Adenovirus RIDβ Subunit Contains a Tyrosine Residue That Is Critical for RID-Mediated Receptor Internalization and Inhibition of Fas- and TRAIL-Induced Apoptosis (original) (raw)
Abstract
The adenovirus-encoded receptor internalization and degradation (RID) protein (previously named E3-10.4K/14.5K), which is composed of RIDα and RIDβ subunits, down-regulates a number of cell surface receptors in the tumor necrosis factor (TNF) receptor superfamily, namely Fas, TRAIL receptor 1, and TRAIL receptor 2. Down-regulation of these “death” receptors protects adenovirus-infected cells from apoptosis induced by the death receptor ligands Fas ligand and TRAIL. RID also down-regulates certain tyrosine kinase cell surface receptors, especially the epidermal growth factor receptor (EGFR). RID-mediated Fas and EGFR down-regulation occurs via endocytosis of the receptors into endosomes followed by transport to and degradation within lysosomes. However, the molecular interactions underlying this function of RID are unknown. To investigate the molecular determinants of RIDβ that are involved in receptor down-regulation, mutations within the cytoplasmic tail of RIDβ were constructed and the mutant proteins were analyzed for their capacity to internalize and degrade Fas and EGFR and to protect cells from death receptor ligand-induced apoptosis. The results demonstrated the critical nature of a tyrosine residue near the RIDβ C terminus; mutation of this residue to alanine abolished RID function. Mutating the tyrosine to phenylalanine did not abolish the function of RID, arguing that phosphorylation of the tyrosine is not required for function. These data suggest that this tyrosine residue forms part of a tyrosine-based sorting signal (Yxxφ). Additional mutations that target another potential sorting motif and several possible protein-protein interaction motifs had no discernible effect on RID function. It was also demonstrated that mutation of serine 116 to alanine eliminated phosphorylation of RIDβ but did not affect any of the functions of RID that were examined. These results suggest a model in which the tyrosine-based sorting signal in RID plays a role in RID's ability to down-regulate receptors.
Host-virus interactions are characterized by a struggle in which the host tries to protect itself against infection while the virus attempts to thwart host defenses. Chief among the host's defenses are the innate and adaptive arms of the immune system. However, viruses have evolved numerous mechanisms to evade the host immune system. Among the known immune evasion mechanisms are (i) interference with major histocompatibility complex (MHC) class I antigen presentation, (ii) synthesis of cytokine receptor mimics, (iii) secretion of viral cytokines that mimic or antagonize cellular cytokines, (iv) suppression of immune cell activity, and (v) down-regulation of cell surface death receptors required for death receptor ligand-induced apoptosis (reviewed in references 2 and 60). Adenoviruses (Ads) in particular expend a great deal of their resources to prevent death receptor-mediated apoptosis (32, 48, 78).
Binding of a “death” ligand in the tumor necrosis factor (TNF) family (e.g., TNF, Fas ligand, and TRAIL) to its cognate death receptor (TNF receptor 1 [TNFR1], Fas, and TRAIL receptors 1 and 2, respectively) triggers events that may ultimately lead to destruction of the cell via apoptosis. Although incompletely understood, the molecular mechanisms underlying these events involve complex protein-protein interactions that result in a cascade of caspase-mediated proteolytic cleavages (reviewed in reference 46). Many of the initial protein-protein interactions occur through two specific binding domains termed the death domain (DD) and the death effector domain (DED). Upon ligand engagement with and subsequent trimerization of Fas, the cytoplasmic domain of Fas recruits Fas-associated death domain protein (FADD) via the DD present in both proteins (9, 19). In turn, the death effector domain present in FADD and procaspase 8 interact (8, 52), resulting in autoproteolytic cleavage of procaspase 8 to produce active caspase 8 (53). Activation of the caspase cleavage cascade ensues, with the outcome being cellular apoptosis. TNF binding to TNFR1 causes a similar cascade of events, except that FADD binds indirectly to TNFR1, using TNFR1-associated death domain protein (TRADD) as a bridge (33). These proteins associate via their DDs (33). The DD also mediates interaction of receptor-interacting protein (RIP) with the TNFR1-TRADD complex (33, 68, 70).
Ad types 2 and 5 (Ad2 and Ad5, respectively) encode at least five proteins within the early region 3 (E3) transcription unit that are involved in evasion of the host immune response (32, 48, 78). In cases where the molecular mechanism of action of these Ad-encoded proteins has been studied in detail, they function by binding to and modulating the activity of cellular proteins, thus protecting Ad-infected cells from the host immune response. E3-gp19K is a type I integral membrane protein that is localized to the endoplasmic reticulum (ER) as a consequence of an ER retrieval signal located in the cytoplasmic portion of the protein (34, 54, 58). MHC class I molecules bind to E3-gp19K and are retained in the ER, thus preventing MHC class I-mediated cell surface presentation of peptides and cytotoxic T-cell killing of infected cells (3, 4, 13, 14, 62). In addition, E3-gp19K binds TAP (transporter associated with antigen processing) and blocks the interaction between MHC class I antigens and TAP (6).
Another E3 protein, E3-14.7K, prevents TNF-mediated cytolysis of infected cells and release of arachidonic acid (AA) (25, 26, 31, 36, 41, 81). It has also been reported that E3-14.7K can block apoptosis initiated through the Fas pathway (18). E3-14.7K is a cytoplasmic protein and has been shown to bind three cellular proteins termed 14.7K-interacting protein (FIP)-1, -2, and -3 (43-45). The three FIPs in turn interact with other cellular proteins that are involved in nucleus-cytoplasm trafficking and cell cycle control (47), membrane trafficking and morphogenesis (29), and activation of the NF-κB signal transduction pathway (80). In this manner, E3-14.7K may affect several different cellular pathways.
Ad receptor internalization and degradation (RID) protein protects infected cells by at least two different mechanisms: (i) internalization and degradation of cell surface receptors, and (ii) prevention of cytoplasmic phospholipase A2 (cPLA2)-mediated release of AA. RID directs the internalization and degradation of some members of both the TNFR superfamily and the tyrosine kinase family of receptors. Among the receptors reported to be down-regulated are Fas (22, 66, 71), TRAIL receptor 1 (5, 75), TRAIL receptor 2 (5), and epidermal growth factor receptor (EGFR) (16, 74). The mechanism of RID-induced degradation of Fas has been shown to involve trafficking of the internalized receptors through endosomes to the lysosome, where Fas is degraded (71). TRAIL receptor 2 down-regulation was shown to require RID and another E3 protein named E3-6.7K (5). One research group has reported that RID is necessary and sufficient for down-regulation of TRAIL receptor 1 (75), whereas another group reported that E3-6.7K is also required for this function (5). A third group has reported that, independent of other Ad proteins, E3-6.7K protects cells from apoptosis mediated by Fas, TNFR, and TRAIL receptors (51). Furthermore, E3-6.7K was shown to maintain ER Ca2+ homeostasis and protect cells from thapsigargin-induced apoptosis (51).
RID also has effects on pathways induced by activation of TNFR by TNF. As mentioned above, RID blocks the release of AA mediated by TNF (36). The inhibition mediated by RID and by E3-14.7K are independent of one another (36). RID-mediated inhibition involves blockage of the membrane translocation of cPLA2 and subsequent release of AA, a precursor to the potent proinflammatory eicosinoid family of molecules (21). It was recently reported that RID also blocks activation of NF-κB through an as-yet-undefined mechanism (23).
