Role of the Hemagglutinin-Neuraminidase Protein in the Mechanism of Paramyxovirus-Cell Membrane Fusion (original) (raw)
Abstract
Paramyxovirus infects cells by initially attaching to a sialic acid-containing cellular receptor and subsequently fusing with the plasma membrane of the cells. Hemagglutinin-neuraminidase (HN) protein, which is responsible for virus attachment, interacts with the fusion protein in a virus type-specific manner to induce efficient membrane fusion. To elucidate the mechanism of HN-promoted membrane fusion, we characterized a series of Newcastle disease virus HN proteins whose surface residues were mutated. Fusion promotion activity was substantially altered in only the HN proteins with a mutation in the first or sixth β sheet. These regions overlap the large hydrophobic surface of HN; thus, the hydrophobic surface may contain the fusion promotion domain. Furthermore, a comparison of the HN structure crystallized alone or in complex with 2-deoxy-2,3-dehydro-_N_-acetylneuraminic acid revealed substantial conformational changes in several loops within or near the hydrophobic surface. Our results suggest that the binding of HN protein to the receptor induces the conformational change of residues near the hydrophobic surface of HN protein and that this change triggers the activation of the F protein, which initiates membrane fusion.
Viral envelope glycoproteins bind to specific cellular receptors and initiate fusion with the cell membrane, which allows the penetration of the viral genome into host cells. These two functions, binding and fusion, are mediated by one or multiple envelope glycoproteins. Influenza virus hemagglutinin, vesicular stomatitis virus G protein, and retrovirus envelope proteins are well-characterized viral glycoproteins that participate in attachment and fusion. Exposure to low pH after receptor binding triggers the conformational change of the glycoproteins, a process that is essential for membrane fusion (11, 22). In contrast, paramyxoviruses have two glycoproteins that mediate these functions: hemagglutinin-neuraminidase (HN) or hemagglutinin is responsible for binding, and the fusion (F) protein induces fusion (16). With the exception of some strains of simian virus 5 (SV5), paramyxovirus HN or hemagglutinin is generally required for F-protein-induced membrane fusion (14, 15).
Most paramyxovirus F proteins require HN protein from the homologous virus to induce membrane fusion, and substantial membrane fusion is observed only when HN and F proteins of the same virus or closely related viruses are expressed in the same cells (2, 12, 13, 17). Although F protein of some of the SV5 strains and mutant F protein of Newcastle disease virus (NDV) can induce cell fusion by themselves, coexpression of virus type-specific HN proteins substantially enhances the viruses' fusion activity (12, 21). These findings indicate that in addition to its attachment function, HN protein interacts with a virus type-specific F protein to bring about efficient membrane fusion. Results of coimmunoprecipitation experiments have suggested that there is a physical association between virus type-specific HN and F proteins (HN-F) at the cell surface (9, 24, 32). Furthermore, experiments using chimeric and mutant HN proteins have indicated that both the stalk and head regions of HN protein are involved in the specific interaction with the F protein that is required for fusion promotion (2, 3, 10, 28, 30). The occurrence of membrane fusion at neutral pH indicates that virus-cell fusion takes place at the surface of the cell. The conformational change of HN protein, which may be triggered by receptor binding, sends a signal to the F protein to initiate membrane fusion (15). Although HN′s crucial role in the process of infection is well established, the mechanism by which HN protein participates in membrane fusion is unknown.
