In vivo and in vitro interaction between human transcription factor MOK2 and nuclear lamin A/C (original) (raw)
Abstract
The human and murine MOK2 proteins are factors able to recognize both DNA and RNA through their zinc finger motifs. This dual affinity of MOK2 suggests that MOK2 might be involved in transcription and post-transcriptional regulation of MOK2 target genes. The IRBP gene contains two MOK2-binding elements, a complete 18 bp MOK2-binding site located in intron 2 and the essential core MOK2-binding site (8 bp of conserved 3′-half-site) located in the IRBP promoter. We have demonstrated that MOK2 can bind to the 8 bp present in the IRBP promoter and repress transcription from this promoter by competing with the CRX activator for DNA binding. In this study, we identify a novel interaction between lamin A/C and hsMOK2 by using the yeast two-hybrid system. The interaction, which was confirmed by GST pull-down assays and co-immunolocalization studies in vivo, requires the N-terminal acidic domain of hsMOK2 and the coiled 2 domain of lamin A/C. Furthermore, we show that a fraction of hsMOK2 protein is associated with the nuclear matrix. We therefore suggest that hsMOK2 interactions with lamin A/C and the nuclear matrix may be important for its ability to repress transcription.
INTRODUCTION
The human and murine MOK2 orthologous genes, which are preferentially expressed in brain and testis tissues, encode two different Krüppel/TFIIIA-related zinc finger proteins (1,2). Although both proteins are clearly similar in that they contain seven tandem zinc finger motifs and a five amino acid extension at their C-terminal ends, the human protein contains three additional tandemly arranged zinc finger motifs and a non-finger acidic domain of 173 amino acids in its N-terminus. Alternative splicing of hsMOK2 also results in an isoform that contains a smaller N-terminal acidic domain of 76 amino acids (hsMOK2Δ).
The human and murine MOK2 proteins can recognize both DNA and RNA through their zinc finger motifs (3). Electron microscopy and specific RNA homopolymer binding activity showed clearly that the murine and human MOK2 proteins are RNA-binding proteins that associate mainly with nuclear RNP components, including nucleoli and extranucleolar structures. The murine and human MOK2 proteins have been shown to bind the same 18 bp specific sequence in duplex DNA. Binding site selection with whole genomic PCR revealed that DNA-binding sites are localized in intron sequences. These results and the dual affinity of MOK2 for RNA and DNA suggest that MOK2 may be involved in transcription and post-transcriptional regulation of MOK2 target genes. Our studies identified eight putative hsMOK2 target genes, of which four, IRBP, PAX3, STEP and _hRXR_α (4–7), were already known. Initially, we showed that MOK2 negatively modulates expression of the interphotoreceptor retinoid-binding protein gene, IRBP (8). IRBP is exclusively expressed in retinal photoreceptor cells and in a sub-group of pinealocytes, where it is thought to be involved in the visual cycle of the vertebrate retina (9–11). The IRBP gene contains two MOK2-binding elements, a complete 18 bp MOK2-binding site located in intron 2 and the essential core MOK2-binding site (8 bp of the conserved 3′-half-site) located in the IRBP promoter. The results demonstrated that MOK2 could bind to the 8 bp sequence present in the IRBP promoter and repress transcription from this promoter, when transiently overexpressed in retinoblastoma Weri-RB1 cells. In the IRBP promoter, the TAAAGGCT MOK2-binding site overlaps with the photoreceptor-specific Crx-binding element (CRXE), suggesting that MOK2 represses transcription by competing with the cone–rod homeobox protein (CRX) for DNA binding and decreasing transcriptional activation through CRX. We have shown that Mok2 is expressed early in mouse embryonic development and in adult brain, suggesting that Mok2 might play an important role in development, and particularly in neuronal development.
The arrangement of the 8 bp essential core MOK2-binding site in the promoter and the 18 bp MOK2-binding site in the intron is reminiscent of that of another potential MOK2 target gene, Pax3. In this gene, the 18 bp MOK2-binding site is located in the last intron. A search for MOK2-binding sites in the proximal promoter region of human Pax3 reveals the presence of a TAAAAGGCT sequence that could possibly bind MOK2. Therefore, MOK2 might regulate the transcriptional activity of its target genes at different levels. One of the central questions regarding MOK2 relates to the mechanisms that control its dual binding activity. One model postulates that certain interaction partners of MOK2 might influence its activity.
To identify the cellular proteins that interact with human hsMOK2, we employed the yeast two-hybrid system to screen a HeLa cDNA library. We identified lamin A/C protein as a novel interaction partner of hsMOK2 and confirmed the interaction through a variety of experimental methods. We demonstrate direct interaction between the N-terminal acidic domain of hsMOK2 and the coiled 2 domain of lamin A/C. This interaction can also take place in vivo in eukaryotic HeLa cells. Furthermore, we show that a significant fraction of hsMOK2 protein is associated with the nuclear matrix.
MATERIALS AND METHODS
Plasmid constructs
All plasmids generated for this study were verified by DNA sequencing. For the yeast two-hybrid assay, the full-length coding region of hsMOK2, the non-finger acidic domain of hsMOK2 (nucleotides encoding amino acids 1–173) and the finger domain of hsMOK2 (nucleotides encoding amino acids 177–458) were generated from the pBhsMOK2 vector (8) by PCR using 5′ primers containing an _Eco_RI site and 3′ primers containing a _Sal_I site. After digestion with restriction enzyme, the three PCR products were cloned in-frame into the pLexA vector (BD Clontech) to generate, respectively, pLex-hsMOK2, pLex-NH2 and pLex-finger fused to a LexA DNA binding domain.