RID consists of a complex of RIDα (formerly E3-10.4K) and RIDβ (formerly E3-14.5K) subunits (72-74). The complex is localized predominantly to the plasma membrane in Ad-infected cells or in cells cotransfected with plasmids that express the individual proteins (30, 69, 71). Neither subunit when expressed by itself can reach the cell surface; instead, the proteins are localized predominantly in the Golgi (RIDα expression only) or the ER and Golgi (RIDβ expression only) (69, 71). RIDα is a small integral membrane protein that exists as either a full-length form or a proteolytically processed form in which the signal sequence is cleaved off (37, 73). Both forms adopt a type I orientation in the membrane and form a complex with each other through a disulfide linkage (30, 37). RIDβ is a type I integral membrane protein and is posttranslationally modified by O glycosylation and by phosphorylation of a serine residue within the cytoplasmic domain of the protein (38-40).
The mechanism of action of RID and the functional significance of RIDβ phosphorylation are unknown. However, it is noteworthy that all Ad-encoded immune function modulators studied to date exert their effects through interactions with host cell proteins (32, 48, 78). Analysis of the primary amino acid sequence of RIDβ revealed several motifs within the cytoplasmic tail that could potentially mediate protein-protein interactions. These motifs included a tyrosine-based sorting motif (Yxxφ) (35), a C-terminal acidic motif (61, 77), and three core Src homology 3 (SH3) ligand motifs (PxxP) (17). In addition, this region contains two serine residues, either or both of which could be the site of RIDβ phosphorylation. In order to dissect the molecular mechanisms by which RID functions and to define the site(s) of RIDβ phosphorylation, site-specific point mutations that eliminated these motifs were introduced into the cytoplasmic tail of RIDβ. The mutant proteins were assessed for their ability to form a complex with RIDα, become phosphorylated, internalize and degrade cell surface receptors, and protect cells from death receptor-mediated apoptosis. The results demonstrate the critical nature of tyrosine 124 and suggest that RID may function through a tyrosine-based sorting motif.
MATERIALS AND METHODS
Cells.
COS7 (African Green monkey kidney), HeLa (human cervical carcinoma), and A549 (human lung carcinoma) cells were grown in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Antibodies.
The use of rabbit anti-RIDβ and rabbit anti-RIDα antisera for immunofluorescence and Western blot analysis was described previously (69, 74). Fas was detected with a rabbit anti-peptide antibody used at a 1:1,000 dilution (antibody C-20; Santa Cruz Biotechnology, Santa Cruz, Calif.) or with the ZB4 mouse monoclonal antibody (Panvera, Madison, Wis.) used at a 1:100 dilution. A hybridoma cell line expressing the mouse monoclonal antibody [clone 200-3-G6-4 (20.4)] directed against the low-affinity nerve growth factor receptor (LANGFR) was obtained from the American Type Culture Collection. Tissue culture supernatants containing this antibody were used undiluted. EGFR was detected with a rabbit antipeptide antiserum (74) or with a mouse monoclonal antibody (528; Santa Cruz Biotechnology) used at a dilution of 1:100. For indirect immunofluorescence studies, AlexaFluor 594 conjugated to goat anti-mouse immunoglobulin G (IgG) and AlexaFluor 488 conjugated to goat anti-rabbit IgG were purchased from Molecular Probes (Eugene, Oreg.) and used at a dilution of 1:500. For Western blot studies, goat anti-rabbit immunoglobulins (IgG, IgA, and IgM) conjugated to horseradish peroxidase were purchased from Cappel/ICN (Costa Mesa, Calif.) and used at a dilution of 1:4,000. Fas-mediated apoptosis was induced with the CH-11 Fas agonist mouse monoclonal antibody (Panvera). To detect LANGFR by indirect immunofluorescence when CH-11 was used for induction of apoptosis, a goat anti-mouse-IgG1-specific antibody coupled to AlexaFluor 594 was used (Molecular Probes).
Construction of mutants.
The primers used in constructing the RIDβ cytoplasmic tail mutant genes are shown in Table 1 and were synthesized by Life Science Technologies (Bethesda, Md.) or Operon Technologies, Inc. (Alameda, Calif.). Mutations were introduced by either a one-step or a two-step PCR method (Table 2). In the one-step PCR method, the desired mutation was incorporated into a primer encompassing the 3′ end of the RIDβ gene. The mutagenic primer and a 5′ primer consisting of the 5′ end of the RIDβ gene were used in a PCR containing the wild-type RIDβ gene as the template. In the two-step PCR method, a mega primer was synthesized using the mutagenic primer paired with a primer encompassing the 3′ end of the RIDβ gene. Following gel purification, the mega primer was used as the 3′ primer in a second round of PCR along with a 5′ primer encompassing the 5′ end of the RIDβ gene. Each reaction mixture contained 0.5 mM concentrations of each deoxynucleoside triphosphate (dNTP), 0.1 to 1.0 μg of template DNA, a 1.0 μM concentration of each primer, 2.5 U of Pfu polymerase (Stratagene, La Jolla, Calif.), and 1× Pfu polymerase buffer (supplied by the manufacturer). Cycling conditions were as follows: a single denaturation step at 97°C for 10 min, addition of Pfu polymerase, 30 cycles of denaturation, annealing, and extension (1 min at 94°C, 1 min at 55°C, and 2 min at 72°C), and a single extension step at 72°C for 10 min. The final PCR products were digested with _Eco_RI and cloned into the _Eco_RI site of the eukaryotic expression vector pMT2 via standard molecular cloning techniques. To facilitate screening for the presence of the mutation, some primers were designed such that a new restriction enzyme site was introduced along with the desired mutation. All mutants were sequenced using an ABI 310 sequencer to confirm the sequence of the mutant gene and to determine the orientation of the gene within the vector.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| 5′RIDβb | 5′GGATAAGAATTCTTTAATTATGAAATTTACTGTGAC 3′ |
| 3′RIDβb | 5′GGATAAGAATTCAGATCTAGGGTGTCAGTC3′ |
| 5′pMT2 | 5′CCACTTTGCCTTTCTCTCCACAGG3′ |
| 3′pMT2 | 5′TAAACAAGTTCTCTAGAGTCGACGGC3′ |
| P112A-4c | 5′CCAATCAGGCGCGCCCACCTTCTCCCACC3′ |
| P114Ac | 5′CAGCCTCGCGCGCCTTCTCC3′ |
| S116Ad | 5′CGCCCACCGGCGCCCACCCCC3′ |
| S123A-4 | 5′CCACTGAAATCGCCTACTTTAATCTAACAGGAGG3′ |
| Y124A | 5′CCACTGAAATAAGCGCCTTTAATCTAACAGGAGG3′ |
| Y124F-2 | 5′CCACTGAAATAAGCTTCTTTAATCTAACAGGAGG3′ |
| L127Ab | 5′CCATAAGAATTCAGATCTAGGGTGTCAGTCATCTCCTCCTGTTGCATTAAAGTAGC3′ |
| DD-AAb,d | 5′CCATAAGAATTCAGATCTAGGGTGTCAGGCGGCGCCTCCTGTTAG3′ |
TABLE 2.
