Membrane Interactions of the Tick-Borne Encephalitis Virus Fusion Protein E at Low pH (original) (raw)
Abstract
Membrane fusion of the flavivirus tick-borne encephalitis virus is triggered by the mildly acidic pH of the endosome and is mediated by envelope protein E, a class II viral fusion protein. The low-pH trigger induces an oligomeric rearrangement in which the subunits of the native E homodimers dissociate and the monomeric subunits then reassociate into homotrimers. Here we provide evidence that membrane binding is mediated by the intermediate monomeric form of E, generated by low-pH-induced dissociation of the dimer. Liposome coflotation experiments revealed that association with target membranes occurred only when liposomes were present at the time of acidification, whereas pretreating virions at low pH in the absence of membranes resulted in the loss of their ability to stably attach to liposomes. With the cleavable cross-linker ethylene glycolbis(succinimidylsuccinate), it was shown that a truncated soluble form of the E protein (sE) could bind to membranes only when the dimers were free to dissociate at low pH, and binding could be blocked by a monoclonal antibody that recognizes the fusion peptide, which is at the distal tip of the E monomer but is buried in the native dimer. Surprisingly, analysis of the membrane-associated sE proteins revealed that they had formed trimers. This was unexpected because this protein lacks a sequence element in the C-terminal stem-anchor region, which was shown to be essential for trimerization in the absence of a target membrane. It can therefore be concluded that the formation of a trimeric form of sE is facilitated by membrane binding. Its stability is apparently maintained by contacts between the ectodomains only and is not dependent on sequence elements in the stem-anchor region as previously assumed.
Enveloped viruses have evolved different but conceptually related mechanisms to fuse their membranes with cellular membranes during entry into cells. The process is controlled by viral surface glycoproteins that undergo triggered conformational changes required for mediating fusion (16, 30).
At least two different classes of viral fusion proteins can be distinguished (21). Class I is represented by orthomyxo-, retro-, paramyxo-, and filoviruses. Their fusion proteins mature by proteolytic cleavage of a precursor protein, yielding a membrane-anchored subunit with an amino-terminal or amino-proximal fusion peptide. Application of the fusion trigger (receptor binding or low pH) results in the formation of a characteristic trimeric postfusion structure with a triple-stranded coiled coil at its core (6, 17, 27, 30). Class II fusion proteins are not proteolytically cleaved and have internal rather than amino-terminal fusion peptides. They are synthesized as a complex with a second membrane glycoprotein, and the activation of the fusogenic potential involves the cleavage of this accessory protein (11, 12, 18). X-ray crystallography of two class II fusion proteins, the E protein of the flavivirus tick-borne encephalitis (TBE) virus (24) and the E1 protein of the alphavirus Semliki Forest virus (SFV) (21), has revealed a common overall fold for these proteins, which are structurally unrelated to class I viral fusion proteins, for which the influenza virus hemagglutinin is the prototype (5, 31).
The E protein of TBE virus forms flat head-to-tail homodimers (24) that are oriented parallel to the viral membrane and organized in an icosahedral lattice (9), another characteristic that flaviviruses have in common with alphaviruses (21, 23). At the mildly acidic pH at which membrane fusion occurs, the virion surface undergoes a concerted rearrangement involving a dissociation of the native E homodimers and a subsequent irreversible reorganization into a trimeric form (2, 29).
Full-length E dimers isolated by treating virions with detergent have been shown to undergo the low-pH-driven dimer-trimer transition in solution, but truncated forms lacking the membrane anchor and connecting stem region undergo the dissociation step without trimerizing. It has been proposed, based on deletion mapping, that a predicted α-helix extending from amino acids 401 to 413 in the proximal stem region is involved in trimer formation (3, 29), but its exact role in this process is not yet clear.
Although the low-pH-induced rearrangements of the TBE virus E protein have been studied fairly extensively, most studies so far have been carried out in the absence of membranes. In the present work, we used in vitro experiments to relate these changes to the first step of the fusion process, the binding of the E protein to a target membrane. We provide evidence that the low-pH-induced dissociation of the E homodimer during the dimer-to-trimer transition is necessary for the initial interactions of the virus with its target membrane and that, contrary to earlier predictions, membrane-bound E proteins can form stable homotrimers even when the stem-anchor region has been removed.