A recombinant pGEX-ΔlaminC plasmid was obtained by inserting the blunt-ended _Xho_I–_Sma_I fragment of pGAD-ΔlaminC plasmid into _Sma_I-treated pGEX-2T (Amersham Pharmacia Biotech). The _Xho_I–_Sma_I fragment from pGAD-ΔlaminC contains nucleotides 516–1851 of lamin C cDNA (amino acids 128–572, accession no. M13451). pGEX-laminA/C-coil1BΔ (nucleotides encoding amino acids 128– 218), pGEX-laminA/C-coil2 (nucleotides encoding amino acids 243–387) and pGEX-laminA/C-tail (nucleotides encoding amino acids 384–566) were generated from pGEX-ΔlaminC by PCR using 5′ primers and 3′ primers containing a _Bam_HI site and an _Eco_RI site, respectively. After digestion with _Eco_RI and _Bam_HI, the products were cloned into the corresponding sites of the pGEX-3X vector (Amersham Pharmacia Biotech). The pCMV-GST-ΔlaminC vector was obtained by inserting the blunt-ended _Eco_RI fragment from pGEX-ΔlaminC into the pCMV-GST vector (12) digested by _Sma_I. The pBNH2hsMOK2 vector was obtained by inserting the _Bam_HI–_Eco_RI fragment from pBhsMOK2 into the corresponding sites of the pBluescript KS+ vector (Stratagene). The recombinant pCMV-GST-NH2hsMOK2 was obtained by inserting the blunt-ended _Eco_RI–_Sal_I fragment of pLex-NH2 into the _Pst_I/DNA polymerase-treated pCMV-GST vector. The recombinant pCMV-hsMOK2 and pCMV-hsMOK2Δ vectors were previously described (3,8).
Yeast two-hybrid screen
pLex-hsMOK2, pLex-NH2 and pLex-finger expression plasmids were transformed into the yeast L40 strain [MATa his3_Δ_200 trp1-901 leu2-3112 ade2 LYS2::(4lexAop-HIS3) URA3::(8lexAop-lacZ)_GAL4_] using a standard lithium acetate transformation procedure. Plasmids were stably maintained by selection for the pLexA selection marker TRP1, which allows yeast growth in the absence of tryptophan. L40 yeast cells transformed with pLex-hsMOK2, pLex-NH2 or pLex-finger were then tested for activation of the reporter gene LacZ. pLex-hsMOK2 and pLex-finger constructs alone or paired with the empty vector (pGAD-GH) did not activate the LacZ reporter gene. Very low activation of the LacZ reporter gene was detected with the pLex-NH2 construct. A human HeLa S3 Matchmaker cDNA library (BD Clontech), constructed in the pGAD-GH vector expressing the GAL4 activation domain fusion protein, was transformed into L40 containing the pLex-hsMOK2 construct. Transformants were selected by growing them on selective minimal media lacking histidine, leucine and tryptophan, but containing 10 mM 3-aminotriazole (3-AT) (Sigma) at 30°C for 4 days. Colonies that grew in this selective minimal media condition were scored for β-galactosidase activity by the filter assay method. Filters were incubated in β-galactosidase assay buffer at 30°C for 90 min. Bait loss was carried out by successive inoculation into media lacking leucine. Plasmids were isolated from positive colonies and transformed into Escherichia coli TG1 for plasmid amplification. The rescued pGAD plasmids containing the library inserts were transformed back into L40, either with the empty pLexA vector or with pLexA-hsMOK2, to verify that the interaction depended on the presence of both hsMOK2 and the candidate insert in pGAD-GH. The cDNA inserts were further characterized by sequencing them on an ABI sequence analyzer and searching for gene sequence similarity in the GenBank™ database with the program BLAST.
Cell lines and antibodies
HeLa cells were grown in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) supplemented with 10% fetal calf serum. Affinity purified rabbit polyclonal anti-hsMOK2 antibody was obtained as described previously (8). Mouse monoclonal anti-GST antibody clone 1D10 was purchased from Euromedex. Mouse monoclonal anti-lamin A/C(636) was purchased from Santa Cruz Biotechnology. Rhodamine (TRITC)-conjugated goat anti-mouse IgG, CY2™-conjugated goat anti-rabbit IgG, peroxidase-conjugated goat anti-rabbit IgG and peroxidase-conjugated goat anti-mouse IgG were purchased from Jackson Immunoresearch Laboratories.
Purification of GST fusion proteins, in vitro protein synthesis and pull-down assay
GST–Δlamin C, GST–lamin A/C coil1BΔ, GST–lamin A/C coil2 and GST–lamin A/C tail were produced in E.coli strain TG1. Protein expression was induced by adding 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 5 h at 30°C. GST fusion proteins were prepared as described by Frangioni and Neel (13) and purified on glutathione–agarose gels (Sigma). The purity and quantity of the recombinant proteins were determined by examining SDS–PAGE gels stained with Coomassie blue.
pBNH2hsMOK2 was translated in a 50 µl reaction of [35S]Met/Cys (ICN) mixture using the TNT coupled transcription–translation system according to the manufacturer’s instructions (Promega). An aliquot of the reaction was then resolved by SDS–PAGE to verify the quality of the synthesized product. For nuclear extracts, HeLa cells were plated at 106 cells on a 100 mm Petri dish for 24 h prior to transfection with 15 µg of recombinant plasmid by the calcium phosphate method as described previously (14). Twenty-four hours after transfection, nuclear extracts were prepared according to Dignam (15) and dialyzed against buffer containing 25 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 10 µM ZnSO4 and 0.1% Nonidet P-40 at 4°C for 4 h. After dialysis and centrifugation, the nuclear extract was stored at –80°C. The protein concentration was determined by the Coomassie blue protein assay (Pierce).
GST pull-down experiments were performed using 10 µg of GST fusion protein bound to glutathione beads and 10 µl of in vitro synthesized 35S-labeled protein or 20 µg of nuclear proteins from transfected HeLa cells, in a total volume of 400 µl of binding buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 10 µM ZnSO4, 0.1% Nonidet P-40). After 4 h at 4°C, the beads were extensively washed with binding buffer, resuspended in SDS sample buffer (16) and separated by SDS–PAGE. The radiolabeled proteins were visualized by autoradiography after treatment with 16% sodium salicylate. Nuclear bound proteins from HeLa cells were analyzed by immunoblotting using the Supersignal West Pico Chemiluminescent Signal kit (Pierce). The binding was visualized by exposing the blots to Kodak BioMax MR film.