Strategies used for construction of mutants
| Mutant | Strategy | Step 1 | Step 2 | Template | ||
|---|---|---|---|---|---|---|
| 5′ primer | 3′ primer | 5′ primer | 3′ primer | |||
| P112A | Two step | P112A-4 | 3′pMT2 | 5′pMT2 | P112A mega | pMT2-RIDβ |
| P114A | Two step | P114A | 3′RIDβ | 5′RIDβ | P114A mega | pMT2-RIDβ |
| S116A | Two step | S116A | 3′RIDβ | 5′RIDβ | S116A mega | pMT2-RIDβ |
| S123A | Two step | S123A-4 | 3′pMT2 | 5′pMT2 | S123A mega | pMT2-RIDβ |
| S116A/S123A | Two step | S123A-4 | 3′pMT2 | 5′pMT2 | S/S mega | pMT2-RIDβ S116A |
| Y124A | Two step | Y124A | 3′pMT2 | 5′pMT2 | Y124A mega | pMT2-RIDβ |
| Y124F | Two step | Y124F-2 | 3′pMT2 | 5′pMT2 | Y124F mega | pMT2-RIDβ |
| L127A | One step | 5′RIDβ | L127A | NAa | NA | pMT2-RIDβ |
| D131A/D132A | One step | 5′RIDβ | DD-AA | NA | NA | pMT2-RIDβ |
Transfection.
For all transfections, cells were plated in 35-mm dishes at a density of 1 × 105 to 2 × 105 cells/dish 1 day before transfection. HeLa and A549 cells were transfected with plasmid DNAs using FuGene 6 (Roche, Indianapolis, Ind.) according to the manufacturer's recommended procedure. COS7 cells were transfected with FuGene 6 or DEAE-dextran. Briefly, plasmid DNAs were added to 0.5 ml of serum-free DMEM. Fresh DEAE-dextran (0.8 mg/ml in serum-free DMEM containing 0.1 M Tris [pH 7.5]) was prepared and filter sterilized. An equal volume of DEAE-dextran solution was added to the diluted DNA, and this mixture was added dropwise to cells that had been washed with and were incubating in serum-free DMEM. After incubation at 37°C for 3 h, the medium was replaced with serum-free DMEM containing 150 μM chloroquine and the dishes were incubated at 37°C for 4 h. Cells were washed once with serum-free DMEM and then incubated for up to 48 h in complete DMEM. The amount of plasmid used for most experiments was as follows: 0.5 μg of pMT2-RIDα per dish and 0.1 μg of pMT2-RIDβ per dish. For the apoptosis assay, pMT2-RIDα, pMT2-RIDβ, and pMT2-6.7K were all used at 1.0 μg per dish, while pCDNA3.1Z-ΔLANGFR (63) was used at 0.1 μg per dish.
Cell labeling and immunoprecipitation.
COS7 cells transfected by the FuGene method were labeled with [35S]methionine and [35S]cysteine at 24 h posttransfection. After being washed once in 35S-labeling medium (methionine-free, cysteine-free, serum-free DMEM), cells were incubated in this medium at 37°C for 30 min. The medium was then replaced with 35S-labeling medium containing 50 μCi of [35S]cysteine (Perkin-Elmer Life Sciences, Boston, Mass.) and 50 μCi of Easy Tag Express [35S] protein labeling mix (Perkin-Elmer Life Sciences), and the cells were incubated at 37°C for 6 h. Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and then frozen at −80°C.
COS7 cells transfected by the DEAE-dextran method were labeled with inorganic [32P]phosphate at 26 h posttransfection. After being washed once in 32P-labeling medium (phosphate-free, serum-free DMEM), cells were incubated in this medium at 37°C for 1 h. The medium was then replaced with 32P-labeling medium containing 250 μCi of inorganic [32P]phosphate (Perkin-Elmer Life Sciences), and the cells were incubated at 37°C for 3 h. Cells were washed three times with ice-cold PBS and then frozen at −80°C.
Thawed cells were scraped into 0.1 ml of lysis buffer (10 mM Tris [pH 7.4]-0.4% [wt/vol] deoxycholic acid-66 mM EDTA-1.0% NP-40), and cytoplasmic lysates were prepared by centrifugation at 16,000 × g for 5 min at 4°C. The protein concentration of each lysate was determined using the Bio-Rad DC protein assay. Labeled RIDβ was immunoprecipitated from 50 to 75 μg of cytoplasmic lysate with rabbit anti-RIDβ antiserum (74). Immunoprecipitates separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were visualized by fluorography (Entensify; Perkin-Elmer Life Sciences) of the dried gels.
Western blotting.
Lysates of transfected cells were prepared and the protein concentration was determined as described above. Ten to 25 μg of protein from each cell lysate was separated by SDS-PAGE and then transferred to polyvinylidene difluoride membranes by using a semidry blotting apparatus (Bio-Rad, Hercules, Calif.). Transfers were performed in transfer buffer (48 mM Tris-39 mM glycine-20% [vol/vol] methanol [pH 9.2]) for 30 min at 2.5 mA/cm2. Membranes were blocked for at least 30 min at 4°C in rinse buffer (137 mM NaCl-1.47 mM KH2PO4-8.1 mM Na2HPO4-2.68 mM KCl-0.05% [vol/vol] Tween 20) containing 5% (wt/vol) nonfat dry milk. Blots were incubated for 60 min with primary antibody, washed five times in 50 ml of rinse buffer, incubated for 30 min with secondary antibody, washed as above, incubated with LumiGlo chemiluminescent reagents according to the manufacturer's instructions (Kirkegard & Perry Laboratories, Gaithersburg, Md.), and exposed to BioMax MR film (Kodak, Rochester, N.Y.). All incubations were performed at room temperature (RT). Antibodies were diluted in rinse buffer containing 5% (wt/vol) nonfat dry milk.
Receptor degradation assay.
COS7 cells were transfected by the DEAE-dextran method. To analyze RID-mediated degradation of Fas, cells were transfected with 1.0 μg of pCDNA3.1-Fas (a generous gift from V. Dixit) in addition to the vectors expressing RIDα and RIDβ. Alternatively, degradation of EGFR was examined in cells transfected with a plasmid expressing EGFR under control of the cytomegalovirus promoter (pRT3ER; a generous gift from Marsha R. Rosner). At 24 h posttransfection, cells were treated with cycloheximide (25 μg/ml) for 4 h at 37°C. Cells were lysed and Fas or EGFR was detected by Western blotting.
Indirect immunofluorescence.
Indirect immunofluorescence was performed as described previously (69). Briefly, cells were plated on glass coverslips in 35-mm dishes and then transfected the following day using the DEAE-dextran method. At 36 h posttransfection, coverslips were washed once with PBS and then fixed for 10 min at −20°C in methanol containing 0.25 μg of 4′,6-diamidino-2-phenylindole (DAPI)/ml. Coverslips were then washed once with methanol at −20°C and three times with PBS. Cells were incubated in a humidified chamber at 37°C for 60 min with rabbit anti-RIDβ antiserum and a mouse monoclonal antibody against either Fas or EGFR. After washing three times in PBS for 10 min at RT, coverslips were incubated with the appropriate fluorophore-conjugated secondary antibodies in a humidified chamber at 37°C for 30 min. After washing as described above, coverslips were mounted and sealed on glass microscope slides and viewed with a Nikon Optiphot microscope equipped with epifluorescence. Digital images were captured with a Nikon DMX1200 digital camera and ACT-1 software (Nikon).