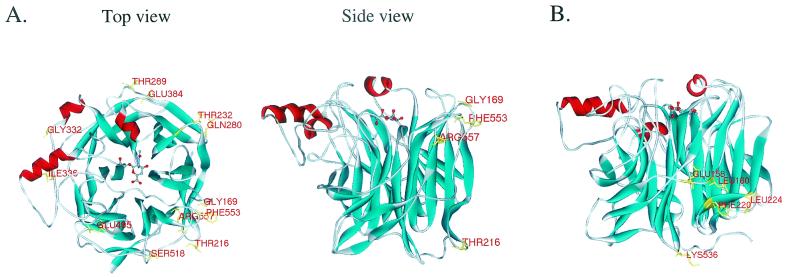
We recently crystallized the protease-cleaved soluble form of HN protein and reported its atomic structure (8). This structure has provided insights into the biological activities of the multifunctional HN protein and has allowed us to characterize structure-function relationships in detail. On the basis of the three-dimensional (3D) structure of the NDV HN, we substituted alanine for various residues on the surface of HN protein (Fig. 1A) to determine whether a particular surface region is involved in fusion promotion. NDV HN cDNA in expression vector pCAGGS (26) was mutated using the Transformer Site-Directed Mutagenesis Kit (Clontech). The mutant HNs were expressed in transfected HeLa T4+ cells, and their biological activities (neuraminidase [NA], hemadsorption [HAD], and fusion promotion) were determined. Cell surface expression of the mutant HNs were determined by enzyme-linked immunosorbent assay using a cocktail of HN-specific monoclonal antibodies (N1, N3, N6, and N7) as described previously (7). High levels of each mutant HN were expressed; the transfection efficiency (25 to 30%) and overall levels of each mutant expression did not differ substantially from that of wild-type HN (Table 1). To measure NA activity of mutant HN proteins, transfected cells were incubated in 0.2 M phosphate buffer (pH 5.9) containing _N_-acetylneuraminyl-lactose for 2 h at 37°C, and the amount of sialic acid in the solution was determined as reported previously (2). In most HN mutants, the NA activity was not significantly affected. Only the Phe553Ala mutation reduced the NA activity to about 30% of that of the wild-type HN; the NA activities of all other mutants were 78 to 123% of that of wild-type HN (Table 1). HAD activity was assayed by incubating the transfected cells with 2% (vol/vol) chicken red blood cells (RBCs) in phosphate-buffered saline on ice for 30 min. Unadsorbed RBCs were removed by repeated gentle washing with phosphate-buffered saline, bound RBCs were lysed by adding TNE buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA), and clarified supernatant was measured at a wavelength of 545 nm (7, 18). Although most of the mutations had little effect on HAD activity, there were some exceptions. The Phe553Ala and Arg557Ala mutations reduced HAD activity to approximately 20 to 30% of wild-type HN values. We expected this reduction in HAD activity, because residues 553 and 557 are located on or near the loop between the second and third strands of the sixth β sheet, which is close to the binding pocket of HN protein (Fig. 1A).
FIG. 1.
3D structure of NDV HN protein and locations of mutated residues. The mutations we characterized in the first (A) and second (B) series of experiments are shown in the structure of HN-Neu5Ac2en complex. The side view in panel A shows the locations of mutations that affected fusion promotion activity in the first series of analysis. The structures were generated using WebLab ViewerPro 3.5 (Molecular Simulations Inc.).
TABLE 1.
Biological activities of mutant HNs
| HN | Expressiona | HADb | NAb | Fusionc |
|---|---|---|---|---|
| WT | 100.0 | 100.0 | 100.0 | 22.1 ± 4.0 |
| Mutants | ||||
| G169A | 107.1 ± 27.8 | 87.9 ± 20.0 | 79.1 ± 9.1 | 36.0 ± 2.7 |
| T216A | 93.7 ± 6.3 | 53.6 ± 9.7 | 122.7 ± 8.2 | 1.9 ± 1.8 |
| T232A | 98.5 ± 0.2 | 113.3 ± 8.3 | 78.4 ± 14.1 | 15.6 ± 2.3 |
| Q280A | 107.7 ± 3.5 | 95.5 ± 10.7 | 87.6 ± 8.0 | 20.4 ± 3.3 |
| T289A | 114.8 ± 9.9 | 103.7 ± 13.0 | 99.7 ± 2.8 | 19.5 ± 2.1 |
| G332A | 103.1 ± 17.1 | 109.1 ± 21.3 | 92.8 ± 13.6 | 21.3 ± 3.1 |
| I336A | 104.3 ± 12.2 | 93.5 ± 18.8 | 107.1 ± 7.6 | 20.4 ± 5.5 |
| E384A | 118.1 ± 24.3 | 88.1 ± 5.9 | 99.0 ± 5.8 | 20.8 ± 2.9 |
| E495A | 113.4 ± 3.9 | 117.3 ± 19.6 | 113.4 ± 9.2 | 22.9 ± 3.0 |
| S518A | 136.2 ± 19.4 | 101.7 ± 18.6 | 79.9 ± 3.5 | 16.7 ± 2.2 |
| F553A | 93.7 ± 25.9 | 28.0 ± 3.1 | 30.3 ± 5.8 | 2.6 ± 1.1 |