Immunofluorescence microscopy
HeLa cells were plated at 105 cells on a 35 mm Petri dish containing a glass coverslip for 24 h prior to transfection with 2 µg of expression vector by the calcium phosphate method as described previously (14). Twenty-four hours after transfection, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min and then permeabilized with 0.3% Triton X-100 in PBS for 15 min at room temperature (RT). Alternatively, cells were treated with –20°C methanol for 3 min. The cells were incubated with the primary antibody anti-hsMOK2, anti-GST or anti-lamin A/C, followed by incubation with the secondary antibody. The incubations were done for 1 h each and were carried out sequentially (with washes in PBS after each step), as this gave optimal labeling. The cellular DNA was labeled with 0.05% 4,6-diamidino-2-phenylindole (DAPI). The slides were mounted in antifadent AF1/glycerol/PBS mounting medium (Citifluor Ltd, London, UK). Immunofluorescence microscopy was performed using a Leica DMR microscope (Leica, Heidelberg, Germany) and an APOCHROMAT 63 × 1.32 oil immersion objective. Photographs were taken using a Micromax (Princeton Instruments) CCD camera and Metaview (Universal Imaging Corp.) software. For confocal analysis, images were sequentially collected by confocal laser scanning on a Leica TCS-NT/SP, equipped with an air-cooled argon–krypton mixed gas laser and an APOCHROMAT 63 × 1.3 2 oil immersion objective. Z series were generated by collecting a stack consisting of 10–12 optical sections using a step size of 0.284 µm in the Z direction. Images were processed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Cellular fractionation and nuclear matrix isolation
For electrophoretic analysis, subcellular fractionation and nuclear matrix isolation were performed as described by Buckler-White et al. (17) with slight modifications. In brief, transfected HeLa cells were sequentially extracted with: 1% Triton X-100 in TMS buffer (50 mM Tris–HCl pH 7.4, 5 mM MgSO4, 250 mM sucrose) for 5 min at RT (fraction 1); 50 U/ml RNase-free DNase RQ1 (Promega) in TMS at 37°C for 30 min and adding ammonium sulfate to a final concentration of 0.25 M (fraction 2); 2 M NaCl in TM buffer (10 mM Tris–HCl pH 7.4, 0.02 mM MgSO4) twice for 15 min at RT (fraction 3); 50 µg/ml RNase A in TM for 15 min at RT (fraction 4). The final pellet containing the nuclear matrix (fraction 5) was resuspended in SDS sample buffer (16) and boiled for 5 min. The protein concentrations were determined by the Amido Schwartz assay (18) and the proteins were separated by SDS–PAGE. hsMOK2 and truncated proteins were revealed by immunoblotting using the Supersignal West Pico Chemiluminescent Signal kit (Pierce). The blots were visualized using a Fluor-S Max MultiImager with Quantity One software (Bio-Rad).
For in situ fractionation, cells grown on polylysine- coated coverslips were washed three times with ice-cold PBS. After washing, the cells were sequentially extracted using the method described above with slight modifications. In these fractionation experiments, we used 0.5% Triton X-100 instead of 1% and added 0.5% Triton X-100 in all digestion buffers (TMS and TM) as described in the currently used protocol of He et al. (19). For these experiments, the DNase treatment was done at RT. Monolayers of extracted cells at different steps of the nuclear matrix preparation were fixed with –20°C methanol for 3 min and then processed for indirect immunofluorescence microscopy as described above.
RESULTS
hsMOK2 interacts with lamin C in yeast
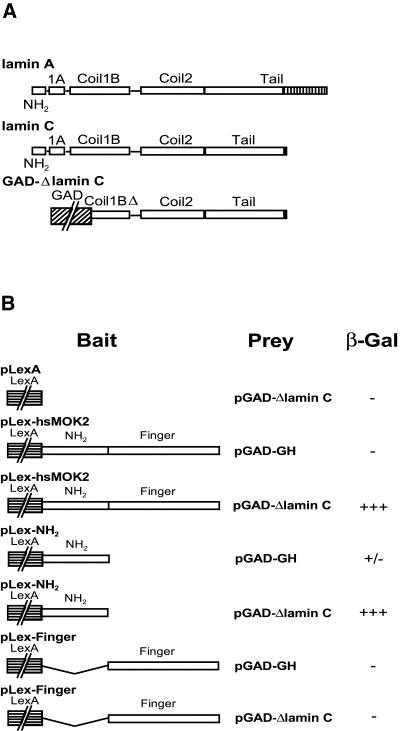
To identify interaction partners of hsMOK2 that might be involved in regulating hsMOK2 functions, we performed a two-hybrid yeast screen. We generated as bait a vector that expressed a fusion protein of the LexA binding domain and the full-length coding region of hsMOK2. The pLex-hsMOK2 plasmid did not autoactivate HIS3 or LacZ reporter genes upon transformation into L40 yeast. The pLex-hsMOK2 bait was used to screen a commercial human HeLa S3 Matchmaker cDNA library fused to the GaL4 activation domain in yeast L40 cells. After screening 5 × 106 transformants, 43 yeast colonies that fitted the criteria for interaction between bait and prey proteins (i.e. positive for both histidine growth and β-galactosidase activity) were obtained. The 43 clones were sequenced and the sequences were used to screen GenBank, which yielded nine unique sequences. Eight of these await further investigation. The ninth sequence encoded the C-terminal region of lamin C protein (amino acids 128–572; Fig. 1A) and was selected for further study.
Figure 1.
Identification of lamin C as an interaction partner of hsMOK2 by the yeast two-hybrid system and identification of the hsMOK2 interaction domain. (A) Schematic representation of human lamins A and C. Lamins A and C are identical in sequence except that lamin C has a unique 6 amino acid extension at its C-terminus (black box), while lamin A has a 98 amino acid extension (stripped box). Schematic representation of the pGAD-ΔlaminC library vector identified from a HeLa cDNA library on the basis of its ability to activate the LacZ reporter gene in the presence of the LexA binding domain/hsMOK2 hybrid. (B) Constructs expressing full-length or the indicated domains of hsMOK2 and the human polypeptide Δlamin C were co-transformed into yeast. The specificity of the interaction between bait and prey was determined by estimating the degree of color development after incubating the filter for 90 min in the filter lift β-galactosidase assay as described in Materials and Methods. +++, high blue color development; +/–, very low blue color development; –, no color development.
To determine which region of hsMOK2 interacts with lamin C, we co-transformed the library pGAD-ΔlaminC vector with pLex containing either one of the two different domains of hsMOK2, the non-finger acidic domain (pLex-NH2) or the finger domain (pLex-finger) into the yeast strain L40 and again performed β-galactosidase assays. The results revealed that Δlamin C interacts only with the N-terminal acidic domain of hsMOK2 (Fig. 1B). No interaction was found with the finger domain.
