Expression of Simian Immunodeficiency Virus nef in Immune Cells of Transgenic Mice Leads to a Severe AIDS-Like Disease (original) (raw)
Abstract
In order to study the functions of simian immunodeficiency virus (SIV) Nef in vivo in a small-animal model, we constructed transgenic (Tg) mice expressing the SIVmac239 nef gene in the natural target cells of the virus under the control of the human CD4 gene promoter (CD4C). These CD4C/SHIV-_nef_SIV Tg mice develop a severe AIDS-like disease, with manifestations including premature death, failure to thrive or weight loss, wasting, thymic atrophy, an especially low number of peripheral CD8+ T cells as well as a low number of peripheral CD4+ T cells, diarrhea, splenomegaly, and kidney (interstitial nephritis, segmental glomerulosclerosis), lung (lymphocytic interstitial pneumonitis), and heart disease. In addition, these Tg mice fail to mount a class-switched antibody response after immunization with ovalbumin, they produce anti-DNA autoantibodies, and some of them develop Pneumocystis carinii lung infections. All these results suggest a generalized Nef-induced immunodeficiency. The low numbers of peripheral CD8+ and CD4+ T cells are likely to reflect a thymic defect and may be similar to the DiGeorge-like “thymic defect” immunophenotype described for a subgroup of human immunodeficiency virus type 1-infected children. Therefore, it appears that SIV Nef alone expressed in mice, in appropriate cell types and at sufficient levels, can elicit many of the phenotypes of simian and human AIDS. These Tg mice should be instrumental in studying the pathogenesis of SIV Nef-induced phenotypes.
nef is an accessory gene present in the genomes of human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV). A functional SIV or HIV-1 Nef is critical for many of the same in vitro and in vivo phenotypes induced by these viruses (for reviews, see references 15, 34, 52, 55, and 68). In vivo, infection of rhesus monkeys with a SIVmac239 in which nef had been deleted caused a chronic infection with low viral loads and rare progression to AIDS (41). In addition, rhesus monkeys infected with a SIVmac239 clone containing a premature stop codon in nef were found to mutate rapidly to nef open reading frame (ORF) revertants, indicating a strong selective pressure, in vivo, for an open, functional form of nef to generate a pathogenic SIV (41). In agreement with these observations, humans infected with a form of HIV-1 in which nef was deleted have maintained low viral loads for more than a decade (16, 42).
In vitro, both SIV Nef and HIV-1 Nef have been found to be involved in the downregulation of cell surface expression of CD4 (1, 6, 9, 22, 24, 54) and of major histocompatibility complex (MHC) class I molecules on infected human cells (59) and to enhance virion infectivity (2, 13, 15, 28, 41, 63). In addition, SIV and HIV-1 Nef associate with a serine/threonine kinase, the Nef-associated kinase (NAK), which is a member of the p21-activated kinase (PAK) family (4, 57).
Although SIV and HIV-1 Nef proteins appear to be functionally similar, they are nonetheless distinct molecules with clear differences. (i) SIV Nef is larger than HIV-1 Nef (43), and the molecules share little amino acid homology. The most homologous regions are the N-terminal myristylation region and a highly conserved core region (43). (ii) The genomic organizations of SIV and HIV-1 nef are distinct: SIV nef coding sequences overlap with env coding sequences, while HIV-1 nef coding sequences do not. (iii) SIV Nef harbors SH2 binding domains at its N terminus, which are not present in HIV-1 Nef (21). (iv) In contrast to HIV-1 Nef, SIV Nef is unable to downregulate mouse CD4 cell surface expression (22, 23), whereas they have both been found to be able to downregulate the mouse CD8 cell surface marker in vitro (22). (v) SIV Nef binds poorly to the Hck SH3 domain, while HIV-1 Nef binds tightly to it (14, 27). (vi) SIV Nef has been reported to activate Hck and Lck, while HIV-1 Nef inhibits Lck kinase (27). (vii) In contrast to SIV and HIV-2 Nef, HIV-1 Nef does not interact with the zeta chain of the T-cell receptor (8, 37, 58). (viii) To downregulate the human cell surface CD4 receptor, SIV Nef uses either two tyrosine motifs or a leucine-based motif whereas HIV-1 Nef uses a leucine-based motif only (1, 12, 24, 38). (ix) HIV-1 Nef contacts the AP-2 complex via its C terminus, while SIV Nef binds to it via its N terminus (49). (x) Although HIV-1 Nef can replace SIV Nef and generate infectious viruses, such chimeric viruses (simian-human immunodeficiency virus [SHIV]) produce high virus loads and AIDS less consistently than the parental SIV in infected rhesus macaques (3, 39, 43, 50, 61).
We have previously reported a novel in vivo assay for the nef gene of HIV-1 in transgenic (Tg) mice (30). These Tg mice, expressing Nef in the natural target cells of the virus (immature CD4+ CD8+ T cells, mature CD4+ T cells, macrophages, and dendritic cells) under the control of the regulatory sequences of the human CD4 gene (CD4C), developed a severe disease with multiple phenotypes (premature death, wasting, diarrhea, atrophy of the lymphoid organs, progressive and preferential loss of CD4+ T cells, lymphocytic interstitial pneumonitis [LIP], interstitial nephritis, segmental glomerulosclerosis [29, 30], and cardiac disease [D. G. Kay, P. Yue, Z. Hanna, S. Jothy, E. Tremblay, and P. Jolicoeur, submitted for publication]) resembling those reported for AIDS. More recently, we reported that these CD4C/HIV Tg mice exhibit impaired germinal-center formation and produce elevated levels of autoantibodies (auto-Ab) (53). The appearance of this AIDS-like disease in CD4C/HIV Tg mice strongly suggested that at least some of the HIV-1 Nef effectors were conserved in mice and humans. Since SIV and HIV-1 Nef molecules are structurally distinct and potentially interact with different effectors, we explored whether SIV Nef would have pathogenic effects similar to those of HIV-1 Nef in Tg mice.
MATERIALS AND METHODS
Transgene construction and generation of Tg mice.
The HIV-1 nef ORF in the CD4C/HIVMutG construct (29, 30) was replaced with the SIVmac239 nef ORF. The SIVmac239 cloned DNA (26) was obtained through the AIDS Research and Reference Reagent Program of the Division of AIDS, National Institute of Allergy and Infectious Diseases. The SIV nef was amplified with two oligonucleotides in order to generate an _Mlu_I site at the 5′ end and a _Not_I site at the 3′ end (oligonucleotides GACGCGTCTACAATATGGGTGGAG and AGCGGCCGCTGTTTCAGCGATTT, respectively). The PCR product was confirmed by sequencing in both orientations. This amplified fragment was then inserted into the _Mlu_I-_Not_I site created in the CD4C/HIV cassette described previously (29, 30), thus replacing the HIV-1NL4-3 nef. Transgene DNA was isolated from the vector by _Aat_II digestion and purified to produce Tg mice as described previously (29, 30). These mice were bred as heterozygotes on the C3H background. Age-matched non-Tg littermates were used as controls.
Tg and non-Tg mice were maintained in a specific pathogen-free (SPF) animal facility. Nine non-Tg CD1 mice were housed as sentinels in each of the same SPF rooms as the Tg and non-Tg experimental animals. These mice were assigned to two cages, and each cage was placed on one of the cage racks in each SPF room. A bedding sample from every cage of each rack was placed in the relevant sentinel cage. Every three months, the sentinels were tested serologically; once a year, they were tested serologically and bacteriologically and were examined for the presence of parasites.
Serological evaluation (Scan Plus [SPM]; performed by the Rodent Health Surveillance Program, University of Miami) revealed no infection with the following agents: mouse hepatitis virus, Sendai virus, Mycoplasma pulmonis or Mycoplasma arthritidis, pneumonia virus of mice, minute virus of mice, Theiler's murine encephalomyelitis virus (GDVII), rotavirus (EDIM), and lymphocytic choriomeningitis virus. The mice were also negative for the presence of several respiratory and enteric bacteria (they were screened for more than 80 pathogenic and nonpathogenic bacteria as well as more than 35 species of fungi). The serological test was performed by standard enzyme-linked immunosorbent assay (ELISA) procedures. Reagents were individually prepared in tissue culture and titrated for optimal reactivity. Samples were run with known negative and positive controls. To be considered positive, the sample's reading (optical density) had to be double the reading of the negative sample. All positive or questionable samples were confirmed by immunofluorescence assay and/or outside reference laboratory. Cultures were placed on different media including MacConkey agar, Columbia agar, Trypticase soy agar, Hekton E, and chocolate. They were incubated at 37°C and reviewed by licensed medical technologists. Follow-up tests included Gram staining, various differential biochemical tests, and a commercially available kit called Api (Biomerieux, Marcy l'Etoile, France).
RNA purification and Northern blot analysis.
RNAs from different tissues were isolated by using Trizol (Gibco BRL), and 10 μg from each sample was electrophoresed on formaldehyde agarose gels and processed for hybridization by using a mixture of three 32P-labeled probes, two of which were the 2.1-kbp _Hin_dIII (nucleotides [nt] 6026 to 8131) and 400-bp _Bam_HI-_Mlu_I (nt 8401 to 8800) fragments of the HIV-1NL4-3 genome, while the third was the 820-bp _Mlu_I-_Not_I fragment (nt 9320 to 10140) of the SIVmac239 genome.
Assessment of transgene expression by ISH.
In situ hybridization (ISH) was performed on paraformaldehyde perfusion-fixed, paraffin-embedded tissues and on sorted peripheral CD4+ T cells plated by cytospotting, by using a transgene-specific 35S-UTP-labeled antisense probe and control sense RNA probes, as described previously (29, 30). Tissues from non-Tg control animals hybridized with antisense probes, as well as tissues from Tg animal hybridized with sense probes, failed to exhibit any specific hybridization signal.
Protein extraction and Western blot analysis.
Proteins were extracted by lysing the cells or tissues in 20 μl of radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% sodium deoxycholate, 1% NP-40, and 0.1% sodium dodecyl sulfate [SDS]) containing inhibitors (aprotinin [2 μg/ml], pepstatin [1 μg/ml], _N_α-_p_-tosyl-l-lysine chloromethyl ketone [TLCK; 50 μg/ml], leupeptin [2 μg/ml], sodium orthovanadate [0.37 μg/ml], sodium fluoride [0.04 μg/ml], and phenylmethylsulfonyl fluoride [100 μg/ml], all from Sigma) by using pellet pestles (Fisher). Lysates were left on ice for 30 min and centrifuged for 15 min at 4°C. Supernatants were collected, and proteins were quantitated by using a MicroBCA assay (Sigma). Proteins (100 μg) were loaded on 10% polyacrylamide gels and, following migration, were transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore) by using a semidry transfer apparatus (Schleicher & Schuell). A monoclonal antibody to SIVmac251 Nef protein (17.2) (no. 2659; lot 295016) was obtained from Kai Krohn and Vladimir Ovod through the AIDS Research and Reference Reagent Program and was used at a 1:1,000 dilution. A peroxidase-coupled secondary antibody against mouse immunoglobulins (Ig) (DAKO) was used at a 1:10,000 dilution. Proteins were detected by using the Renaissance enhanced chemiluminescent substrate (NEN, Life Science Products). To confirm equal protein loading, membranes were first stripped of antibodies by washing for 30 min at 55°C in a solution containing 2% SDS, 74.4 mM Tris-HCl (pH 6.8), and 0.7% β-mercaptoethanol and then blocked with 3% bovine serum albumin before being reprobed with an anti-actin antibody (1:800; Sigma) followed by peroxidase-coupled anti-rabbit Ig (1:10,000; Sigma) as the secondary antibody.
