Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates (original) (raw)
Abstract
We investigated the evolution of the families of LINE-1 (L1) retrotransposons that have amplified in the human lineage since the origin of primates. We identified two phases in the evolution of L1. From ∼70 million years ago (Mya) until ∼40 Mya, three distinct L1 lineages were simultaneously active in the genome of ancestral primates. In contrast, during the last 40 million years (Myr), i.e., during the evolution of anthropoid primates, a single lineage of families has evolved and amplified. We found that novel (i.e., unrelated) regulatory regions (5′UTR) have been frequently recruited during the evolution of L1, whereas the two open-reading frames (ORF1 and ORF2) have remained relatively conserved. We found that L1 families coexisted and formed independently evolving L1 lineages only when they had different 5′UTRs. We propose that L1 families with different 5′UTR can coexist because they don't rely on the same host-encoded factors for their transcription and therefore do not compete with each other. The most prolific L1 families (families L1PA8 to L1PA3) amplified between 40 and 12 Mya. This period of high activity corresponds to an episode of adaptive evolution in a segment of ORF1. The correlation between the high activity of L1 families and adaptive evolution could result from the coevolution of L1 and a host-encoded repressor of L1 activity.
LINE-1 (L1) retrotransposons (Fig. 1A) constitute the most abundant family of autonomously replicating retroelements in mammals, and their continuous amplification over the last ∼170 million years (Myr) has had a profound impact on the organization and function of mammalian genomes (Smit 1996; Lander et al. 2001; Kazazian 2004). L1 elements replicate via an RNA intermediate that is copied into genomic DNA at the site of insertion (Luan et al. 1993; Luan and Eickbush 1995; Cost et al. 2002). This mechanism of replication is not very efficient and generates mostly defective copies that are truncated at their 5′ end. These copies can be classified into families of hundreds to thousands of elements based on the shared nucleotide differences they inherit from their common progenitor (or group of closely related progenitors). Because the vast majority of L1 inserts are pseudogenes, they accumulate mutations at the neutral rate (Voliva et al. 1984; Hardies et al. 1986; Pascale et al. 1993; Boissinot et al. 2000). Consequently, older families are more divergent than younger ones. Phylogenetic analyses of L1 families in murine rodents and in primates (see Furano 2000, and references therein) have shown that, over the long-term, a single lineage of L1 families amplifies and evolves, one family replacing its predecessor as the dominant family. This mode of evolution is exemplified in human, where a single lineage of families amplified over the last 25 Myr (Smit et al. 1995; Boissinot and Furano 2001). Families of closely related variants can occasionally coexist for short periods of time (Cabot et al. 1997; Boissinot et al. 2000) until one family prevails and dominates the replicative process. The reason(s) why multiple lineages rarely coexist in modern mammals remains unknown, but it has been suggested that competition between L1 families, possibly for a limiting host factor, could account for this pattern of evolution (Casavant and Hardies 1994; Cabot et al. 1997).
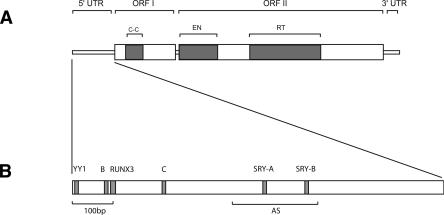
Figure 1.
(A) Structure of a modern human full-length element. A full-length element is 6 Kb long and contains a 5′ untranslated region (5′UTR), two open-reading frames (ORFI and ORFII), and a 3′UTR. The 5′UTR has a regulatory function (Swergold 1990; Minakami et al. 1992). ORFI encodes a protein with nucleic acid-binding properties that can also act as a nucleic acid chaperone (Martin et al. 2000; Martin and Bushman 2001). ORFIp also contains a coiled-coil domain (C-C) that mediates interaction of ORFIp with itself (Martin et al. 2000). ORFII encodes a protein with endonuclease (EN) (Feng et al. 1996) and reverse transcriptase (RT) activity (Mathias et al. 1991). The 3′UTR contains a conserved poly-purine tract (Howell and Usdin 1997). Genomic copies of L1 are typically flanked by an A-rich tail at their 3′ end. (B) Functional motifs in the 5′UTR of a modern L1 element (L1PA1). The first 100 bp (100bp) of the 5′UTR was shown to be critical for transcription (Swergold 1990). The 5′UTR contains a YY1 binding site that plays an important role in transcription initiation (Athanikar et al. 2004), a functional RUNX3 binding site (Yang et al. 2003), two functional SRY-related transcription factor binding sites (SRY-A and SRY-B) (Tchenio et al. 2000), and two cellular factor-binding motifs (B and C) (Minakami et al. 1992). The 5′UTR also contains an antisense promoter (AS) between positions 400 and 600 that can drive transcription of adjacent cellular genes (Speek 2001).
Although L1 seems to have been continuously active since marsupials and placental mammals diverged (Burton et al. 1986), the rate of L1 amplification, and presumably the impact of L1 activity on genomes, has changed over evolutionary times. In murine rodents (Pascale et al. 1993; Verneau et al. 1998) and in primates (Smit et al. 1995; Liu et al. 2003; Boissinot et al. 2004), bursts of amplification alternate with periods of low activity, and in the human lineage, the rate of L1 amplification seems to have slowly decreased over the last 25 Myr (Lander et al. 2001). Correlations between bursts of amplification and evolutionary radiations (Pascale et al. 1990) suggest that the history of populations, especially the occurrence of population bottlenecks (Mathews et al. 2003), could affect the dynamics of L1 amplification. However, this alone could not explain the large variations in replicative success observed between L1 families, and it was recently suggested that positive or negative interactions of a host factor with L1 replicative machinery could be responsible for the episodic nature of L1 amplification (Pascale et al. 1993; Furano 2000; Boissinot and Furano 2001; Furano et al. 2004;).
Previous studies on the evolution of L1 retrotransposons in human have focused on the last 25 Myr (Boissinot et al. 2000; Sheen et al. 2000; Boissinot and Furano 2001; Myers et al. 2002; Ovchinnikov et al. 2002), while a study reaching back about 150 Myr relied mostly on the analysis of 3′ ends, since over 90% of L1 elements are 5′truncated (Smit et al. 1995). Here we examine the molecular evolution and the tempo of amplification of the L1 families that amplified in the human genome during primate evolution over a period of some 70 Myr (Goodman et al. 1998). We derived and analyzed full-length consensus sequences for most of the families that emerged from the L1MA6 family (following the nomenclature of Smit et al. 1995). We found that ancestral primate genomes contained several distinct L1 lineages that amplified and evolved simultaneously for as long as 30 Myr. In contrast, a single lineage of L1 families amplified over the last 40 Myr of human evolution. Interestingly, it seems that distinct L1 families coexisted over long periods of time only when they had different 5′UTR. We propose that L1 families with different 5′UTR can coexist because they exploit different transcriptional niches.