Apoptosis assay.
HeLa or A549 cells were plated on poly-l-lysine-treated coverslips and transfected in duplicate the following day using the FuGene method. The plasmid pCDNA3.1Z-ΔLANGFR was used as a marker for transfected cells. This plasmid expresses a truncated version of LANGFR in which the entire cytoplasmic domain has been deleted (a kind gift from Clay Smith). To assess the ability of RID to protect cells from Fas-mediated apoptosis, duplicate coverslips were treated for 5.5 h at 37°C with cycloheximide (25 μg/ml) alone or in combination with the CH-11 Fas agonist monoclonal antibody (1 μg/ml) starting at 24 h posttransfection. Alternatively, to measure protection from TRAIL-mediated apoptosis, transfected cells were treated with cycloheximide with or without TRAIL (20 ng/ml) for 3.5 h at 37°C (75). At the end of the treatment period, cells were washed once with serum-free DMEM, fixed for 10 min at RT with 3.7% paraformaldehyde in PBS, air dried at RT for 10 min, permeabilized for 6 min at −20°C with methanol-DAPI, and rinsed for 2 min at −20°C with methanol. Cells were rehydrated with PBS, and the expression of ΔLANGFR was assessed by indirect immunofluorescence. The level of apoptosis in transfected cells was determined by evaluating the morphology of DAPI-stained nuclei in ΔLANGFR-positive cells. The percentage of apoptotic cells was calculated as follows: (number of apoptotic-positive ΔLANGFR-positive cells/total number of ΔLANGFR-positive cells) × 100. The percentage of apoptotic cells induced specifically by TRAIL or Fas agonist treatment was calculated as follows: (percentage of apoptotic cellscycloheximide + TRAIL or Fas agonist) − (percentage of apoptotic cellscycloheximide only). More than 1,000 ΔLANGFR-positive cells were counted for each condition tested.
RESULTS
Construction and biochemical characterization of RIDβ mutants.
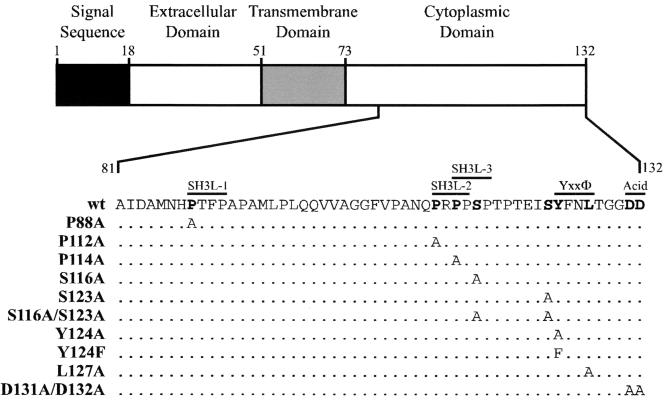
Given that RID can internalize members of at least two distinct, unrelated receptor families (the TNFR superfamily and tyrosine kinase family) and that many Ad proteins function by binding to cellular proteins, it is likely that rather than binding directly to its target receptors RID acts by binding to a cellular adaptor molecule(s). With this in mind, the Ad5 RIDβ amino acid sequence was analyzed for motifs that might mediate interaction with a cellular protein(s). Three such potential protein-protein interaction motifs were identified in the RIDβ cytoplasmic tail (Fig. 1). First, a potential Yxxφ (Y is a tyrosine, x is any amino acid, and φ is a hydrophobic amino acid with a bulky side chain) sorting motif was identified near the C terminus of the protein. This type of tyrosine-based motif is known to mediate binding to the medium (μ) chains of adaptor protein (AP) complexes (56, 67). AP complexes thus link clathrin-coated vesicles with cargo proteins that contain the Yxxφ motif (35). Second, a possible acidic sorting motif was identified at the C terminus. This motif has been shown to enhance the activity of the Yxxφ motif and may itself act as a sorting motif (61, 77). Third, three potential SH3 domain ligand motifs of the form PxxP (P is proline and x is any amino acid) were also detected. This motif allows binding to proteins containing an SH3 domain (17).
FIG. 1.
Ad5 RIDβ mutations used in this study. A schematic representation of the domain structure of RIDβ is shown. The expanded portion shows the deduced amino acid sequence of part of the cytoplasmic tail domain. The amino acid sequence of the same part of RIDβ is shown for each mutant that was constructed. Amino acids that were mutated are shown in bold. Amino acids unchanged from the wild-type sequence are represented by a dot. Potential protein-protein interaction domains are shown above the wild-type sequence.
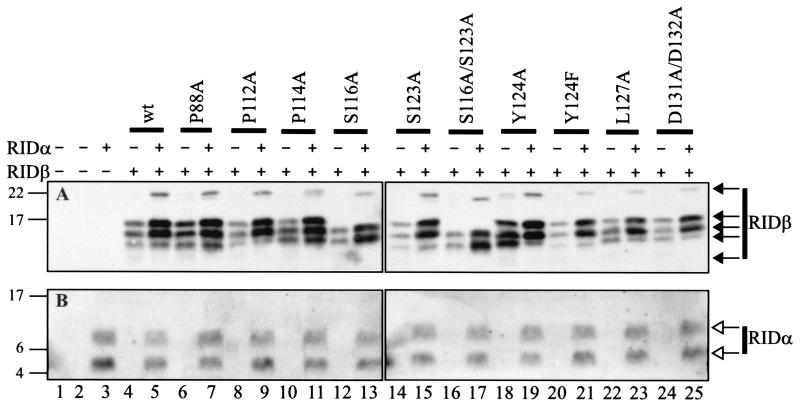
Key amino acid residues within each potential motif were mutated to an alanine residue in order to test the functional significance of these motifs (Fig. 1). The stability and processing of each mutant protein was addressed by examining its synthesis in COS7 cells transiently transfected with a plasmid that expresses wild-type or mutant forms of RIDβ. Some samples were also transfected with a plasmid that expresses RIDα. Wild-type RIDβ produced the characteristic quadruplet of bands in the absence of RIDα (note that the fastest-migrating band was only clearly visible upon a longer exposure) (Fig. 2A, lane 4). These represent the mature and partially processed immature forms of RIDβ (72). RIDα migrated as two characteristic bands (Fig. 2B). The upper band is an uncleaved primary translation product and the lower band is a form that has been cleaved between residues 22 and 23 (37, 73). In the presence of RIDα, the fastest-migrating RIDβ band diminished in intensity and a new band at about 22 kDa appeared (Fig. 2A, lane 5). The presence of RIDα also increased the steady-state level of RIDβ, perhaps by enhancing its stability.
FIG. 2.