Interaction in vitro between the N-terminal acidic domain of hsMOK2 and the coiled 2 domain of lamin A/C
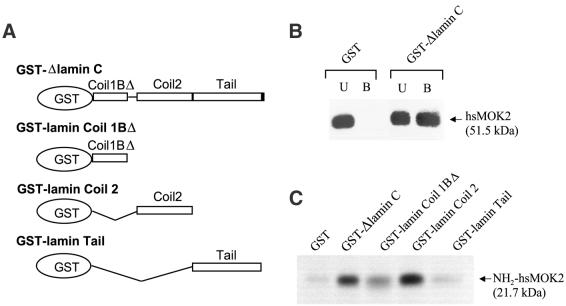
GST pull-down experiments, employing full-length proteins and truncation mutants thereof, were conducted to verify the observations made in yeast and to map the respective interaction domains of hsMOK2 and lamin C. First, GST fused to Δlamin C (GST–Δlamin C, amino acids 128–572; Fig. 2A) (20) was expressed in E.coli and immobilized on glutathione–agarose beads, which were then incubated with hsMOK2 transfected HeLa cell nuclear extracts. The results show that hsMOK2 specifically interacted with immobilized GST–Δlamin C but not with GST alone (Fig. 2B). Next, GST fusion proteins containing different regions of lamin C were bound to glutathione–agarose beads and incubated with the in vitro translated 35S-labeled N-terminal acidic domain of hsMOK2. Consistent with the yeast two-hybrid screen, the N-terminal domain of hsMOK2 was found to interact with Δlamin C (Fig. 2C). As lamin C is identical to lamin A except for 6 amino acids at its C-terminus, the different regions used to locate the interaction domain are present in both proteins (Figs 1A and 2A). The coil 2 domain of lamin A/C, from amino acid 243 to 387, showed the same binding activity as Δlamin C (amino acids 128–572), while the coil 1BΔ domain (amino acids 128–218) and the tail domain (amino acids 384–566) showed no excess activity in comparison with the negative GST control (Fig. 2C). Therefore, the coiled 2 domain of lamin A/C (amino acids 243–387) mediates hsMOK2 binding.
Figure 2.
GST pull-down of hsMOK2 by lamin A/C. (A) Schematic representation of the human polypeptide Δlamin C and the three fragments of human lamin A/C expressed in bacteria as fusions with GST. (B) Nuclear extracts (20 µg) from HeLa cells transfected with the expression vector hsMOK2 were incubated with an equal amount (10 µg) of GST or GST–Δlamin C bound to glutathione. Unbound (U) and bound (B) proteins were separated by SDS–PAGE and visualized by immunoblotting with affinity purified anti-hsMOK2 antibody. (C) In vitro 35S-labeled NH2hsMOK2 was incubated with an equal amount (10 µg) of recombinant GST fusion proteins containing the full-length or one of the three different segments of lamin A/C or GST alone bound to glutathione beads. After thoroughly washing the beads, the bound proteins were eluted in SDS sample buffer, resolved by SDS–PAGE and visualized by autoradiography after treatment with 16% sodium salicylate.
hsMOK2 and lamin A/C interact in vivo
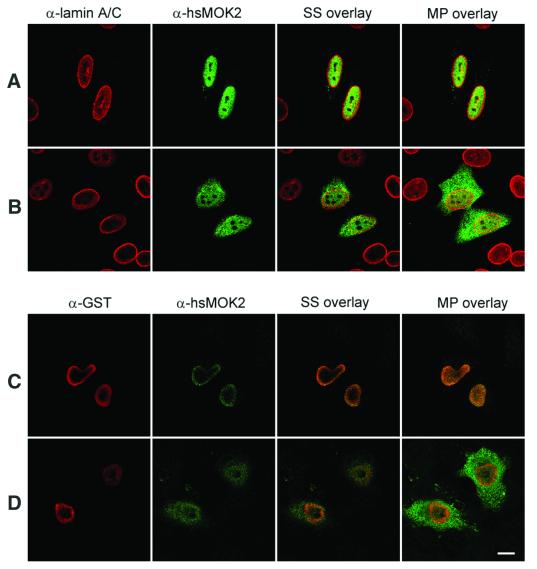
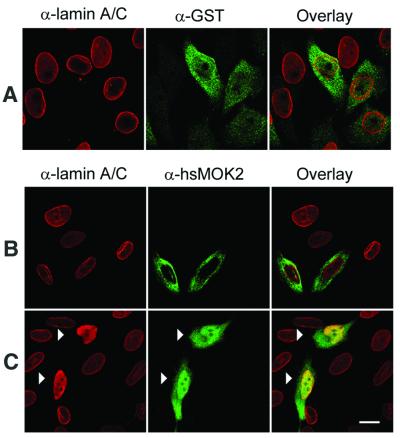
To pursue the interaction between hsMOK2 and lamin A/C in vivo, we performed immunofluorescence studies in transfected HeLa cells. We chose to examine this question in transfected cells, because hsMOK2 protein is below the level required for detection by antibodies, as shown in Figures 3, 4 and 6, where any labeling was found in untransfected cells, and in Arranz et al. (3). We previously demonstrated by in situ immunodetection and electron microscopy that endogenous human and murine MOK2 proteins are clearly localized in the nucleus (3). We first examined the subcellular localization of hsMOK2 and compared it with that of lamin A/C. HeLa cells transfected with the expression vector CMV-hsMOK2 were fixed, permeabilized, stained with the relevant antibodies and then visualized by confocal laser microscopy. We obtained similar results when methanol was employed instead of paraformaldehyde for fixation (data not shown). Staining of the cells with anti-lamin A/C antibody revealed staining of the nuclear periphery together with some staining of the nuclear interior (Fig. 3A, red), as reported by others (21–23). As previously observed, hsMOK2 exhibits a homogeneous nuclear staining pattern in all transfected cells (Fig. 3A, green). Expression of the N-terminal acidic domain of hsMOK2 showed both cytoplasmic and nuclear staining (Fig. 3B, green). The small N-terminal acidic domain (21.7 kDa), which lacks a nuclear localization sequence, is probably passively distributed throughout the cytosol and the nucleus. To observe the effect of lamin A/C on the localization of the hsMOK2 protein or on the localization of the N-terminal acidic domain, the GST–Δlamin C expression vector was co-transfected into human HeLa cells. Co-transfection of hsMOK2 and GST–Δlamin C demonstrated a predominant staining of the nuclear periphery for hsMOK2 in the majority of the co-transfected cells as shown by the confocal overlays of the single section scanned in the same optical plane (Fig. 3C, green). This pattern was different from the nuclear distribution of hsMOK2 and suggested that the cellular localization of hsMOK2 changes in the presence of Δlamin C to a more nuclear periphery distribution. Co-transfection of the N-terminal acidic domain of hsMOK2 and GST–Δlamin C did not appreciably change the cellular localization of this domain as shown by the confocal overlays of the single section scanned in the same optical plane (Fig. 3D). To avoid the passive passage of the small N-terminal acidic domain into the nucleus, we fused it to GST in the eukaryotic expression vector CMV-GST. Expression of the small GST protein (27.9 kDa) showed a similar cytoplasmic and nuclear staining as observed with the small N-terminal acidic domain of hsMOK2 (Fig. 4A, green). Transfection with the vector expressing GST–NH2hsMOK2 alone demonstrated a predominant cytoplasmic staining pattern in all transfected cells (Fig. 4B, green), indicating that the large fusion protein (46.3 kDa) prevented passive diffusion of the NH2hsMOK2 domain and GST protein into the nucleus. Co-transfection with the vector expressing GST–NH2hsMOK2 with the vector expressing GST–Δlamin C substantially changed this exclusively cytoplasmic localization: in all co-transfected cells, GST–NH2hsMOK2 can be seen both in the nucleus and in the cytoplasm (Fig. 4C, green). The changed localization of hsMOK2 and the nuclear translocation of GST–NH2hsMOK2 fusion proteins in the presence of GST–Δlamin C indicate that hsMOK2 and lamin A/C can also interact in living cells.