Cell type specificity of transgene expression.
Single-cell suspensions of thymuses, peripheral lymph nodes (LN), mesenteric LN, and spleens from Tg mice and non-Tg age-matched controls were prepared by meshing the organs in RPMI medium (5% inactivated fetal bovine serum [FBSI]). Red blood cells were lysed by incubation in Gay's solution for 5 min on ice. The cells were washed once, counted, resuspended in fluorescence-activated cell sorter (FACS) blocking solution (phosphate-buffered saline [PBS] containing 20% FBSI and 0.02% azide), and left on ice for 1 h. Afterwards, cells were washed and resuspended in FACS buffer (PBS containing 2% FBSI and 0.01% azide) to be stained with the following mixture of antibodies: a phycoerythrin (PE)-coupled anti-mouse CD8α monoclonal antibody (Ly2), a fluorescein isothiocyanate (FITC)-coupled anti-mouse αβTcR monoclonal antibody, and a biotin-coupled anti-mouse CD4 monoclonal antibody (all from Cedarlane) followed by streptavidin-allophycocyanin (APC) (Pharmingen). Staining was also performed with a PE-coupled anti-mouse B220 (Ly5) monoclonal antibody and an FITC-coupled anti-mouse IgM monoclonal antibody (both from Cedarlane). After being stained by incubation for 45 min on ice, cells were resuspended in FACS buffer at a concentration of 15 × 106 cells/ml. The cells were then sorted to obtain B220+ IgM+ B cells, CD4+ TcRαβ+ T cells, and CD8+ TcRαβ+ T cells from Tg and non-Tg mice by using a cell sorter (Moflo; Cytomation). Macrophages from Tg and non-Tg mice were also obtained by peritoneal lavage using a total of 10 ml of cold RPMI medium (10% FBSI-0.35 mM 2-mercaptoethanol). Recovered cells were washed three times before being plated on 100-mm petri dishes (Falcon) and incubated overnight at 37°C to allow cell adherence. Macrophages were then harvested with a Costar cell scraper.
Flow cytometry.
Single-cell suspensions of thymuses, spleens, and mesenteric LN were prepared from Tg and non-Tg age-matched littermates. The procedure explained above was followed. The antibodies used in this study were a PE-coupled anti-mouse CD4 monoclonal antibody (L3/T4), a PE-coupled anti-mouse CD8α monoclonal antibody (Ly2), an FITC-coupled anti-mouse CD8α monoclonal antibody (Ly2), an FITC-coupled anti-mouse αβTcR monoclonal antibody, an FITC-coupled anti-mouse CD44 monoclonal antibody, an FITC-coupled anti-mouse B220 (Ly5) monoclonal antibody, a PE-coupled anti-mouse CD11b Mac-1 monoclonal antibody, an FITC-coupled anti-mouse Thy1.2 monoclonal antibody (all from Cederlane Laboratories), and an FITC-coupled anti-mouse CD69 monoclonal antibody (Pharmingen Canada). After staining, the cells were resuspended in FACS buffer at a concentration of 106 cells in 300 μl. Flow cytometric analyses were performed with a FACScan (Becton Dickinson), and cells taking up propidium iodide (final concentration, 5 μg/ml) were excluded by electronic gating. FACS data were analyzed by using Windows Multiple Document Interface for Flow Cytometry (WinMDI), version 2.8.
Microscopic analysis.
Mice were sacrificed by CO2 asphyxiation, and the organs (thymus, heart, liver, kidneys, spleen, and gut) were immediately collected and immersion-fixed in 3.7% formaldehyde (Fisher) in PBS (1×). Organs to be assessed were embedded in parraffin, sectioned at a thickness of 5 μm, and stained with hematoxylin and eosin, as described previously (29). Tg and non-Tg tissues examined by Gram staining (detecting both gram-positive and gram-negative bacteria) and by Grocott staining were chosen at random. Tissues were examined by two investigators (D.G.K. and S.J.).
Preparation of single-cell suspensions and CD4+ T-cell purification.
Peripheral (axillary, inguinal, popliteal, and brachial) LN from Tg and non-Tg littermates were collected, and single-cell suspensions were prepared by gently disrupting the organ with a syringe plunger, pipetting several times up and down with a Pasteur pipette, and filtering through nylon mesh. Cells were incubated for 1 h at 37°C in a humidified 5% CO2 incubator to remove adherent cells (dendritic cells and macrophages). After incubation, nonadherent cells were harvested, counted, and resuspended (107 cells/ml) in PBS without fetal bovine serum, as described elsewhere (33). CD4+ T-cell purification was performed by a procedure that we published previously (33). In brief, the cells were purified by using sheep anti-rat antibody-coated magnetic beads (Dynabeads) after a preincubation with an antibody cocktail of hybridoma supernatants. The cocktail contained rat anti-B220 (RA36B2), a kind gift from R. Coffman (DNAX Research Institute of Cellular and Molecular Biology, Palo Alto, Calif.), and rat anti-MHC-II (M5-114), and rat anti-CD8 (53.6.78) (both from the American Type Culture Collection, Manassas, Va.). The purity of CD4+ TcRαβ+ cells as analyzed by flow cytometry was ≥96% for the non-Tg controls.
CFSE fluorescent dye labeling and cell division assay.
CFSE (5- and 6-carboxyfluorescein diacetate succinimidyl ester) was purchased from Molecular Probes Inc. (Eugene, Oreg.). The LN single-cell suspension was labeled by adding CFSE at a final concentration of 1 μM, followed by incubation at room temperature for 10 min. The reaction was stopped by addition of complete medium (Iscove's modified Dulbecco medium; Gibco BRL, Life Technologies) supplemented with 5% FBSI (Gibco), 50 μM 2-mercaptoethanol (Sigma), penicillin (100 U/ml), and streptomycin (100 U/ml). The cells were washed twice and cultured in anti-CD3 (5 μg/ml)-coated flat-bottom 96-well plates at 105 cells/well in 200 μl of complete medium for 3 days. After the 3-day culture, cells were harvested and stained for FACS, as described above. CFSE profiles were read on live CD4+ or CD8+ T cells, as described previously (33).
Detection of DNA-specific Ig.
Ig were detected by standard ELISA procedures using flat-bottom plates (Nalge Nunc International) coated with sonicated double-stranded human placenta DNA (200 to 300 bp; 4 μg/ml), essentially as described previously (53). Horseradish peroxidase (HRP)-coupled goat anti-mouse IgM (1:500) and IgG1 (1:1,000) (Southern Biotechnology Associates) were used to reveal isotypes. Ortho-phenylene diamine (Sigma) was used as a substrate.
Immunization of mice and detection of OVA-specific Ig.
Female Tg and non-Tg mice were used between the ages of 8 and 12 weeks. Mice were immunized with alum-precipitated ovalbumin (OVA; 300 μg administered subcutaneously, 100 μg in the neck and 50 μg per paw; Sigma). To obtain a secondary response, mice were given the same immunization 14 days after the primary injection. Mice were either sacrificed 10 days after the primary injection or 5 days after the secondary immunization, as previously described (53). OVA-specific Ig were detected by standard ELISA procedures, as previously published (53), using flat-bottom plates (Nalge Nunc International) coated with OVA (1 μg/ml; Sigma). HRP-coupled goat anti-mouse IgM (1:500), IgG1 (1:1,000), and IgG2b (1:1,000) and biotin-coupled goat anti-mouse IgG2a (1:300) followed by streptavidin-HRP (1:1,000) (Southern Biotechnology Associates) were used to reveal isotypes. OPD (Sigma) was used as a substrate.
Statistical analysis.
Statistical analysis was performed with Microsoft Excel using an unpaired, two-tailed Student's t test with equal variance. All results are means ± standard deviation.
RESULTS
Construction of CD4C/SHIV-_nef_SIV Tg mice.
To construct SIV Nef-expressing Tg mice, nef coding sequences of SIVmac239 were inserted into the NL4-3 HIV-1MutG genome, replacing the HIV-1 nef (Fig. 1). In this HIVMutG genome, the ORFs of all other known HIV-1 genes (gag, pol, env, vif, vpu, rev, tat, and vpr) were interrupted (30). This SHIV-_nef_SIV DNA fragment was ligated downstream of the CD4C regulatory sequences of the human CD4 gene (Fig. 1). These CD4C sequences have been shown to drive expression of surrogate genes, including HIV-1, in CD4+ CD8− and CD4+ CD8+ thymocytes, as well as in peripheral CD4+ T cells and in cells of the macrophage/dendritic lineages of Tg mice (29-32).
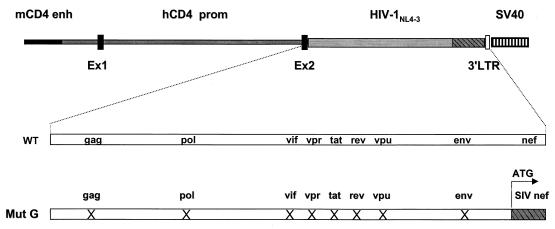
FIG. 1.
Structure of the CD4C/SHIV-_nef_SIV transgene. The mouse CD4 enhancer (mCD4 enh), the human CD4 promoter (hCD4 prom), the HIV-1NL4-3 mutant G genome (Mut G), and the polyadenylation sequences from simian virus 40 (SV40) were ligated. Ex1 and Ex2 are the first two untranslated exons of the hCD4 gene; 3′ LTR is part of the 3′ LTR of the HIV-1 genome. WT, wild type. The symbol X means that the ORF of the indicated HIV-1 gene was interrupted.
Five founders (F) carrying this CD4C/SHIV-_nef_SIV transgene (F55541, F57691, F57693, F57690, and F61217) were produced. Southern blot analysis indicated that the structure of the transgene appeared to be grossly intact (data not shown). All founders showed a lower-than-expected frequency of Tg progeny (Table 1), suggesting that these founders were mosaic. Independent Tg lines could be established with all founders. Mice from the two founders F57690 and F61217 were found by Northern blot analysis to be negative for transgene expression (data not shown). Therefore, no further analysis of mice from these lines was carried out. Mice from three founders (F57691, F55541, and F57693) were studied. Progeny mice were genotyped and routinely examined for signs of disease.
TABLE 1.
Characteristics of CD4C/SHIV-_nef_SIV founders
| Founder | Sexa | No. of Tg mice born/total no. of mice born (%) | Mosaicism | Expression level (according to Western blotting)b | Line established |
|---|---|---|---|---|---|
| F57690 | F | 6/42 (14) | Yes | None | Yes |
| F61217 | M | 5/35 (14) | Yes | None | Yes |
| F55541 | F | 25/76 (33) | Yes | High | Yes |
| F57691 | F | 5/54 (9) | Yes | Very high | Yes |
| F57693 | F | 20/77 (26) | Yes | Medium high | Yes |
Transgene expression in CD4C/SHIV-_nef_SIV Tg mice.