Mutant RIDβ proteins are expressed at the same level as wild-type RIDβ in transiently transfected cells. Lysates were prepared from COS7 cells at 28 h posttransfection and were subjected to Western blot analysis using anti-RIDβ (A) or anti-RIDα (B) antiserum. The expression plasmids included in each transfection are indicated above the blots. The identity of the RIDβ gene included in the transfection is also indicated above the blots. The position and molecular mass in kilodaltons of the protein standards are shown to the left of each blot. RIDβ-specific (solid arrows) and RIDα-specific (open arrows) bands are indicated to the right of each blot.
All of the mutant proteins were stably expressed, since their level of expression, either in the absence or presence of RIDα, was similar to that of wild-type RIDβ (Fig. 2A). Furthermore, none of the mutations in RIDβ affected the level of expression of RIDα when it was coexpressed in the same cells (Fig. 2B). Most of the mutant proteins showed a pattern of bands similar to that of wild-type RIDβ, indicating that no defects in processing were evident. The two exceptions were S116A RIDβ and S116A/S123A RIDβ, in which the upper and lower bands in the quadruplet were absent (Fig. 2A, lanes 12, 13, 16, and 17). In addition, the highest-molecular-weight form seen in the presence of RIDα migrated slightly faster than its wild-type counterpart (Fig. 2A, lanes 13 and 17). These defects in processing may result from a change in phosphorylation status of RIDβ, since both mutants contain the S116A mutation and RIDβ is known to be phosphorylated on either serine 116 or serine 123 (40).
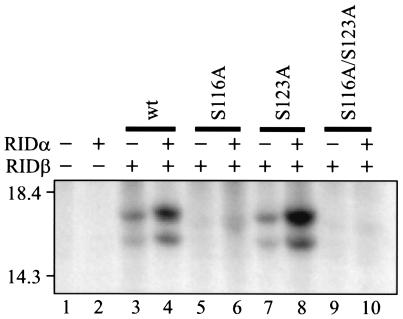
RIDβ is phosphorylated exclusively on serine 116.
RIDβ has been reported to be phosphorylated on a serine residue(s) in the presence of RIDα (40). To test if S116A and S116A/S123A were phosphorylated, these two mutant proteins, as well as S123A and wild-type RIDβ, were labeled with inorganic [32P]phosphate in COS7 cells transiently transfected with plasmids that express these proteins. Samples immunoprecipitated with anti-RIDβ antibody were visualized by fluorography following SDS-PAGE. Two phosphorylated forms were present in cells expressing wild-type RIDβ, regardless of whether RIDα was present (Fig. 3, lanes 3 and 4). A similar result was obtained for the S123A mutant protein (Fig. 3, lanes 7 and 8). However, no phosphorylated forms of RIDβ were detected in cells expressing either S116A or S116A/S123A (Fig. 3, lanes 5, 6, 9, and 10). These results demonstrate that RIDβ expressed by transient transfection contains only a single site for phosphorylation and that the site is located at serine 116. In addition, the presence of two phosphorylated forms of RIDβ corresponds to the disappearance of two bands for the S116A and S116A/S123A mutants (Fig. 2).
FIG. 3.
Phosphorylation status of wild-type and serine mutant proteins of RIDβ. COS7 cells were transfected with different combinations of plasmids and labeled with inorganic [32P]phosphate for 3 h at 27 h posttransfection. A portion of each lysate was immunoprecipitated with RIDβ-specific antiserum. Immunoprecipitates were subjected to SDS-PAGE, and the labeled proteins were visualized by fluorography of the dried gel. The expression plasmids included in each transfection are indicated above the gel by a + symbol. The identity of the RIDβ gene included in the transfection is also indicated above the gel. The position and molecular mass in kilodaltons of the protein standards are shown to the left of the gel.
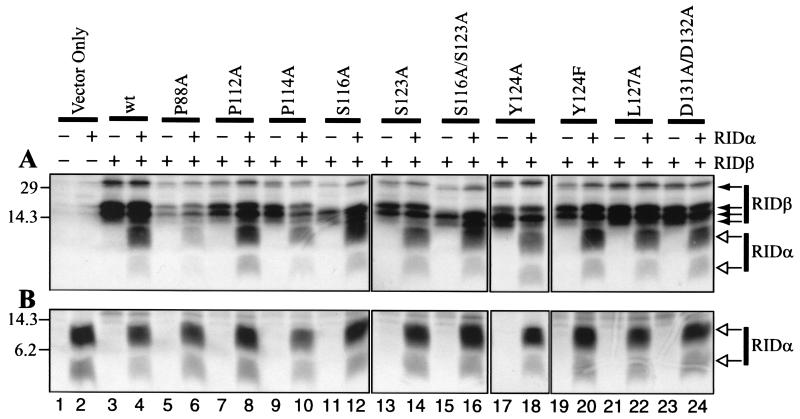
RIDβ mutant proteins are able to form a complex with RIDα.
Formation of a complex between RIDβ and RIDα is necessary for the function of RID, since neither subunit functions by itself (69, 74). Coimmunoprecipitation of RIDα by anti-RIDβ antibodies can be used to test for complex formation, and so this assay was used to assess the capacity of each RIDβ mutant protein to form a complex with RIDα. COS7 cells transiently transfected with plasmids that express RIDα and wild-type or mutant forms of RIDβ were labeled with [35S]methionine and [35S]cysteine. Cell lysates immunoprecipitated with an anti-RIDβ antibody were analyzed by SDS-PAGE and fluorography. The results showed that the wild type and all mutant forms of RIDβ are able to coimmunoprecipitate RIDα when it is coexpressed with RIDβ (Fig. 4A, even-numbered lanes except for lane 2). Nearly all of the mutant proteins coimmunoprecipitated an amount of RIDα similar to the amount immunoprecipitated by wild-type RIDβ protein. The decreased amount of RIDα coimmunoprecipitated by the P88A mutant protein was likely due to the decreased level of expression of P88A RIDβ compared to wild-type RIDβ and the other mutant proteins (Fig. 4A, lanes 5 and 6). This conclusion was confirmed by additional experiments in which the level of RIDα coimmunoprecipitated by P88A RIDβ was equivalent to that seen for coimmunoprecipitation by wild-type RIDβ (data not shown). As a comparison, when RIDα was not coexpressed only RIDβ was immunoprecipitated (Fig. 4A, odd-numbered lanes except for lane 1). Figure 4B shows that RIDα was expressed and labeled equally well in all transfections. These data suggest that all of the mutant proteins can form a complex with RIDα comparable to that of wild-type RIDβ.
FIG. 4.
Mutant forms of RIDβ interact with RIDα, as shown by coimmunoprecipitation. COS7 cells were transfected with different combinations of plasmids and labeled with [35S]cysteine and [35S]methionine for 6 h at 24.5 h posttransfection. A portion of each lysate was immunoprecipitated with RIDβ-specific (A) or RIDα-specific (B) antipeptide antiserum. Immunoprecipitates were subjected to SDS-PAGE, and the labeled proteins were visualized by fluorography of the dried gels. The expression plasmids included in each transfection are indicated above the gels by a + symbol. The identity of the RIDβ gene included in the transfection is also indicated above the gels. The position and molecular mass in kilodaltons of the protein standards are shown to the left of each gel. RIDβ-specific (solid arrows) and RIDα-specific (open arrows) bands are indicated to the right of each gel.
RIDβ mutant proteins localize properly within cellular membranes.
