Biological Significance of a Human Enterovirus B-Specific RNA Element in the 3′ Nontranslated Region (original) (raw)
Abstract
The secondary structures predicted for the enteroviral 3′ nontranslated region (3′NTR) all seem to indicate a conformation consisting of two (X and Y) hairpin structures. The higher-order RNA structure of the 3′NTR appears to exist as an intramolecular kissing interaction between the loops of these two hairpin structures. The enterovirus B-like subgroup possesses an additional stem-loop structure, domain Z, which is not present in the poliovirus-like enteroviruses. It has been suggested that the Z domain originated from a burst of short sequence repetitions (E. V. Pilipenko, S. V. Maslova, A. N. Sinyakov, and V. I. Agol, Nucleic Acids Res. 20:1739-1745, 1992). However, no functional features have yet been ascribed to this enterovirus B-like-specific RNA element in the 3′NTR. In this study, we tested the functional characteristics and biological significance of domain Z. A mutant of the cardiovirulent coxsackievirus group B3 strain Nancy which completely lacked the Z domain and which therefore acquired enterovirus C-like secondary structures exhibited a wild-type growth phenotype, as determined by single-cycle growth analysis with BGM cells. This result proves that the Z domain is virtually dispensable for viral growth in tissue cultures. Partial distortion of the Z domain structure resulted in a disabled virus with reduced growth kinetics, probably due to alternative conformations of the overall structure of the domain. Infection of mice showed that the recombinant coxsackievirus group B3 mutant which completely lacked the Z domain was less virulent. Pancreatic tissues from mice infected with wild-type virus and recombinant virus were equally affected. However, the heart tissue from mice infected with the recombinant virus showed only slight signs of myocarditis. These results suggest that the enterovirus B-like-specific Z domain plays a role in coxsackievirus-induced pathogenesis.
Enteroviruses are members of the picornavirus family, a large and diverse group of small RNA viruses. According to the present classification (10), the enterovirus genus comprises the following species: poliovirus, human enterovirus A (HEV-A) (coxackie A viruses and enterovirus 71), HEV-B (coxsackie B viruses, echoviruses, coxsackie A9 virus, and enteroviruses 69 and 73), HEV-C (coxsackie A viruses), HEV-D (enteroviruses 68 and 70), and at least three animal enterovirus species (bovine, simian, and porcine enteroviruses). They all contain a genome of approximately 7,500 bases and positive [(+)]-strand polarity. After infection of the host cell, the genome is translated in a cap-independent manner into a single polyprotein, which is subsequently processed by virus-encoded proteases into the structural capsid proteins and the nonstructural proteins, which are mainly involved in the replication of the virus (36).
The coding region is flanked by 5 and 3′ nontranslated regions (5′NTR and 3′NTR, respectively). The highly structured 5′NTR contains the internal ribosome entry site directing cap-independent viral translation initiation (25). The cloverleaf structure at the 5′ end of this region is a multifunctional _cis_-acting replication element which interacts with viral and cellular proteins to form a ribonucleoprotein complex. The cloverleaf structure is involved in (i) mediation of the switch from viral translation to replication (4), (ii) initiation of viral negative [(−)]-strand RNA synthesis by induction of a circular conformation of the genome (7), (iii) uridylylation of the viral protein VPg (15), and (iv) enhancement of the translation efficiency of the internal ribosome entry site region (38). The viral RNA-dependent RNA polymerase 3D catalyzes both (+)-strand and (−)-strand RNA syntheses. The initiation of (−)-strand and (+)-strand RNA syntheses commences at the 3′ ends of the respective (+)-strand and (−)-strand. Since the 3′ ends of the enteroviral complementary strands are dissimilar, the replication machinery should be able to recognize two different types of _cis_-acting elements to initiate (−)-strand and (+)-strand syntheses (1).
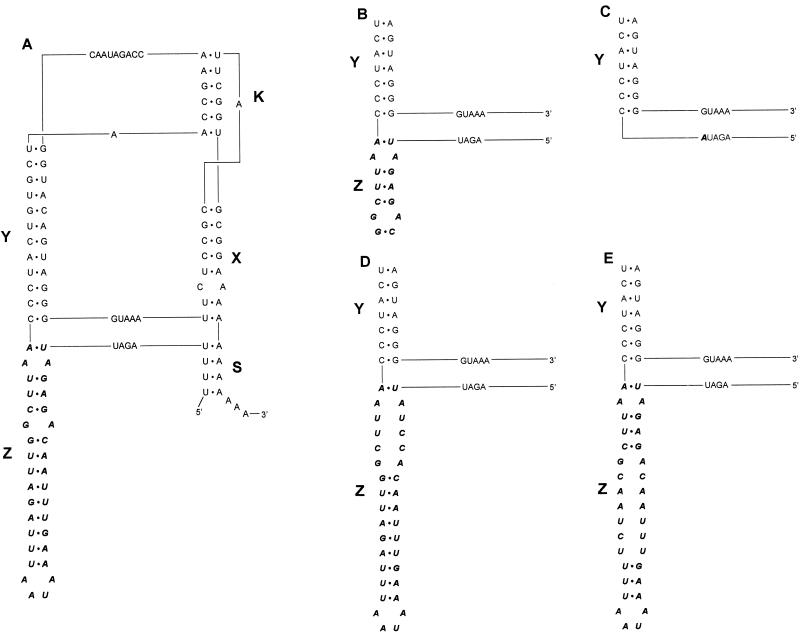
The enteroviral genome is a single-stranded RNA molecule containing a genetically encoded poly(A) tail at its 3′ terminus. Obviously, this ubiquitous homopolymeric stretch cannot serve to direct the RNA-dependent RNA polymerase to the 3′NTR to initiate viral (−)-strand synthesis. Since the viral replication machinery is known to exhibit a remarkable specificity for virus-specific RNAs, it is natural to assume that specified structures should contain heteropolymeric elements conserved among related viruses. The enteroviral 3′NTR folds into two common stem-loop structures (domains X and Y) in which the genetically encoded poly(A) tract is partly included (see Fig. 2A) (26). It was previously shown that sequences within the loop structure of domain X base pair with complementary sequences in the loop structure of domain Y to form a tertiary intramolecular kissing interaction in all enteroviruses analyzed thus far (20, 23, 27, 35). The kissing domain (K domain) can be stacked with domains X and S to form one tertiary superhelical domain connected to the Y domain by a small bridge of 5 nucleotides (see Fig. 2A). The spatial interdependence of the two helical domains, X and Y, is determined by the kissing interaction (21). This interaction may be involved in the specific recognition of ligands (proteins and nucleic acids) which play a role in the regulation of the initiation of viral (−)-strand RNA synthesis (30). The 3′NTR of HEV-B, however, possesses an additional stem-loop structure, the Z domain, which can be stacked with the Y domain to form a second superhelical domain. Pilipenko et al. (26) previously proposed that the entire Z domain could be regarded as being composed of three direct incomplete repeats.