MATERIALS AND METHODS
Virus growth and purification.
TBE virus strain Neudoerfl was grown in primary chicken embryo cells, harvested 48 h after infection, and purified by 2 cycles of sucrose density gradient centrifugation as described elsewhere (13).
Preparation of sE dimers.
Truncated soluble E dimers (sE dimers) were generated by limited trypsin digestion of purified virions at 0°C. Residual particles were removed by ultracentrifugation, and the purification of the sE dimers was performed by anion-exchange chromatography as described previously (14).
Liposomes.
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (Avanti Polar Lipids, Inc., Birmingham, Ala.) and 1-cholesterol (Sigma) were mixed in a molar ratio of 1:1:1 from stock solutions in chloroform. The mixture was dried to a thin film with a rotary evaporator and then dried further in high vacuum for at least 2 h. The lipid film was hydrated in 10 mM triethanolamine (pH 8.0)-140 mM NaCl and subjected to 5 cycles of freeze-thawing, followed by 21 cycles of extrusion through two 200-nm-pore-size polycarbonate membranes with a Liposofast syringe-type extruder (Avestin, Ottawa, Canada).
Coflotation of virions with liposomes.
Virions were mixed with liposomes in a ratio of 1 μg of E protein to 300 nmol of lipid and incubated for 5 min at 37°C. The mixture was acidified with 150 mM morpholineethanesulfonic acid (MES), incubated for 10 min at pH 5.5 and 37°C, back-neutralized with 150 mM triethanolamine, adjusted to a final volume of 2 ml of 20% (wt/wt) sucrose in TAN buffer (0.05 M triethanolamine [pH 8.0], 0.1 M NaCl), and layered onto a 1-ml 50% (wt/wt) sucrose cushion. The gradients were completed with a layer of 1 ml of 5% (wt/wt) sucrose. Centrifugation was carried out for 2 h at 50,000 rpm at 4°C in a Beckman SW55 rotor, and fractions of 200 μl were collected by upward displacement with an ISCO model 640 fraction collector. The amount of E protein in each fraction was determined by a quantitative four-layer enzyme-linked immunosorbent assay after denaturation of the samples with 0.4% sodium dodecyl sulfate (SDS) (15).
Coflotation of sE dimers with liposomes.
sE dimers were mixed with liposomes in a ratio of 1 μg of sE protein to 15 nmol of lipid and incubated for 5 min at 37°C. The samples were acidified with 300 mM MES, incubated for 30 min at 37°C at pH 5.5, back-neutralized, and adjusted to a final volume of 0.4 ml of 20% (wt/wt) sucrose as described above. The 0.4-ml sE protein-liposome mixture was applied to a 50% cushion and overlaid with 1.6 ml of 15% (wt/wt) sucrose and 1 ml of 5% (wt/wt) sucrose. Acidic step gradients were made in the same way with MES buffer (0.05 M MES [pH 5.5], 0.1 M NaCl) instead of TAN buffer (pH 8.0). Centrifugation was carried out as described above for coflotation of virus with liposomes. For the inhibition of coflotation by monoclonal antibodies (MAbs), the sE protein (final concentration, 50 μg/ml) was incubated with E protein-specific MAbs (final concentration, 100 μg/ml) for 30 min at pH 8.0 or 5.5 and room temperature before the addition of liposomes.
Chemical cross-linking of sE dimers with EGS and cleavage of EGS with hydroxylamine.
For coflotation experiments with cross-linked sE dimers, the samples were incubated for 30 min at room temperature in the presence of 1 mM ethylene glycolbis(succinimidylsuccinate) (EGS) (Pierce). The reaction was stopped by the addition of Tris-HCl (pH 7.5) to a final concentration of 62.5 mM Tris. Cleavage of the cross-linker was achieved by incubation of the samples in the presence of 1 M hydroxylamine-HCl (Pierce) at pH 8.5 for 3 h at 37°C. The efficiency of cross-linking and cleavage was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (22). Before the samples were used for coflotation with liposomes, hydroxylamine was washed out by membrane filtration (Ultrafree 30K; Millipore Corp, Bedford, Mass.) to a final concentration less than 0.3 mM.