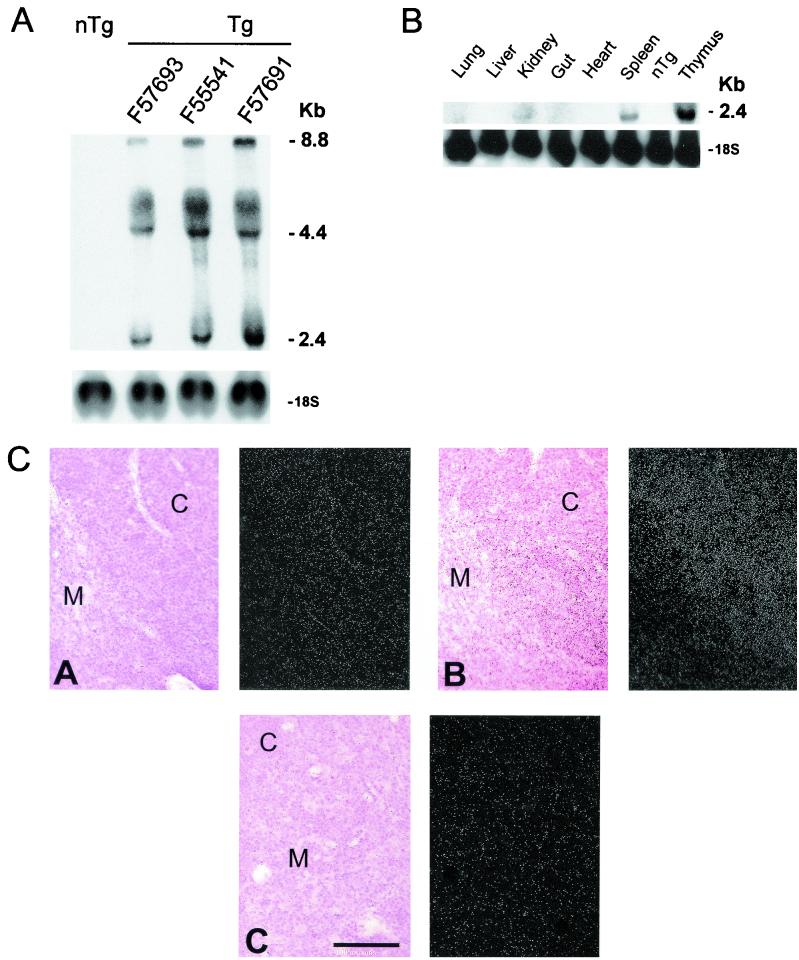
Transgene expression was first evaluated by Northern blot analysis. This analysis revealed the three main transcripts of the HIV-1 genome (the 8.8-kb full-length, 4.3-kb _env_-specific, and 2.0-kb multiply spliced transcript) at high levels in the thymus (Fig. 2A). Weaker transgene expression was detected in the spleen, lungs, gut, and kidneys (Fig. 2B), most likely reflecting expression in CD4+ T cells and/or in resident macrophages and dendritic cells, as previously documented (29, 30). No transgene expression was detected in the heart or liver (Fig. 2B). Expression was highest in mice from founder F57691, moderately high in mice from founder F55541, and lowest in mice from founder F57693 (F57691 > F55541 > F57693).
FIG. 2.
Expression of SIVmac239 nef RNA in CD4C/SHIV-_nef_SIV Tg mice. (A and B) Northern blot analysis of RNA from thymuses of mice from Tg founders F57691, F55541, and F57693 (A) and from organs of mice from founder F57691 (B). Organs from non-Tg littermates were used as controls. RNAs were hybridized with 32P-labeled transgene-specific probes. Filters were then washed and rehybridized with the 18S ribosomal RNA-specific probe. (C) ISH analysis. Thymuses from Tg (panels CA and CB) or non-Tg (panel CC) mice were probed with sense (panel CA) or antisense (panels CB and CC) transgene-specific riboprobes. Left and right sides of each panel show tissues in bright- and dark-field illumination, respectively. A strong and specific hybridization signal was detected in panel CB, showing higher expression in cortical (C) than in medullary (M) thymocytes. Bar, 100 μM. Counterstain, hematoxylin and eosin.
Assessment of transgene expression by ISH revealed that thymocytes from both the cortex and the medulla expressed the transgene, with a higher level of expression detected in the cortex (Fig. 2C). Transgene-expressing cells were also detected in the T-cell zones of the spleen and mesenteric LN as well as in FACS-sorted, purified peripheral CD4+ T cells (data not shown). In addition, mononuclear cells in the lamina propria of the intestine and infiltrating the interstitium of the kidney expressed the transgene (data not shown). No specific hybridization signal was detected in tissues from Tg mice exposed to a sense probe or in tissues from non-Tg mice exposed to an antisense probe (Fig. 2C and data not shown).
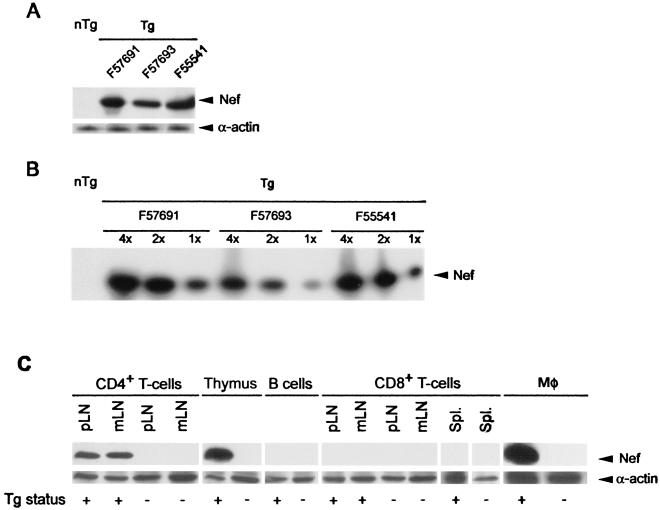
SIV Nef protein expression was also analyzed in these Tg mice by Western blotting. Medium-high to very high expression of the SIV Nef protein could be detected in Tg mice from the three founders expressing Tg RNA (F57691 > F55541 > F57693), while no SIV Nef protein could be detected in control non-Tg mice (Fig. 3A and B). A similar experiment performed on sorted cells showed that Nef expression was detectable in CD4+ CD8+ thymocytes, CD4+ TcRαβ+ T cells from the peripheral lymphoid organs, and peritoneal macrophages of Tg mice, but not in B220+ IgM+ B cells or CD8+ TcRαβ+ T cells of Tg mice, or in any of these populations of non-Tg mice (Fig. 3C).
FIG. 3.
Expression of SIVmac239 Nef protein in CD4C/SHIV-_nef_SIV Tg mice. (A) Total protein extracts (100 μg) from thymuses of 1-month-old Tg lines (from founders F57691, F55541, and F57693) and non-Tg (nTg) littermates were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting with a monoclonal antibody specific to SIV Nef. Equal loading was confirmed by probing with an anti-actin antibody. (B) Semiquantitative analysis of SIVmac239 Nef protein. Thymic extracts from mice from Tg lines were serially diluted and compared to each other for the level of Nef expression. Relative amounts of protein are shown above lanes; the well indicated by “4×” contains four times as much protein as the well indicated by “1×”. (C) Analysis of Nef expression in specific cell populations. CD4+ TcRαβ+ (CD4+) T cells and CD8+ TcRαβ+ (CD8+) T cells from spleens (Spl.) or peripheral (p) or mesenteric (m) LN, B220+ IgM+ B cells from spleens, double-positive CD4+ CD8+ thymocytes (thymus), and peritoneal macrophages (Mφ) from Tg and non-Tg mice were obtained by cell sorting (or plating in the case of macrophages), and cell extracts were processed for Western blotting with an anti-Nef monoclonal antibody. Blots were stripped of bound antibodies and reprobed with anti-actin antibodies to assess the amount of loaded protein.
Together, these results indicate a faithful and expected pattern of expression for these CD4C regulatory elements. This pattern of expression was indistinguishable from that observed previously for CD4C/HIVWT and CD4C/HIVMut Tg mice (29, 30).
Clinical phenotypes of CD4C/SHIV-_nef_SIV Tg mice.
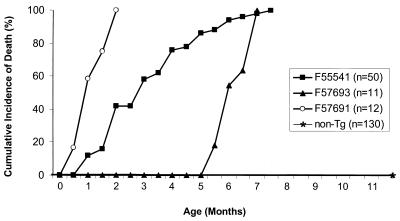
Mice from the three expressor lines (F55541, F57691, and F57693) developed a fatal disease similar to those described for CD4C/HIVWT and CD4C/HIVMut Tg mice (29, 30). Disease progression varied between founders; it was much faster in high-expressing mice from the F57691 line (Fig. 4; Table 1), confirming an earlier observation with HIV-1 that disease latency was strongly correlated with levels of Nef expression (30). All Tg progeny (n = 12) from the F57691 line died of severe disease within 45 to 60 days. The founder itself (F57691) died at 17 months, of the same disease as its Tg offspring. This long latency most likely reflects its mosaicism. At necropsy, it was found to have splenomegaly, kidney disease, lung disease, and an enlarged heart. It also had hepatomegaly and a perianal tumor whose cell composition, characterized by FACS, was found to be mostly B cells (B220+ CD4− CD8−) (data not shown). Similarly, the mosaic founders F57693 and F55541 died later than their progeny: at 17 versus 8 months and 12 versus 4 months, respectively.
FIG. 4.
Cumulative incidence of death among CD4C/SHIV-_nef_SIV Tg mice. Each point represents the percentage of Tg mice which were found to be moribund or dead at a specific time in each group. n, number of animals in each group.
Mice from the three expressor lines developed a similar disease except that the latency period was longer for mice from founders F55541 and F57693. Early on, mice from the latter two lines had no detectable clinical signs of disease. As they aged, they became ill. Most Tg mice from founders F55541 (85%), F57691 (86%), and F57693 (84%) exhibited a failure to thrive; they were approximately two-thirds the size of their non-Tg littermates: (body weights, 27.8 ± 7.7 g for non-Tg mice [n = 8]; 19.1 ± 4.7 g for F55541 Tg mice [n = 5]; 19.6 ± 12.5 g for F57691 Tg mice [n = 3]; and 32.6 ± 11.3 g for F57693 Tg mice [n = 3]). Tg mice from all lines (94%) became hypoactive and weak, showed ruffled hair, and developed a severe wasting syndrome (27 of 32 [84%]), sometimes accompanied by edema (6 of 32 [19%]) or diarrhea (40%). Non-Tg littermates housed in the same cages as the Tg mice did not develop any disease, indicating that these phenotypes were specific to the transgene.
Pathological assessment of CD4C/SHIV-_nef_SIV Tg mice.
At necropsy, macroscopic observation confirmed the presence of wasting and edema and revealed phenotypes of enlarged heart (13 of 32 [41%]), splenomegaly (17 of 32 [53%]), thymic atrophy (31 of 32 [97%]), and kidney disease (25 of 32 [78%]) in Tg mice from the three expressor lines (Table 2). Again, no pathological changes were observed for non-Tg mice. Moreover, no bacteria (except in the intestinal lumen) were detected by Gram staining in the kidneys, lungs, livers, and hearts of Tg (n = 9) and non-Tg (n = 7) mice randomly chosen during this study (data not shown). Generally, when a gross pathological change was seen in one organ, other organs of the same mice were also affected.
TABLE 2.