Figure 3.
Alteration of the subcellular localization of hsMOK2 by co-transfection with Δlamin C. HeLa cells were transfected with CMV-hsMOK2 (A) or CMV-NH2hsMOK2 (B) or co-transfected with CMV-hsMOK2 and CMV-GST-ΔlaminC (C) or CMV-NH2hsMOK2 and CMV-GST-ΔlaminC (D). Thirty-six hours after transfection, the cells were fixed, permeabilized and double stained sequentially with anti-lamin A/C and anti-hsMOK2 antibodies (A and B) or with anti-GST and anti-hsMOK2 antibodies (C and D). Cells were examined by confocal scanning laser microscopy as described in Materials and Methods. Images correspond to a single confocal section though the middle of the nucleus scanned in the same optical plane. SS overlay corresponds to the single confocal section shown and MP overlay corresponds to maximum projection of all optical sections. In overlay, the co-localization of rhodamine-labeled and fluorescein-labeled structures gives a yellow color. Bar, 10 µm.
Figure 4.
Nuclear translocation of the N-terminal acidic domain of hsMOK2 expressed as a fusion with GST. HeLa cells were transfected with CMV-GST (A) or CMV-GST-NH2hsMOK2 (B) or co-transfected with CMV-GST-NH2hsMOK2 and CMV-GST-ΔlaminC (C). Thirty-six hours after transfection, the cells were fixed, permeabilized and double stained with the indicated antibodies. Cells were examined by confocal scanning laser microscopy as described in Materials and Methods. Images represent a maximum projection of ten 0.284 µm optical sections. In overlay, the co-localization of rhodamine-labeled and fluorescein-labeled structures gives a yellow color. The arrows in (C) show the cells co-transfected with both plasmids. Bar, 10 µm.
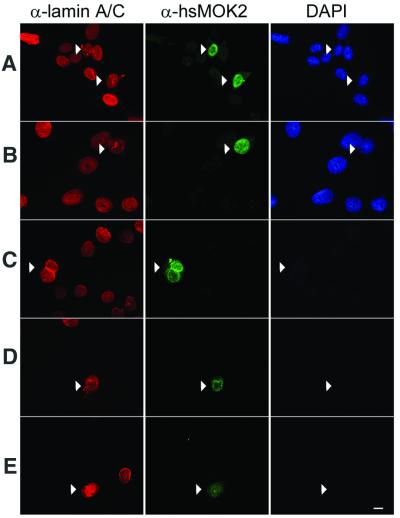
Figure 6.
Immunolocalization of hsMOK2 protein in transfected cells following in situ sequential fractionation. HeLa cells were transfected by CMV-hsMOK2 vector. Cells were untreated (A) or submitted to in situ sequential extraction with 0.5% Triton X-100 (B), DNase I at RT (C), 2 M NaCl (D) and RNase A (E) before fixation. Double immunolabeling was performed as described in Materials and Methods using monoclonal anti-lamin A/C and polyclonal anti-hsMOK2 antibodies. All the immunofluorescence photographs were acquired at the same exposure time. Images were printed at the same range of intensities for each color to facilitate comparisons except the image corresponding to untreated cells probed with anti-hsMOK2 antibody (A, green). At the same range of intensities, the image of untreated cells for hsMOK2 was saturated, which is consistent with the observation that a significant fraction of hsMOK2 is released with soluble proteins in western blotting (Fig. 5A). For each field of cells analyzed, DAPI staining is shown. Bar, 10 µm.
The transcription factor hsMOK2 is associated with the nuclear matrix
Hozak et al. (22) showed that lamin A forms part of a diffuse skeleton throughout the nuclear interior in HeLa cells. We have shown previously by electron microscopy that the murine and human MOK2 proteins are mainly associated with nuclear RNP components, including nucleoli and extranucleolar structures. Previously, hnRNP proteins have been shown to be associated with the nuclear matrix (24). These results suggest that MOK2 may bind to the nuclear matrix. To investigate this possibility, we isolated the nuclear matrix by sequential extraction of HeLa cells transfected with the eukaryotic expression vector CMV-hsMOK2, CMV-hsMOK2Δ or CMV-NH2hsMOK2. In the first fractionation step, soluble proteins are removed by extraction with 1% Triton X-100. Chromatin proteins are then released by DNase I digestion and extraction with 0.25 M ammonium sulfate. After washing with 2 M NaCl and RNase A digestion, the last fraction is composed of structural nuclear matrix proteins and nuclear matrix-associated proteins. Supernatants from each step and the final nuclear matrix pellet were analyzed by SDS–PAGE and immunoblotting. The fractionation procedure was controlled by immunoblotting with antibody directed against lamin A/C, two nuclear matrix-associated proteins. In our extracts, lamin A and C proteins were detected as expected in the nuclear matrix fraction, but also in the high salt wash (Fig. 5A, lanes 3 and 5). It has been observed that lamin A and C are not only found in nuclear lamina but also in the nucleoplasm (22,25). Lamin A and C released by 2 M NaCl in our fractionation protocol could correspond to lamin A and C present in the nucleoplasm. As shown in Figure 5A, a sizeable portion of hsMOK2 was released in the 1% Triton X-100 soluble fraction and the chromatin proteins while the remainder was tightly associated with the nuclear matrix proteins. No release of hsMOK2 was observed after RNase A treatment (Fig. 5A, lane 4). These results suggest that the association of hsMOK2 with the nuclear matrix was independent of RNA components of the matrix that were accessible to RNase A digestion. Two times more hsMOK2Δ isoform truncated for the N-terminal acidic domain was detected in the nuclear matrix fraction while the majority of N-terminal acidic domain (∼97%) was released in the Triton X-100 soluble fraction (Fig. 5B and C). These results suggest that the zinc finger domain is required for efficient targeting of hsMOK2 to the nuclear matrix.