FIG. 2.
Tertiary structure models displaying the Z domain mutants. The tertiary structure models represent wild-type CVB3 (A), CVB3 Z domain deletion mutants (B and C), and CVB3 Z domain distortion mutants (D and E). The HEV-B 3′NTR folds into a conformation containing three stem-loop structures (20, 27) designated X, Y, and Z. Also, a helical structure, designated S, can be formed that closes the 3′NTR. This domain results from an interaction between the most distal U stretch of the coding region and the poly(A) tail. An interaction between the loops of the X and Y domains results in a higher-order RNA structure termed the K domain. The HEV-B-specific domain Z is depicted in italic characters.
The Z domain is phylogenetically well conserved, which argues for a biological and even an enterovirus B-like-specific significant function (Fig. 1). To investigate the biological significance of the Z domain, we constructed a coxsackievirus group B3 (CVB3) mutant lacking the entire Z domain (CVB3-ΔZ) and CVB3 mutants in which the Z domain was distorted. The recombinant viruses obtained were analyzed for their growth properties in vitro, and the pathogenicity of the recombinant CVB3-ΔZ mutant was determined in vivo.
FIG. 1.
Alignment of HEV-B-specific Z domains. The sequence homology for the HEV-B Z domains is highlighted in grey. The consensus sequence is depicted at the bottom. CV, coxsackievirus; SVDV, swine vesicular disease virus; Echo, echovirus; EV, enterovirus.
MATERIALS AND METHODS
Cells and viruses.
Virus propagations and RNA transfections were performed with BGM cells grown in minimal essential medium supplemented with 10% fetal bovine serum (GIBCO). After transfection, minimal essential medium containing 10% fetal bovine serum was added to the BGM cells. Virus yields were determined by end-point titration with eight replicates of serial 10-fold dilutions in 96-well plates containing BGM cell monolayers (34). The 50% tissue culture infective dose (TCID50) was calculated as described by Reed and Muench (29).
Site-directed mutagenesis.
A full-length copy of the DNA of cardiovirulent CVB3 (pCB3/T7) was cloned behind a T7 RNA polymerase promoter (12). For oligonucleotide-directed mutagenesis, CVB3 was subcloned into phagemid pALTER-1 (20), and mutations were introduced by using an Altered Sites in vitro mutagenesis system (Promega) according to the recommendations of the manufacturer. Synthetic oligonucleotides (Biolegio, Malden, The Netherlands) were used to introduce site-specific mutations (Table 1). The mutated fragments were cloned into the infectious cDNA clone, and the nucleotide sequence of the mutant cDNA was verified by sequence analysis as previously described (20).
TABLE 1.
Mutations and oligonucleotide sequences
| Mutation | Oligonucleotide sequence |
|---|---|
| Partial Z- domain deletion | 5′-CCGTTATCTGGTTCGGTTAGCACAGTAGGGTTAAGCCGTCTCTAATCTAAAAG GAGTCCAACCACTTCCTGCG-3′ |
| CV B3-ΔZ mutation | 5′-CCGTTATCTGGTTCGGTTAGCACAGTAGGGTATCTAAAAGGAGTCCAACC ACTTCCTGCG-3′ |
| Upper distortion of the CVB3 Z domain | 5′-GTTAAGCCAATCTAAATTATTTCAAATTAAGGATAATCTAAAAGGAGTCCAAC CACTTCCTGCGTA-3′ |
| Lower distortion of the CVB3 Z domain | 5′-TCTGGTTCGGTTAGCACAGTAGGGTTAAGCGTTAGAAAATTATTTCAAATTG TCTCTAATCTAAAA-3′ |
Transfection of cells with RNA transcripts.
Wild-type and mutant pCB3/T7-derived plasmids were linearized by digestion with _Sal_I and transcribed in vitro with T7 RNA polymerase as described previously (20). Cells were transfected in duplicate with 4 μg of RNA by the DEAE-dextran method (20). The cells were grown at 36°C. The cells were incubated until the cytopathic effect (CPE) was complete. Upon CPE completion, the cultures were subjected to three cycles of freezing and thawing, and the viruses were stored at −80°C.
Single-cycle growth analysis.
Confluent BGM cell monolayers were infected with virus at a multiplicity of infection of 1 TCID50 per cell and grown at 33, 36, and 39°C for 2, 4, 6, and 8 h (20). Each growth curve was determined three times at each different temperature. Viruses were released by three successive cycles of freezing and thawing, and virus yields were determined with BGM cells by end-point titration with eight replicates of serial 10-fold dilutions in 96-well plates (34). The TCID50 was calculated as described by Reed and Muench (29).
Sequence analysis of mutant viruses.
Viral RNA extraction, cDNA synthesis, and PCR amplification with a poly(T) primer and a primer located in the 3D-coding region of the CVB3 genome (5′-GTTGTTTGACCCTCCCCGCG-3′) were performed as described previously (20). The resulting PCR products were purified from low-melting-temperature agarose, and the nucleotide sequence of the 3′NTR was determined by sequence analysis as described previously (20).
Mouse experiment design.