Incidence of disease in CD4C/SHIV-_nef_SIV Tg mice
| Pathology observeda | No. with trait/total no. studiedb | |||||||
|---|---|---|---|---|---|---|---|---|
| F55541 | F57691 | F57693 | Totald | |||||
| Tg | nTgc | Tg | nTg | Tg | nTg | Tg | nTg | |
| Hypoactivity and ruffled hair | 18/20 | 0/51 | 7/7 | 0/49 | 5/5 | 0/57 | 30/32 | 0/157 |
| Small body size | 17/20 | 0/51 | 6/7 | 0/49 | 4/5 | 0/57 | 27/32 | 0/157 |
| Wasting | 17/20 | 0/51 | 6/7 | 0/49 | 4/5 | 0/57 | 27/32 | 0/157 |
| Small or absent thymus | 19/20 | 0/51 | 7/7 | 0/49 | 5/5 | 0/57 | 31/32 | 0/157 |
| Splenomegaly | 14/20 | 0/51 | 1/7 | 0/49 | 2/5 | 0/57 | 17/32 | 0/157 |
| Small and/or mottled kidney | 17/20 | 0/51 | 5/7 | 0/49 | 3/5 | 0/57 | 25/32 | 0/157 |
| Edema | 5/20 | 0/51 | 1/7 | 0/49 | 0/5 | 0/57 | 6/32 | 0/157 |
| Enlarged heart | 8/20 | 0/51 | 4/7 | 0/49 | 1/5 | 0/57 | 13/32 | 0/157 |
(i) Lymphoid organs. (a) Thymus.
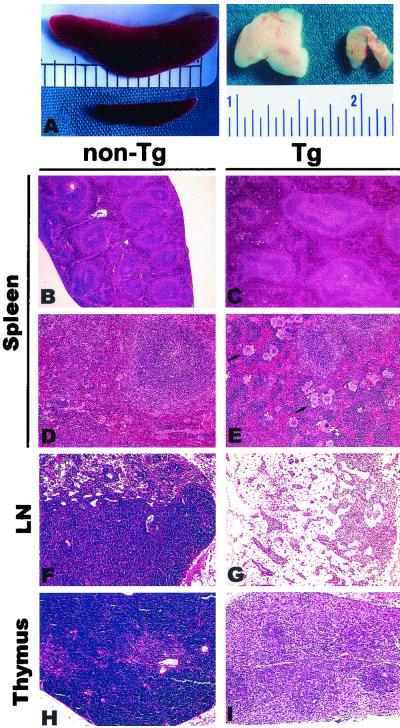
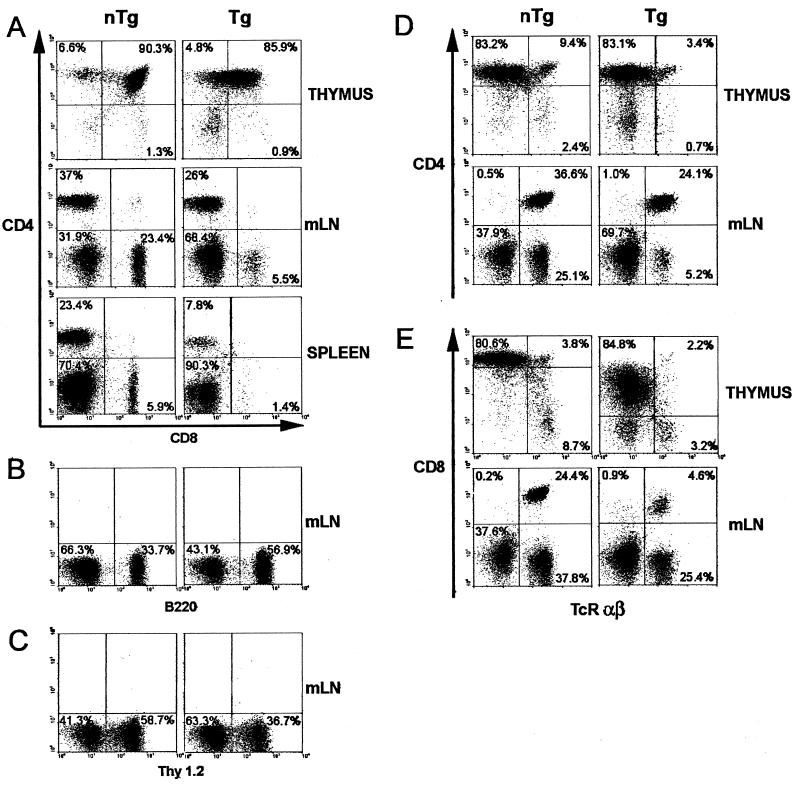
The majority of Tg mice (31 of 32 [97%]) exhibited gross thymic atrophy compared to age-matched non-Tg controls. This atrophy could already be observed at birth (data not shown) or in very young Tg mice (Fig. 5A), which showed as much as a 20-fold decrease in the number of thymocytes at the age of 21 days (Table 3). Histological assessment of Tg thymuses showed a generalized hypocellularity and a loss of the corticomedullary junction (Fig. 5I). FACS analysis, performed on total remaining thymocytes of Tg mice from founders F57693 and F55541 (Fig. 6; Table 4), revealed a dramatic downregulation of CD8 cell surface expression and downregulation of CD4 cell surface expression to a lesser extent. The relative percentages and absolute numbers of CD4+ CD8+ and single-positive thymocytes (CD4+ CD8− or CD4− CD8+) were decreased in Tg mice compared to non-Tg controls (Fig. 6A), whereas relative percentages of Thy1.2+ cells (Table 4) were less affected. There were also decreases in the percentages and absolute numbers of TcRαβHi cells (Fig. 6D and E), which represent the most mature stages in thymocyte development. These results suggested that Tg thymocytes are skewed toward an immature phenotype.
FIG. 5.
Pathology in lymphoid organs of CD4C/SHIV-_nef_SIV Tg mice. (A) Macroscopic view of the spleen and thymus. Splenomegaly is seen in the Tg mouse (top) relative to the non-Tg mouse (bottom), and thymic atrophy is seen in the Tg mouse (right) relative to the non-Tg mouse (left). (B through E) Histology of the spleen. Note the splenomegaly and the altered architecture of the white pulp of the Tg mouse, and note that both enlarged white and red pulp areas contribute to the splenomegaly (C). An apparent increase in megakaryocyte numbers (arrows) is seen in the spleen of a Tg animal (E) relative to those in a non-Tg animal (D). (F and G) LN. Extensive depletion of the T-cell zone and hypocellularity of the B-cell zone are seen in mesenteric LN of a Tg animal (G) compared to non-Tg LN (F). (H and I) Thymus. A Tg thymus (I) exhibits hypocellularity and loss of the corticomedullary architecture, compared to a non-Tg thymus (H). Counterstain, hematoxylin and eosin. Magnifications, ×2.5 (A), ×9 (B and C), and ×36 (D through I).
TABLE 3.
Quantitation of cells of lymphoid organs of control and CD4C/SHIV-_nef_SIV Tg mice
| Mice | Cells (106)a in: | ||
|---|---|---|---|
| Thymus | Spleen | Mesenteric LN | |
| Non-Tg (n = 19) | 118 ± 67.5 | 43.7 ± 17.2 | 9.4 ± 2.0 |
| Tg F55541 (n = 10) | 5.99 ± 2.9*** | 146.0 ± 5.7* | 9.5 ± 5.7 |
| Tg F57693 (n = 6) | 12.2 ± 6.8** | 80 ± 17 | 6.8 ± 2.0 |
| Tg F27367 (n = 7) | 117 ± 47.9 | ND | ND |
FIG. 6.
Cytofluorometric analysis of thymocytes, splenocytes, and mesenteric-LN (mLN) lymphocytes of young CD4C/SHIV-_nef_SIV Tg mice. Thymocytes, splenocytes, and mesenteric-LN lymphocytes from a representative Tg mouse (F57693) and a non-Tg control mouse were analyzed by flow cytometry. (A) Two-color analysis for the expression of CD4 and CD8. (B and C) One-color analysis for the expression of B220 (B) and Thy 1.2 (C). (D and E) Two-color analysis for the expression of CD4 and TcRαβ (D) as well as CD8 and TcRαβ (E). Percentages of cells found in each quadrant are shown; 104 live cells were analyzed.
TABLE 4.
Thymic cell surface marker analysis of CD4C/SHIV-_nef_SIV Tg mice
| Mice | No. of mice | Cell populationa (absolute no. of cells, 106) | Mean fluorescence | |||||
|---|---|---|---|---|---|---|---|---|
| Thy 1.2+ | CD4+CD8+ | CD4+CD8− | CD4−CD8+ | Thy 1.2 | CD4 | CD8 | ||
| Non-Tg | 19 | 94.46 ± 2.2 | 88.9 ± 2.1 | 10.7 ± 1.6 | 4.1 ± 2.0 | 407 ± 125 | 1,224 ± 332 | 1,691 ± 927 |
| (113 ± 64.9) | (106 ± 60.7) | (9.7 ± 7.1) | (4.2 ± 3.3) | |||||
| Tg (F55541) | 10 | 75.95 ± 13.9*** | 33.1 ± 19.4***b | 3.6 ± 0.6*** | 3.0 ± 0.9* | 498 ± 148 | 812 ± 335** | 469 ± 214*** |
| (18.1 ± 1.9)*** | (5.0 ± 5.9)*** | (0.25 ± 0.06)* | (0.59 ± 0.06)* | |||||
| Tg (F57693) | 3 | 92.3 ± 1.7 | 78.4 ± 6.5*** | 5.7 ± 0.9*** | 1.1 ± 0.6* | 283 ± 165 | 517 ± 33** | 359 ± 187* |
| (14.2 ± 3.3)* | (12.2 ± 3.7)* | (0.9 ± 0.09) | (0.2 ± 0.1) |
(b) Peripheral lymphoid organs.
Tg mesenteric LN were usually of normal size, as determined by the recovery of similar numbers of cells from Tg and non-Tg mesenteric LN (Table 3). However, the peripheral LN were usually severely atrophied (data not shown). Consistent with this observation, histological observation of Tg peripheral LN showed low cell density in the deep cortical zone, with a less frequently observed hypocellularity of the B-cell zone (Fig. 5G). FACS analysis performed on the mesenteric LN of Tg mice from founders F55541 and F57693 also showed decreases in both the relative percentages and absolute numbers of CD4+ TcRαβ+ T cells (Fig. 6D and Table 5). Strikingly, even greater decreases were observed in both percentages and absolute cell numbers of CD8+ TcRαβ+ T cells of Tg mice (Fig. 6E). These low numbers of peripheral CD4+ and CD8+ T cells were apparent early in life, and there was no subsequent dramatic loss of these cell populations as the disease progressed. As noted for total thymocytes, expression of both CD4 and CD8 cell surface markers was low, but the CD8 cell surface marker was, again, more affected (Fig. 6A). A decrease in the percentages of Thy1.2+ cells (Fig. 6C) and increases of B220+ cells (Fig. 6B) and Mac1+ cells (data not shown) were also noted in Tg mice.
TABLE 5.