Results
A total of 7046 full-length (FL) L1 elements was identified. The number of FL elements we were able to identify varied considerably, from 11 elements in families L1PA12 and L1MA5 to 1333 in family L1PA3. The number of FL L1PA17, L1PB4, and L1MA4 elements that could be aligned was so small that it was not possible to derive an accurate FL consensus sequence, and only partial consensus were derived. In large families, only a subset of FL elements was aligned. We found that it was very difficult to adequately align family L1PA13 because it contains two subsets of elements with nonhomologous 5′UTR. Therefore, these two subsets of L1PA13 elements (called L1PA13A and L1PA13B) were aligned separately. Alignments are available on request and FL consensus sequences are available online (Supplemental material 1).
Phylogenetic analysis and tempo of amplification of L1 families
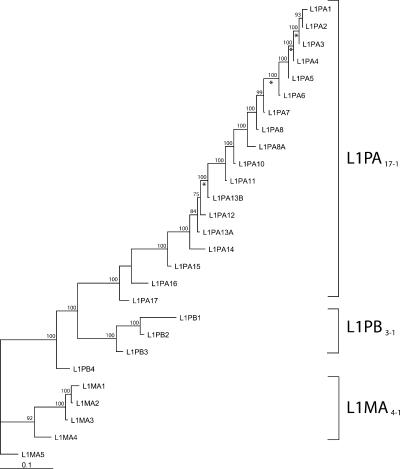
Because the 5′UTR of some families are not homologous (see below), phylogenetic trees were built using ORF1 and ORF2 sequences. Figure 2 shows a maximum likelihood phylogeny of L1 consensus sequences based on ORF1 and ORF2 sequences. The neighbor-joining and maximum-parsimony methods produced very similar trees. This phylogenetic analysis supports previous studies based only on the 3′ extremity of L1 (Smit et al. 1995). Three well-supported lineages appear on the tree, i.e., the L1MA4-1 lineage (consisting of families L1MA4 to L1MA1), the L1PB3-1 lineage (families L1PB3 to L1PB1), and the L1PA17-1 lineage (families L1PA17 to L1PA1). The topology of the tree indicates that these three distinct lineages were simultaneously active and evolved in parallel in ancestral primate genomes. Ultimately, the L1MA4-1 and L1PB3-1 lineages became extinct and only the L1PA17-1 lineage persisted until modern times. In contrast to the diversity of active lineages that existed in ancestral genomes, the 18 L1PA families have evolved as a single lineage during most of primate evolution, one family replacing its predecessor as the dominant family until it was replaced by a younger one (Fig. 2).
Figure 2.
Phylogeny of L1 consensus sequences. This maximum likelihood tree is based on the consensus sequences of the ORF1 and ORF2 of 27 L1 families. The numbers above the nodes indicate the percentages of time the labeled node was present in 1000 bootstrap replicates of the data. Asterisks indicate branches on which the free-ratio model assigned estimates of ω > 1.
We determined the age of each family by estimating the average pairwise divergence between elements. Ages presented in Table 1 are based on an intermediate calibration between the lower and higher divergence rates described in the Methods. The age of the L1 families analyzed here ranged from more than 70 Myr for the oldest families to 3 Myr for the currently active L1PA1 family. Therefore, our analysis covers the entire evolution of human L1 families since before the origin of primates 63 million years ago (Mya). The family ages are consistent with their distribution in primate genomes. For instance, the L1PA5 family that amplified ∼20 Mya is, as expected, absent from the baboon genome and sites that are occupied by an L1PA8 insertion (∼41-Myr-old) in the human genome also contain a L1PA8 insertion in the baboon genome. Some L1PA7 inserts were present in both the human and baboon genomes, while others were absent from the baboon genome. This suggests that the L1PA7 family (∼31.4-Myr-old) amplified before and after the split between Old World monkeys and the human/ape lineage (∼25 Mya). Similarly, we found that some L1MA2 inserts were found in all primate species, while others were absent from the lemur genome. This is not surprising, as the L1MA2 family amplified (∼66 Myr) around the time of separation of the lemur lineage (∼63 Myr). Using the higher divergence rate, we calculated that the oldest families in the three lineages defined above amplified at least 70-74 Mya. As the last active family in lineage L1PB3-1 amplified about 46 Mya, the L1PA17-1 and L1PB3-1 lineages coexisted for at least 24 Myr in ancestral primate genomes and probably more, since they were already well differentiated when they began amplifying. Although these estimates are rough because of variations in the rate of sequence evolution, they are rather conservative because they are based on the average age of the families. Indeed, we found that the L1PB1 family didn't suddenly become extinct, but instead, remained active until 40 Mya (P. Warburton and S. Boissinot, unpubl.). Therefore, it is likely that the L1PA17-1 and L1PB3-1 lineages coexisted for as long as 30 Myr. Using similar calculations, we inferred that the L1MA4-1 lineage coexisted with the L1PA17-1 and L1PB3-1 lineages for at least 11 Myr. Therefore, it seems that L1 diversity has been limited to a single lineage, the L1PA lineage, only during the last ∼40 Myr of primate evolution, i.e., during the evolution of anthropoid primates.
Table 1.
Copy number, divergence, and age of L1 families based on their 3′ extremity
| Lineage | Family | Genomic copy number | Number of sequences aligned | Length of sequences aligned (bp) | Average pairwise divergence (% ± S.E.)a | Average divergence from consensus (% ± S.E.)a | Age (Myr)b | Chimpanzee 6Myr | Baboon 25Myr | Lemur 63Myr |
|---|---|---|---|---|---|---|---|---|---|---|