All animal procedures were officially approved in accordance with the German Animal Protection Law. To evaluate the influence of virus dose on lethality and the severity of myocarditis, groups of six female BALB/c mice were inoculated with 102, 103, 104, 105, and 106 TCID50s of cDNA-generated wild-type CVB3 and cDNA-generated CVB3-ΔZ. In a parallel experiment, groups of six A.CA mice were infected with 2 × 102, 2 × 103, 2 × 104, and 2 × 105 TCID50s of cDNA-generated wild-type CVB3 and cDNA-generated CVB3-ΔZ. The body weights of the mice were estimated daily. Either at day 14 or at day 30 postinfection (p.i.), the mice were exsanguinated under ether anesthesia; specimens of heart, pancreas, spleen, and blood were collected. The organ weights of the specimens were determined, and pancreas and heart tissues were used for histologic evaluations. The sera were checked for CVB3-specific antibodies by an enzyme-linked immunosorbent assay (ELISA).
Histologic analyses.
Heart and pancreas tissues were fixed in 6% formalin solution for at least 24 h and mounted in paraffin. For histologic examinations, 5-μm sections were stained with hematoxylin and eosin (HE). At least three adjacent sections were examined microscopically for the presence of lesions. For pancreas tissue, the necrosis of the exocrine parenchyma was scored according to the degree of damage (scores: 0, no lesions; 1, ≤10% lesions; 2, 10 to 40% lesions; 3, 40 to 80% lesions; and 4, 80 to 100% lesions). For heart tissue, histologic evidence of myocarditis and inflammation was classified in terms of the degree of cellular infiltration, myocardial cell necrosis, extent of calcification, and fibrosis and was scored on a scale of 0 to 4, as follows. Score 0 indicates no lesions. Score 1 (slight) describes rare groups of two or three mononuclear cells displacing myocytes, the presence of one or two small lesions (focal myocytic degeneration and/or necrosis; calcification of necrotic myocytes), and an insignificant reaction of interstitial connective tissue. Score 2 (mild) describes larger and more frequent mononuclear cell aggregates without confluence between adjacent foci, the presence of several small lesions scattered throughout the myocardium, and an increase in interstitial connective tissue with foci. Score 3 (medium) describes larger and more frequent mononuclear cell infiltrates with confluent foci present, multiple small lesions or several large lesions, and areas of myocyte disappearance replaced by dense stellate scars with tendril-like extensions into the surrounding myocardium. Score 4 (severe) describes many large foci of inflammatory cells, multiple large lesions, accentuated and heavily collagenized focal scars, and thickening of unscarred reticulum fibers.
Statistical comparisons were carried out with Microsoft Excel and Student's t test.
ELISA.
For the ELISA, microtiter plates were coated with CVB3-specific polyclonal rabbit antiserum (K3 anti CVB3; Institute for Virology, Jena, Germany) (0.1 μg per well [100 μl]) diluted in carbonate buffer (pH 9.6) at 37°C for 3 h. After incubation, the plates were washed with phosphate-buffered saline (PBS) and 0.05% Tween 20 and blocked with 200 μl of 1% bovine serum albumin in PBS at 37°C for 30 min. The plates were then washed and incubated with 50 μl of purified virus antigen (CVB3 after HeLa cell passages; precipitated in 1,1,2-trichlorotrifluoroethane; log10 TCID50, ≥8.0) or control antigen (noninfected HeLa cells precipitated in 1,1,2-trichlorofluoroethane) diluted in PBS-Tween 20 overnight at 4°C. The plates were then washed and incubated for 1 h at 37°C with 50 μl of mouse serum samples diluted 1:10,000 (immunoglobulin M [IgM]) or 1:5,000 (IgG and IgM) in PBS containing 1% bovine serum albumin. After additional washing, 100 μl of an appropriate dilution of peroxidase-labeled goat anti-mouse immunoglobulin (either IgG plus IgM or IgM) was added and incubated for 2 h at 37°C. After washing, the amount of peroxidase was determined by the addition of 200 μl of _o_-phenylenediamine dihydrochloride as a substrate, and the plates were incubated for 30 min in the dark at room temperature. The enzyme-substrate reaction was stopped with 50 μl of 2 M H2SO4 per well and measured at 490 nm. Each assay included reference positive serum. To establish the cutoff level for a positive value, sera from noninfected control mice were diluted and tested in the assay. The mean optical density value plus 2 standard deviations was determined, and an optical density value greater than the corresponding cutoff value indicated a positive reaction.
RESULTS
Structural requirements of the Z domain in vitro.
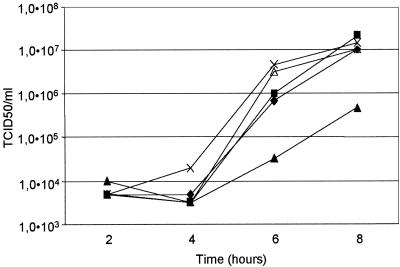
The Z domain is a specific well-conserved structural hairpin (Fig. 1 and 2A) which is most pronounced in the HEV-B species. Similar truncated domains may be present in other human and animal enteroviruses (39). So far, no function has been attributed to this putative secondary structure. Also, no evidence was found that the acquisition of this domain was accomplished through duplications of extended segments. It was suggested that the Z domain could have originated from a burst of short repetitions (26). To investigate the structural requirement of the Z domain for CVB3 replication, we used a deletion mutant in which only the upper part of the Z domain was retained (Fig. 2B) and a mutant which contained a complete deletion of the Z domain (Fig. 2C) and which therefore acquired a poliovirus-like genotype (CVB3-ΔZ). The mutations were verified by sequence analysis and introduced into the infectious CVB3 cDNA clone pCB3/T7. To study the effect of Z domain deletions on virus growth, BGM cells were transfected with RNA transcripts of the engineered constructs. CPE was observed upon transfection of both deletion mutants. Sequence analysis of the viruses obtained showed retention of the introduced mutations with no reversions in the 3′NTR. The growth characteristics of the viruses obtained were further analyzed by single-cycle growth analysis at 33, 36, and 39°C. Both mutant viruses exhibited growth characteristics similar to those of the wild-type virus (Fig. 3).
FIG. 3.
Single-cycle growth curves for the wild type and the Z domain mutants. BGM cell monolayers were infected with wild-type virus (♦) and mutant viruses (▪, upper distortion; ▴, lower distortion; ×, entire Z domain deletion; ▵, partial Z domain deletion) at a multiplicity of infection of 1 TCID50. The cells were grown at 36°C for 2, 4, 6, and 8 h. Virus titers were determined as described in Materials and Methods. Growth curves at 33 and 39°C showed similar growth kinetics.