Spleen and mesenteric LN cell surface marker analysis of CD4C/SHIV-_nef_SIV Tg mice
| Organ(s) analyzed | Mice | No. of mice | Cell populationa (absolute no. of cells, 106) | Mean fluorescence | |||||
|---|---|---|---|---|---|---|---|---|---|
| Thy 1.2+ | CD4+ | CD8+ | B220+ | Thy 1.2 | CD4 | CD8 | |||
| Spleen | Non-Tg | 6 | 31.0 ± 12.2 (19.0 ± 14.3) | 19.6 ± 4.2 (13.5 ± 7.9) | 8.3 ± 5.0 (4.3 ± 2.1) | 55.7 ± 7.2 (23.1 ± 12.8) | 260 ± 153 | 931 ± 482 | 1,439 ± 901 |
| Spleen | Tg (F55541) | 8 | 7.7 ± 1.8***b (3.3 ± 2.0)* | 4.5 ± 0.9*** (3.63 ± 3.3)* | 0.7 ± 0.4** (0.8 ± 0.7)** | 66.0 ± 7.7* (60.0 ± 40.0)* | 146 ± 53 | 144 ± 116* | 188 ± 69* |
| Spleen | Tg (F57693) | 7 | 13.3 ± 3.3 (10.5 ± 0.7) | 6.6 ± 2.5*** (3.4 ± 4.1)* | 0.9 ± 0.8** (0.5 ± 0.8)** | 68.0 ± 6.4** (23.3 ± 19.0) | 231 ± 48 | 279 ± 114* | 140 ± 89* |
| LN | Non-Tg | 6 | 63.6 ± 8.4 (5.57 ± 1.5) | 53.2 ± 10.4 (4.8 ± 1.6) | 20.0 ± 3.9 (1.8 ± 0.4) | 24.7 ± 7.6 (2.1 ± 0.60) | 178 ± 180 | 569 ± 130 | 380 ± 92 |
| LN | Tg (F55541) | 8 | 19.8 ± 6.5*** (1.7 ± 0.9)** | 13.2 ± 5.7*** (1.1 ± 0.5)** | 1.8 ± 0.9*** (0.2 ± 0.2)*** | 66.5 ± 17.3** (7.0 ± 5.8) | 133 ± 91 | 247 ± 207* | 94 ± 30*** |
| LN | Tg (F57693) | 3 | 46.3 ± 13.5 (3.3 ± 1.9) | 35.9 ± 14.0 (2.6 ± 1.7) | 5.4 ± 0.1** (0.4 ± 0.09)** | 47.9 ± 12.7* (3.2 ± 0.07) | 222 ± 30 | 451 ± 206 | 171 ± 30* |
A significant fraction (17 of 32[53%]) of Tg mice had splenomegaly (spleen weights, 0.12 ± 0.04 g for non-Tg mice [n = 8]; 0.29 ± 0.19 g for F55541 Tg mice [n = 5]; 0.36 ± 0.04 g for F57691 Tg mice [n = 3]; 0.19 ± 0.03 g for F57693 Tg mice [n = 3]) (Fig. 5A; Tables 2 and 3). The spleens of Tg mice were often enlarged, with greater areas of both red and white pulp participating in the enlargement (Fig. 5C). Several Tg spleens exhibited increased numbers of megakaryocytes (Fig. 5E). FACS analysis of splenocytes from mice from both F55541 and F57693 founders showed changes similar to those observed for Tg mesenteric LN (Fig. 6A and Table 5). This analysis revealed low numbers of CD4+ and CD8+ T cells, with low and very low levels of CD4 and CD8 cell surface expression, respectively (Fig. 6A; Table 5). In addition, accumulation of high percentages of B cells and macrophages, as found in mesenteric LN, and an increase in the level of erythroid CD71+ Ter119+ progenitors were observed (data not shown). Further analysis showed that a CD4− CD8− CD3+ population was absent in the LN of Tg mice (data not shown), indicating that the low number of CD8+ T cells in peripheral lymphoid organs truly reflected their absence rather than simply the inability to detect the CD8 cell surface molecule. No difference in the expression of the CD44 or CD69 cell surface marker was found on T lymphocytes from the thymuses, spleens, and mesenteric LN of Tg versus non-Tg mice (data not shown). Together, these results demonstrate an early and severe thymic defect in these Tg mice, as well as profound changes in the cell populations of the peripheral lymphoid organs.
(ii) Kidneys.
In most Tg mice (25 of 32 [78%]), the kidneys were markedly atrophic and paler, with an irregular surface, compared to those of controls. Some mice (6 of 32 [19%]) exhibited edema, which is seen in renal failure.
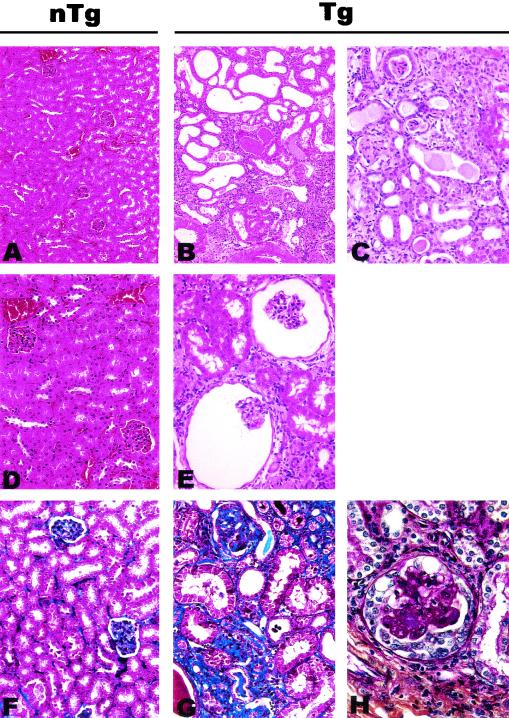
The histopathological changes observed in the kidneys consisted of interstitial nephritis, fibrosis, atrophy with dilation, and cystic changes (Fig. 7B and C). More-extensive glomerular changes were observed in the CD4C/SHIV-_nef_SIV Tg mice than in their CD4C/HIV Tg mouse counterparts (30). Thus, expansion of Bowman's space, with occasional atrophy of the podocyte cell layer (Fig. 7E), and segmental glomerulosclerosis were observed (Fig. 7G and H). These histopathological findings are consistent with those of a chronic tubulo-interstitial nephropathy.
FIG. 7.
Pathology in kidneys of CD4C/SHIV-_nef_SIV Tg mice. Shown are non-Tg (nTg) (A, D, and F) and Tg (B, C, E, G, and H) kidneys at low (A and B), intermediate (C), or high (D through H) power. Kidneys of Tg mice exhibit tubular atrophy and dilation with cystic changes and interstitial nephritis. Note expansion of Bowman's space (E). Compared to a non-Tg control mouse kidney (F), the renal cortex of a Tg mouse (G) has focal areas of interstitial fibrosis (blue). Sclerosis is also present in the glomerulus in the upper part of panel G. Occasional glomeruli in the kidneys of Tg mice have segmental hyaline obliteration of capillary loops and cellular crescents (H). Trichrome stain (F and G), periodic acid-Schiff stain (H), and hematoxylin and eosin (A through E) were used. Magnifications, ×43 (A and B), ×85 (C through G), and ×170 (H).
(iii) Lungs.
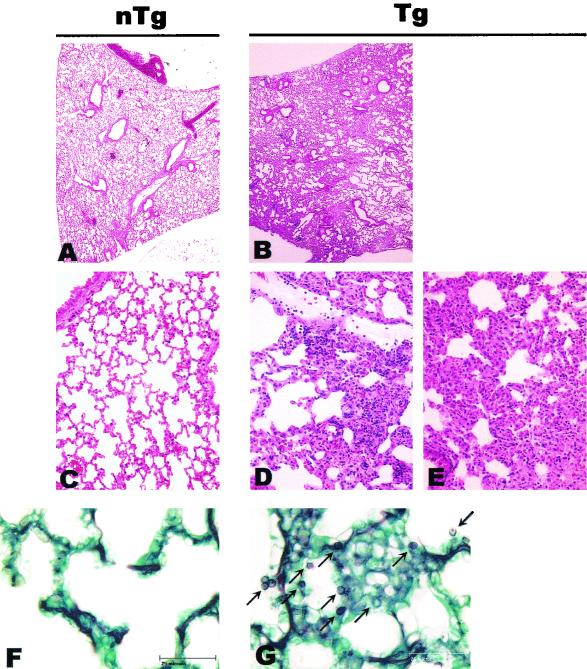
No gross changes in the lungs were noticed upon dissection of Tg mice compared to their non-Tg littermates. However, histological examination revealed that several Tg animals exhibited a LIP which was sometimes extensive (Fig. 8B, D, and E). Tg lungs (56%; n = 9), but not control non-Tg lungs, were positive for Pneumocystis carinii (22% had about 100 to 1,000 organisms, and 34% had only a few [up to 100]) (Fig. 8F and G), suggesting a state of severe immunodeficiency. Since some of the Tg animals still exhibited LIP without any signs of P. carinii infection, it is likely that LIP develops as a consequence of Nef expression, rather than being caused by P. carinii infection.
FIG. 8.
Pathology in lungs of CD4C/SHIV-_nef_SIV Tg mice. Shown are non-Tg (A, C, and F) or Tg (B, D, E, and G) lungs at low (A and B) and intermediate (C through E) power. LIP was observed in Tg animals (B, D, and E). Note the widespread nature of the pathology seen in panel B. Panels F and G show methamine silver (Grocott)-stained tissues from non-Tg and Tg animals, respectively. Note the presence of P. carinii particles in the lung from the Tg mouse (arrows in panel G). Counterstain, hematoxylin and eosin (A through E). Magnifications, ×11 (A and B) and ×85 (C through E). Bar in panels F and G, 25 μm.
(iv) Heart.
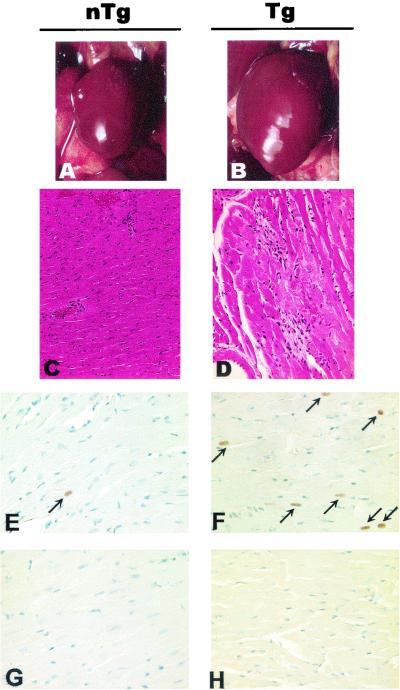
In about 34% (13 of 38) of Tg mice, the heart was obviously enlarged (Fig. 9B; Table 2). The pathology was multifocal in nature, consisting of areas of myocytolysis and myocarditis (Fig. 9D). Despite the enlargement of the hearts, no clear evidence of widespread cardiomyocyte hypertrophy was seen. When observed, hypertrophic fibers were isolated as single or small clusters of two to three fibers. No clear evidence of heart dilation could be documented histologically.
FIG. 9.
Pathology in hearts of CD4C/SHIV-_nef_SIV Tg mice. Control non-Tg (nTg) (A, C, E, and G) and Tg (B, D, F, and H) mice were studied. (A and B) Macroscopic images of hearts in situ. Note the enlargement of the Tg heart (B) compared to the non-Tg heart (A). (C and D) Histology of hearts. Myocytolysis and myocarditis are observed in the left ventricle of a Tg heart (D) but not in that of a non-Tg heart (C). (E through H) Detection of Ig deposition. Heart sections were probed with anti-mouse Ig (E and F) or with anti-rat Ig as a negative control (G and H). Increased frequency of anti-nuclear auto-Ab deposition (arrows) is observed in a Tg heart (F) compared to a non-Tg heart (E), when sections are probed with anti-mouse Ig. Counterstain, hematoxylin and eosin (C and D) or hematoxylin (E through H). Magnifications, ×2.5 (A and B), ×72 (C and D), and ×145 (E through H).