| R557A | 91.4 ± 12.8 | 22.5 ± 1.5 | 80.9 ± 10.2 | 1.0 ± 0.9 |
| E158A | 69.7 ± 12.2 | 48.4 ± 16.1 | 116.7 ± 5.1 | 1.7 ± 0.8 |
| L160A | 84.5 ± 14.9 | 20.3 ± 8.4 | 129.7 ± 11.0 | 2.8 ± 0.2 |
| F220A | 79.7 ± 2.9 | 21.8 ± 5.8 | 87.2 ± 16.8 | 2.1 ± 1.3 |
| L224A | 74.3 ± 12.6 | 67.6 ± 13.4 | 102.9 ± 11.3 | 2.4 ± 1.6 |
| K536A | 69.2 ± 7.7 | 77.4 ± 6.0 | 85.9 ± 3.1 | 3.6 ± 1.6 |
We evaluated the fusion promotion activity of each mutant HN in HeLa T4+ cells that had been transfected with mutant HN expression plasmids and pCAGGS-NDVF, a plasmid from which the NDV F protein is expressed (7). Of the 12 mutants we first examined, eight induced levels of syncytium formation that were similar to those induced by wild-type HN and F protein (Table 1). In contrast, the HN containing the Gly169Ala mutation displayed a level of activity that was 60% greater than that of wild-type HN and F protein. Mutations at Thr216, Phe553, and Arg557 abolished the fusion promotion activity. The 3D structure of NDV HN protein consists of a six-bladed β-propeller fold typical of NA molecules (Fig. 1A) (8, 29). Interestingly, the mutations that affected the fusion promotion activity are located on the loops of the first and sixth β sheets of HN protein, suggesting that the fusion promotion domain resides within this particular region of the protein.
We analyzed the electrostatic potential of the surface of HN protein to determine whether the regions that contain the mutant HNs (first and sixth β sheets) that altered fusion promotion activity also demonstrated unique surface properties. Our analysis revealed that the region containing the first and sixth β sheets is largely hydrophobic (Fig. 2B). In fact, two isolated HN monomers formed a dimer by interacting with each other's hydrophobic site in the asymmetric unit of the crystal (Fig. 2A) (8). These results suggest that the large hydrophobic area on the surface of HN protein plays an important role in fusion promotion.
FIG. 2.
The fusion promotion domain is located at the large hydrophobic surface of the HN protein. (A) The surface area of HN involved in the dimer interface of the HN-Neu5Ac2en complex is yellow. The area involved in the dimer interface of HN alone is blue. The dimer interface area involved in both forms of crystals is green. (B) GRASP software was used to calculate the electrostatic potential of the surface of HN. Areas of positive potential (blue) and areas of negative potential (red) are shown. (C) Residues that affected the fusion promotion activity of HN are green, and those that did not affect it are purple. All of the structures in panels A to C are shown in the same orientation.
To test the hypothesis that the hydrophobic surface of HN protein has fusion promotion activity, we created five mutant HNs in which residues within or near the large hydrophobic area (Glu158, Leu160, Phe220, Leu224, and Lys536) were replaced by alanine residues (Fig. 1B). The NA activity of the mutant HN protein was similar to that of the wild-type HN (86 to 130% of wild-type HN levels) (Table 1). In contrast, the HAD activity varied among the HN mutations in the hydrophobic region. Leu224Ala and Lys536Ala were located far from the binding pocket and did not markedly affect HAD activity (68 and 77% of HAD levels of wild-type HN, respectively); however, Glu158Ala reduced HAD activity to levels that were 50% of that of wild-type HN, and Leu160Ala or Phe220Ala demonstrated HAD activity that was only 20% of that of wild-type HN. All mutations within or near the hydrophobic region (Fig. 2C) eliminated fusion promotion activity (Table 1). It is noteworthy that mutant HNs containing Leu224Ala or Lys536Ala retained NA and HAD activities that were similar to those of wild-type HN but lost the fusion promotion activity. These results further indicate the importance of the hydrophobic area in the fusion promotion activity of HN protein.