The structural requirements of the Z domain for the maintenance of the overall structure of the CVB3 3′NTR were examined by the construction of two additional mutants in which either the upper part or the lower part of the Z domain was disrupted (Fig. 2D and E). Again, CPE was observed after transfection, and sequence analysis of the obtained viruses revealed no reversions.
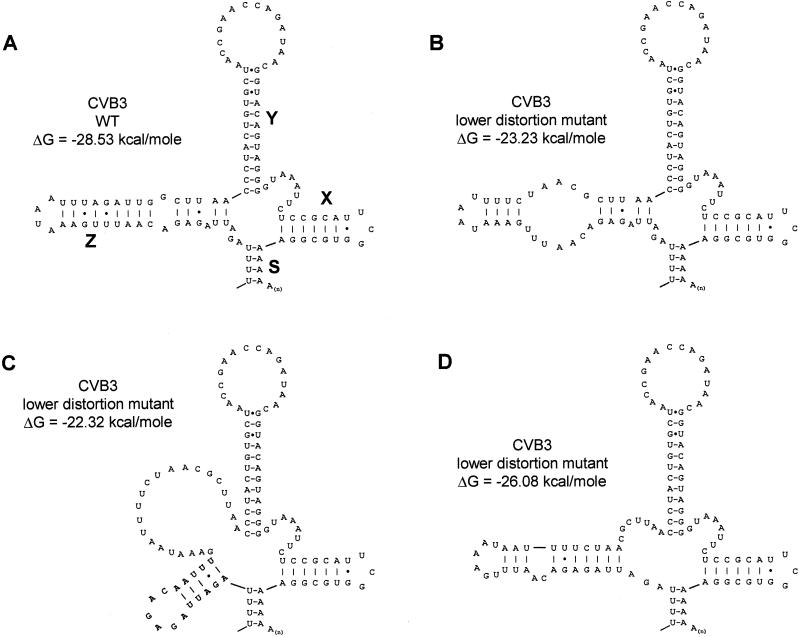
Single-cycle growth curves for BGM cells revealed that distortion of the upper part of the Z domain (Fig. 2D) resulted in a virus with growth characteristics similar to those of wild-type virus in BGM cells (Fig. 3). Distortion of the lower part of the Z domain (Fig. 2E), however, produced a disabled virus with a yield about 5% that of the wild-type virus at 8 h p.i. at all different temperatures (Fig. 3). M-Fold predictions (41) showed that disrupting the lower part of the Z domain resulted in alternative conformations of this domain, thereby altering the helical structure and terminal loop sequence (Fig. 4). These alternative configurations were not observed in the upper distortion mutant. These results could explain the negative effect of the lower distortion mutant on viral replication in BGM cells.
FIG. 4.
M-Fold structure predictions. Predicted RNA secondary structures for wild-type (WT) CVB3 (A) and the lower distortion mutant (B to D) are shown. Distortion of the Z domain could result in alternative conformations of the stem-loop structure with significantly reduced free energies (41).
Structural requirements of the Z domain in vivo.
To investigate whether virus-induced lethality was influenced by the deletion of the Z domain and/or the virus dose, BALB/c mice were inoculated with 104 and 105 TCID50s of cDNA-generated wild-type CVB3 and 104, 105, and 106 TCID50s of cDNA-generated CVB3-ΔZ. Mice were observed up to day 30 p.i. As shown in Fig. 5, a number of mice died during the acute phase of infection (from day 7 to day 17). While all animals infected with 105 TCID50s of wild-type virus died within 13 days, 50% of the mice infected with 105 TCID50s of CVB3-ΔZ survived. Even when inoculated with 106 TCID50s of CVB3-ΔZ, 35% of the infected mice still survived. The number of surviving mice increased when lower virus doses were used for infection. All surviving animals showed a loss of body weight (means, 5 g for wild-type CVB3-infected mice and 3.4 g for CVB3-ΔZ-infected mice) (Fig. 6A) and were seropositive, indicating CVB3-induced disease. Noninfected control mice were seronegative and showed an increase in body weight (mean, 1 g) (Fig. 6A).
FIG. 5.
Lethality of wild-type CVB3 and mutant CVB3-ΔZ for BALB/c mice. Groups of six female BALB/c mice were each infected with 104 (thin dashed line) and 105 (thin solid line) TCID50s of cDNA-generated wild-type CVB3 and 104 (dotted line), 105 (thick dashed line), and 106 (thick solid line) TCID50s of cDNA-generated mutant CVB3-ΔZ. The survival rates were determined daily.
FIG. 6.
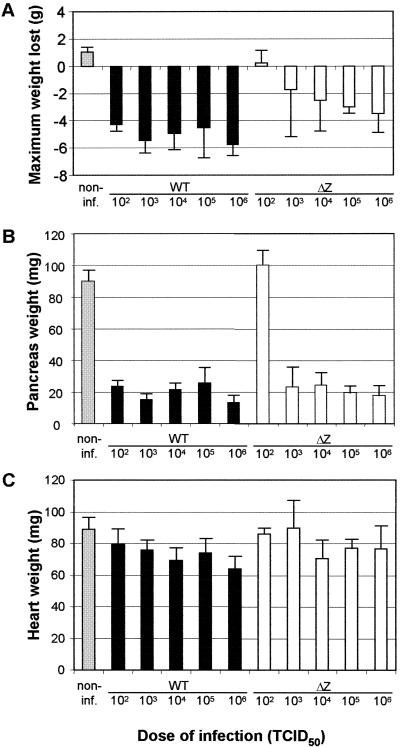
Body and organ weights. The data indicate the maximum weight loss during the observation period of 14 days (A) and the pancreas (B) and heart (C) weights at the end of the experiment. The body weights of all mice were determined daily. Black bars, wild-type (WT) CVB3; white bars, mutant CVB3-ΔZ; grey bars, noninfected (non-inf.) animals. Data are the means and standard deviations for three to five values.
Influence of virus dose on severity of disease.