| L1PA | L1PA1 | 1028 | 198 | 532 | 0.98 ± 0.16 | 0.59 ± 0.13 | 3.1 (2.3–3.9) | – | – | – |
| L1PA2 | 4067 | 199 | 520 | 2.40 ± 0.17 | 1.24 ± 0.10 | 7.6 (5.6–9.6) | + | – | – | |
| L1PA3 | 8712 | 200 | 521 | 3.96 ± 0.28 | 2.21 ± 0.20 | 12.5 (9.2–15.8 | + | – | – | |
| L1PA4 | 9936 | 165 | 515 | 5.69 ± 0.28 | 3.02 ± 0.19 | 18.0 (13.2–22.8) | + | – | – | |
| L1PA5 | 9446 | 195 | 515 | 6.47 ± 0.25 | 3.39 ± 0.18 | 20.4 (15.0–25.9) | + | – | – | |
| L1PA6 | 4798 | 195 | 510 | 8.49 ± 0.31 | 4.44 ± 0.21 | 26.8 (19.7–34.0) | + | ? | – | |
| L1PA7 | 9863 | 174 | 495 | 9.95 ± 0.32 | 5.16 ± 0.20 | 31.4 (23.0–39.8) | + | +/– | – | |
| L1PA8 | 6672 | 196 | 530 | 12.96 ± 0.45 | 6.92 ± 0.31 | 40.9 (30.0–51.8) | + | + | – | |
| L1PA8A | 1474 | 184 | 520 | 13.20 ± 0.44 | 6.87 ± 0.26 | 41.7 (30.6–52.8) | + | + | – | |
| L1PA10 | 4827 | 181 | 455 | 14.68 ± 0.54 | 7.65 ± 0.31 | 46.4 (34.0–58.7) | + | + | – | |
| L1PA11 | 3047 | 189 | 480 | 16.88 ± 0.49 | 8.82 ± 0.30 | 53.3 (39.1–67.5) | + | + | – | |
| L1PA13B | 3114 | 63 | 579 | 18.93 ± 0.58 | 9.78 ± 0.32 | 59.8 (43.8–75.7) | + | + | – | |
| L1PA12 | 892 | 78 | 518 | 18.90 ± 0.62 | 9.82 ± 0.35 | 59.7 (43.7–75.6) | + | + | – | |
| L1PA13A | 4671 | 94 | 579 | 18.98 ± 0.64 | 10.07 ± 0.39 | 59.9 (43.9–75.9) | + | + | – | |
| L1PA14 | 2818 | 161 | 500 | 19.19 ± 0.60 | 10.14 ± 0.34 | 60.6 (44.4–76.8) | + | + | – | |
| L1PA15 | 5951 | 153 | 470 | 22.33 ± 0.70 | 11.74 ± 0.39 | 70.5 (51.7–89.3) | + | + | + | |
| L1PA16 | 9430 | 87 | 520 | 25.23 ± 0.67 | 13.27 ± 0.39 | 79.7 (58.4–100.9) | + | + | + | |
| L1PA17 | 3309 | 39 | 490 | 32.01 ± 1.18 | 17.17 ± 0.70 | 101.1 (74.1–128.0) | + | + | + | |
| L1PB | L1PB1 | 7412 | 124 | 540 | 14.80 ± 0.54 | 8.07 ± 0.38 | 46.7 (34.3–59.2) | + | + | – |
| L1PB2 | 1759 | 118 | 541 | 18.43 ± 0.62 | 9.63 ± 0.36 | 58.2 (42.7–73.7) | + | + | – | |
| L1PB3 | 2073 | 110 | 631 | 23.29 ± 0.75 | 12.39 ± 0.45 | 73.5 (53.9–93.2) | + | + | + | |
| L1PB4 | 5454 | 35 | 398 | 30.39 ± 1.11 | 16.50 ± 0.68 | 96.0 (70.3–121.6) | + | + | + | |
| L1MA | L1MA1 | 2528 | 55 | 530 | 19.52 ± 0.56 | 10.04 ± 0.30 | 61.6 (45.2–78.1) | + | + | – |
| L1MA2 | 4237 | 70 | 542 | 20.85 ± 0.61 | 10.88 ± 0.35 | 65.8 (48.3–83.4) | + | + | +/– | |
| L1MA3 | 5129 | 70 | 524 | 21.56 ± 0.73 | 11.39 ± 0.43 | 68.1 (49.9–86.2) | + | + | + | |
| L1MA4 | 6859 | 41 | 511 | 32.41 ± 1.02 | 17.85 ± 0.63 | 102.3 (75.0–129.6) | + | + | + | |
| L1MA5 | 2886 | 40 | 390 | 31.23 ± 1.17 | 16.78 ± 0.65 | 98.6 (72.3–124.9) | + | + | + |
Table 1 also shows that L1 families differ dramatically in their replicative success, as indicated by their different copy number. Family L1PA12 is the smallest one, with ∼900 copies. In contrast, five families have more than 8000 copies (L1PA16, L1PA7, L1PA5, L1PA4, and L1PA3). Four of the most active L1 families amplified between 40 and 12 Mya, indicating a period of high L1 activity just after the split between Old World and New World primates that occurred ∼40 Mya. While a single lineage was responsible for this increased activity, the simultaneous activity of several lineages or families in ancestral primate genomes did not produce a higher rate of L1 amplification. For instance, between 55 and 65 Mya, three lineages (L1MA4-1, L1PB3-1, and L1PA17-1) were simultaneously active, and four clearly differentiated L1PA families (L1PA12, L1PA13A, L1PA13B, and L1PA14) amplified at about the same time (∼60 Mya), but the number of L1 copies produced by all of these concurrently active families is similar to the activity of the sole L1PA lineage over the same length of time during the last 40 Myr. This observation suggests that L1 amplification might be in some way limited, either because L1 transposition requires a limiting host factor, or because there is a limit to the number of new inserts a genome can tolerate.
Repeated recruitment of 5′UTR sequences
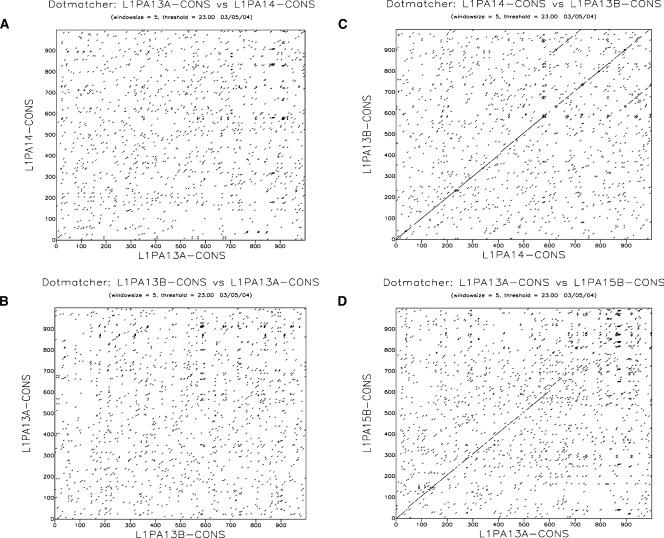
The comparison of the family-specific consensus sequences revealed that the 5′UTR of some L1 families are not homologous. For instance, the 5′UTR of families L1PA13B and L1PA14 were homologous, but neither was homologous to the 5′UTR of the L1PA13A family (Fig. 3). This cannot be explained by a high rate of evolution of the 5′UTR, because these three families are of similar age (Table 1) and because we were able to detect some similarity between the L1PA13A 5′UTR and the even older L1PA15 5′UTR. After comparing all L1 families by dot-plots, we were able to identify seven different types of 5′UTR with no or very little similarity to each other, except over the first ∼50 bp of the 5′UTR and over the 20 bp adjacent to the start-codon of ORF1. As we relied on RepeatMasker to identify full-length elements, it is plausible that some additional types of 5′UTRs are present in the genome, but are not detected by Repeat-Masker. Because we failed to find any intermediate 5′UTR sequences between these seven types, we can infer that these 5′UTRs did not evolve from each other by the accumulation of mutations, but instead, that L1 lineages acquired radically different 5′UTR several times.
Figure 3.
L1 families of similar ages have unrelated 5′UTR sequences. This dot plot analysis, based on the first 1000 bp of the 5′UTR, shows that the 5′UTR of family L1PA13A is unrelated to the 5′UTR of families L1PA14 (A) and L1PA13B (B). In contrast, families L1PA14 and L1PA13B (C) and families L1PA13A and L1PA15 (D) are relatively similar.