Figure 5.
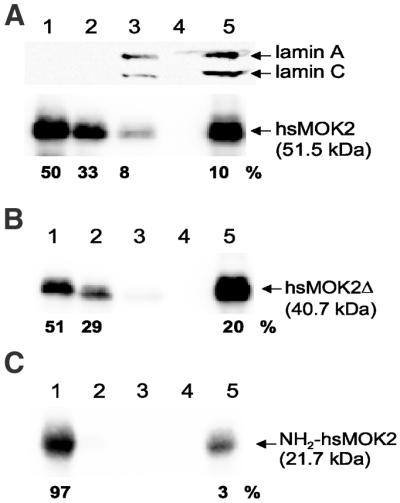
A fraction of hsMOK2 protein is associated with nuclear matrix. HeLa cells transfected with CMV-hsMOK2 (A), CMV-hsMOK2Δ (B) or CMV-NH2hsMOK2 (C) vectors were sequentially extracted as described in Materials and Methods. The various protein fractions correspond to 1% Triton X-100 (fraction 1), DNase I at 37°C and 0.25 M (NH4)2SO4 (fraction 2), 2 M NaCl (fraction 3), RNase A (fraction 4) and, finally, the pellet containing nuclear matrix (fraction 5) resuspended in SDS sample buffer. Four micrograms of proteins from fractions 1–4 and 20 µg of proteins from fraction 5, corresponding to the nuclear matrix, were subjected to SDS–PAGE and western blot analysis. Probing was realized with polyclonal anti-hsMOK2 antibody to detect hsMOK2 and truncated proteins or monoclonal anti-lamin A/C(636) antibody to detect the two nuclear matrix lamins A and C. Western blots were visualized with a Fluor-S Max MultiImager and quantified with Quantity One software (Bio-Rad). The percentages indicated below each panel were normalized at 4 µg of proteins. The molecular weights of the proteins are indicated on the right.
To further investigate the association of hsMOK2 with the nuclear matrix, monolayers of HeLa cells transfected with hsMOK2 were submitted to in situ sequential fractionation. In these fractionation experiments, we have reduced the concentration of Triton X-100 to 0.5% as described in the currently used protocol of He et al. (19) because we lost too many cells when 1% Triton X-100 was used in the first step. As seen in Figure 6, a significant fraction of hsMOK2 remained associated with the nuclear matrix structure after extraction with detergent (Fig. 6B), DNase treatment and low salt wash (Fig. 6C), 2 M high salt wash (Fig. 6D) and RNase treatment (Fig. 6E). The efficiency of chromatin digestion was indicated by the disappearance of the DAPI stain (Fig. 6C–E), while the integrity of the nuclear matrix was verified by lamin A/C antibody (Fig. 6A–E). After DNA digestion and chromatin elution with 0.25 M ammonium sulfate, the residual structures were barely visible in phase contrast microscopy (not shown). The results obtained by these in situ fractionations confirmed the results obtained by the above biochemical subcellular fractionations.
DISCUSSION
We used the yeast two-hybrid screen to identify novel protein partners for the transcription factor hsMOK2. Here, we present evidence to show that lamin C binds directly to the N-terminal acidic domain of hsMOK2. This interaction was confirmed by GST pull-down assays, using both in vitro and in vivo produced hsMOK2. In mammalian somatic cells, there exist two major A-type lamins, termed lamins A and C, which are alternatively spliced products of the same gene. Both proteins are identical for the first 566 amino acids, but contain unique C-terminal extensions after this common region (20,26). In lamin A, the C-terminal extension consists of 98 amino acids while in lamin C it comprises six residues (26). These isotypes are usually expressed in approximately equal amounts in differentiated somatic cells but their relative expression can differ in some tumors (27). We demonstrated that the N-terminal acidic domain of hsMOK2 interacts with the region between amino acids 243 and 387 of lamin A/C. This small region corresponds to the coil 2 domain localized within the C-terminal half of the central α-helical rod of lamin A/C (20). Thus, it is possible that both types of lamin can interact with hsMOK2 through an identical sequence present in the conserved coil 2 domain. Interestingly, the retinoblastoma (Rb) protein has also been found to bind to the coil 2 domain of lamin A/C (28), suggesting that the two different transcriptional repressor hsMOK2 and Rb factors compete for binding to the same small region of lamin A/C.
Further, we showed that the physical interaction between hsMOK2 and lamin A/C can also take place in vivo in eukaryotic HeLa cells. The in vivo interaction between hsMOK2 and lamin A/C has been demonstrated by the change in nuclear localization of hsMOK2 and by the nuclear translocation of the N-terminal acidic domain observed in cells co-transfected with Δlamin C. Nuclear translocation of the N-terminal acidic domain of hsMOK2 showed that in vivo this domain is also responsible for lamin A/C binding. We have chosen to examine the in vivo interaction in transfected cells because the endogenous MOK2 proteins are difficult to assess due to the very low expression level. In order to visualize the endogenous MOK2 proteins and to improve the subnuclear localization, we have previously used in situ immunodetection and electron microscopy methods (3). In human HeLa and murine L cells, which are only weakly labeled, we have found that the MOK2 proteins were associated with perichromatin fibrils and clusters of interchromatin granules, two structures involved in transcription and processing of pre-mRNA (29,30). We also showed that HeLa cells overexpressing human hsMOK2 protein did not exhibit any alteration in nuclear structure and that the general distribution of labeled nuclear RNP components was conserved (3). It has also been reported that lamin A co-localizes with RNP (21). hnRNP proteins are known to be associated with the nuclear matrix (24). These results suggest that MOK2 may bind to the nuclear matrix. Here, we have shown that a significant fraction of hsMOK2 protein is associated with the nuclear matrix prepared from hsMOK2-transfected cells. Since RNase treatment did not affect the fraction of hsMOK2 associated with the nuclear matrix, we suspected that this attachment was independent of the hsMOK2 RNA binding property and instead involved protein–protein interactions. Lamins are major components of the nuclear matrix (31,32). A potential mechanism for tethering hsMOK2 to the nuclear matrix would be its interaction with the nuclear matrix structural protein lamin A/C. Our results suggest that the zinc finger domain of hsMOK2 is required for targeting to the nuclear matrix. This domain is different from the domain involved in lamin A/C binding. Therefore, the association of hsMOK2 with nuclear matrix might involve other proteins. Nuclear targeting requiring a zinc finger domain was also reported for the RNA-binding protein ZNF74 (33). In contrast, the association of the transcription factors YY1 (34) and AKAP95 (35) with the nuclear matrix is independent of their zinc finger DNA-binding domains.