In order to down-regulate cell surface receptors, RID must localize to the plasma membrane. However, as shown previously, cell surface localization of RID requires coexpression of RIDα and RIDβ (74). In the absence of the other subunit, each subunit is severely retarded in reaching the cell surface, resulting in predominant localization to both the ER and Golgi (RIDβ) (69) or to the Golgi (RIDα) (71). Subcellular localization of mutant RIDβ proteins was evaluated by indirect immunofluorescence in COS7 cells transiently transfected with plasmids that express mutant forms of RIDβ. In the absence of RIDα expression, transport of wild-type RIDβ to the cell surface did not occur, as indicated by the localization of RIDβ to the ER and Golgi (Fig. 5E and reference 69). For each mutant, the cellular localization pattern in the absence of RIDα expression was similar to that of wild-type RIDβ (Fig. 5I, M, Q, U, Y, CC, GG, KK, OO, and SS). As shown in Fig. 4, RIDα and wild-type RIDβ coexpression resulted in formation of a complex and, as expected, RIDβ was observed in the plasma membrane (Fig. 5G). All of the mutant RIDβ proteins were also localized predominantly to the cell surface in the presence of RIDα (Fig. 5K, O, S, W, AA, EE, II, MM, QQ, and UU). Together, these data indicate that none of the mutations compromised the ability of RIDβ to localize correctly.
FIG. 5.
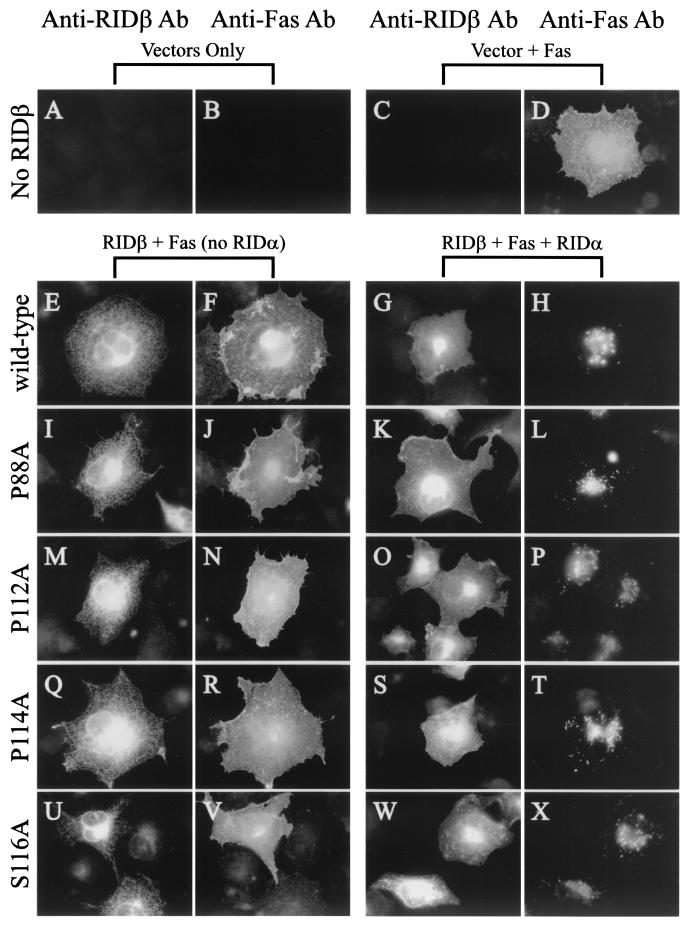
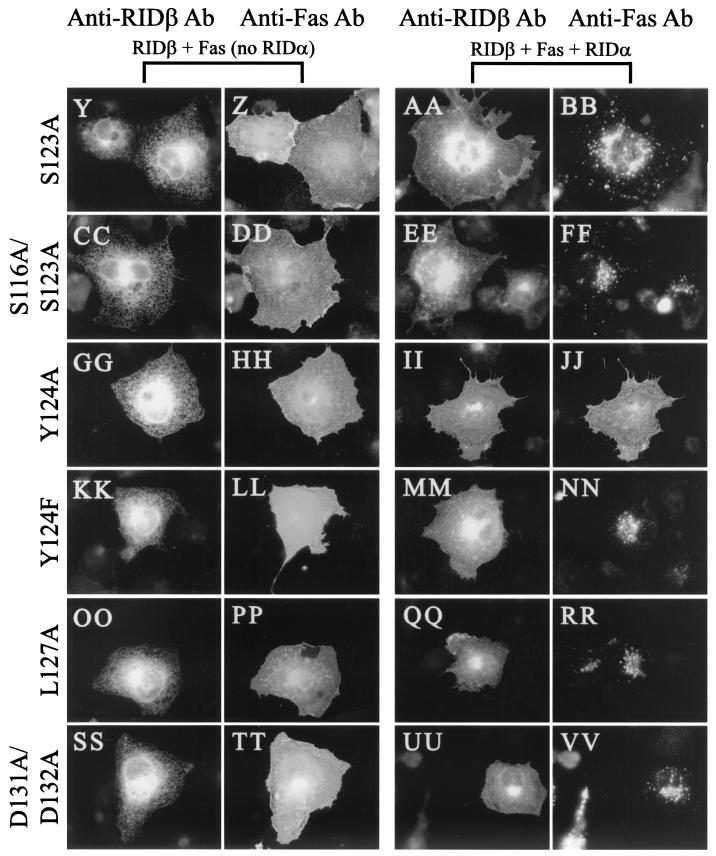
Y124A RIDβ is defective for internalization of Fas. COS7 cells on coverslips were transfected with various combinations of pCDNA3.1-Fas, pMT2-RIDα, and pMT2-RIDβ. At 24 h posttransfection, cells were fixed and immunostained for RIDβ and Fas. RIDβ was detected with a RIDβ-specific rabbit antiserum and AlexaFluor 488-conjugated goat anti-rabbit IgG. Fas was detected with the ZB4 mouse monoclonal antibody and AlexaFluor 594 conjugated to goat anti-mouse IgG. The plasmids included in the transfection are indicated above the panels. The primary antibody used for immunostaining is indicated above each column of panels.
Only the Y124A mutant protein is defective in internalization and degradation of cell surface receptors.
Since all of the mutant proteins were stably expressed, were capable of forming a complex with RIDα, and localized properly, the ability of each mutant protein to internalize cell surface receptors was assessed. Fas was localized to the cell surface in COS7 cells transfected with just the Fas expression plasmid (Fig. 5D). Coexpression of Fas and either wild-type or mutant forms of RIDβ resulted in no discernible alteration in the cell surface localization of Fas (Fig. 5F, J, N, R, V, Z, DD, HH, LL, PP, and TT). Coexpression of Fas, RIDα, and wild-type RIDβ resulted in clearance of Fas from the cell surface and relocalization of Fas to intracellular structures, presumably late endosomes and lysosomes (Fig. 5H and reference 71). Most of the mutant RIDβ proteins were also capable of internalizing Fas in a manner similar to that of wild-type RIDβ (Fig. 5L, P, T, X, BB, FF, NN, RR, and VV). The one exception was the Y124A mutant protein, which did not affect the cell surface localization of Fas (Fig. 5JJ). Since phenylalanine can substitute for tyrosine in some Yxxφ sorting motifs (15, 50, 76), it was of particular interest that the Y124F mutant protein was capable of down-regulating Fas (Fig. 5NN). We conclude that only Y124A RIDβ is defective in internalizing Fas.