Sedimentation analysis.
To analyze the oligomeric structure of E proteins that coflotated with the liposomes, the corresponding fractions were solubilized with 2% Triton X-100 for 1 h at room temperature. Then the samples were applied to 7-to-20% (wt/wt) continuous sucrose gradients in TAN buffer (pH 8.0) or MES buffer (pH 5.5) containing 0.1% Triton X-100, and centrifugation was carried out for 20 h at 38,000 rpm at 15°C in a Beckman SW40 rotor. The amount of E protein in the fractions was determined by a quantitative enzyme-linked immunosorbent assay as described above. As controls, whole virions or sE dimers were subjected to the same low-pH treatment as described for the coflotation assay, but in the absence of liposomes.
Chemical cross-linking with DMS, SDS-PAGE, and immunoblotting.
The E-protein-containing fractions from the sedimentation analyses were subjected to cross-linking with dimethylsuberimidate (DMS) as described earlier (2). The cross-linked samples were separated by electrophoresis on SDS-5% polyacrylamide gels with a phosphate-buffered system (22), blotted onto polyvinylidene difluoride membranes (Bio-Rad) with a Bio-Rad Trans-Blot semidry transfer cell, and detected and visualized immunoenzymatically as described previously (25).
RESULTS
Interactions of virions with liposomes.
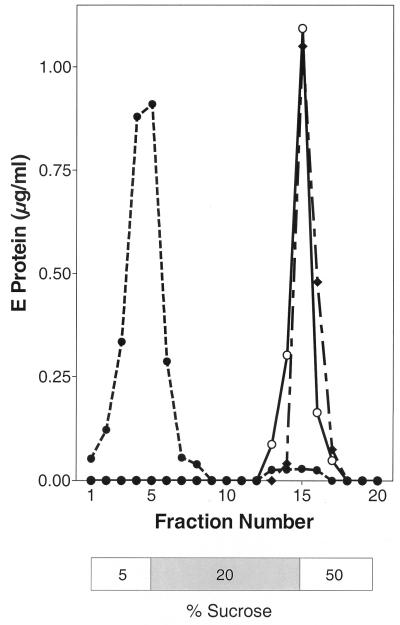
In order to investigate the interactions of TBE virus with target membranes at an acidic pH, we studied the association with liposomes by coflotation analysis. Native virions were exposed to pH 5.5 in the presence of liposomes, back-neutralized, and subjected to centrifugation in sucrose step gradients at pH 8.0. As a control, the virions were kept at pH 8.0 during the whole procedure. After centrifugation, the E proteins from native virions treated at low pH in the presence of target membranes were found at the top of the gradient together with the liposomes, whereas no association was observed in controls incubated at pH 8.0, indicating that this association was completely dependent on low pH (Fig. 1).
FIG. 1.
Liposome coflotation assay with three different samples of TBE virus. Native virions were incubated with liposomes for 10 min at pH 8.0 (○), incubated with liposomes for 10 min at pH 5.5 and readjusted to pH 8.0 (•), or preincubated for 10 min at pH 5.5 without a target membrane, back-neutralized, incubated for 10 min at pH 5.5 in the presence of liposomes, and readjusted to pH 8.0 (⧫). These samples were then analyzed by sucrose step gradient centrifugation at pH 8.0. The positions of the sucrose layers are shown below the curves; the 20% layer contains the virus-liposome mixture before centrifugation.
When virions were preexposed for 10 min to pH 5.5 in the absence of a target membrane, back-neutralized, mixed with liposomes, and then subjected to the same treatment as above, they were unable to stably associate with liposomes and instead sedimented to the 20-to-50% interface of the gradient, similar to the pH 8.0 control (Fig. 1). This indicates that the target membrane must be present at the time of acidification for membrane association to occur. The same results were obtained when samples were not back-neutralized and the coflotation analysis was done at pH 5.5 (data not shown).