Nuclear lamins were initially identified as major components of the nuclear lamina which underlies the inner nuclear membrane (36,37). Actually, there is strong evidence that lamins are not restricted to the nuclear periphery. The nuclear lamins have also been found in the nucleoplasm and may be important constituents of the nucleoskeleton (21–23,38). It has been suggested that internal populations of lamin A and lamin C might anchor or organize components of the nuclear matrix (39). An involvement of lamins A/C in transcription has been supported by the finding that Rb protein binds to lamins A/C in vivo and in vitro (28,40,41). Our results also support an involvement of lamins A/C in transcription. MOK2 proteins recognize both DNA and RNA through their zinc finger motifs. This dual affinity and the subnuclear localization of these proteins have suggested that MOK2 might play a role in transcription, as well as in post-transcriptional regulation processes, of its target genes. One of its target genes, the IRBP gene, contains two MOK2-binding elements, a complete 18 bp MOK2-binding site located in intron 2 and the essential core MOK2-binding site (8 bp of the conserved 3′-half-site) located in the IRBP promoter. We have previously demonstrated that MOK2 can bind to the 8 bp present in the IRBP promoter and repress transcription from this promoter by competing with the activator CRX for DNA binding (8). The particular arrangement of two MOK2-binding sites observed in the human IRBP and PAX3 genes suggests that hsMOK2 could repress transcription through different mechanisms. Actually, we do not know the role of the 18 bp MOK2-binding site present in intron 2 of IRBP. This site could allow MOK2 to repress transcription by blocking transcriptional elongation. Interest ingly, it has been suggested that a negative regulatory element affecting mRNA elongation might be involved in controlling IRBP gene expression during fetal retinal development (42). It is possible that the binding of hsMOK2 to one or two of these target sequences and its interaction with lamin A/C may be involved in the transcriptional repression of the IRBP gene through the formation of a specific highly ordered chromatin structure. This structural assembly may be associated with the nuclear lamina or the nuclear matrix and function as a global mechanism to repress transcription of hsMOK2 target genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Isabelle Tratner for precious advice concerning the yeast two-hybrid system. We also thank Patrick Hughes and Patrick Anglard for critical reading of the manuscript. This research was supported by grants from Fondation Raymonde et Guy Strittmatter and the association Retina France.
REFERENCES
- 1.Ernoult-Lange M., Kress,M. and Hamer,D. (1990) A gene that encodes a protein consisting solely of zinc finger domains is preferentially expressed in transformed mouse cells. Mol. Cell. Biol., 10, 418–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernoult-Lange M., Arranz,V., Leconiat,M., Berger,R. and Kress,M. (1995) Human and mouse Kruppel-like (MOK2) orthologue genes encode two different zinc finger proteins. J. Mol. Evol., 41, 784–794. [DOI] [PubMed] [Google Scholar]
- 3.Arranz V., Harper,F., Florentin,Y., Puvion,E., Kress,M. and Ernoult-Lange,M. (1997) Human and mouse MOK2 proteins are associated with nuclear ribonucleoprotein components and bind specifically to RNA and DNA through their zinc finger domains. Mol. Cell. Biol., 17, 2116–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong S.L. and Bridges,C.D. (1990) Interstitial retinol-binding protein: purification, characterization, molecular cloning and sequence. Methods Enzymol., 189, 207–213. [DOI] [PubMed] [Google Scholar]
- 5.Stoykova A. and Gruss,P. (1994) Roles of Pax-genes in developing and adult brain as suggested by expression patterns. J. Neurosci., 14, 1395–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Luna,J., Lombroso,P.J. and Francke,U. (1995) Molecular cloning of the human homolog of a striatum-enriched phosphatase (STEP) gene and chromosomal mapping of the human and murine loci. Genomics, 28, 442–449. [DOI] [PubMed] [Google Scholar]
- 7.Mangelsdorf D.J., Borgmeyer,U., Heyman,R.A., Zhou,J.Y., Ong,E.S., Oro,A.E., Kakizuka,A. and Evans,R.M. (1992) Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev., 6, 329–344. [DOI] [PubMed] [Google Scholar]
- 8.Arranz V., Dreuillet,C., Crisanti,P., Tillit,J., Kress,M. and Ernoult-Lange,M. (2001) The zinc finger transcription factor, MOK2, negatively modulates expression of the interphotoreceptor retinoid-binding protein (IRBP) gene. J. Biol. Chem., 276, 11963–11969. [DOI] [PubMed] [Google Scholar]
- 9.Chader G. (1989) Interphotoreceptor retinoid-binding protein (IRBP): a model protein for molecular biological and clinically relevant studies. Friedenwald lecture. Invest. Ophthalmol. Vis. Sci., 30, 7–22. [PubMed] [Google Scholar]
- 10.Liou G., Geng,L. and Baehr,W. (1991) Interphotoreceptor retinoid-binding protein: biochemistry and molecular biology. Prog. Clin. Biol. Res., 362, 115–137. [PubMed] [Google Scholar]
- 11.Pepperberg D.R., Okajima,T.L., Wiggert,B., Ripps,H., Crouch,R.K. and Chader,G.J. (1993) Interphotoreceptor retinoid-binding protein (IRBP). Molecular biology and physiological role in the visual cycle of rhodopsin. Mol. Neurobiol., 7, 61–85. [DOI] [PubMed] [Google Scholar]
- 12.Tsai R.Y.L. and Reed,R.R. (1997) Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J. Neurosci., 17, 4159–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frangioni J.V. and Neel,B.G. (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem., 210, 179–187. [DOI] [PubMed] [Google Scholar]