Presence of auto-Ab and lack of T-cell help in CD4C/SHIV-_nef_SIV Tg mice.
We recently reported that CD4C/HIV Tg mice, which develop an AIDS-like disease very similar to the disease found in CD4C/SHIV-_nef_SIV Tg mice, show B-cell activation, impaired germinal-center formation, and Ig class switching and produce auto-Ab (53). Similar assays were performed to determine whether CD4C/SHIV-_nef_SIV Tg mice show the same defects.
First, peripheral T cells (total or purified CD4+ T cells) from Tg and non-Tg mice were compared for their proliferation capacities in vitro, following anti-CD3 stimulation, by using CFSE labeling, as previously described (33). No major differences were noted between the groups, except for an increase of the proliferation capacities of both CD4+ and CD8+ T cells in some Tg mice (data not shown).
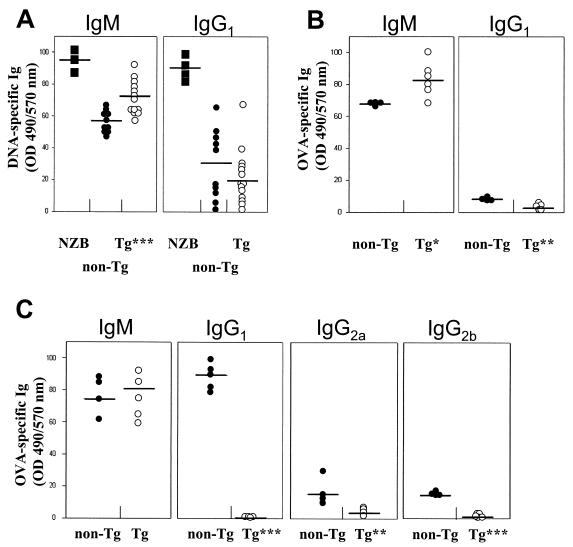
Then the sera of diseased Tg animals (i.e., those exhibiting thymic atrophy, kidney disease, and large spleens) were assessed for the presence of anti-DNA auto-Ab. These Tg sera were found to have higher levels of anti-DNA IgM and lower levels of anti-DNA IgG than non-Tg sera (Fig. 10A). The presence of auto-Ab of other specificities, and specifically for another tissue, was also studied by testing for decoration of heart sections with endogenous Ig, using tissue staining with an anti-Ig antibody. This analysis revealed deposition of markedly high levels of anti-nuclear Ig in the hearts of a significant percentage of Tg mice (43%; n = 7) compared to non-Tg mice (Fig. 9F). Finally, preliminary data did not show any difference between Tg and non-Tg mice in the number of CD5+ B cells (data not shown). Therefore, these results indicated the presence of autoimmune responses in these Tg mice.
FIG. 10.
Presence of anti-DNA antibodies and reduced OVA-specific Ig isotype switching in CD4C/SHIV-_nef_SIV Tg mice. (A) Anti-DNA IgM and IgG1 antibody levels in sera of nonimmunized non-Tg and Tg mice were measured by ELISA. The data shown were normalized to the DNA reactivity observed with reference sera from NZB mice (arbitrarily established as 100%) (filled squares). Each circle represents the serum of one mouse at the end point of the titration curve. Bars, means from five different experiments. (B and C) OVA-specific antibody levels in sera of non-Tg and Tg mice taken 10 days following a primary immunization (B) and 5 days following a secondary immunization (C). Each circle represents the serum of one mouse at the end point of the ELISA titration curve. Bars, means from three different experiments. P values by Student's t test: ∗, P < 0.05; ∗∗, P < 0.001; ∗∗∗, P < 0.0001. OD490, optical density at 490 nm.
The higher ratio of IgM to IgG anti-DNA antibody levels found in these Tg mice relative to their non-Tg control littermates also suggested an impaired Ig isotype class-switching ability. This phenomenon was studied further after immunization with OVA. Sera of OVA-immunized mice (Tg and non-Tg) were tested for the presence of OVA-specific Ig. Ten days postinjection (primary response), Tg mice had lower levels of OVA-specific IgG1 and higher levels of OVA-specific IgM than non-Tg mice (Fig. 10B). Similarly, after a secondary immunization, the levels of OVA-specific IgG1, IgG2a, and IgG2b were much lower in Tg than in non-Tg mice, while OVA-specific IgM levels showed less difference (Fig. 10C), confirming a severe incapacity of T-cell help-dependent Ig class switching in Tg mice.
DISCUSSION
SIV Nef induces a fatal AIDS-like disease in Tg mice.
This report shows that mice expressing the SIVmac239 nef gene under the regulation of the CD4C promoter (CD4C/SHIV-_nef_SIV) develop a severe AIDS-like disease. This disease is characterized by premature death, wasting, failure to thrive, kidney disease (interstitial nephritis and glomerulosclerosis), LIP, cardiac disease, thymic atrophy, splenomegaly, and low T-cell numbers both in the thymus and in peripheral lymphoid organs. As previously discussed extensively (29, 30, 53), all these phenotypes, including the cardiac disease phenotype (Kay et al., submitted) and except those of the lymphoid organs (see below), are very similar to those induced by the expression of HIV-1 _ne_f in Tg mice and resemble human AIDS, especially pediatric AIDS. Since an allelic construct (CD4C/HIVMutH) not expressing Nef was found to be unable to induce disease in Tg mice (30), it appears that SIV Nef is mainly responsible for inducing these phenotypes.
This AIDS-like disease of CD4C/SHIV-_nef_SIV Tg mice is also very similar to simian AIDS (SAIDS) (17, 18). Premature death is frequent in SAIDS (7, 10, 40). Wasting disease and failure to thrive, in the absence of detectable secondary pathogens, has also been described in SAIDS (35, 66); such wasting is usually correlated with a high incidence of diarrhea (7, 36), often caused by pathogenic bacteria or protozoa. In contrast, wasting without diarrhea was frequent in CD4C/SHIV-_nef_SIV Tg mice, most likely because they were housed in an SPF facility. Kidney (5, 7, 25, 65) and lung (25, 51) diseases have also been described in SAIDS. Similarly, cardiac disease is found in SIV-infected primates (60). Finally, thymic atrophy with preferential loss of CD4+ CD8+ immature thymocytes and CD4+ CD8− mature thymocytes, correlating with the progression of the infection, has been reported in SAIDS (46, 47, 70). In addition, the loss of both CD4+ and CD8+ peripheral T cells late in the disease is typical of SAIDS (10).
The similarity of the diseases developing in SIV or HIV-1 _nef_-expressing Tg mice (29, 30) indicates a strong functional resemblance between SIV and HIV-1 Nef. The functional relatedness of the two molecules has been noted previously in several assays, both in vitro (down-modulation of human CD4 [1, 6, 9, 22, 24, 54] and of human MHC class I cell surface molecules [59]; enhanced viral replication [2, 13, 15, 28, 41, 63]) and in vivo, where Nef is required for high virus loads in SIV-infected primates (41) and in HIV-1-infected humans (16, 42). Our data extend these observations by demonstrating the functional resemblance of SIV and HIV-1 Nef in an additional in vivo assay, in another species (mouse), and show that this functional similarity is independent of virus replication or of an immune response to the virus. In addition, the present results extend to the nef gene of another lentivirus (SIV) our previous findings that HIV-1 nef alone (CD4C/HIVMutG) is sufficient to induce an AIDS-like disease in mice (30).
Development of SIV _nef_-induced disease in Tg mice correlates with the level of its expression.
The cell type specificity of SIV nef transgene expression determined by the CD4C promoter in Tg mice is likely to contribute to the resemblance of the phenotype to human and simian AIDS. Indeed, other Tg mice expressing only HIV Nef under the regulation of the long terminal repeat (LTR) or of other T-cell-specific promoter/enhancer elements (11, 19, 20, 48, 62) or expressing SIV Nef under the regulation of the cytomegalovirus promoter (45) do not exhibit the same multiorgan syndrome as that observed in CD4C/HIVMutG (30) or CD4C/SHIV-_nef_SIV Tg mice. The CD4C promoter has been reported previously to drive the expression of surrogate genes in mature and immature T cells and in cells of the macrophage/dendritic lineages of Tg mice (29-32). The expression of the transgene in mice seems to be following the tissue specificity of this promoter, thus targeting the same cell populations affected by the virus in natural infections.
Interestingly, the development of disease in CD4C/SHIV-_nef_SIV Tg mice seems to be influenced not only by the promoter specificity but also by the level of SIVmac239 nef expression. Transgene expression in mice from the lines investigated correlated well with disease latency (F57691 ≦ F55541 ≦ F57693). In addition, disease progression seems clearly influenced by the number of target cells expressing SIV Nef. Indeed, the three founder mice which were mosaic for transgene expression, and which likely had a lower number of cells expressing the transgene, developed the disease much later than their nonmosaic offspring. Therefore, the Nef viral load in specific target cells appears to be important for disease progression in these CD4C/SHIV-_nef_SIV Tg mice. This biological parameter may also be quite critical in human and simian AIDS and may be partly reflected in the plasma viral load, which is known to correlate well with disease progression (64, 67, 69). We have previously observed a similar effect of HIV-1 Nef levels on disease progression in CD4C/HIV Tg mice (30).
Some phenotypes distinguish SIV from HIV-1 Nef expression in Tg mice.
Although the disease arising in Tg mice expressing SIV nef is very similar to that of Tg mice expressing HIV-1 nef (29, 30), heart enlargement was observed at a higher penetrance and kidney glomerular pathology was greater in CD4C/SHIV-_nef_SIV mice than in CD4C/HIV Tg mice. Moreover, three immune phenotypes are virtually unique to CD4C/SHIV-_nef_SIV Tg mice: splenomegaly, early thymic atrophy, and early onset of low numbers of peripheral T cells.
Splenomegaly is frequent in CD4C/SHIV-_nef_SIV mice, while it is rare in CD4C/HIVWT or CD4C/HIVMutG Tg mice, where spleen atrophy is more frequently observed (29, 30). Splenomegaly has previously been reported for other Tg mice expressing HIV-1 Nef under the regulation of a T-cell-specific promoter/enhancer element (48). Splenomegaly has also been reported upon infection of rhesus macaques with SIVmac239/YEnef (21, 56) or SIVsmmPBj14 (40), two particularly virulent alleles of Nef, and with SIVmac251 (64) or SIV/Delta (7). It has been observed, as well, upon infection of pigtailed macaques with SIVagm9063 (36). The enlarged spleens in CD4C/SHIV-_nef_SIV Tg mice show a low number of T cells, accumulation of high percentages of B cells and macrophages, and the presence of a large number of megakaryocytes (Fig. 5E) and erythroid CD71+ Ter119+ progenitors (data not shown). This phenotype is associated with the white appearance of the long bones (in contrast to their reddish color in the non-Tg controls) (data not shown), suggesting the possibility of a bone marrow (BM) defect in the production of cells of the erythroid lineage. Therefore, this splenomegaly may in part reflect extramedullary hematopoiesis, common in mice. Detailed investigation of hematopoietic precursors in spleens and in BM will be needed to determine whether a BM defect with compensatory splenic extramedullary hematopoiesis exists in these Tg mice.