Binding of truncated soluble E proteins (sE) to liposomes.
Coflotation experiments using virions whose membranes had been metabolically labeled with fluorescent lipids (8) revealed that not only had the virions in the top fractions attached to the liposomes but also, as expected, membrane fusion had occurred (data not shown). Therefore, to experimentally uncouple the attachment step from the viral membrane fusion event, we carried out similar coflotation experiments with sE instead of whole virions. The sE protein, which was used previously for structure determination by X-ray crystallography (24), is a homodimer that lacks the approximately 100 C-terminal amino acids comprising the stem-anchor region (14). It was shown in an earlier study that the sE dimers, when acidified in the absence of membranes, dissociate into monomers without forming trimers and that back-neutralization restores the original dimeric state (29).
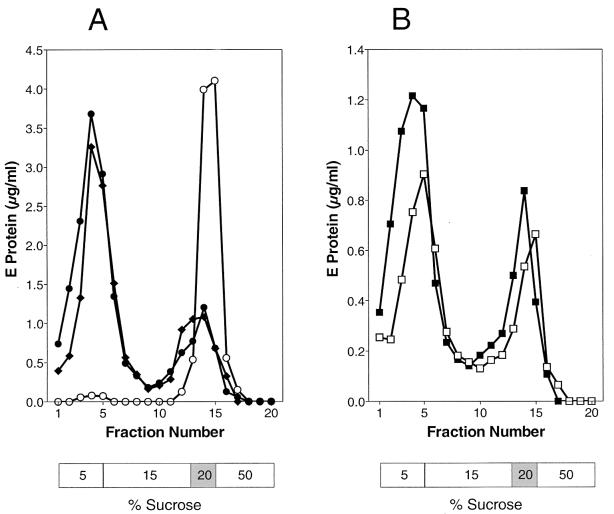
sE dimers were incubated at pH 8.0 or 5.5 in the presence of liposomes and subjected to centrifugation in step gradients at the corresponding pH. As shown in Fig. 2A, the dimeric sE proteins were unable to bind to liposomes at pH 8.0, but at pH 5.5 about 70% of the isolated sE proteins were able to stably attach to liposomes. When the centrifugation step was carried out at pH 8.0 rather than pH 5.5, no difference in the coflotation of sE was observed (Fig. 2A), indicating that the binding was irreversible.
FIG. 2.
Coflotation with liposomes of sE protein (A) and sE that had already undergone a cycle of pH-dependent dissociation and reassociation (B). (A) sE was incubated with liposomes at pH 8.0 for 30 min and analyzed by sucrose step gradient centrifugation at pH 8.0 (○), incubated with liposomes at pH 5.5 for 30 min and analyzed by sucrose step gradient centrifugation at pH 5.5 (⧫), or incubated with liposomes at pH 5.5 for 30 min, back-neutralized, and then subjected to centrifugation in sucrose step gradients at pH 8.0 (•). (B) sE was pretreated for 10 min at pH 5.5 without liposomes (dissociation), back-neutralized (reassociation), incubated for 30 min at pH 5.5 in the presence of liposomes, and analyzed by sucrose step gradient centrifugation at pH 8.0 (□); alternatively, sE was incubated with liposomes at pH 5.5 for 30 min, back-neutralized, and then subjected to centrifugation in sucrose step gradients at pH 8.0 (▪). The positions of the sucrose layers are shown below the curves; the 20% layer contains the sE-liposome mixture before centrifugation.
In the case of whole virions, preincubation at low pH induces an irreversible trimerization of E (2) and in the previous experiments led to a complete loss of the capacity to bind to liposomes. The sE protein, on the other hand, reverts to the dimeric form when back-neutralized (29). To see whether this reassociation also restores membrane binding activity, a set of experiments similar to those described above (Fig. 1) was carried out with sE dimers that had already been preexposed to low pH and then back-neutralized. In striking contrast to what was observed with whole virions, the preexposure of the sE dimer for 10 min to pH 5.5 did not destroy its membrane-binding potential. When these samples were reacidified in the presence of liposomes, approximately the same percentage of membrane-associated sE proteins was observed as with samples that had not been pretreated (Fig. 2B).