The course of CVB3 infection was also studied with the mouse model. Upon intraperitoneal infection, the pancreas is one of the organs strongly affected by the virus. After replication in the exocrine parenchyma of the pancreas, the virus is spread via viremia to other organs, including the heart and spleen. During infection with highly myocarditic virus variants, mice usually exhibit a significant loss of body weight (up to 40%) (data not shown), which is the consequence of pancreas destruction. However, cDNA-generated CVB3 strain Nancy is considerably less virulent, as expressed by a less pronounced maximum body weight loss. Figure 6A shows that infection of BALB/c mice with cDNA-generated wild-type CVB3 induced a body weight loss ranging from 4 to 6 g at 102 to 106 TCID50s. CVB3-ΔZ induced a dose-dependent maximum weight loss: all mice infected with 102 TCID50 of this mutant exhibited a slight increase in body weight (mean, 0.2 g), while infections with higher doses of this mutant resulted in stepwise increasing weight loss up to 4 g (Fig. 6A).
The destruction of the pancreatic tissue is summarized in Fig. 6B and 7A. With wild-type CVB3, all infected mice showed a reduction in pancreas weight up to 80% (Fig. 6B). The histologic grades commonly used to describe pancreas destruction were greater than 3 (Fig. 7A). With CVB3-ΔZ, at least 103 TCID50s were necessary to establish an infection. Accordingly, no detectable histopathologic changes and a mean pancreas weight of 100 mg were observed in mice infected with 102 TCID50s of CVB3-ΔZ. At higher doses (103 to 106 TCID50s), the pancreas weight was reduced up to 80% and histopathologic changes scored greater than 3 as well.
FIG. 7.
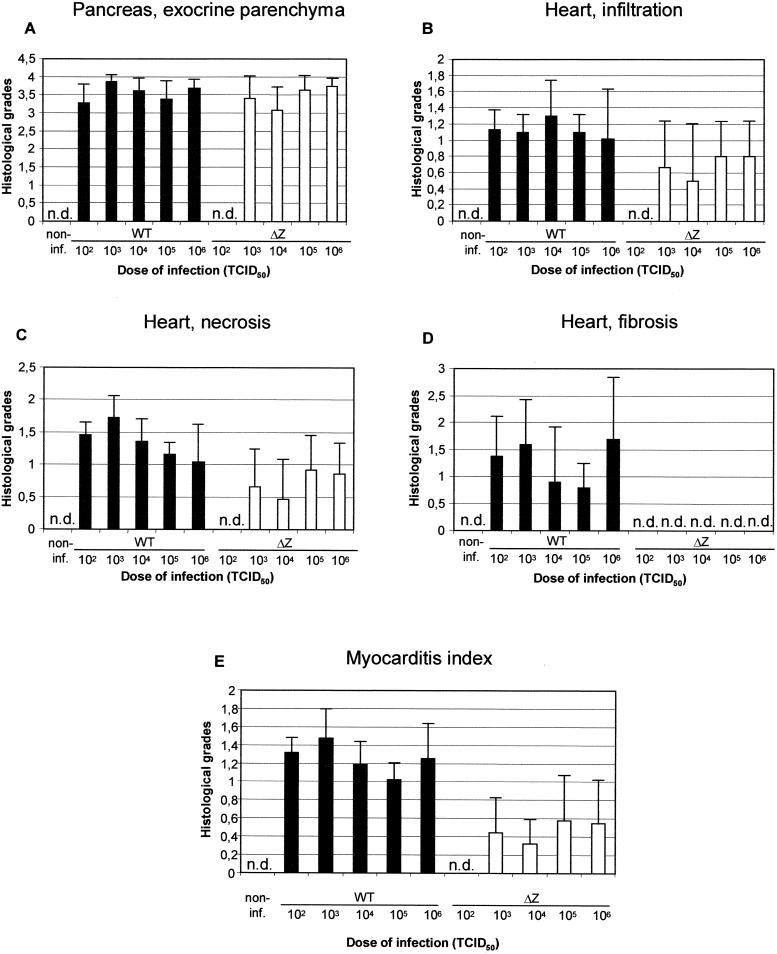
Histologic analyses. The histologic grades of pancreas destruction (A) and heart lesions (B, C, and D) were scored as described in Materials and Methods. The myocarditis index (E) is the mean of the values for cellular infiltration (B), necrosis (C), and fibrosis (D). Data are the means and standard deviations for three to five values. n.d., not detectable; non-inf., noninfected.
At day 14 p.i., no heart lesions were macroscopically visible and the heart weights of infected mice did not differ significantly from those of noninfected animals (Fig. 6C). However, histopathologic changes were evident (Fig. 7B to E and 8). Mice infected with wild-type CVB3 exhibited mild symptoms of myocarditis, i.e., mononuclear cell infiltration, myocyte necrosis, and fibrotic alterations (Fig. 8A and B). The myocarditis index, which represents the mean of the histopathologic scores of cellular infiltration, necrosis, and fibrosis, ranged from 1 to 1.5 (Fig. 7E) and was significantly higher than that in mice infected with CVB3-ΔZ. BALB/c mice infected with CVB3-ΔZ showed only slight myocarditis, i.e., single-myocyte necrosis adjacent to blood vessels and few infiltrating lymphocytes, and had no detectable fibrotic alterations (Fig. 8C and D). The myocarditis index ranged from 0.3 to 0.6. The replication pattern for CVB3-ΔZ-infected BALB/c mice indicated an abortive infection. Noninfected control animals showed no symptoms of myocarditis (data not shown).
FIG. 8.