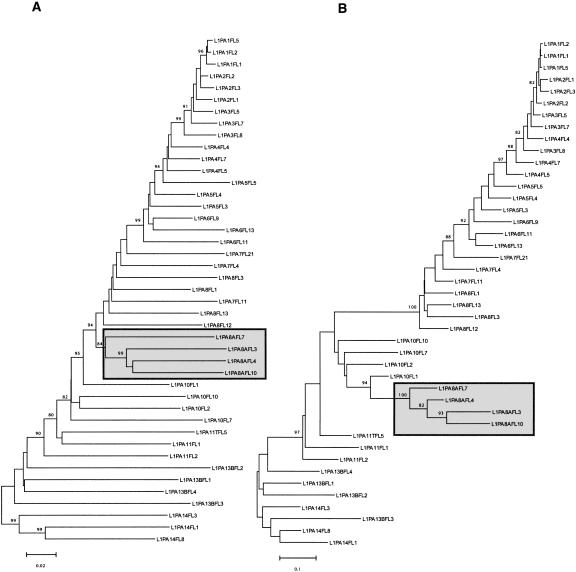
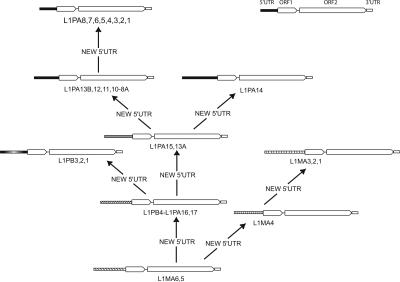
Figure 4 shows two trees built using the same FL length elements. Tree A is based on the 3′ 2000 bp, while tree B is based on the 5′ 300 bp of the same elements. For the most part, tree A shows a gradual evolution of L1 families, whereas tree B shows a discontinuity during the evolution of the 5′UTR. This discontinuity (between families L1PA8A and L1PA8) most likely corresponds to an additional replacement of 5′UTR with a divergent but homologous 5′UTR sequence in family L1PA8. This discontinuity is also discernable on the tree based on the 3′ end where the L1PA8A family forms a separate lineage that seems to have coexisted for some time with the L1PA8 family. Figure 5 shows a possible scenario based on the phylogenetic analysis on Figure 2. The 5′UTR of family L1MA5 is similar to the 5′UTR of the L1MA6 (data not shown) and therefore corresponds to the ancestral state. It was then replaced at least eight times over the last 70 Myr of primate evolution. In fact, more replacements probably occurred because we found some small subsets of FL elements that had clearly different 5′UTR, but their number in the genome was too small to build an accurate consensus. The acquisition of the modern type of 5′UTR occurred in family L1PA13B, and variations of this type of 5′UTR have been retained in all subsequently amplifying families, including the currently active L1PA1 family (Supplemental material 2). All of the lineages that coexisted within the genome of ancestral primates, between 70 and 40 Mya, had different 5′UTR, whereas the single lineage that amplified during the last ∼40 Myr retained the same type of 5′UTR. In conclusion, our analysis suggests that the recruitment of novel 5′UTRs is a common phenomenon that occurred as many as eight times over the last 70 Myr of primate evolution.
Figure 4.
Phylogeny of L1 genomic sequences. These trees were built using the neighbor-joining method based on Kimura's two-parameter distances. These two trees were built using the same full-length elements but using different regions of L1. Tree A was built using the 3′ end of the elements (2000 bp) and tree B was built using the 5′ end of the elements (300 bp). Only bootstrap values > 80 are shown. The gray boxes indicate that the L1PA8A family is forming a distinct lineage (see text).
Figure 5.
L1 lineages have frequently recruited novel 5′UTR sequences. This scenario was inferred from the phylogenetic tree in Figure 2. An arrow indicates the acquisition of a new 5′UTR. The 5′UTR sequences are drawn proportionally to their size. As the L1PA14 family is nested within the L1PA13A family (data not shown), it is likely that the modern type of 5′UTR was probably recruited independently by the L1PA14 and L1PA13B families.
Despite their lack of homology, all of these different types of 5′UTR presumably had the ability to drive the transcription of L1. We examined the level of conservation of motifs that have been shown experimentally to be functionally important. Those functional domains and motifs are shown on Figure 1B. Only the first 54 bp of the 5′UTR shows some similarity between 5′UTR types. The only feature common to all types is the presence of a Yin Yang 1 (YY1) binding site between positions 21 and 13 on the antisense strand (Becker et al. 1993). The conservation of the YY1 site is consistent with the important role of YY1 in directing L1 transcription initiation (Athanikar et al. 2004). In contrast, the functional RUNX3 transcription-factor binding site (Yang et al. 2003), the two factor-binding motifs (motifs B and C in Minakami et al. 1992), the two functional SRY-related transcription factor sites (Tchenio et al. 2000) and the antisense promoter region (Speek 2001) are all specific acquisition of the L1PA lineage. The segments corresponding to the RUNX-3 binding site and motifs B and C can be found in families L1PA1-L1PA14, but the motifs themselves are not particularly conserved in families older than L1PA8. The two SRY-binding sites are not conserved in families older than L1PA7. The antisense promoter is located in a region of the 5′UTR that has undergone a large number of large indels. Indeed, the antisense promoter region is conserved only among families L1PA6-L1PA1. We searched for potential transcription-factor binding sites in older families using the TFSEARCH engine at http://www.cbrc.jp/research/db/TFSEARCH.html and found several good candidates (data not shown). However, the biological significance of these putative binding sites is unclear and needs to be assessed experimentally. Altogether, these observations indicate that, except for the presence of an YY1 site close to the transcription initiation site, the different types of 5′UTR don't share any obvious common motifs. This suggests that the transcription of elements belonging to ancestral L1 families required different host-encoded factors and that the regulation of transcription probably differed between simultaneously active families and lineages.
Evolution of ORF1
We examined in more detail the evolution of ORF1 because its coiled-coil domain has undergone an episode of adaptive evolution during the evolution of families L1PA5-L1PA3 (Boissinot and Furano 2001). This analysis was limited to the five most recent L1 families and we extend it here to older families. Because consensus sequences of the oldest families contained a number of ambiguities in the coiled-coil domain, we were able to analyze only the 17 most recent L1PA families (from L1PA16 to L1PA1). The PLATO analysis identified a region starting at position 12 and ending at position 396 that has evolved significantly faster than the entire ORF1 (Z = 20.66, P < 001, sliding window size = 5). Modifying the parameters of the analysis identified the same region and yielded similar Z values that were all significant. This region has undergone a large number of amino acid substitutions and three in-frame indels. We tested whether the high rate of amino acid replacement was caused by positive selection as it was previously reported (Boissinot and Furano 2001). First, we determined whether the ratio ω has changed over time. The free-ratio model (allows ω to vary across lineages) fit the data significantly better than the one-ratio model (2ΔlnL = 58.86, _P_ < 0.01, df = 31), indicating that the nature and/or strength of the selection that acted on this fast-evolving region has changed over evolutionary times. The free-ratio model assigned estimates of ω > 1 on several branches of the tree (identified by an asterisk in Fig. 2), and the estimates of ω it produced are very similar to those calculated by the method of Yang and Nielsen (2000) presented in Table 2. Table 2 shows that the ratio ω has varied considerably during the evolution of the L1PA lineages. During the evolution of the ancestral L1PA16 through L1PA8 families (with the exception of the branch leading to L1PA13B) the ratio ω has remained relatively low (from 0.24 to 0.74). In contrast, during the evolution of families L1PA8 through L1PA3, ω has been consistently high, with values higher than 0.8, and in some cases, significantly higher than 1, suggesting that positive selection (i.e., selection in favor of amino-acid changes) has affected the evolution of this region of ORF1. Although the criteria ω > 1 is usually used as evidence for positive selection, this criteria is also overly stringent, and it is more likely that the high values of ω from L1PA8 to L1PA3 correspond to a single episode of positive selection that started ∼40 Mya and ended ∼12 Mya. In comparison with our earlier study (Boissinot and Furano 2001), we found that positive selection might not be limited to the coiled-coil domain, but also affect the evolution of the sequence upstream of it (Table 2).