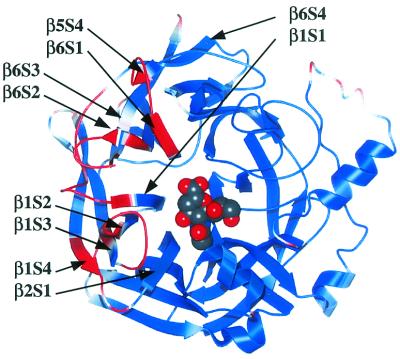
We crystallized soluble HN protein alone or in complex with the inhibitor 2-deoxy-2,3-dehydro-_N_-acetylneuraminic acid (Neu5Ac2en) and solved the structures (8, 27). Ligand-free HN formed orthorhombic crystals at pH 4.6, and HN-Neu5Ac2en complexes formed hexagonal crystals at pH 6.5. A comparison of the two crystals revealed marked structural changes of several loops within or near the large hydrophobic surface (Fig. 3). The chief structural changes were found in the loops between the second and third strands of the sixth β sheet (β6S2-β6S3), between β1S2 and β1S3, between β1S4 and β2S1, between β6S4 and β1S1, and between β5S4 and β6S1 (Fig. 3). Interestingly, these structural changes lie between the binding/NA active pocket and the large hydrophobic area that is essential for fusion promotion activity. The results of our biochemical and structural analyses suggest that as HN binds to its receptor, the loop near the large hydrophobic surface undergoes an important conformational change that triggers the activation of F protein, the mediator of membrane fusion.
FIG. 3.
Receptor binding-induced conformational change of HN protein. A space-filling model of a monomer of HN-Neu5Ac2en depicts the protein colored according to the root mean square deviation of the α-carbon atoms between the two crystal forms of HN. The red regions are those regions that move more than 0.5 Å and show that the primary conformational change is restricted to a small region connecting the sialic acid-binding/hydrolysis site to the large hydrophobic surface, which is to the left of the image.
HN protein mediates receptor binding and promotes membrane fusion through a specific interaction with F protein. We investigated fusion promotion activity by characterizing NDV HN mutants and comparing their biological activities to those of wild-type HN protein. Fusion promotion activity was significantly altered in only those mutant HNs that contained a substituted alanine residue in the first or sixth β sheet. Our current results agree with previous findings that identified residues that are important for fusion promotion activity of different paramyxovirus HN proteins. Bousse et al. previously reported that the Asn242Lys mutation in the HN of human parainfluenza virus type 1 (hPIV1) markedly enhances fusion promotion (3). Residue 242 of hPIV1 HN corresponds to residue 220 of NDV HN and is therefore presumed to be located on the hydrophobic surface. As we showed in this study, the Phe220Ala mutation abolished the fusion promotion activity of NDV HN. Another study using chimeric HN proteins of human parainfluenza virus type 2 (hPIV2) and SV41 revealed that regions I (the stalk domain) and II (residues 148 to 209 in the globular head) specifically interact with the F protein to induce membrane fusion (31). Residues 148 to 209 in hPIV2 HN correspond to residues 154 to 215 of NDV HN and are presumed to be part of the first three strands of the first β sheet. Together, these findings indicate that the hydrophobic surface of HN protein is involved in fusion promotion.