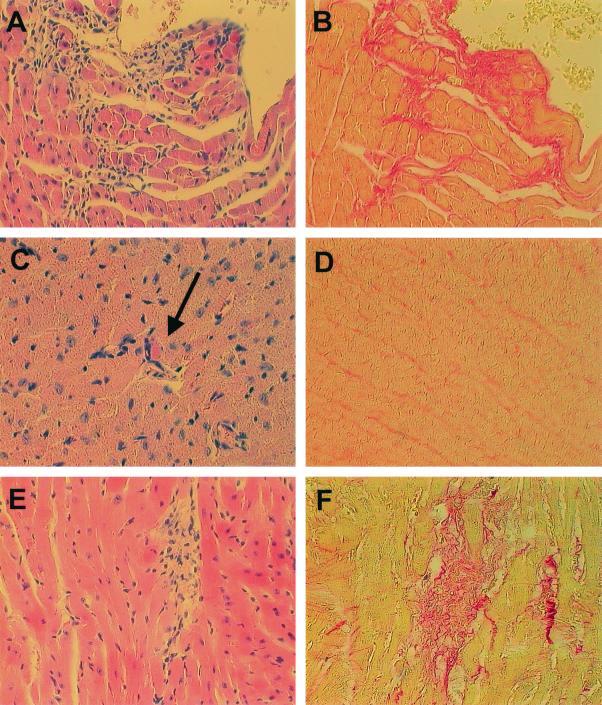
Histologic lesions in the hearts of virus-infected mice. BALB/c (A, B, C, and D) and A.CA (E and F) mice were infected with wild-type CVB3 (A and B) and CVB3-ΔZ (C, D, E, and F). At day 14 p.i., the mice were sacrificed and the hearts were removed for histologic examination. (A) An HE-stained section of a wild-type CVB3-infected mouse (myocarditis score, 1.6) shows numerous infiltrating lymphocytes (original magnification, ×200). (B) Considerable fibrosis in the same region of the heart is evident in a sirius red-stained section (original magnification, ×200). (C) An HE-stained section of a CVB3-ΔZ-infected mouse (myocarditis score, 0.7) shows one of few single necrotic myocytes (arrow) characterized by attraction of lymphocytes, loss of its association with the tissue, and destruction of intracellular fibrillar structures (original magnification, ∼×315). (D) No fibrosis can be detected in the same region of the heart in a sirius red-stained section (original magnification, ×200). (E and F) Lymphocyte infiltration (E) and fibrotic lesions (F) in the heart of an A.CA mouse infected with CVB3-ΔZ (myocarditis score, 1.4) (original magnifications, ∼×315).
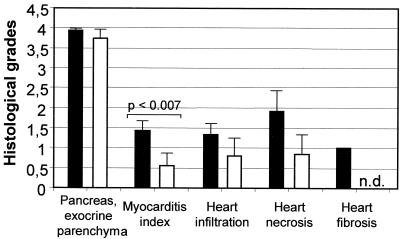
To demonstrate the dependence of virus-induced myocarditis on the host genetic background, the histopathologic scores of A.CA mice infected with 2 × 102 TCID50s of CVB3-ΔZ and BALB/c mice infected with 106 TCID50s of CVB3-ΔZ were compared. As shown in Fig. 8C to F and 9, the lesions of the hearts of CVB3-ΔZ-infected mice were generally more pronounced in A.CA mice than in BALB/c mice. The myocarditic lesions of CVB3-ΔZ-infected A.CA mice resembled those of wild-type virus-infected mice and indicated productive replication in the heart.
FIG. 9.
Dependence of CVB3-ΔZ-induced heart damage on host genetic background. A.CA mice (black bars) and BALB/c mice (white bars) were infected with 2 × 102 TCID50s and 106 TCID50s of CVB3-ΔZ, respectively. The histologic grades of pancreas destruction and heart lesions at day 14 p.i. were scored as described in Materials and Methods. The myocarditis index is the mean of the lesion scores for infiltration, necrosis, and fibrosis of the heart. Data are the means and standard deviations for three to five values. n.d., not detectable.
DISCUSSION
The enteroviral 3′NTR is a highly conserved heteropolymeric domain which contains two hairpin structures, the X and Y domains, both involved in the formation of a higher-order RNA structure, representing the intramolecular kissing interaction (20, 27). The precise function of the 3′NTR in viral replication still remains obscure. Point mutations introduced to destabilize the kissing interaction resulted in a lethal phenotype; therefore, the 3′NTR seemed of vital importance for viral replication (20, 35). However, mutants encompassing a complete deletion of the enteroviral 3′NTR appeared to be viable, albeit severely hampered in virus growth; therefore, the 3′NTR seemed to be dispensable for viral replication (31; unpublished results).
Agol et al. (1) suggested that the enteroviral 3′NTR is probably involved in the regulation of viral replication efficiency, possibly along different metabolic pathways. For example, regulation can be mediated by the efficient positioning of _cis_-acting replication element VPgpUpU (24) on the poly(A) tail to initiate viral (−)-strand synthesis and/or the interaction of helical elements within the 3′NTR with protein components of the replication machinery. The presence of an additional helical element in the 3′NTR, termed the Z domain, exemplifies the evolution of the 3′NTR into a number of foldings characteristic of each enterovirus species. The large Z domain may reflect an adaptation of HEV-B to the intracellular environment to regulate viral replication. Surprisingly, complete and partial genomic deletion mutants of the CVB3 Z domain did not impair virus growth or reduce replication kinetics in vitro. Likewise, a poliovirus mutant which harbored a complete CVB3 Z domain insertion into the poliovirus 3′NTR, thus generating an enterovirus B-like poliovirus construct, had wild-type poliovirus growth characteristics (unpublished results). This result proves that the Z domain is virtually dispensable for virus growth in tissue cultures. Disruption of the lower part of the Z domain structure resulted in a mutant virus with reduced growth characteristics compared to those of wild-type CVB3. Assuming that the overall tertiary structures of the 3′NTR, i.e., the formation of the kissing element (domain K) and the formation of putative helix S, are not disturbed (Fig. 2), the nucleotide substitutions in the lower distortion mutant (Fig. 4B) result in a significant decrease in free energy (−28.53 kcal/mol for wild-type CVB3 and −23.23 kcal/mol for the lower distortion mutant). Alternatively, configurations of the Z domain with altered helical structures and terminal loop sequences may have comparable free energies (Fig. 4C) or may be favored (Fig. 4D). As these alternative configurations were not observed in the upper distortion mutant, the changes in the Z domain structure could serve to explain the negative effect of the lower distortion mutant on replication in BGM cells.
In many respects, experimental CVB3 infection in the mouse results in lesions that resemble certain enterovirus-induced diseases in humans, such as myocarditis and pancreatitis (reviewed in references 28 and 37). Depending on the host genetic background and the virus strain used for experimental infection, CVB3 induces acute or chronic myocarditis which may heal or lead to death. Two pathogenic mechanisms of enteroviral heart disease have been described: (i) virus-induced dysfunction and cytolysis of infected myocytes and (ii) destruction of myocytes by a virus-stimulated immune process (8, 19). In an attempt to analyze the effect of the CVB3-ΔZ mutation on viral virulence, we used the immunologically well-characterized BALB/c strain, which was previously described as a model for acute CVB3-induced myocarditis with a low probability for the development of ongoing myocarditis. This mouse strain is characterized by a pronounced Th1 response and high-level antibody production (5, 6, 9). For comparison of the virulence of CVB3-ΔZ with that of the wild-type virus, histologic lesion scores for BALB/c mice were compared with those for A.CA mice. The latter mouse strain was used as a model for chronic myocarditis (11).