Table 2.
Maximum likelihood estimates of ω for different regions of L1
| ORF1 (5′) non-CCa | Coiled-coil domainb | ORF1 (5′)c | ORF1 (3′)d | ORF2 | |
|---|---|---|---|---|---|
| L1PA2 => L1PA1 | 0.329 | NA | 0.296 | 0.335 | 0.217 |
| L1PA3 => L1PA2 | 0.161 | 0.740 | 0.445 | 0.083 | 0.277 |
| L1PA4 => L1PA3 | 0.312 | 4.435 | 2.291 | 0.658 | 0.094 |
| L1PA5 => L1PA4 | 0.926 | ∞ | 4.469 | 0.654 | 0.289 |
| L1PA6 => L1PA5 | 0.430 | 2.906 | 0.809 | 0.116 | 0.159 |
| L1PA7 => L1PA6 | 1.293 | 1.428 | 1.342 | 0.162 | 0.207 |
| L1PA8 => L1PA7 | 0.901 | 0.872 | 0.843 | 0.221 | 0.373 |
| L1PA8A => L1PA8 | 0.253 | 0.324 | 0.259 | 0.213 | 0.276 |
| L1PA10 => L1PA8A | 1.193 | 0.288 | 0.417 | 0.229 | 0.130 |
| L1PA11 => L1PA10 | 0.166 | 1.307 | 0.738 | 0.400 | 0.183 |
| L1PA13B => L1PA11 | 0.623 | 0.512 | 0.523 | 0.209 | 0.210 |
| L1PA12 => L1PA13B | 1.999 | 0.901 | 1.253 | 0.189 | 0.191 |
| L1PA13A => L1PA12 | 0.154 | 0.282 | 0.242 | 0.191 | 0.270 |
| L1PA14 => L1PA13A | 0.357 | 0.410 | 0.384 | 0.258 | 0.181 |
| L1PA15 => L1PA14 | 0.479 | 0.519 | 0.515 | 0.230 | 0.189 |
| L1PA16 => L1PA15 | 0.637 | 0.150 | 0.266 | 0.164 | 0.236 |
Discussion
In this study, we have analyzed the evolution of the L1 families that have amplified in the human genome since before the origin of primates until the present time. We distinguished two major phases in the evolution of L1. During the last ∼40 Myr, a single lineage of L1 families has dominated the replicative process. In the short term, distinct L1 families have occasionally been concurrently active (e.g., L1PA8A and L1PA8) (Table 2; Fig. 4), but eventually only one persisted and in the long term, a single lineage is observed. In contrast, until ∼40 Mya, several clearly distinct L1 lineages coexisted and were simultaneously active in the genome of ancestral primates for as long as 30 Myr. The long-term coexistence of several lineages remains the exception in extant mammals. Phylogenetic analyses in mice, rats, and primates (Martin et al. 1985; Hardies et al. 1986; Pascale et al. 1990, 1993; Furano et al. 1994; Boissinot and Furano 2001; Boissinot et al. 2004) have shown that in most mammalian species investigated so far a single lineage of L1 families is present. Although the past coexistence of L1 lineages has also been detected in the genome of New World monkeys (Boissinot et al. 2004) and rabbits (Price et al. 1992), the long-term persistence of more than one L1 lineage in a modern mammal has been described only in deer mice (Casavant et al. 1996).
The reason(s) why L1 lineages rarely coexist for extended periods of time remains unclear. It has previously been suggested that L1 families engage in some sort of competition, possibly for some host factors, until one attains replicative supremacy (Casavant and Hardies 1994; Cabot et al. 1997). Our analysis revealed that the most significant difference between coexisting families was in the 5′UTR. Indeed, the families of elements that evolved into the three main lineages (L1MA4-1, L1PB3-1, and L1PA17-1) had completely different (i.e., unrelated) 5′UTRs. In contrast, ORF2 and most of ORF1 remained relatively conserved at the amino acid level. As the different types of primate-specific 5′UTRs don't share any obvious common motifs, except a YY1-binding site, it is likely that the transcription of elements with different 5′UTR required different host-encoded factors. Presumably, these L1 elements could be simultaneously active because they didn't rely on the same limiting host-factors for their transcription and were not competing with each other. Similarly, coexistence of distinct L1PA families (e.g., L1PA13A and L1PA14, L1PA8A and L1PA8) occurred only between families of elements that have different 5′UTRs. A similar situation can be found in the genome of mice, which contains three distinct and simultaneously active L1 families (L1MdA, Tf and Gf) (Goodier et al. 2001; Mears and Hutchison III 2001). These three murine families have very different 5′UTRs and the 5′UTR of family L1MdA is unrelated to the 5′UTR of families Tf and Gf (Adey et al. 1994; Goodier et al. 2001; Mears and Hutchison III 2001). These different examples suggest that distinct L1 families can coexist if they have unrelated 5′UTRs. Therefore, we propose that the absence of competition for transcription factors between elements with different 5′UTRs allows them to coexist in mammalian genomes for extended periods of evolutionary times. This long-term coexistence results in the formation of independently evolving lineages such as the L1MA4-1, L1PB3-1, and L1PA17-1 lineages. Coexistence between different L1 families would eventually end when a novel family acquires a 5′UTR, which is more efficient at driving transcription than other L1 families, and therefore dominates the replicative process. This scenario implies that there is a second level of competition between L1 families, possibly for a host factor required for L1 transposition. Interestingly, the extinction of the L1PB3-1 lineage (∼40 Mya) roughly corresponds to the acquisition of the modern type of 5′UTR that occurred just before a period of intense amplification (families L1PA8-L1PA3). Adey et al. (1994) also noted that the acquisition of the A type of 5′UTR in mice correlated with the demise of a number of ancestral L1 families with different 5′UTRs.
Our analysis revealed that L1 families have frequently recruited novel 5′UTRs in the human lineage. A similar observation has been made in mouse, where L1 families acquired novel 5′UTRs at least twice in the past 5-6 Myr (Adey et al. 1994; Furano 2000). The lack of homology between primates, mouse, rat, and rabbit 5′UTRs also suggests that the acquisition of novel 5′UTRs in mammals is a fundamental feature of L1 evolution (Scott et al. 1987; Wincker et al. 1987; Furano et al. 1988; Padgett et al. 1988; Jubier-Maurin et al. 1992; Schichman et al. 1993; Adey et al. 1994; Furano 2000). The acquisition of novel regulatory sequences is probably facilitated by the ability of the L1 reverse transcriptase to switch templates during the target-site primed reverse transcription reaction (Martin et al. 2005). Strand-switching seems to occur frequently as exemplified by the numerous cases of chimeric L1 elements (Hayward et al. 1997; Saxton and Martin 1998; Buzdin et al. 2002, 2003) and constitutes a source of variation on which selection can act. It is also possible that novel regulatory sequences were acquired by gene conversion. However, this seems relatively unlikely because conversion events between L1 elements are very rare in humans (Boissinot et al. 2001; Myers et al. 2002). Whatever the mechanism of recruitment, the acquisition of a novel 5′UTR could have been advantageous if elements with a new type of 5′UTR did not compete for host-factor with the bulk of L1 elements (and therefore occupied a different transcriptional niche) or if it allowed elements to bypass host repression of transcription, or both.