Effect of cross-linking on liposome binding.
The experiments described above suggest that the monomeric form of E is required for membrane interactions. To examine whether subunit dissociation is indeed a prerequisite for the association with target membranes, we carried out coflotation experiments with sE dimers that had been pretreated with the cleavable cross-linker EGS to prevent dissociation.
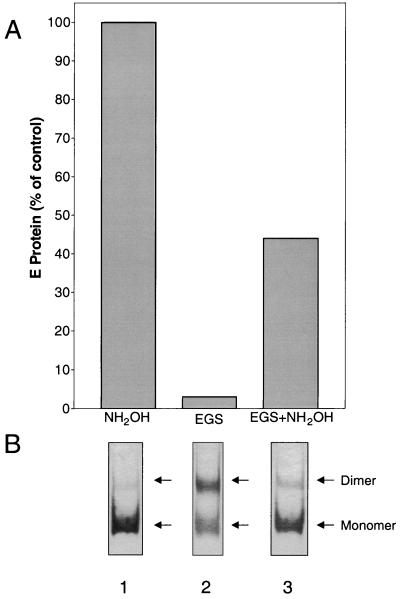
The results of these experiments are shown in Fig. 3. Treatment with EGS led to intersubunit cross-linking, as revealed by a strong band corresponding to a dimer in SDS-PAGE (Fig. 3B, lane 2). These cross-linked dimers were found to be strongly impaired in their ability to associate with liposomes (Fig. 3A). However, partial cleavage of the cross-linker with hydroxylamine, as demonstrated by a weakening of the dimer band and a corresponding increase in the intensity of the monomer band (Fig. 3B, lane 3), restored the ability of the sE proteins to interact with liposomes at low pH to 40 to 45% of control levels. This demonstrates that the ability of the dimers to dissociate is essential for forming stable interactions with the target membrane.
FIG. 3.
Effect of cross-linking with EGS on liposome binding. (A) Percentage of sE protein associated with liposomes after treatment with EGS and/or hydroxylamine compared to untreated controls. (B) SDS-PAGE of cross-linked samples. Lane 1, sE proteins not subjected to cross-linking but treated with the amount of hydroxylamine used to cleave the cross-linker (lane 3); lane 2, sE proteins cross-linked with EGS; lane 3, sE proteins cross-linked with EGS and then treated with hydroxylamine to cleave the cross-linker. The data are representative of four experiments.
Blocking of liposome binding by a fusion peptide-specific antibody.
It is likely that the stable association of the sE proteins with liposomes is the result of specific interactions between the membrane and the internal fusion peptide, which would be predicted to become exposed when the sE dimer dissociates (1). To obtain additional evidence as to whether the interactions of sE with liposomes occur via the fusion peptide, sE dimers were incubated with a MAb (A1) that specifically recognizes this region (1). Liposomes were then added, and the samples were acidified, back-neutralized, and analyzed by coflotation as described above. As shown in Fig. 4, MAb A1 completely inhibited the binding of sE dimers to liposomes, in contrast to a control MAb (C5) that binds to another region of the E protein (10). To exclude the possibility that A1 inhibits binding indirectly by obstructing the dissociation of the dimer, we also preincubated the sE proteins at low pH to dissociate them before adding MAb A1. As shown in Fig. 4, the antibody was able to block low-pH-induced liposome binding under these conditions as well.
FIG. 4.
Inhibition of liposome binding by a fusion peptide-specific MAb (A1). sE proteins were preincubated with MAb A1 at pH 8.0, acidified in the presence of liposomes, back-neutralized, and analyzed by step gradient centrifugation at pH 8.0 (○). Alternatively, sE proteins were pretreated at pH 5.5 and then incubated with MAb A1. After the addition of liposomes at the same pH, these samples were analyzed by step gradient centrifugation at pH 5.5 (•). As a control, sE proteins were incubated with MAb C5 at pH 8.0, acidified in the presence of liposomes, back-neutralized, and analyzed by step gradient centrifugation at pH 8.0 (⧫).