Another intriguing and unexpected phenotype observed in these CD4C/SHIV-_nef_SIV Tg mice is the early appearance of low numbers of peripheral T cells in combination with low expression of the CD8 cell surface marker. Such a CD8 phenotype is highly reproducible, is seen even in young mice, and is observed in the three founder lines studied, ruling out an integration effect. This CD8 phenotype was not seen in CD4C/HIVWT or CD4C/HIVMut Tg mice (29, 30), which are, in contrast to CD4C/SHIV-_nef_SIV Tg mice, born with close-to-normal numbers of thymocytes and peripheral T cells and which rather show a progressive and preferential loss of CD4+ T cells. Only at the end stage of the disease do they show, as in human AIDS, a loss of CD8+ T cells (29, 30).
This dramatic low expression of the CD8 and, to a lesser extent, the CD4 cell surface marker in CD4C/SHIV-_nef_SIV Tg mice confirms earlier in vitro data showing that SIV Nef can downregulate the mouse CD8, but not the CD4, cell surface marker (22). Similar but less-marked low expression of the CD8 cell surface molecule has been observed previously, in vivo, in HIV Nef-expressing Tg mice (11, 48, 62). The very low cell surface expression of CD8 in CD4+ CD8+ thymocytes may reflect a downregulation caused by a strong interaction of Nef with the CD8 molecule, which would be expected to disrupt T-cell differentiation in the thymus and consequently to lead to the generation of an abnormal peripheral CD8+ T-cell pool. Consistent with this observation is the observation that peripheral CD8+ T cells are few in number and show very low CD8 cell surface expression, while not expressing the transgene at detectable levels.
Other findings also suggest that the profound changes in the cell populations of the peripheral lymphoid organs, including the low numbers of both CD4+ and CD8+ peripheral T cells in CD4C/SHIV-_nef_SIV Tg mice, most likely reflect a defect in production of these cells in the thymus rather than their rapid loss. CD4C/SHIV-_nef_SIV Tg mice are born with smaller thymuses than non-Tg controls. Moreover, at the age of 21 days, CD4C/SHIV-_nef_SIV Tg mice show a 20-fold-lower number of thymocytes than age-matched non-Tg controls. These data strongly suggest that CD4C/SHIV-_nef_SIV Tg mice are born with a severe thymic defect. In fact, this observation of early thymic atrophy, coupled with our preliminary ontogeny studies and fetal liver transplantation experiments, confirms the thymus defect hypothesis (M.-C. Simard, P. Chrobak, D. G. Kay, Z. Hanna, and P. Jolicoeur, unpublished data). A similar phenotype, consisting of a transient and reversible (62) or a more chronic (11, 48) suppression of the number of single-positive CD8+ T cells, has been reported previously for Tg mice expressing HIV-1 Nef under the regulation of T-cell-specific enhancer/promoter elements. In contrast, CD4C/HIVMutG Tg mice do not exhibit such a phenotype and are born with near-normal numbers of thymocytes and peripheral CD4+ T cells, which are progressively lost as the disease progresses (29, 30). Loss of CD8+ T cells together with loss of CD4+ T cells is also common in the advanced stage of SAIDS but is usually not observed in the early stage of the disease. Therefore, this remarkable low number of peripheral CD8+ T cells and of CD4+ T cells is unique to Tg mice expressing SIV Nef. This phenotype is reminiscent of the DiGeorge-like immunophenotype described for some children infected with HIV-1, who harbor low numbers of both CD4+ and CD8+ T cells and are thought to have an HIV-1 induced thymic defect (44).
The underlying cellular and molecular bases of this early thymic atrophy, dramatic low expression of the CD8 cell surface marker, and low numbers of CD4+ and CD8+ peripheral T cells are unclear. This phenotype could reflect subtle changes in transgene expression undetectable by the methods we used. We believe this to be unlikely, however, for three reasons: (i) this phenotype was observed in three distinct founder lines, thus ruling out an integration (positional) effect; (ii) no such variation has been observed in 19 independent founder lines of CD4C/HIV Tg mice expressing wild-type HIV-1 Nef (strain NL4-3), all of which develop very similar phenotypes consisting of a progressive and preferential loss of peripheral CD4+ T cells and an increase in CD8+ T-cells (29, 30); (iii) alternatively, and most likely, this phenotype could reflect either functional differences between HIV-1 and SIV Nef or allelic variations. Future comparison of different HIV-1 and SIV nef alleles in the same in vivo Tg assay should help to determine which of these possibilities is the most likely.
Therefore, the overall disease observed in these SIV _nef_-expressing Tg mice is remarkably similar to human and simian AIDS and especially to a subgroup of pediatric AIDS. This Tg model represents a novel in vivo assay for SIV Nef which is likely to be instrumental in structure-function studies as well as for understanding the pathogenesis of this disease.
Acknowledgments
This work was supported by grants to P.J. from the Medical Research Council of Canada. Marie-Chantal Simard is the recipient of a Canadian Institutes of Health Research Ph.D. scholarship.
We gratefully acknowledge the excellent technical assistance of Benoît Laganière, Ève-Lyne Thivierge, Chunyan Hu, Karina Lamarre, Ginette Massé, Émilie Gauthier, and Viorica Lascau. We thank Nathalie Tessier and Éric Massé of the Cytofluorometry Core Facility for their support. We also thank Michel Robillard and Quinzhang Zhu of the Transgenic Mouse Core Facility, Christian Charbonneau and Hélène Lienard of Photography, Claire Crevier of Histology, and Stéphane Matte of the SPF Core Facility for their excellent work. We are grateful to Johanne Poudrier for helpful discussions and to Patrick Vincent for help in the cell-sorting experiments.
REFERENCES
- 1.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76**:**853-864. [DOI] [PubMed] [Google Scholar]
- 2.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69**:**5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, L., Z. Du, A. Y. Howe, S. Czajak, and R. C. Desrosiers. 1999. Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing nef of human immunodeficiency virus type 1. J. Virol. 73**:**5814-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander, L., Z. Du, M. Rosenzweig, J. U. Jung, and R. C. Desrosiers. 1997. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J. Virol. 71**:**6094-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alpers, C. E., C. C. Tsai, K. L. Hudkins, Y. Cui, L. Kuller, R. E. Benveniste, J. M. Ward, and W. R. Morton. 1997. Focal segmental glomerulosclerosis in primates infected with a simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 13**:**413-424. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, S., D. C. Shugars, R. Swanstrom, and J. V. Garcia. 1993. Nef from primary isolates of human immunodeficiency virus type 1 suppresses surface CD4 expression in human and mouse T cells. J. Virol. 67**:**4923-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baskin, G. B., M. Murphey-Corb, E. A. Watson, and L. N. Martin. 1988. Necropsy findings in rhesus monkeys experimentally infected with cultured simian immunodeficiency virus (SIV)/delta. Vet. Pathol. 25**:**456-467. [DOI] [PubMed] [Google Scholar]
- 8.Bell, I., C. Ashman, J. Maughan, E. Hooker, F. Cook, and T. A. Reinhart. 1998. Association of simian immunodeficiency virus Nef with the T-cell receptor (TCR) zeta chain leads to TCR down-modulation. J. Gen. Virol. 79**:**2717-2727. [DOI] [PubMed] [Google Scholar]
- 9.Benson, R. E., A. Sanfridson, J. S. Ottinger, C. Doyle, and B. R. Cullen. 1993. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral superinfection. J. Exp. Med. 177**:**1561-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benveniste, R. E., W. R. Morton, E. A. Clark, C. C. Tsai, H. D. Ochs, J. M. Ward, L. Kuller, W. B. Knott, R. W. Hill, M. J. Gale, et al. 1988. Inoculation of baboons and macaques with simian immunodeficiency virus/Mne, a primate lentivirus closely related to human immunodeficiency virus type 2. J. Virol. 62**:**2091-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady, H. J., D. J. Pennington, C. G. Miles, and E. A. Dzierzak. 1993. CD4 cell surface downregulation in HIV-1 Nef transgenic mice is a consequence of intracellular sequestration. EMBO J. 12**:**4923-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bresnahan, P. A., W. Yonemoto, and W. C. Greene. 1999. Cutting edge: SIV Nef protein utilizes both leucine- and tyrosine-based protein sorting pathways for downregulation of CD4. J. Immunol. 163**:**2977-2981. [PubMed] [Google Scholar]
- 13.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. Fitch, D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68**:**2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collette, Y., S. Arold, C. Picard, K. Janvier, S. Benichou, R. Benarous, D. Olive, and C. Dumas. 2000. HIV-2 and SIV Nef proteins target different Src family SH3 domains than does HIV-1 Nef because of a triple amino acid substitution. J. Biol. Chem. 275**:**4171-4176. [DOI] [PubMed] [Google Scholar]
- 15.Cullen, B. R. 1994. The role of Nef in the replication cycle of the human and simian immunodeficiency viruses. Virology 205**:**1-6. [DOI] [PubMed] [Google Scholar]
- 16.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, et al. 1995. Genomic structure of an attenuated quasispecies of HIV-1 from a blood transfusion donor and recipients. Science 270**:**988-991. [DOI] [PubMed] [Google Scholar]
- 17.Desrosiers, R. C., and N. L. Letvin. 1987. Animal models for acquired immunodeficiency syndrome. Rev. Infect. Dis. 9**:**438-446. [DOI] [PubMed] [Google Scholar]
- 18.Desrosiers, R. C., and D. J. Ringler. 1989. Use of simian immunodeficiency viruses for AIDS research. Intervirology 30**:**301-312. [DOI] [PubMed] [Google Scholar]
- 19.Dickie, P. 2000. Nef modulation of HIV type 1 gene expression and cytopathicity in tissues of HIV transgenic mice. AIDS Res. Hum. Retrovir. 16**:**777-790. [DOI] [PubMed] [Google Scholar]
- 20.Dickie, P., F. Ramsdell, A. L. Notkins, and S. Venkatesan. 1993. Spontaneous and inducible epidermal hyperplasia in transgenic mice expressing HIV-1 Nef. Virology 197**:**431-438. [DOI] [PubMed] [Google Scholar]
- 21.Du, Z., S. M. Lang, V. G. Sasseville, A. Lackner, P. O. Ilyinskii, M. D. Daniel, J. U. Jung, and R. C. Desrosiers. 1995. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell 82**:**665-674. [DOI] [PubMed] [Google Scholar]
- 22.Foster, J. L., S. J. Anderson, A. L. Frazier, and J. V. Garcia. 1994. Specific suppression of human CD4 surface expression by Nef from the pathogenic simian immunodeficiency virus SIVmac239open. Virology 201**:**373-379. [DOI] [PubMed] [Google Scholar]
- 23.Garcia, J. V., and J. L. Foster. 1996. Structural and functional correlates between HIV-1 and SIV Nef isolates. Virology 226**:**161-166. [DOI] [PubMed] [Google Scholar]
- 24.Garcia, J. V., and A. D. Miller. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by Nef. Nature 350**:**508-511. [DOI] [PubMed] [Google Scholar]