Following intraperitoneal infection of the mice, virus was detectable in the pancreas, liver, and feces as early as 12 h p.i. The virus strongly affected the pancreas. The disease was characterized by lysis of the acinar cells and severe inflammation, while pancreatic ducts and the islets of Langerhans appeared normal (22). We found that in BALB/c mice, infection of the pancreas led to significant weight loss, which was the result of almost complete destruction of the exocrine parenchyma. Compared to wild-type CVB3, CVB3-ΔZ was clearly less virulent: it required 10-fold higher doses (at least 103 TCID50s) than the wild-type virus to induce the generation of CVB3-specific antibodies (as indicated by seroconversion; data not shown) and to replicate in the pancreas. The replication of CVB3-ΔZ in the hearts of BALB/c mice appears to be abortive, as tiny lesions were observed only in the vicinity of capillaries (Fig. 8C). There were no indications of cell-to-cell spread, as is usually seen in wild-type CVB3-infected mice. Lesion scores in the heart were less severe and, most significantly, no fibrosis was induced. Moreover, CVB3-ΔZ-infected BALB/c mice exhibited significantly reduced maximum loss of body weight during the course of infection and a higher survival rate. A.CA mice developed more severe heart disease. This finding was expressed as higher lethality (data not shown) and as a significantly higher myocarditis index. Unlike BALB/c mice, A.CA mice did not exhibit restricted CVB3-ΔZ replication in the heart but showed productive infection (Fig. 8E and F). This finding could be the result of a difference in local immunity due to the different genetic backgrounds. More elaborate investigations are required to answer these questions.
Deletion of the Z domain in CVB3 did not result in alternative growth characteristics in BGM cells, implying that this specific domain has no apparent function in vitro. Surprisingly, BALB/c and AC.A mice infected with CVB3-ΔZ showed a reduction in virulence, implying an in vivo function for the Z domain. Tracy et al. (32) stated that the ability of CVB3 to cause pancreatitis and myocarditis is due to its capability for inducing high titers in the pancreas and not to its persistence. The reduced myocarditis index of CVB3-ΔZ may be due to (i) reduced replication levels in the pancreas, resulting in virus titers which are too low to induce severe myocarditis, or to (ii) normal replication levels in the pancreas but a reduced ability to replicate in the heart, possibly due to an altered tropism resulting in more rapid clearance of mutant virus than of wild-type CVB3 from the heart. No determination could be made as to whether CVB3-ΔZ was less able to induce dysfunction and cytolysis of infected myocytes or less able to cause destruction of myocytes by a virus-stimulated immune process. This discrepancy between wild-type growth kinetics in tissue cultures and reduced virulence has also been described for different mengovirus mutants (3, 18) and flavivirus mutants (17). Flavivirus mutants containing deletions in the highly conserved core element of the 3′NTR showed no reduction in viral growth kinetics in vitro but proved to be attenuated when inoculated in mice. Also, mengovirus mutants containing either a shortened poly(C) tract in the 5′NTR or a stem-loop I deletion in the 3′NTR showed growth characteristics similar to those of wild-type mengovirus in vitro but were clearly less virulent in mice (3). The fact that many animals survived an inoculation of the mengovirus stem-loop I deletion mutant led to the suggestion that the 3′NTR may play a role in cell tropism or induction of the host immune system. These suggestions are in line with our results indicating that a specific domain in the CVB3 3′NTR plays a role in viral virulence as well. Strong conservation of the Z domain throughout enterovirus B-like species argues for this stem-loop structure being a potential host cell protein recognition site.
In previous reports, the molecular basis of CVB3 attenuation was attributed to mutations of capsid protein VP2 (13) and the 5′NTR (14, 33). Although the major attenuating mutations of the Sabine vaccine strains of poliovirus have also been mapped to the 5′NTR (16), the significance of this genomic region in CVB3 attenuation is still under debate (2, 40). In this respect, deletion of a putative secondary structure of the 3′NTR introduces another genomic region which could serve as a possible candidate for the development of a safe vaccine strain.
Acknowledgments
Ingrid Merkle and Mark J. M. van Ooij contributed equally to this work.
We thank Birgit Meiβner, Birgit Schikowski, and Sabine Wachsmuth for excellent technical assistance.
This research was supported by grants from the Council of Chemical Sciences of The Netherlands Organization for Scientific Research (NWO-CW 98008) and the European Communities (INTAS 2012).
REFERENCES
- 1.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62**:**129-147. [DOI] [PubMed] [Google Scholar]
- 2.Chapman, N. M., J. R. Romero, M. A. Pallansch, and S. Tracy. 1997. Sites other than nucleotide 234 determine cardiovirulence in natural isolates of coxsackievirus B3. J. Med. Virol. 52**:**258-261. [PubMed] [Google Scholar]
- 3.Duque, H., and A. C. Palmenberg. 2001. Phenotypic characterization of three phylogenetically conserved stem-loop motifs in the mengovirus 3′ untranslated region. J. Virol. 75**:**3111-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA-virus. Genes Dev. 12**:**2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henke, A., S. Huber, A. Stelzner, and J. L. Whitton. 1995. The role of CD8+ T lymphocytes in coxsackievirus B3-induced myocarditis. J. Virol. 69**:**6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henke, A., R. Zell, G. Ehrlich, and A. Stelzner. 2001. Expression of immunoregulatory cytokines by recombinant coxsackievirus B3 variants confers protection against virus-caused myocarditis. J. Virol. 75**:**8187-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularisation through a protein-protein bridge. Mol. Cell 7**:**581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber, S. A. 1992. Viral myocarditis—a tale of two diseases. Lab. Investig. 66**:**1-3. [PubMed] [Google Scholar]
- 9.Huber, S. A., and B. Pfaeffle. 1994. Differential Th1 and Th2 cell responses in male and female BALB/c mice infected with coxsackievirus group B type 3. J. Virol. 68**:**5126-5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-673. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, Inc., New York, N.Y.
