity. The fusion transcripts might function in various levels, either by creating newly functioning proteins, or by changing the regulation of pre-existing proteins. The function of each fusion event, as well as the mechanism enabling such fusion and the actual impact of this phenomenon on the human genome, remain to be elucidated.
Methods
Computational search
Human ESTs and cDNAs were obtained from NCBI GenBank version 136 (June 2003; http://www.ncbi.nlm.nih.gov/Genbank/) and aligned to the human genome build 33 (April 2003; http://www.ncbi.nlm.nih.gov/genome/guide/human/) using the LEADS clustering and assembly software as described previously (Sorek et al. 2002). Briefly, the software cleans the expressed sequences from vectors and immunoglobulins, masking them for repeats and low-complexity regions. It then aligns the expressed sequences to the genome, taking alternative splicing into account, and clusters overlapping expressed sequences into “clusters” that represent genes or partial genes. Clusters were separated to sense/antisense clusters using the “Antisensor” algorithm as described in Yelin et. al. (2003).
“Complete CDS” annotation of RNA sequences was obtained from the “DEFINITION” field in the GenBank sequence records. In each cluster, overlapping complete CDS cDNA sequences that aligned fully to the genome were grouped together. Each group was referred to as a gene. Gene boundaries were extended in cases where ESTs suggested longer UTRs than present in the RNA. In clusters containing more than one gene, connecting sequences were identified. Connecting sequences were required to have canonical splice sites at the fusion junction, and to share at least one splice site with each of the two separate genes.
For the “alignment artifacts” filtering, each exon in the connecting sequences was aligned to the cDNA sequences of both connected genes. Sequences with exons aligned to both genes were discarded. For the manual filtration of fusion events, we used the following information from the UCSC genome browser: (1) occurrence of CpG islands before both genes; (2) existence of SWISS-PROT annotations for both genes; (3) existence of ORF, 5′ and 3′ UTRs for both genes.
For gene distance calculation, known RefSeqs were localized to the genome using the UCSC genome browser annotations (Karolchik et al. 2003). The distance between each gene pair was calculated from the most downstream hit of the upstream gene to the most upstream hit of the downstream gene. Only distances up to 400 kb were considered, as this is the maximum intron length allowed by the LEADS software.
To calculate possible NMD of transcripts, the fused transcript was first assembled using the upstream and downstream RefSeqs connected by the fusing EST. In this transcript, premature stop codon was searched according to the rule of 55 nucleotides or more upstream to the last exon–exon junction.
For the “evolution of protein complexes” analysis, annotations of genes were downloaded from the “RefSeq Summary” field in UCSC genome browser and from the comments fields in SWISS-PROT. Processed pseudogenes indicating on TIC were systematically searched in the database of >8000 processed pseudogenes compiled by Zhang et al (2003).
Experimental validation of fusion events
Total RNA was isolated from a variety of human tissues (kidney, liver, brain, ovary, white blood cells [WBC], testis, prostate, spleen, heart, breast, pancreas, lung, colon, bladder, thymus, Farage B lymphocyte cell line [CRL-2360, ATCC], K562 cell line [CCL-243, ATCC], JURKAT T lymphocyte cell line [TIB-152, ATCC], and HepG2 liver cell line [HB-8065, ATCC]) by Tri-Reagent (MRC) according to the manufacturer's instructions. First-strand cDNAs were prepared using SuperScript II reverse transcriptase (Invitrogen) primed with random hexamers and oligo dT's (Invitrogen). RT–PCR reactions were performed in 50-μL reactions encompassing 1 μL of RT in the presence of 2 mM dNTPs, 10 pmol of primers, 2.5 units of SUPER-THERMO polymerase, and subjected to 28–35 amplification cycles. Reactions were designed to include a forward primer from the upstream gene and two reverse primers, one from the last exon of the upstream gene and one from the internal exon of the downstream gene, in order to identify both the existence of a wild-type RNA and the fused RNA (primers appear in Supplemental Table S2). RT–PCR products were isolated from agarose gels and verified by sequencing.