In the current models of virus-induced membrane fusion, activation of the fusion protein induces a conformational change that involves the two heptad repeat domains near the fusion peptide and transmembrane (TM) regions. This conformational change is believed to direct membrane fusion by inserting the fusion peptide into the target membrane and by bringing the fusion peptides into close proximity to the TM anchors to merge the two membranes (11, 22). In the case of influenza virus hemagglutinin, exposing the protein to low pH triggers a series of conformational changes that lead to the exposure and insertion of the fusion peptide into the target membrane and the subsequent formation of helical bundles between the two heptad repeat domains. The formation of these bundles is essential for merging the two membranes (4, 5, 22). In contrast, most retroviruses use a pH-independent entry mechanism (11, 23). Receptor binding induces conformational changes in the envelope protein that drive fusion at the plasma membrane. A recent study of avian leukosis virus (ALV), however, suggested that the entry mechanism of this retrovirus requires a low-pH step that acts downstream of receptor binding (19). Mothes and colleagues (19) showed that receptor binding converts a pH-insensitive envelope glycoprotein to a form that is responsive to low pH; therefore, both receptor binding and exposure to low pH are required for membrane fusion of ALV. Exposure to low pH is not required for paramyxovirus membrane fusion. Unlike influenza virus, paramyxoviruses penetrate cells by fusing their viral envelope with the plasma membrane of the target cell (16), and the HN-F-specific interaction is required for F-protein-induced membrane fusion. An initial event, such as receptor binding, may induce a conformational change in HN that triggers additional conformational changes in the F protein and thus activates it (16). The conformational change of the F protein that activates it should be triggered after the attachment of the virus to the target cells so that the virus envelope and the cell membrane fuse to initiate infection.
Our structural data and results of functional analysis of mutant NDV HNs support the hypothesis that receptor binding induces the conformational change that further triggers the activation of F protein. A comparison of the structure of HN alone with that of the HN-Neu5Ac2en complex revealed crucial changes in the positions of several loops between the binding/NA active pocket and the hydrophobic surface (Fig. 3). Some conserved residues in the binding/NA active site of HN, such as Arg174 (which lies on β1S1) and Lys236 (which lies on β1S2) underwent substantial conformational changes in the HN-Neu5Ac2en complex (Fig. 3) (8). Arg174 and Lys236 are linked to the surface residues that constitute the hydrophobic area, and conformational changes of Arg 174 and Lys236 appear to influence the translocation of the hydrophobic surface residues that also undergo conformational changes, as described above (Fig. 3). It is noteworthy that significant conformational change was limited to the area between the binding pocket and the hydrophobic surface that includes the fusion promotion domain. These structural findings suggest that binding to the sialic acid-containing receptor induces key conformational changes of several loops near the hydrophobic surface and that these changes trigger the activation of the F protein to initiate membrane fusion.
Studies of the HN-F interaction of various paramyxoviruses revealed that the specificity of the interaction is conferred by the globular head and predicted stalk region of HN protein (10, 28, 31). The HN stalk contains a potential heptad repeat region, which probably forms an α-helix (25). Mutation of the heptad repeats resulted in a substantial reduction of fusion promotion activity, a result that supports the theory that HN associates with the F protein via its heptad repeat region in the stalk (25). The two heptad repeat regions of F protein, which are located near the fusion peptide and TM domains, refold into helical bundles. The result of this refolding is that the fusion peptide comes into close proximity to the TM domains (1). Peptides derived from these heptad repeat regions specifically inhibit fusion, a finding that shows that the formation of the helical bundles is essential for merging two membranes (20). Therefore, the predicted heptad repeat in the stalk region of HN may specifically interact with the heptad repeat in the stalk region of the F protein to maintain the F protein in a metastable, native state. Receptor binding and the following conformational change in the HN protein may result in the dissociation of the specific HN-F interaction in the stalk region, and this dissociation may allow the fusion peptide of F to relocate, form the helical bundles, and merge the two membranes.