L1 families show considerable variation in copy numbers, suggesting large differences in replicative success. The most intense period of L1 activity involved families L1PA8-L1PA3 and lasted from ∼40 Mya to ∼12 Mya. The amplification of these very successful families is responsible for the amplification of the bulk of the _Alu_Y elements and of many processed pseudogenes (Batzer and Deininger 2002; Ohshima et al. 2003) and accounts for the larger genome size of anthropoid primates (e.g., monkeys, apes, and human) compared with prosimian primates (e.g., lemurs and galagos) which split from the anthropoid lineage 63 Mya (Liu et al. 2003). The start of the period of high L1 activity, 40 Mya, coincides with two events that could explain the replicative success of families L1PA8-L1PA3, i.e., the acquisition of a novel 5′UTR by family L1PA8 and the extinction of the L1PB1 family. These two events might not be independent, as the extinction of the L1PB1 family could have been caused by the recruitment of a new regulatory sequence (see above). First, the acquisition of a very efficient promoter (i.e., a promoter that produces more L1 transcripts) could by itself cause an increase in retrotransposition. Such a novel promoter could have either bypassed host repression or been more efficient at recruiting host transcription factors required for L1 expression. Second, it is possible that elements belonging to families L1PB1 and L1PA8 competed with each other for an additional host factor necessary for retrotransposition (Casavant and Hardies 1994; Cabot et al. 1997). The extinction of the L1PB1 family, caused either by the accumulation of inactivating mutations or by host repression, could have relieved L1PA8 elements from any competition and allowed the L1PA8-L1PA3 families to amplify at a higher rate. In the absence of more experimental data, it is not possible to determine whether one or both of these hypotheses is correct. Whatever the reason for the replicative success of families L1PA8-L1PA3, an important event occurred ∼40 Mya that has dramatically affected the pattern of evolution and amplification of L1 and, consequently, the evolution of primate genomes.
We determined that the 5′end of ORF1 (from nucleotide 12 to 396) underwent an episode of positive selection that occurred during the evolution of families L1PA8-L1PA3. In contrast, this region has remained remarkably conserved during the evolution of older (from families L1PA16-L1PA8, with the exception of family L1PA13B) and younger (L1PA2 and L1PA1) families. This suggests that the strength or nature of the selective pressure that has driven the rapid evolution of this region has changed over time. It was recently proposed that positive selection in ORF1 could reflect an adaptation of L1 to its hosts (Boissinot and Furano 2001; Furano et al. 2004). More specifically, we and others suggested that adaptive evolution in ORF1 reflects L1 adaptation to a host-encoded repressor of L1 replication or to a rapidly evolving host factor necessary for L1 replication. This model predicts that adaptive evolution in ORF1 should correlate with the rate of amplification of L1 because a high level of L1 activity creates the selective environment for the host to reduce L1 replication and for L1 to preserve its activity. This scenario fits with our present finding, i.e., the episode of positive selection corresponds to the time of activity of the most successfully replicative L1 families (from families L1PA8-L1PA3).
Methods
Collection and alignment of full-length elements
Full-length elements belonging to the L1 families that amplified since before the origin of primates (families L1MA5-L1MA1, L1PA17-L1PA1, and L1PB4-L1PB1) were collected by searching table RepeatMasker (assembly of April 2003) at http://genome.ucsc.edu for all L1 elements longer than 6 Kb. The classification of each element was confirmed by phylogenetic analysis (see below) and by comparing their 3′UTR to the consensus sequences published by Smit et al. (1995) and available from Repbase (at http://www.girinst.org/). Full-length elements belonging to the same family were then aligned to each other using CLUSTALW (Thompson et al. 1994) and a consensus sequence was derived for each subset of elements. Consensus sequences can be understood as the best possible reconstruction of the progenitors at the origin of a family. Mutations in the highly mutable CpG dinucleotides were eliminated from the consensus except when they corresponded to fixed differences among families. For the five most recent families (L1PA5-L1PA1) we used the consensus derived in Boissinot and Furano (2001). Alignments were performed and visualized using the BioEdit platform of program (Hall 1999). The consensus sequences obtained for each family were then aligned to each other. When necessary, the dot-plot method (at http://bioweb.pasteur.fr/seqanal/interfaces/dotmatcher.html) was used to search for homology between sequences that couldn't be easily aligned using CLUSTALW.
Analysis of full-length elements
Phylogenetic trees of L1 genomic copies were built using the neighbor joining (NJ) method (Saitou and Nei 1987) based on Kimura 2-parameters' distances (Kimura 1980), and the robustness of the trees was assessed using a bootstrap procedure. Phylogenetic relationships among consensus sequences were analyzed by NJ, maximum parsimony, and maximum likelihood (ML). NJ and maximum parsimony trees were calculated using the MEGA 2.1 program package (Kumar et al. 2001) and ML analyses were performed using the PAML 3.0 program package (Yang 2000).
To detect variations in the rate of evolution (either caused by recombination or selection) along L1 sequences, we used the program PLATO (Grassly and Holmes 1997). Using a ML tree based on the entire L1 sequence as the null hypothesis, PLATO uses a sliding window through which deviations from the ML-generated branch lengths are calculated, thereby identifying regions that differ in evolutionary rate from the complete sequence.
We examined the possibility that positive selection has played a role in the evolution of L1 by estimating the ratio of nonsynonymous to synonymous substitution rate (ω) within a phylogenetic context. A ratio significantly > 1 indicates that nonsynonymous substitutions have been reaching fixation faster than synonymous substitutions and is indicative of positive selection. First, we examined whether ω varied during the evolution of L1 families. This was tested by comparing a model that assumes a constant ω (M0 model in Yang et al. 2000) with one that allows ω to vary across the branches of the ML tree (free-ratio model in Yang 1998). The double of the log-likelihood difference between the two models is compared with a χ2 distribution with degrees of freedom equal to the number of branches on the ML tree minus 1. The free-ratio model also permits one to determine on which branch of the tree positive selection has occured (i.e., branches for which ω > 1). However, because this model is parameter-rich, the estimates of ω it produces are unlikely to be accurate. Therefore, pairwise comparisons of ω were also performed using the model of Yang and Nielsen (2000). All of these calculations were performed using the PAML 3.0 program package (Yang 2000).