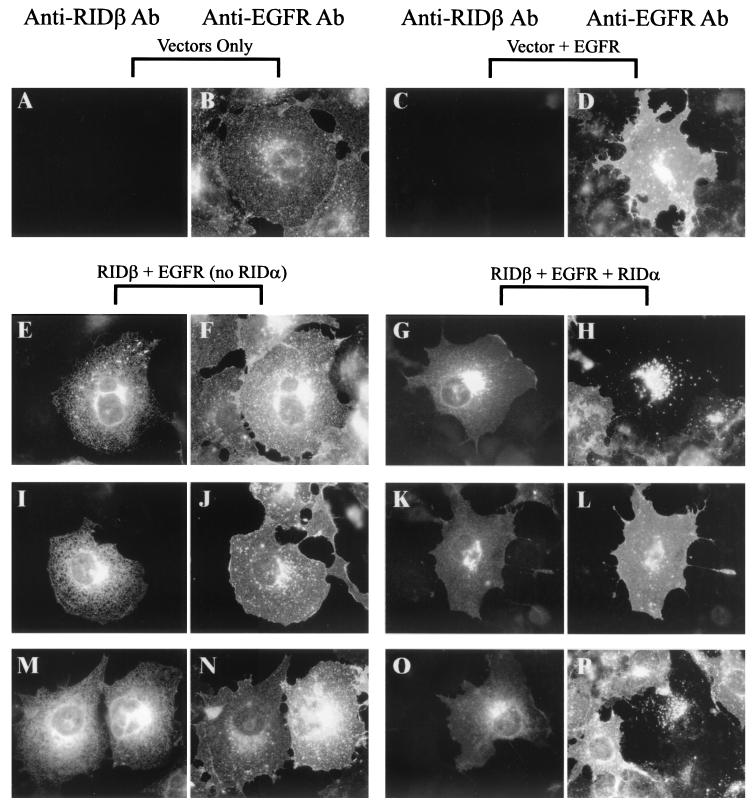
To determine whether the defect in the Y124A mutant was specific for internalization of Fas, internalization of EGFR was also examined by indirect immunofluorescence in COS7 cells transiently transfected with plasmids that express wild-type or mutant forms of RIDβ. Endogenous simian EGFR reacted with the monoclonal antibody against EGFR that was used (Fig. 6B), but the EGFR signal was much greater when a plasmid that expresses human EGFR was transfected into the cells (Fig. 6D). As was the case with Fas, EGFR cell surface localization was not affected by expression of wild-type RIDβ only (Fig. 6F). However, when wild-type RIDβ and RIDα were coexpressed, EGFR was cleared from the cell surface and found predominantly in intracellular vesicles (Fig. 6H). Expression of Y124A or Y124F by themselves did not alter the localization of EGFR (Fig. 6J and N). Coexpression of RIDα with these two mutant proteins showed that Y124A was defective in EGFR internalization, but Y124F behaved like wild-type RIDβ (Fig. 6L and P, respectively). The remaining mutant proteins were also tested for their ability to internalize EGFR, and all of them behaved like wild-type RIDβ (data not shown). In transiently transfected A549 cells, all the mutant proteins had the same phenotype with respect to Fas and EGFR internalization, indicating that the phenotype observed for each mutant protein was not specific to COS7 cells (data not shown).
FIG. 6.
Y124A RIDβ is defective for internalization of EGFR. COS7 cells on coverslips were transfected with various combinations of an EGFR expression plasmid (pRT3ER), pMT2-RIDα, and pMT2-RIDβ. At 24 h posttransfection, cells were fixed and immunostained for RIDβ and EGFR. RIDβ was detected as described in the legend to Fig. 5. EGFR was detected with the 528 monoclonal antibody and AlexaFluor 594 conjugated to goat anti-mouse IgG. The plasmids included in the transfection are indicated above the panels. The primary antibody used for immunostaining is indicated above each column of panels. Cells were transfected with a plasmid that expresses wild-type RIDβ (E to H), Y124A (I to L), or Y124F RIDβ (M to P).
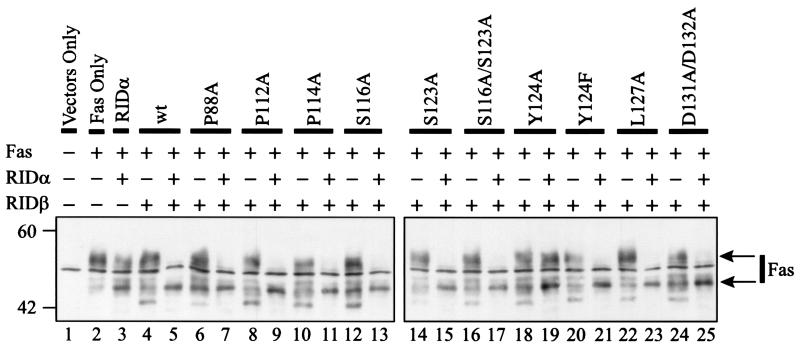
RID not only internalizes Fas but also causes the degradation of Fas in lysosomes (22, 71). Each of the mutant proteins was tested for its ability to degrade Fas. Lysates from transiently transfected COS7 cells were subjected to Western blot analysis to detect Fas. Transient transfection of only a plasmid that expresses Fas resulted in the appearance of two Fas-specific proteins (Fig. 7, compare lanes 1 and 2). The sharp middle band is a background protein inasmuch as it was detected in cells not transfected with the plasmid that expresses Fas (Fig. 7, lane 1). Coexpression of Fas and RIDα resulted in a slight decrease in the intensity of the upper band and an increase in the intensity of the lower band (Fig. 7, lane 3). When wild-type RIDβ and Fas were coexpressed, the upper and lower Fas bands were unaffected but a new faster-migrating band appeared (Fig. 7, lane 4). This band may be a Fas precursor or a breakdown product; further experiments will be required to distinguish between these two possibilities. A similar phenotype was seen with all of the mutant proteins when they were expressed in the absence of RIDα (Fig. 7). Coexpression of wild-type RIDβ, RIDα, and Fas resulted in degradation of Fas, as shown by the near-complete loss of the upper band and an increase in the intensity of the lower band (Fig. 7, lane 5). Most of the mutant proteins, including Y124F RIDβ, were also able to degrade Fas (Fig. 7, lanes 7, 9, 11, 13, 15, 17, 21, 23, and 25). The exception was Y124A RIDβ, which had no effect on the amount of Fas (Fig. 7, lane 19). These data correspond with the inability of Y124A RIDβ to internalize Fas. All mutants were also tested for their capacity to degrade EGFR; the results were similar to those seen for Fas (data not shown).
FIG. 7.
Y124A RIDβ is defective for degradation of Fas. Lysates were prepared from COS7 cells at 28 h posttransfection and were subjected to Western blot analysis using anti-Fas antiserum. The expression plasmids included in each transfection are indicated above the blots. The identity of the RIDβ gene included in the transfection is also indicated above the blots. The position and molecular mass in kilodaltons of the protein standards are shown to the left of the blots. Arrows to the right of the blot indicate Fas-specific bands.