- 25.Gattone, V. H., C. Tian, W. Zhuge, M. Sahni, O. Narayan, and E. B. Stephens. 1998. SIV-associated nephropathy in rhesus macaques infected with lymphocyte-tropic SIVmac239. AIDS Res. Hum. Retrovir. 14**:**1163-1180. [DOI] [PubMed] [Google Scholar]
- 26.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10**:**607-616. [DOI] [PubMed] [Google Scholar]
- 27.Greenway, A. L., H. Dutartre, K. Allen, D. A. McPhee, D. Olive, and Y. Collette. 1999. Simian immunodeficiency virus and human immunodeficiency virus type 1 Nef proteins show distinct patterns and mechanisms of Src kinase activation. J. Virol. 73**:**6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guatelli, J. C. 1997. The positive influence of Nef on viral infectivity. Res. Virol. 148**:**34-37. [DOI] [PubMed] [Google Scholar]
- 29.Hanna, Z., D. G. Kay, M. Cool, S. Jothy, N. Rebai, and P. Jolicoeur. 1998. Transgenic mice expressing human immunodeficiency virus type 1 in immune cells develop a severe AIDS-like disease. J. Virol. 72**:**121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna, Z., D. G. Kay, N. Rebai, A. Guimond, S. Jothy, and P. Jolicoeur. 1998. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell 95**:**163-175. [DOI] [PubMed] [Google Scholar]
- 31.Hanna, Z., N. Rebai, J. Poudrier, and P. Jolicoeur. 2001. Distinct regulatory elements are required for faithful expression of human CD4 in T cells, macrophages and dendritic cells of transgenic mice. Blood 98**:**2275-2278. [DOI] [PubMed] [Google Scholar]
- 32.Hanna, Z., C. Simard, and P. Jolicoeur. 1994. Specific expression of the human CD4 gene in mature CD4+ CD8− and immature CD4+ CD8+ T cells and in macrophages of transgenic mice. Mol. Cell. Biol. 14**:**1084-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanna, Z., X. Weng, D. G. Kay, J. Poudrier, C. Lowell, and P. Jolicoeur. 2001. The pathogenicity of human immunodeficiency virus (HIV) type 1 Nef in CD4C/HIV transgenic mice is abolished by mutation of its SH3-binding domain, and disease development is delayed in the absence of Hck. J. Virol. 75**:**9378-9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris, M. 1996. From negative factor to a critical role in virus pathogenesis: the changing fortunes of Nef. J. Gen. Virol. 77**:**2379-2392. [DOI] [PubMed] [Google Scholar]
- 35.Heise, C., P. Vogel, C. J. Miller, C. H. Halsted, and S. Dandekar. 1993. Simian immunodeficiency virus infection of the gastrointestinal tract of rhesus macaques. Functional, pathological, and morphological changes. Am. J. Pathol. 142**:**1759-1771. [PMC free article] [PubMed] [Google Scholar]
- 36.Hirsch, V. M., G. Dapolito, P. R. Johnson, W. R. Elkins, W. T. London, R. J. G. S. Montali, and C. Brown. 1995. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J. Virol. 69**:**955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howe, A. Y., J. U. Jung, and R. C. Desrosiers. 1998. Zeta chain of the T-cell receptor interacts with Nef of simian immunodeficiency virus and human immunodeficiency virus type 2. J. Virol. 72**:**9827-9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hua, J., and B. R. Cullen. 1997. Human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus Nef use distinct but overlapping target sites for downregulation of cell surface CD4. J. Virol. 71**:**6742-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Igarashi, T., R. Shibata, F. Hasebe, Y. Ami, K. Shinohara, T. Komatsu, C. Stahl-Hennig, H. Petry, G. Hunsmann, T. Kuwata, et al. 1994. Persistent infection with SIVmac chimeric virus having tat, rev, vpu, env and nef of HIV type 1 in macaque monkeys. AIDS Res. Hum. Retrovir. 10**:**1021-1029. [DOI] [PubMed] [Google Scholar]
- 40.Israel, Z. R., G. A. Dean, D. H. Maul, S. P. O'Neil, M. J. Dreitz, J. I. Mullins, P. N. Fultz, and E. A. Hoover. 1993. Early pathogenesis of disease caused by SIVsmmPBj14 molecular clone 1.9 in macaques. AIDS Res. Hum. Retrovir. 9**:**277-286. [DOI] [PubMed] [Google Scholar]
- 41.Kestler, H. W., III, D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65**:**651-662. [DOI] [PubMed] [Google Scholar]
- 42.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332**:**228-232. [DOI] [PubMed] [Google Scholar]
- 43.Kirchhoff, F., J. Munch, S. Carl, N. Stolte, K. Matz-Rensing, D. Fuchs, P. T. Haaft, J. L. Heeney, T. Swigut, J. Skowronski, and C. Stahl-Hennig. 1999. The human immunodeficiency virus type 1 nef gene can to a large extent replace simian immunodeficiency virus nef in vivo. J. Virol. 73**:**8371-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kourtis, A. P., C. Ibegbu, A. J. Nahmias, F. K. Lee, W. S. Clark, M. K. Sawyer, and S. Nesheim. 1996. Early progression of disease in HIV-infected infants with thymus dysfunction. N. Engl. J. Med. 335**:**1431-1436. [DOI] [PubMed] [Google Scholar]
- 45.Larsen, N. B., H. W. Kestler, and J. J. Docherty. 1998. Mice transgenic for simian immunodeficiency virus nef are immunologically compromised. J. Biomed. Science 5**:**260-266. [DOI] [PubMed] [Google Scholar]
- 46.Letvin, N. L., M. D. Daniel, P. K. Sehgal, R. C. Desrosiers, R. D. Hunt, L. M. Waldron, J. J. MacKey, D. K. Schmidt, L. V. Chalifoux, and N. W. King. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230**:**71-73. [DOI] [PubMed] [Google Scholar]
- 47.Li, S. L., E. Kaaya, C. Ordonez, M. Ekman, H. Feichtinger, P. Putkonen, D. Bottiger, G. Biberfeld, and P. Biberfeld. 1995. Thymic immunopathology and progression of SIVsm infection in cynomolgus monkeys. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9**:**1-10. [PubMed] [Google Scholar]
- 48.Lindemann, D., R. Wilhelm, P. Renard, A. Althage, R. Zinkernagel, and J. Mous. 1994. Severe immunodeficiency associated with a human immunodeficiency virus 1 NEF/3′-long terminal repeat transgene. J. Exp. Med. 179**:**797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lock, M., M. E. Greenberg, A. J. Iafrate, T. Swigut, J. Muench, F. Kirchhoff, N. Shohdy, and J. Skowronski. 1999. Two elements target SIV Nef to the AP-2 clathrin adaptor complex, but only one is required for the induction of CD4 endocytosis. EMBO J. 18**:**2722-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mandell, C. P., R. A. Reyes, K. Cho, E. T. Sawai, A. L. Fang, K. A. Schmidt, and P. A. Luciw. 1999. SIV/HIV Nef recombinant virus (SHIVnef) produces simian AIDS in rhesus macaques. Virology 265**:**235-251. [DOI] [PubMed] [Google Scholar]
- 51.Mankowski, J. L., D. L. Carter, J. P. Spelman, M. L. Nealen, K. R. Maughan, L. M. Kirstein, P. J. Didier, R. J. Adams, M. Murphey-Corb, and M. C. Zink. 1998. Pathogenesis of simian immunodeficiency virus pneumonia: an immunopathological response to virus. Am. J. Pathol. 153**:**1123-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peter, F. 1998. HIV nef: the mother of all evil? Immunity 9**:**433-437. [DOI] [PubMed] [Google Scholar]
- 53.Poudrier, J., X. Weng, D. G. Kay, G. Paré, E. L. Calvo, Z. Hanna, M. H. Kosco-Vilbois, and P. Jolicoeur. 2001. The AIDS disease of CD4C/HIV transgenic mice shows impaired germinal centers and autoantibodies and develops in the absence of IFN-γ and IL-6. Immunity 15**:**173-185. [DOI] [PubMed] [Google Scholar]
- 54.Rhee, S., and J. W. Marsh. 1994. Human immunodeficiency virus type 1 Nef-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J. Virol. 68**:**5156-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saksela, K. 1997. HIV-1 Nef and host cell protein kinases. Front. Biosci. 2**:**d606-d618. [DOI] [PubMed] [Google Scholar]
- 56.Sasseville, V. G., Z. Du, L. V. Chalifoux, D. R. Pauley, H. L. Young, P. K. Sehgal, R. C. Desrosiers, and A. A. Lackner. 1996. Induction of lymphocyte proliferation and severe gastrointestinal disease in macaques by a nef gene variant SIVmac239. Am. J. Pathol. 149**:**163-176. [PMC free article] [PubMed] [Google Scholar]
- 57.Sawai, E. T., A. Baur, H. Struble, B. M. Peterlin, J. A. Levy, and C. Cheng-Mayer. 1994. Human immunodeficiency virus type 1 Nef associates with a cellular serine kinase in T lymphocytes. Proc. Natl. Acad. Sci. USA 91**:**1539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaefer, T. M., I. Bell, B. A. Fallert, and T. A. Reinhart. 2000. The T-cell receptor zeta chain contains two homologous domains with which simian immunodeficiency virus Nef interacts and mediates down-modulation. J. Virol. 74**:**3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2**:**338-342. [DOI] [PubMed] [Google Scholar]
- 60.Shannon, R. P., M. A. Simon, M. A. Mathier, Y. J. Geng, S. Mankad, and A. A. Lackner. 2000. Dilated cardiomyopathy associated with simian AIDS in nonhuman primates. Circulation 101**:**185-193. [DOI] [PubMed] [Google Scholar]
- 61.Shibata, R., F. Maldarelli, C. Siemon, T. Matano, M. Parta, G. Miller, T. Fredrickson, and M. A. Martin. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176**:**362-373. [DOI] [PubMed] [Google Scholar]
- 62.Skowronski, J., D. Parks, and R. Mariani. 1993. Altered T cell activation and development in transgenic mice expressing the HIV-1 nef gene. EMBO J. 12**:**703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spina, C. A., T. J. Kwoh, M. Y. Chowers, J. C. Guatelli, and D. Richman. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179**:**115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staprans, S. I., P. J. Dailey, A. Rosenthal, C. Horton, R. M. Grant, N. Lerche, and M. B. Feinberg. 1999. Simian immunodeficiency virus disease course is predicted by the extent of virus replication during primary infection. J. Virol. 73**:**4829-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephens, E. B., C. Tian, Z. Li, O. Narayan, and V. H. Gattone II. 1998. Rhesus macaques infected with macrophage-tropic simian immunodeficiency virus (SIVmacR71/17E) exhibit extensive focal segmental and global glomerulosclerosis. J. Virol. 72**:**8820-8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stone, J. D., C. C. Heise, C. J. Miller, C. H. Halsted, and S. Dandekar. 1994. Development of malabsorption and nutritional complications in simian immunodeficiency virus-infected rhesus macaques. AIDS 8**:**1245-1256. [DOI] [PubMed] [Google Scholar]
- 67.Ten Haaft, P., B. Verstrepen, K. Uberla, B. Rosenwirth, and J. Heeney. 1998. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J. Virol. 72**:**10281-10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trono, D. 1995. HIV accessory proteins: leading roles for the supporting cast. Cell 82**:**189-192. [DOI] [PubMed] [Google Scholar]
- 69.Watson, A., J. Ranchalis, B. Travis, J. McClure, W. Sutton, P. R. Johnson, S. L. Hu, and N. L. Haigwood. 1997. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J. Virol. 71**:**284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wykrzykowska, J. J., M. Rosenzweig, R. S. Veazey, M. A. Simon, K. Halvorsen, R. C. Desrosiers, R. P. Johnson, and A. A. Lackner. 1998. Early regeneration of thymic progenitors in rhesus macaques infected with simian immunodeficiency virus. J. Exp. Med. 187**:**1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]