Estimation of the timing and intensity of amplification
Because L1 elements accumulate mutations at the neutral rate, the age of a family is proportional to the divergence between its members. Thus, we estimated the age of each family by calculating the average pairwise divergence between each element and all of the other elements of the family. For each family, we collected a large number of ∼500-bp sequences corresponding to the 3′ extremity of L1. These sequences were aligned using CLUSTALW. CpG dinucleotides and the highly mutable polypurine tract located in the 3′UTR were removed from the alignment. The level of divergence (and standard error) associated with each family was then calculated using Kimura 2-parameters correction. Because the rate of DNA substitution has recently decreased in the human lineage (Goodman 1961; Goodman et al. 1971; Yi et al. 2002), we converted divergences to time using two different calibrations. First, we used an L1 primate rate of 0.125%/Myr based on human/orangutan comparisons (Boissinot et al. 2000) and human/baboon comparisons (Liu et al. 2003). Because this calibration is likely to overestimate the age of old families, we also applied a calibration of 0.216%/Myr based on a comparison between human and lemur sequences (calibrated using the data of Liu et al. 2003). This calibration reflects more faithfully the faster rate of sequence evolution in ancestral primates. The age of the families was verified by examining their presence (or absence) in the genome of three nonhuman primates for which large amounts of genomic sequences are available (Liu et al. 2003). These three species are the chimpanzee, the baboon, and the lemur, which split from the human lineage 6, 25, and 63 Mya, respectively (Goodman et al. 1998). The replicative success of each family was estimated by determining genomic copy numbers from table RepeatMasker at http://genome.ucsc.edu. Because L1 families have been classified based on their 3′ends, we estimated copy number by counting for each family the number of 3′UTR present in the genome.
Acknowledgments
We thank Anthony Furano, Aron Branscomb, and three anonymous reviewers for their helpful comments on the manuscript.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4001406.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Adey, N.B., Schichman, S.A., Graham, D.K., Peterson, S.N., Edgell, M.H., and Hutchison III, C.A. 1994. Rodent L1 evolution has been driven by a single dominant lineage that has repeatedly acquired new transcriptional regulatory sequences. Mol. Biol. Evol. 11 778-789. [DOI] [PubMed] [Google Scholar]
- Athanikar, J.N., Badge, R.M., and Moran, J.V. 2004. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32 3846-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, M.A. and Deininger, P.L. 2002. Mammalian retroelements. Genome Res. 12 1455-1465. [DOI] [PubMed] [Google Scholar]
- Becker, K.G., Swergold, G.D., Ozato, K., and Thayer, R.E. 1993. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum. Mol. Genet. 2 1697-1702. [DOI] [PubMed] [Google Scholar]
- Boissinot, S. and Furano, A.V. 2001. Adaptive evolution in LINE-1 retrotransposons. Mol. Biol. Evol. 18 2186-2194. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., Chevret, P., and Furano, A.V. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17 915-928. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., Entezam, A., and Furano, A.V. 2001. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 18 926-935. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., Roos, C., and Furano, A.V. 2004. Different rates of LINE-1 (L1) retrotransposon amplification and evolution in New World monkeys. J. Mol. Evol. 58 122-130. [DOI] [PubMed] [Google Scholar]
- Burton, F.H., Loeb, D.D., Voliva, C.F., Martin, S.L., Edgell, M.H., and Hutchison III, C.A. 1986. Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J. Mol. Biol. 187 291-304. [DOI] [PubMed] [Google Scholar]
- Buzdin, A., Ustyugova, S., Gogvadze, E., Vinogradova, T., Lebedev, Y., and Sverdlov, E. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of L1. Genomics 80 402-406. [DOI] [PubMed] [Google Scholar]
- Buzdin, A., Gogvadze, E., Kovalskaya, E., Volchkov, P., Ustyugova, S., Illarionova, A., Fushan, A., Vinogradova, T., and Sverdlov, E. 2003. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 31 385-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabot, E.L., Angeletti, B., Usdin, K., and Furano, A.V. 1997. Rapid evolution of a young L1 (LINE-1) clade in recently speciated Rattus taxa. J. Mol. Evol. 45 412-423. [DOI] [PubMed] [Google Scholar]
- Casavant, N.C. and Hardies, S.C. 1994. The dynamics of murine LINE-1 subfamily amplification. J. Mol. Biol. 241 390-397. [DOI] [PubMed] [Google Scholar]
- Casavant, N.C., Sherman, A.N., and Wichman, H.A. 1996. Two persistent LINE-1 lineages in Peromyscus have unequal rates of evolution. Genetics 142 1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost, G.J., Feng, Q., Jacquier, A., and Boeke, J.D. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21 5899-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q., Moran, J.V., Kazazian, H.H., and Boeke, J.D. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87 905-916. [DOI] [PubMed] [Google Scholar]
- Furano, A.V. 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acid. Res. Mol. Biol. 64 255-294. [DOI] [PubMed] [Google Scholar]
- Furano, A.V., Robb, S.M., and Robb, F.T. 1988. The structure of the regulatory region of the rat L1 (L1Rn, long interspersed repeated) DNA family of transposable elements. Nucleic Acids Res. 16 9215-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano, A.V., Hayward, B.E., Chevret, P., Catzeflis, F., and Usdin, K. 1994. Amplification of the ancient murine Lx family of long interspersed repeated DNA occurred during the murine radiation. J. Mol. Evol. 38 18-27. [DOI] [PubMed] [Google Scholar]
- Furano, A.V., Duvernell, D., and Boissinot, S. 2004. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 20 9-14. [DOI] [PubMed] [Google Scholar]
- Goodier, J.L., Ostertag, E.M., Du, K., and Kazazian Jr., H.H. 2001. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 11 1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M. 1961. The role of immunochemical differences in the phyletic development of human behavior. Hum. Biol. 33 131-162. [PubMed] [Google Scholar]
- Goodman, M., Barnabas, J., Matsuda, G., and Moore, G.W. 1971. Molecular evolution in the descent of man. Nature 233 604-613. [DOI] [PubMed] [Google Scholar]
- Goodman, M., Porter, C.A., Czelusniak, J., Page, S.L., Schneider, H., Shoshani, J., Gunnell, G., and Groves, C.P. 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phyl. Evol. 9 585-598. [DOI] [PubMed] [Google Scholar]
- Grassly, N.C. and Holmes, E.C. 1997. A likelihood method for the detection of selection and recombination using sequence data. Mol. Biol. Evol. 14 239-247. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Series 41 95-98. [Google Scholar]
- Hardies, S.C., Martin, S.L., Voliva, C.F., Hutchison III, C.A., and Edgell, M.H. 1986. An analysis of replacement and synonymous changes in the rodent L1 repeat family. Mol. Biol. Evol. 3 109-125. [DOI] [PubMed] [Google Scholar]
- Hayward, B.E., Zavanelli, M., and Furano, A.V. 1997. Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus. Genetics 146 641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, R. and Usdin, K. 1997. The ability to form intrastrand tetraplexes is an evolutionarily conserved feature of the 3′ end of L1 retrotransposons. Mol. Biol. Evol. 14 144-155. [DOI] [PubMed] [Google Scholar]
- Jubier-Maurin, V., Cuny, G., Laurent, A.-M., Paquereau, L., and Roizes, G. 1992. A new 5′ sequence associated with mouse L1 elements is representative of a major class of L1 termini. Mol. Biol. Evol. 9 41-55. [DOI] [PubMed] [Google Scholar]