Oligomeric structure of liposome-associated E proteins.
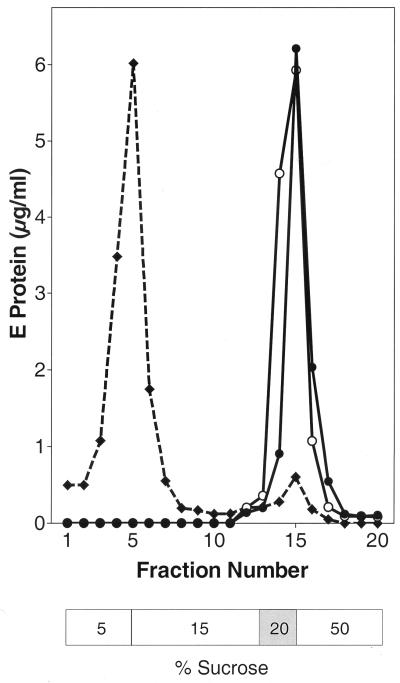
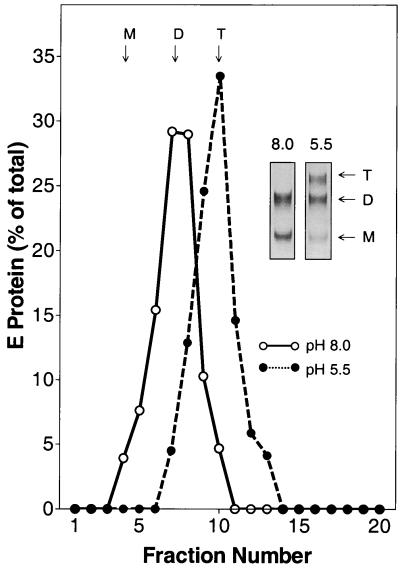
To determine the oligomeric state of full-length viral E or truncated sE after coflotation, the liposome-bound fractions were solubilized with Triton X-100 and subjected to sedimentation analysis (2, 29). As predicted from earlier studies, the solubilized full-length E from virions sedimented entirely in the fractions corresponding to a trimer, and this was confirmed by cross-linking with DMS (Fig. 5).
FIG. 5.
Oligomeric state of full-length E after attachment of TBE virus to liposomes. Whole TBE virions and liposomes were mixed at pH 8.0, exposed to pH 5.5, and subjected to coflotation as shown in Fig. 1. Bound virus was solubilized with 2% Triton X-100 and analyzed by sedimentation in 7 to 20% sucrose gradients containing 0.1% Triton X-100 (•). As a control, virions were incubated with liposomes at pH 8.0 and the E-protein-containing fractions were analyzed in the same way (○). The sedimentation direction is from left to right, and the positions of E monomer (M), dimer (D), and trimer (T) are indicated. (Insets) Cross-linking of the corresponding peak fractions with DMS.
When liposome-associated sE protein like that used for Fig. 2A was analyzed in the same way, we were surprised to find that these proteins were also homotrimeric (Fig. 6A). This was unexpected because earlier studies carried out in the absence of target membranes (and confirmed here in the control in Fig. 6B) had shown that the sE dimer dissociates but does not form trimers at low pH (29) and that trimerization in solution is dependent on the presence of a portion of the stem region that is absent in the sE protein (2). The stem region is therefore not absolutely required for trimer formation if the E ectodomains are bound to a target membrane.
FIG. 6.
Sedimentation analysis of sE protein after incubation at low pH in the presence (A) and absence (B) of liposomes. (A) Liposome-bound sE proteins (Fig. 2) were back-neutralized, solubilized with 2% Triton X-100, and centrifuged into 7-to-20% sucrose gradients in TAN buffer (pH 8.0) containing 0.1% Triton X-100 (•). As a control we used sE protein that had been incubated with liposomes at pH 8.0 (○). (Insets) SDS-PAGE of samples from the corresponding peak fractions cross-linked with DMS. (B) Samples were incubated at pH 5.5 (•) or pH 8.0 (○) in the absence of liposomes, solubilized with Triton X-100, and centrifuged into 7-to-20% sucrose gradients (containing 0.1% Triton X-100) at the corresponding pH. The sedimentation direction is from left to right, and the positions of E monomer (M), dimer (D), and trimer (T) are indicated.