- 11.Klingel, K., C. Hohenadl, A. Canu, M. Albrecht, M. Seemann, G. Mall, and R. Kandolf. 1992. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc. Natl. Acad. Sci. USA 89**:**314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klump, W. M., I. Bergmann, B. C. Müller, D. Ameis, and R. Kandolf. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 64**:**1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowlton, K. U., E. S. Jeon, N. Berkley, R. Wessely, and S. A. Huber. 1996. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 70**:**7811-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, C., E. Maull, N. Chapman, S. Tracy, and C. Gauntt. 1997. Genomic regions of coxsackievirus B3 associated with cardiovirulence. J. Med. Virol. 52**:**341-347. [PubMed] [Google Scholar]
- 15.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and initiation of negative-strand RNA synthesis. J. Virol. 75**:**10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macadam, A. J., G. Ferguson, J. Burlison, D. Stone, R. Skuce, J. W. Almond, and P. D. Minor. 1992. Correlation of RNA secondary structure and attenuation of Sabin vaccine strains of poliovirus in tissue culture. Virology 189**:**415-422. [DOI] [PubMed] [Google Scholar]
- 17.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 72**:**2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, L. R., G. M. Duke, J. E. Osorio, D. J. Hall, and A. C. Palmenberg. 1996. Mutational analysis of the mengovirus poly(C) tract and surrounding heteropolymeric sequences. J. Virol. 70**:**2027-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McManus, B. M., L. H. Chow, J. E. Wilson, D. R. Anderson, J. M. Gulizia, C. J. Gauntt, K. Klingel, B. W. Beisel, and R. Kandolf. 1993. Direct myocardial injury by enterovirus: a central role in the evolution of murine myocarditis. Clin. Immunol. Immunopathol. 68**:**159-169. [DOI] [PubMed] [Google Scholar]
- 20.Melchers, W. J. G., J. G. J. Hoenderop, H. J. Bruins Slot, C. W. A. Pleij, E. V. Pilipenko, V. I. Agol, and J. M. D. Galama. 1997. Kissing of two predominant hairpin loops in the coxsackievirus B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71**:**686-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melchers, W. J. G., J. M. J. E. Bakkers, H. J. Bruins Slot, J. M. D. Galama, V. I. Agol, and E. V. Pilipenko. 2000. Cross-talk between orientation-dependent recognition determinants of a complex control RNA-element, the enterovirus _ori_R. RNA 6**:**976-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mena, I., C. Fischer, J. R. Gebhard, C. M. Perry, S. Harkins, and J. L. Whitton. 2000. Coxsackievirus infection of the pancreas: evaluation of receptor expression, pathogenesis, and immunopathology. Virology 271**:**276-288. [DOI] [PubMed] [Google Scholar]
- 23.Mirmomeni, M. H., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71**:**2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74**:**10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334**:**320-325. [DOI] [PubMed] [Google Scholar]
- 26.Pilipenko, E. V., S. V. Maslova, A. N. Sinyakov, and V. I. Agol. 1992. Towards identification of _cis_-acting elements involved in replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 20**:**1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilipenko, E. V., K. V. Poperechny, S. V. Maslova, W. J. G. Melchers, H. J. Bruins Slot, and V. I. Agol. 1996. _cis_-element, _ori_R, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing’) interactions. EMBO J. 15**:**5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsingh, A. I. 1997. Coxsackieviruses and pancreatitis. Frontiers Biosci. 2**:**53-62. [DOI] [PubMed] [Google Scholar]
- 29.Reed, L. J., and H. A. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27**:**493. [Google Scholar]
- 30.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69**:**7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71**:**8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tracy, S., K. Höfling, S. Pirruccello, P. H. Lane, S. M. Reyna, and C. J. Gauntt. 2000. Group B coxsackievirus myocarditis and pancreatitis: connection between viral virulence phenotypes in mice. J. Med. Virol. 62**:**70-81. [DOI] [PubMed] [Google Scholar]
- 33.Tu, Z., N. M. Chapman, G. Hufnagel, S. Tracy, J. R. Romero, W. H. Barry, L. Zhao, K. Currey, and B. Shapiro. 1995. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5′ nontranslated region. J. Virol. 69**:**4607-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Kuppeveld, F. J. M., J. G. Hoenderop, R. L. Smeets, P. H. Willems, H. B. Dijkman, J. M. D. Galama, and W. J. G. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16**:**3519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, J., J. M. J. E. Bakkers, J. M. D. Galama, H. J. Bruins Slot, E. V. Pilipenko, V. I. Agol, and W. J. G. Melchers. 1999. Structural requirements of the higher order RNA kissing element in the enteroviral 3′UTR. Nucleic Acids Res. 27**:**485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wimmer, E., C. U. T. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27**:**353-436. [DOI] [PubMed] [Google Scholar]
- 37.Woodruff, J. F. 1980. Viral myocarditis. Am. J. Pathol. 101**:**427-479. [PMC free article] [PubMed] [Google Scholar]
- 38.Zell, R., K. Sidigi, A. Henke, J. Schmidt-Brauns, E. Hoey, S. Martin, and A. Stelzner. 1999. Functional features of the bovine enterovirus 5′-non-translated region. J. Gen. Virol. 80**:**2299-2309. [DOI] [PubMed] [Google Scholar]
- 39.Zell, R., and A. Stelzner. 1997. Application of genome sequence information to the classification of bovine enteroviruses: the importance 5′- and 3′-nontranslated regions. Virus Res. 51**:**213-229. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, H. Y., G. E. Yousef, L. Cunningham, N. W. Blake, X. OuYang, T. A. Bayston, R. Kandolf, and L. C. Archard. 1993. Attenuation of a reactivated cardiovirulent coxsackievirus B3: the 5′-nontranslated region does not contain major attenuation determinants. J. Med. Virol. 41**:**129-137. [DOI] [PubMed] [Google Scholar]
- 41.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. NATO ASI Series. Kluwer Academic Publishers, Dordrecht, The Netherlands.