- Kazazian, H.H. 2004. Mobile elements: Drivers of genome evolution. Science 303 1626-1632. [DOI] [PubMed] [Google Scholar]
- Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111-120. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Arizona State University, Tempe, AZ. [DOI] [PubMed]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409 860-921. [DOI] [PubMed] [Google Scholar]
- Liu, G., Zhao, S., Bailey, J.A., Sahinalp, S.C., Alkan, C., Tuzun, E., Green, E.D., and Eichler, E.E. 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13 358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, D.D. and Eickbush, T.H. 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 15 3882-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, D.D., Korman, M.H., Jakubczak, J.L., and Eickbush, T.H. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 72 595-605. [DOI] [PubMed] [Google Scholar]
- Martin, S.L. and Bushman, F.D. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21 467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S.L., Voliva, C.F., Hardies, S.C., Edgell, M.H., and Hutchison III, C.A. 1985. Tempo and mode of concerted evolution in the L1 repeat family of mice. Mol. Biol. Evol. 2 127-140. [DOI] [PubMed] [Google Scholar]
- Martin, S.L., Li, J., and Weisz, J.A. 2000. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 304 11-20. [DOI] [PubMed] [Google Scholar]
- Martin, S.L., Li, W.-H.P., Furano, A.V., and Boissinot, S. 2005. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet. Genome Res. 110 223-228. [DOI] [PubMed] [Google Scholar]
- Mathews, L.M., Chi, S.Y., Greenberg, N., Ovchinnikov, I., and Swergold, G.D. 2003. Large differences between LINE-1 amplification rates in the human and chimpanzee lineages. Am. J. Hum. Genet. 72 739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias, S.L., Scott, A.F., Kazazian, H.H., Boeke, J.D., and Gabriel, A. 1991. Reverse transcriptase encoded by a human transposable element. Science 254 1808-1810. [DOI] [PubMed] [Google Scholar]
- Mears, M.L. and Hutchison III, C.A. 2001. The evolution of modern lineages of mouse L1 elements. J. Mol. Evol. 52 51-62. [DOI] [PubMed] [Google Scholar]
- Minakami, R., Kurose, K., Etoh, K., Furuhata, Y., Hattori, M., and Sakaki, Y. 1992. Identification of an internal _cis_-element essential for the human L1 transcription and a nuclear factor(s) binding to the element. Nucleic Acids Res. 20 3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, J.S., Vincent, B.J., Udall, H., Watkins, W.S., Morrish, T.A., Kilroy, G.E., Swergold, G.D., Henke, J., Henke, L., Moran, J.V., et al. 2002. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 71 312-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima, K., Hattori, M., Yada, T., Gojobori, T., Sakaki, Y., and Okada, N. 2003. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 4 R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov, I., Rubin, A., and Swergold, G.D. 2002. Tracing the LINEs of human evolution. Proc. Natl. Acad. Sci. 99 10522-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett, R.W., Hutchison III, C.A. and Edgell, M.H. 1988. The F-type 5′ motif of mouse L1 elements: A major class of L1 termini similar to the A-type in organization but unrelated in sequence. Nucleic Acids Res. 16 739-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale, E., Valle, E., and Furano, A.V. 1990. Amplification of an ancestral mammalian L1 family of long interspersed repeated DNA occurred just before the murine radiation. Proc. Natl. Acad. Sci. 87 9481-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale, E., Liu, C., Valle, E., Usdin, K., and Furano, A.V. 1993. The evolution of long interspersed repeated DNA (L1, LINE 1) as revealed by the analysis of an ancient rodent L1 DNA family. J. Mol. Evol. 36 9-20. [DOI] [PubMed] [Google Scholar]
- Price, D.K., Ayres, J.A., Pasqualone, D., Cabell, C.H., Miller, W., and Hardison, R.C. 1992. The 5′ ends of LINE1 repeats in rabbit DNA define subfamilies and reveal a short sequence conserved between rabbits and humans. Genomics 14 320-331. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406-425. [DOI] [PubMed] [Google Scholar]
- Saxton, J.A. and Martin, S.L. 1998. Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J. Mol. Biol. 280 611-622. [DOI] [PubMed] [Google Scholar]
- Schichman, S.A., Adey, N.B., Edgell, M.H., and Hutchison III, C.A. 1993. L1 A-monomer tandem arrays have expanded during the course of mouse L1 evolution. Mol. Biol. Evol. 10 552-570. [DOI] [PubMed] [Google Scholar]
- Scott, A.F., Schmeckpeper, B.J., Abdelrazik, M., Comey, C.T., O'Hara, B., Rossiter, J.P., Cooley, T., Heath, P., Smith, K.D., and Margolet, L. 1987. Origin of the human L1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1 113-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, F.M., Sherry, S.T., Risch, G.M., Robichaux, M., Nasidze, I., Stoneking, M., Batzer, M.A., and Swergold, G.D. 2000. Reading between the LINEs: Human genomic variation induced by LINE-1 retrotransposition. Genome Res. 10 1496-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A.F. 1996. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6 743-748. [DOI] [PubMed] [Google Scholar]
- Smit, A.F., Toth, G., Riggs, A.D., and Jurka, J. 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246 401-417. [DOI] [PubMed] [Google Scholar]
- Speek, M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21 1973-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold, G.D. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10 6718-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio, T., Casella, J.F., and Heidmann, T. 2000. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 28 411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, G.D., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verneau, O., Catzeflis, F., and Furano, A.V. 1998. Determining and dating recent rodent speciation events by using L1 (LINE-1) retrotransposons. Proc. Natl. Acad. Sci. 95 11284-11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva, C.F., Martin, S.L., Hutchison III, C.A., and Edgell, M.H. 1984. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J. Mol. Biol. 178 795-813. [DOI] [PubMed] [Google Scholar]
- Wincker, P., Jubier-Maurin, V., and Roizes, G. 1987. Unrelated sequences at the 5′ end of mouse LINE-1 repeated elements define two distinct subfamilies. Nucleic Acids Res. 15 8593-8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15 568-573. [DOI] [PubMed] [Google Scholar]
- ———. 2000. PAML (phylogenetic analysis by maximum likelihood) version 3.0. University College, London.
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17 32-43. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Nielsen, R., Goldman, N., and Krabbe Pedersen, A.-M. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155 431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, N., Zhang, L., Zhang, Y., and Kazazian, H.H. 2003. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 31 4929-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, S., Ellsworth, D.L., and Li, W.H. 2002. Slow molecular clocks in Old World monkeys, apes and humans. Mol. Biol. Evol. 19 1291-1298. [DOI] [PubMed] [Google Scholar]
Web site references
- http://genome.ucsc.edu; human genome database.
- http://www.repeatmasker.org/; online tool for the identification of repetitive sequences.
- http://www.cbrc.jp/research/db/TFSEARCH.html; online tool for the detection of transcription factors binding sites.
- http://www.girinst.org/; database of transposable elements consensus sequences.
- http://bioweb.pasteur.fr/seqanal/interfaces/dotmatcher.html; online tool for dot plot analysis.