In this study, we showed that the hydrophobic surface of HN is involved in the fusion promotion activity of HN protein. The results of our structural analysis of the HN crystallized alone or in complex with Neu5Ac2en suggested that the receptor binding induces a major structural change in the hydrophobic surface of the HN, which may promote membrane fusion. How does the structural change near the hydrophobic area of HN protein activate the F protein? Although isolated HN monomers formed dimers in the crystal through its hydrophobic surface (8), the actual form of HN dimer on the virion is not known. If the hydrophobic region of an HN molecule interacts with the same region of another HN molecule to form a dimer, then when HN binds to a receptor, the association of the two monomers would be affected by the conformational change near the dimer interface. The conformational change of the HN dimer would result in the dislocation of the stalk region of HN, which would release the heptad repeat in the stalk region of the F protein and initiate membrane fusion (Fig. 4A). Another possibility is that two HN molecules align straight up to form a dimer and interact with F trimer through its hydrophobic region (Fig. 4B). If this is the case, the HN-F interaction will prevent the fusion peptide from being exposed to the environment until the virus reaches a target cell. A recent report of the 3D structure of the NDV F protein indicated that this protein is organized into head, neck, and stalk regions (6). The trimeric F protein contains axial and radial channels that perforate the head and head-neck interface. Chen and colleagues (6) proposed that after cleavage of F0 into F1 and F2, the hydrophobic fusion peptide becomes sequestered in a metastable, native state in the radial channel. Therefore, HN protein may directly interact with the F protein through the hydrophobic surface of the molecules to keep the fusion peptide in the radial channel. Receptor binding-induced conformational changes near the hydrophobic region of HN protein would disrupt the specific HN-F physical interaction and release the fusion peptide (Fig. 4B). This process allows the F protein to insert the fusion peptide into the target cell membrane, and additional conformational changes merge the two membranes. In both models, physical interaction between HN and the F proteins prevents the conformational change of the F protein until the HN binds to a cellular receptor. The receptor binding-induced conformational change of HN protein triggers the activation of the F protein to initiate membrane fusion. The fusion peptide inserts into the target membrane, and subsequent formation of a triple-helix coiled-coil of the two heptad repeats merges the two membranes (Fig. 4C).
FIG. 4.
Models of the role of HN protein in membrane fusion. (A) In the first model, HN protein forms a dimer through its hydrophobic site and physically interacts with the fusion (F) protein trimer to form an HN-F complex. HN binding to the receptor triggers a conformational change near the hydrophobic site, which results in the structural change of dimer formation of HN and the dissociation of the HN-F complex. The receptor-binding/NA pocket is shown as a gray spot. (B) In the second model, two HN molecules align to form a dimer. HN protein interacts with the F protein at its hydrophobic surface and prevents the exposure of the fusion peptide to the environment. Receptor binding induces a structural change near the hydrophobic surface of HN, which causes the dissociation of the HN-F complex and releases the fusion peptide to initiate membrane fusion. Yellow spots in the F trimer indicate the locations of radial channels that may sequester the fusion peptide. The hydrophobic surface of the HN is shown in red. (C) In both models shown above, HN protein prevents the conformational change of the fusion peptide (blue arrow) of the F protein until the virus reaches the target cell membrane. Receptor binding and the subsequent conformational change of HN result in the dissociation of the HN-F interaction and the activation of the F protein. The F protein then inserts the fusion peptide into the target membrane and merges the two membranes by forming a coiled-coil structure between the two heptad repeat regions near the fusion peptide and TM domains.
Our structural and functional analyses of NDV HN have provided insight into the multiple functions of this protein, which is essential for viral infection. HN protein is well designed to perform a series of events of initial viral infection. HN cooperates with the F protein of the same type of virus; this cooperation includes a specific interaction and a series of dynamic changes initiated by receptor binding. The specificity and flexibility of HN protein appear to help the F protein induce membrane fusion. Studying the mechanism induced by HN and F will provide clues about ways of preventing paramyxovirus infection.