DISCUSSION
The fusion of TBE virus with membranes is a process that requires a low-pH-triggered reorganization of the proteins of the viral envelope. In mature virions the E proteins form a metastable, presumably icosahedral network of laterally interacting homodimers that are quantitatively and irreversibly converted to a homotrimeric form when exposed to low pH (2). Earlier work (2, 28, 29) provided evidence that this is a two-step process involving a reversible, protonation-dependent dissociation of the dimers followed by an irreversible trimerization step.
The results of this study suggest that specific interactions with the target membrane probably occur after the dissociation step but before the trimerization step. This is based on the observations that (i) preventing the dissociation of the dimer with EGS blocked liposome binding, but binding activity could be restored by cleaving the cross-linker, and (ii) the final trimeric form of E on low-pH-treated virions was no longer able to mediate liposome coflotation at low pH, suggesting that the lipid-binding state is transient.
It is probable that the low-pH-induced dissociation of the E dimer results in the exposure of a specific element in the E protein whose function it is to bind the target membrane. Recently, using site-directed mutagenesis, we showed that a conserved region that includes Leu 107 and lies at the distal tip of each E protein monomer appears to be directly involved in membrane interactions during fusion (1). This “internal fusion peptide” is buried in the dimer interface (24) but would presumably become exposed during the low-pH-induced dissociation step. In this study we show that a MAb recognizing this region is able to block liposome coflotation with the sE protein, supporting the notion that membrane binding is mediated specifically by the internal fusion peptide. This MAb was also able to block fusion of whole virions with liposomes (data not shown). Interestingly, phase-partitioning experiments with Triton X-114 did not reveal a dramatic increase in the overall hydrophobicity of sE at acidic pH (data not shown), suggesting that the exposure of apolar residues in the monomeric low-pH form is rather limited and may involve only specific functional sites.
A surprising result of this study was that sE proteins that had bound to liposomes at low pH were no longer monomeric but instead had been irreversibly converted to stable trimers. This was unexpected because earlier in vitro experiments had shown that the sE protein does not trimerize at low pH in the absence of target membranes (29), and subsequent deletion mapping studies revealed that this was attributable to the lack of a putative α-helix (amino acids 401 to 413) in the proximal stem region (3). It now appears that contacts between the ectodomains are sufficient to stabilize a homotrimeric form of the E protein but that “facilitators”—either the stem-anchor region or target membranes—are required for the trimerization process. Both could be involved in interactions that lead to an increase of the local concentration of correctly aligned monomers and thereby facilitate trimer assembly. The induction of oligomerization upon lipid binding has also been described for other membrane-interacting proteins such as the matrix protein of Ebola virus (26) and several bacterial pore-forming toxins (reviewed in reference 4). Alternatively, the binding of E to the target membrane might induce structural alterations that lead to trimer formation, and these changes could be similar to ones induced by interactions involving the stem regions.
The TBE virus E protein has a number of functional properties that are similar to those of the fusion protein E1 of SFV and other alphaviruses (family Togaviridae) (18). Very recently it was shown that these proteins share a similar overall fold and are probably descended from a common ancestor (21). Unlike the TBE virus E protein, SFV E1 is not homodimeric in its native state but instead exists as a heterodimer with another glycoprotein, E2. The fusion peptide of SFV E1 is at a similar position at the tip of the monomer and is apparently buried in the E1-E2 interface (21). Exposure to low pH results in the dissociation of the heterodimer and irreversible formation of E1 homotrimers (reviewed in reference 18). It therefore appears likely that the fusion mechanism used by alphaviruses is very similar to that used by flaviviruses.