- 14.Arranz V., Kress,M. and Ernoult-Lange,M. (1994) The gene encoding the MOK-2 zinc-finger protein: characterization of its promoter and negative regulation by mouse Alu type-2 repetitive elements. Gene, 149, 293–298. [DOI] [PubMed] [Google Scholar]
- 15.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 17.Buckler-White A.J., Humphrey,G.W. and Pigiet,V. (1980) Association of polyoma T antigen and DNA with the nuclear matrix from lytically infected 3T6 cells. Cell, 22, 37–46. [DOI] [PubMed] [Google Scholar]
- 18.Cheley S. and Bayley,H. (1991) Assaying nanogram amounts of dilute protein. Biotechniques, 10, 730–732. [PubMed] [Google Scholar]
- 19.He D.C., Nickerson,J.A. and Penman,S. (1990) Core filaments of the nuclear matrix. J. Cell Biol., 110, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKeon F.D., Kirschner,M.W. and Caput,D. (1986) Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature, 319, 463–468. [DOI] [PubMed] [Google Scholar]
- 21.Neri L.M., Raymond,Y., Giordano,A., Capitani,S. and Martelli,A.M. (1999) Lamin A is part of the internal nucleoskeleton of human erythroleukemia cells. J. Cell Physiol., 178, 284–295. [DOI] [PubMed] [Google Scholar]
- 22.Hozak P., Sasseville,A.M., Raymond,Y. and Cook,P.R. (1995) Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J. Cell Sci., 108, 635–644. [DOI] [PubMed] [Google Scholar]
- 23.Broers J.L., Machiels,B.M., van Eys,G.J., Kuijpers,H.J., Manders,E.M., van Driel,R. and Ramaekers,F.C. (1999) Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci., 112, 3463–3475. [DOI] [PubMed] [Google Scholar]
- 24.Mattern K.A., van Goethem,R.E., de Jong,L. and van Driel,R. (1997) Major internal nuclear matrix proteins are common to different human cell types. J. Cell. Biochem., 65, 42–52. [DOI] [PubMed] [Google Scholar]
- 25.Bridger J.M., Kill,I.R., O’Farrell,M. and Hutchison,C.J. (1993) Internal lamin structures within G1 nuclei of human dermal fibroblasts. J. Cell Sci., 104, 297–306. [DOI] [PubMed] [Google Scholar]
- 26.Fisher D.Z., Chaudhary,N. and Blobel,G. (1986) cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl Acad. Sci. USA, 83, 6450–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rober R.A., Sauter,H., Weber,K. and Osborn,M. (1990) Cells of the cellular immune and hemopoietic system of the mouse lack lamins A/C: distinction versus other somatic cells. J. Cell Sci., 95, 587–598. [DOI] [PubMed] [Google Scholar]
- 28.Ozaki T., Saijo,M., Murakami,K., Enomoto,H., Taya,Y. and Sakiyama,S. (1994) Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene, 9, 2649–2653. [PubMed] [Google Scholar]
- 29.Visa N., Puvion-Dutilleul,F., Harper,F., Bachellerie,J.P. and Puvion,E. (1993) Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp. Cell Res., 208, 19–34. [DOI] [PubMed] [Google Scholar]
- 30.Visa N., Puvion-Dutilleul,F., Bachellerie,J.P. and Puvion,E. (1993) Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur. J. Cell Biol., 60, 308–321. [PubMed] [Google Scholar]
- 31.Luderus M.E., Vansteensel,B., Chong,L., Sibon,O.C.M., Cremers,F.F.M. and Delange,T. (1996) Structure, subnuclear distribution and nuclear matrix association of the mammalian telomeric complex. J. Cell Biol., 135, 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minguez A. and Moreno Diaz de la Espina,S. (1993) Immunological characterization of lamins in the nuclear matrix of onion cells. J. Cell Sci., 106, 431–439. [DOI] [PubMed] [Google Scholar]
- 33.Grondin B., Bazinet,M. and Aubry,M. (1996) The KRAB zinc finger gene ZNF74 encodes an RNA-binding protein tightly associated with the nuclear matrix. J. Biol. Chem., 271, 15458–15467. [DOI] [PubMed] [Google Scholar]
- 34.Bushmeyer S.M. and Atchison,M.L. (1998) Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J. Cell. Biochem., 68, 484–499. [PubMed] [Google Scholar]
- 35.Akileswaran L., Taraska,J.W., Sayer,J.A., Gettemy,J.M. and Coghlan,V.M. (2001) A-kinase-anchoring protein AKAP95 is targeted to the nuclear matrix and associates with p68 RNA helicase. J. Biol. Chem., 276, 17448–17454. [DOI] [PubMed] [Google Scholar]
- 36.Dwyer N. and Blobel,G. (1976) A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J. Cell Biol., 70, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerace L., Blum,A. and Blobel,G. (1978) Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J. Cell Biol., 79, 546–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jagatheesan G., Thanumalayan,S., Muralikrishna,B., Rangaraj,N., Karande,A.A. and Parnaik,V.K. (1999) Colocalization of intranuclear lamin foci with RNA splicing factors. J. Cell Sci., 112, 4651–4661. [DOI] [PubMed] [Google Scholar]
- 39.Hutchison C.J., Alvarez-Reyes,M. and Vaughan,O.A. (2001) Lamins in disease: why do ubiquitously expressed nuclear envelope proteins give rise to tissue-specific disease phenotypes? J. Cell Sci., 114, 9–19. [DOI] [PubMed] [Google Scholar]
- 40.Shan B., Zhu,X., Chen,P.L., Durfee,T., Yang,Y., Sharp,D. and Lee,W.H. (1992) Molecular cloning of cellular genes encoding retinoblastoma-associated proteins: identification of a gene with properties of the transcription factor E2F. Mol. Cell. Biol., 12, 5620–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mancini M.A., Shan,B., Nickerson,J.A., Penman,S. and Lee,W.H. (1994) The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc. Natl Acad. Sci. USA, 91, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DesJardin L.E., Timmers,A.M. and Hauswirth,W.W. (1993) Transcription of photoreceptor genes during fetal retinal development. Evidence for positive and negative regulation. J. Biol. Chem., 268, 6953–6960. [PubMed] [Google Scholar]