Acknowledgments
We thank D. Schaffer, A. Golubev, A. Haviv, and N. Keren for biocomputatioanl assistance; M. Oz for providing critical resources; G. Naveh for literature assistance; and Z. Levine, U. Nir, K. Savitsky, E. Levanon, D. Milo, S. Pollock, G. Cojocaru, E. Eisenberg, and D. Dahary for fruitful discussions.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4137606.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Communi, D., Suarez-Huerta, N., Dussossoy, D., Savi, P., and Boeynaems, J.M. 2001. Cotranscription and intergenic splicing of human P2Y11 and SSF1 genes. J. Biol. Chem. 276 16561-16566. [DOI] [PubMed] [Google Scholar]
- Cox, P.R., Siddique, T., and Zoghbi, H.Y. 2001. Genomic organization of Tropomodulins 2 and 4 and unusual intergenic and intraexonic splicing of YL-1 and Tropomodulin 4. BMC Genomics 2 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, A.J. and Ouzounis, C.A. 2001. Functional associations of proteins in entire genomes by means of exhaustive detection of gene fusions. Genome Biol. 2 research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears, S., Mathieu, C., Zeleznik-Le, N., Huang, S., Rowley, J.D., and Nucifora, G. 1996. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl. Acad. Sci. 93 1642-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden, M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196 261-282. [DOI] [PubMed] [Google Scholar]
- Gilles, A.M., Presecan, E., Vonica, A., and Lascu, I. 1991. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J. Biol. Chem. 266 8784-8789. [PubMed] [Google Scholar]
- Hardy, R.W. and Wertz, G.W. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72 520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman, R.T., Green, R.E., and Brenner, S.E. 2004. An unappreciated role for RNA surveillance. Genome Biol. 5 R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., Khan, S., Gonzalez, N., O'Neill, B.P., McDonald, K.J., Cooper, B.J., Angel, N.Z., and Hart, D.N. 2003. Hodgkin's lymphoma cell lines express a fusion protein encoded by intergenically spliced mRNA for the multilectin receptor DEC-205 (CD205) and a novel C-type lectin receptor DCL-1. J. Biol. Chem. 278 34035-34041. [DOI] [PubMed] [Google Scholar]
- Kowalski, P.E., Freeman, J.D., and Mager, D.L. 1999. Intergenic splicing between a HERV-H endogenous retrovirus and two adjacent human genes. Genomics 57 371-379. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409 860-921. [DOI] [PubMed] [Google Scholar]
- Magrangeas, F., Pitiot, G., Dubois, S., Bragado-Nilsson, E., Cherel, M., Jobert, S., Lebeau, B., Boisteau, O., Lethe, B., Mallet, J., et al. 1998. Cotranscription and intergenic splicing of human galactose-1-phosphate uridylyltransferase and interleukin-11 receptor α-chain genes generate a fusion mRNA in normal cells. Implication for the production of multidomain proteins during evolution. J. Biol. Chem. 273 16005-16010. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Ng, H.L., Rice, D.W., Yeates, T.O., and Eisenberg, D. 1999. Detecting protein function and protein–protein interactions from genome sequences. Science 285 751-753. [DOI] [PubMed] [Google Scholar]
- Millar, J.K., Christie, S., Semple, C.A., and Porteous, D.J. 2000. Chromosomal location and genomic structure of the human translin-associated factor X gene (TRAX; TSNAX) revealed by intergenic splicing to DISC1, a gene disrupted by a translocation segregating with schizophrenia. Genomics 67 69-77. [DOI] [PubMed] [Google Scholar]
- Pardigol, A., Forssmann, U., Zucht, H.D., Loetscher, P., Schulz-Knappe, P., Baggiolini, M., Forssmann, W.G., and Magert, H.J. 1998. HCC-2, a human chemokine: Gene structure, expression pattern, and biological activity. Proc. Natl. Acad. Sci. 95 6308-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, G., Reymond, A., Dabbouseh, N., Dermitzakis, E.T., Castelo, R., Thomson, T.M., Antonarakis, S.E., and Guigó, R. 2006. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Poulin, F., Brueschke, A., and Sonenberg, N. 2003. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. J. Biol. Chem. 278 52290-52297. [DOI] [PubMed] [Google Scholar]
- Pradet-Balade, B., Medema, J.P., Lopez-Fraga, M., Lozano, J.C., Kolfschoten, G.M., Picard, A., Martinez, A.C., Garcia-Sanz, J.A., and Hahne, M. 2002. An endogenous hybrid mRNA encodes TWE-PRIL, a functional cell surface TWEAK-APRIL fusion protein. EMBO J. 21 5711-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108 501-512. [DOI] [PubMed] [Google Scholar]
- Sorek, R. and Safer, H.M. 2003. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 31 1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Ast, G., and Graur, D. 2002. _Alu_-containing exons are alternatively spliced. Genome Res. 12 1060-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Shemesh, R., Cohen, Y., Basechess, O., Ast, G., and Shamir, R. 2004. A non-EST-based method for exon-skipping prediction. Genome Res. 14 1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, T.M., Lozano, J.J., Loukili, N., Carrio, R., Serras, F., Cormand, B., Valeri, M., Diaz, V.M., Abril, J., Burset, M., et al. 2000. Fusion of the human gene for the polyubiquitination coeffector UEV1 with Kua, a newly identified gene. Genome Res. 10 1743-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya, A.B., Lee, S.H., and DeJong, J. 1999. Identification of a general transcription factor TFIIAα/β homolog selectively expressed in testis. J. Biol. Chem. 274 18040-18048. [DOI] [PubMed] [Google Scholar]
- Veeramachaneni, V., Makalowski, W., Galdzicki, M., Sood, R., and Makalowska, I. 2004. Mammalian overlapping genes: The comparative perspective. Genome Res. 14 280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21 379-386. [DOI] [PubMed] [Google Scholar]
- Yeo, G.W., Van Nostrand, E., Holste, D., Poggio, T., and Burge, C.B. 2005. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 102 2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos, P.G. 1999. RNA molecules containing exons originating from different members of the cytochrome P450 2C gene subfamily (CYP2C) in human epidermis and liver. Nucleic Acids Res. 27 2585-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Harrison, P.M., Liu, Y., and Gerstein, M. 2003. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13 2541-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., Haws, P., and Wu, Q. 2004. Multiple variable first exons: A mechanism for cell- and tissue-specific gene regulation. Genome Res. 14 79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63 405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.ncbi.nlm.nih.gov/Genbank/; NCBI GenBank version 136 (June 2003).
- http://www.ncbi.nlm.nih.gov/genome/guide/human/; human genome build 33 (April 2003).
- http://genome.ucsc.edu/; This site contains the reference sequence and working draft assemblies for a large collection of genomes. It also provides a portal to the ENCODE project.