It was shown earlier that low-pH-induced fusion, but not membrane binding, by SFV can be inhibited by zinc ions (7). Under these conditions the E1-E2 heterodimer was shown to dissociate but not go on to form trimers (7). Furthermore, a nonfusogenic mutant of SFV that was able to dissociate but not to form trimers retained its ability to bind to liposomes at low pH (19). This would suggest that the initial membrane binding step with both TBE virus and SFV requires subunit dissociation but not trimerization. Also, similar to what was observed in this study with the TBE virus sE protein, it has been shown that a C-terminally truncated monomeric form of the SFV E1 protein is able to trimerize at low pH when bound to liposomes, but not when membranes are absent (20).
The class II fusion proteins of alphaviruses and flaviviruses share properties that are quite distinct from those of class I viral fusion proteins, suggesting that these two classes use different mechanisms to achieve membrane fusion. The finding that soluble sE dimers can be converted into a trimeric form by acidification in the presence of liposomes should facilitate the generation of trimeric proteins that might be suitable for structural studies and thereby could provide a basis for investigating the three-dimensional structure of the low-pH form of a class II viral fusion protein.
Acknowledgments
We thank Angela Dohnal and Walter Holzer for excellent technical assistance.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75**:**4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69**:**695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73**:**5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billington, S. J., B. H. Jost, and J. G. Songer. 2000. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol. Lett. 182**:**197-205. [DOI] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371**:**37-43. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93**:**681-684. [DOI] [PubMed] [Google Scholar]
- 7.Corver, J., R. Bron, H. Snippe, C. Kraaijeveld, and J. Wilschut. 1997. Membrane fusion activity of Semliki Forest virus in a liposomal model system: specific inhibition by Zn2+ ions. Virology 238**:**14-21. [DOI] [PubMed] [Google Scholar]
- 8.Corver, J., A. Ortiz, S. L. Allison, J. Schalich, F. X. Heinz, and J. Wilschut. 2000. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269**:**37-46. [DOI] [PubMed] [Google Scholar]
- 9.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7**:**593-602. [DOI] [PubMed] [Google Scholar]
- 10.Guirakhoo, F., F. X. Heinz, and C. Kunz. 1989. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology 169**:**90-99. [DOI] [PubMed] [Google Scholar]
- 11.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55**:**231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinz, F. X., and S. L. Allison. 2001. The machinery for flavivirus fusion with host cell membranes. Curr. Opin. Microbiol. 4**:**450-455. [DOI] [PubMed] [Google Scholar]
- 13.Heinz, F. X., and C. Kunz. 1981. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. J. Gen. Virol. 57**:**263-274. [DOI] [PubMed] [Google Scholar]
- 14.Heinz, F. X., C. W. Mandl, H. Holzmann, C. Kunz, B. A. Harris, F. Rey, and S. C. Harrison. 1991. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol. 65**:**5579-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198**:**109-117. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12**:**627-661. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez, L. D., and J. M. White. 1998. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J. Virol. 72**:**3259-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kielian, M., P. K. Chatterjee, D. L. Gibbons, and Y. E. Lu. 2000. Specific roles for lipids in virus fusion and exit. Examples from the alphaviruses. Subcell. Biochem. 34**:**409-455. [DOI] [PubMed] [Google Scholar]
- 19.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134**:**863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68**:**6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus. An icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105**:**137-148. [DOI] [PubMed] [Google Scholar]
- 22.Maizel, J. V., Jr. 1971. Polyacrylamide gel electrophoresis of viral proteins. Methods Virol. 5**:**179-246. [Google Scholar]
- 23.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105**:**127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375**:**291-298. [DOI] [PubMed] [Google Scholar]
- 25.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70**:**4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scianimanico, S., G. Schoehn, J. Timmins, R. H. Ruigrok, H. D. Klenk, and W. Weissenhorn. 2000. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 19**:**6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95**:**871-874. [DOI] [PubMed] [Google Scholar]
- 28.Stiasny, K., S. L. Allison, C. W. Mandl, and F. X. Heinz. 2001. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 75**:**7392-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiasny, K., S. L. Allison, A. Marchler-Bauer, C. Kunz, and F. X. Heinz. 1996. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 70**:**8142-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16**:**3-9. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289**:**366-373. [DOI] [PubMed] [Google Scholar]