Acknowledgments
This work was supported in part by grants AI-11949 and AI-38956 from the National Institute of Allergy and Infectious Diseases, by Cancer Center Support grant CA-21765 from the National Cancer Institute, by the American Lebanese Syrian Associated Charities (ALSAC), and by grants from the Medical Research Council (United Kingdom) and the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3**:**309-319. [DOI] [PubMed] [Google Scholar]
- 2.Bousse, T., T. Takimoto, W. L. Gorman, T. Takahashi, and A. Portner. 1994. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology 204**:**506-514. [DOI] [PubMed] [Google Scholar]
- 3.Bousse, T., T. Takimoto, and A. Portner. 1995. A single amino acid change enhances the fusion promotion activity of human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. Virology 209**:**654-657. [DOI] [PubMed] [Google Scholar]
- 4.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371**:**37-43. [DOI] [PubMed] [Google Scholar]
- 5.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73**:**823-832. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Cambridge) 9**:**255-266. [DOI] [PubMed] [Google Scholar]
- 7.Connaris, H., T. Takimoto, R. Russell, S. Crennell, I. Moustafa, A. Portner, and G. Taylor. 2002. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 76**:**1816-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7**:**1068-1074. [DOI] [PubMed] [Google Scholar]
- 9.Deng, R., Z. Wang, P. J. Mahon, M. Marinello, A. Mirza, and R. M. Iorio. 1999. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253**:**43-54. [DOI] [PubMed] [Google Scholar]
- 10.Deng, R., Z. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209**:**457-469. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12**:**627-661. [DOI] [PubMed] [Google Scholar]
- 12.Horvath, C. M., R. G. Paterson, M. A. Shaughnessy, R. Wood, and R. A. Lamb. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 66**:**4564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, X. L., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66**:**1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, M., M. Nishio, M. Kawano, S. Kusagawa, H. Komada, Y. Ito, and M. Tsurudome. 1997. Role of a single amino acid at the amino terminus of the simian virus 5 F2 subunit in syncytium formation. J. Virol. 71**:**9855-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197**:**1-11. [DOI] [PubMed] [Google Scholar]
- 16.Lamb, R. A., and D. Kolalofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In B. N. Fields, D. M. Knipe, and A. Kato (ed.), Fundamental virology. Lippincott-Raven, New York, N.Y.
- 17.Malvoisin, E., and T. F. Wild. 1993. Measles virus glycoproteins: studies on the structure and interaction of the haemagglutinin and fusion proteins. J. Gen. Virol. 74**:**2365-2372. [DOI] [PubMed] [Google Scholar]
- 18.Morrison, T. G., and L. W. McGinnes. 1989. Avian cells expressing the Newcastle disease virus HN protein are resistant to NDV infection. Virology 171**:**10-17. [DOI] [PubMed] [Google Scholar]
- 19.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. T. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103**:**679-689. [DOI] [PubMed] [Google Scholar]
- 20.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20**:**4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74**:**5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69**:**531-569. [DOI] [PubMed] [Google Scholar]
- 23.Sodroski, J. G. 1999. HIV-1 entry inhibitors in the side pocket. Cell 99**:**243-246. [DOI] [PubMed] [Google Scholar]
- 24.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71**:**6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone-Hulslander, J., and T. G. Morrison. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73**:**3630-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takimoto, T., T. Bousse, E. C. Coronel, R. A. Scroggs, and A. Portner. 1998. Cytoplasmic domain of Sendai virus HN protein contains a specific sequence required for its incorporation into virions. J. Virol. 72**:**9747-9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takimoto, T., G. L. Taylor, S. J. Crennell, R. A. Scroggs, and A. Portner. 2000. Crystallization of Newcastle disease virus hemagglutinin-neuraminidase glycoprotein. Virology 270**:**208-214. [DOI] [PubMed] [Google Scholar]
- 28.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70**:**6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor, G. 1996. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 6**:**830-837. [DOI] [PubMed] [Google Scholar]
- 30.Tsurudome, M., M. Ito, M. Nishio, M. Kawano, K. Okamoto, S. Kusagawa, H. Komada, and Y. Ito. 1998. Identification of regions on the fusion protein of human parainfluenza virus type 2 which are required for hemagglutinin-neuraminidase proteins to promote cell fusion. J. Gen. Virol. 79**:**279-289. [DOI] [PubMed] [Google Scholar]
- 31.Tsurudome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213**:**190-203. [DOI] [PubMed] [Google Scholar]
- 32.Yao, Q., X. Hu, and R. W. Compans. 1997. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surface. J. Virol. 71**:**650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]