The Y124A mutant protein cannot protect cells from death receptor-mediated apoptosis.
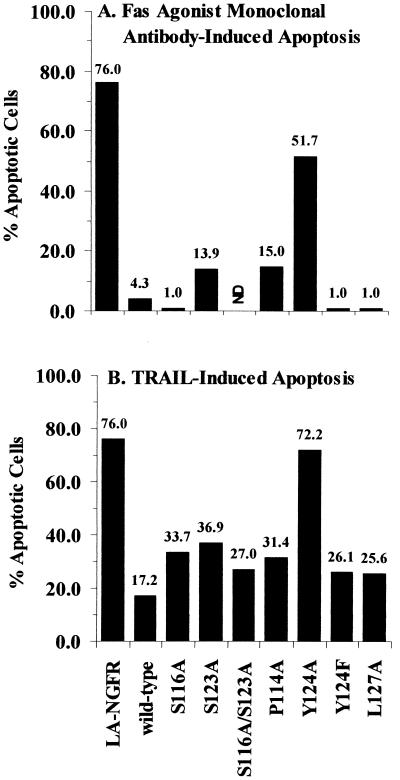
Since RID protects Ad-infected cells from apoptosis triggered by binding of a ligand to its cognate death receptor, some of the mutant proteins were assayed for their ability to block this process. HeLa cells plated on coverslips were transiently cotransfected with plasmids that express a marker protein (LANFGR), RIDα, E3-6.7K, and wild-type or mutant forms of RIDβ. Although not required for RID-mediated internalization and degradation of Fas, the E3-6.7K expression plasmid was included in these experiments since this protein is necessary for RID-mediated down-regulation of TRAIL receptor 2 (5). Apoptosis was induced by treatment with either a Fas agonist monoclonal antibody or TRAIL. LANGFR was detected by indirect immunofluorescence, and nuclear morphology was assessed by DAPI staining. The nuclei of LANGFR-positive cells were scored as being normal or apoptotic. The results shown are representative of two independent experiments. Under the treatment conditions used, about 75% of the LANGFR-positive cells were apoptotic when treated with either the Fas agonist antibody (Fig. 8A) or TRAIL (Fig. 8B). Coexpression of RIDα, E3-6.7K, plus wild-type RIDβ reduced the percentage of apoptotic cells to about 4% in Fas agonist-treated cells and 17% in TRAIL-treated cells. For Fas agonist-treated cells, most of the mutant proteins tested, including Y124F RIDβ, were able to reduce the percentage of apoptotic cells to a level similar to that of wild-type RIDβ. Y124A RIDβ provided only marginal protection of cells from Fas-mediated apoptosis, reducing the level of apoptosis to about 52%. In the presence of wild-type RIDβ TRAIL-induced apoptosis was reduced to about 17%, whereas most mutant proteins reduced the level of apoptosis to 26 to 37% (Fig. 8B). The Y124A mutant was unable to protect cells from TRAIL-mediated apoptosis, suggesting that TRAIL receptor internalization and degradation were impaired in this mutant. Similar results for protection from TRAIL-induced apoptosis were obtained in transiently transfected A549 cells, suggesting that the phenotype of the mutant proteins is not cell-type specific (data not shown). These results showed that, of the mutant proteins tested, only the Y124A mutant protein was defective in its ability to protect cells from death receptor-mediated apoptosis.
FIG. 8.
Y124A RIDβ cannot protect cells from Fas agonist-induced or TRAIL-induced apoptosis. HeLa cells on coverslips transfected with various combinations of plasmids were treated with cycloheximide and either the CH-11 Fas agonist monoclonal antibody (A) or TRAIL (B). Following treatment, cells were fixed and immunostained with a monoclonal antibody directed against LANGFR and AlexaFluor 594 conjugated to goat anti-mouse IgG1 (A) or goat anti-mouse IgG (B). Data are presented as the percentage of transfected cells that were apoptotic (see Materials and Methods for the calculation method) and are representative of two independent experiments.
DISCUSSION
The hypothesis that RID performs its function by interacting with a cellular protein(s) was addressed by constructing and functionally testing site-specific point mutants of the Ad5 RIDβ gene. The mutations target sequences within the cytoplasmic tail of RIDβ that could potentially mediate its molecular mechanism of action and that are part of previously described protein-protein interaction motifs. None of the mutant proteins was grossly defective in synthesis, stability, or subcellular localization (Fig. 2, 4, and 5). However, in functional assays, the mutant Y124A was clearly defective in its ability to internalize and degrade Fas and EGFR (Fig. 5, 6, and 7 and data not shown) and could not protect cells from apoptosis triggered by TRAIL or a Fas agonist antibody (Fig. 8). These results demonstrated that tyrosine 124 is important for RIDβ function. Supporting this idea is the fact that this tyrosine residue is absolutely conserved in all 21 known RIDβ sequences, representing serotypes in all six subgroups of human Ads, present in GenBank (D. L. Lichtenstein and W. S. M. Wold, unpublished observations). These data suggested that tyrosine 124 forms part of a functional Yxxφ motif.
Two types of tyrosine-based motifs have been described that mediate protein-protein interactions and that contain the core sequence Yxxφ, the SH2 ligand motif, and the Yxxφ sorting motif. The SH2 ligand motif binds to proteins containing an SH2 domain (79). Proteins containing the SH2 domain generally function in signal transduction but also function in other pathways (79). The tyrosine-based sorting motif binds to the μ chain of AP complexes and allows incorporation of motif-containing proteins into clathrin-coated vesicles (35). A critical difference between these two tyrosine-based motifs is the requirement for phosphorylation of the tyrosine residue. Biochemical studies demonstrated that interaction between the SH2 domain and its ligand is regulated by the phosphorylation status of the tyrosine residue; efficient binding occurs only when the tyrosine residue is phosphorylated (reviewed in reference 79). Conversely, the Yxxφ sorting motif will only interact with μ chains when the tyrosine residue is not phosphorylated (10, 11, 55, 65). Indeed, the crystal structure of a peptide containing a Yxxφ motif bound to the μ subunit of the AP-2 complex (μ2) suggests that phosphotyrosine would not fit in the μ2 binding pocket (57).
The observation that RIDβ retains its function when tyrosine 124 is mutated to phenylalanine rules out a role for tyrosine phosphorylation in RID function and indicates that this tyrosine residue is not part of an SH2 ligand motif. In support of this idea, previous experiments have shown that wild-type RIDβ is not phosphorylated on tyrosine residues in the presence of RIDα (40). (It should be noted that RIDβ is phosphorylated to a low extent on tyrosine in the absence of RIDα; the functional significance of this phosphorylation is not known [40]).
Our data are in accord with the notion that RIDβ functions by interacting with the cellular sorting machinery through a Yxxφ sorting motif and not by affecting signal transduction pathways. The Yxxφ sorting motif mediates the interaction of membrane proteins with AP complexes by binding to the μ chain of AP complexes (56, 67). AP complexes are membrane-associated cytoplasmic complexes that also bind to clathrin and thus link the cellular sorting machinery with membrane proteins targeted for transport (7, 35). Currently, there are four known types of AP complexes, AP-1 through AP-4 (reviewed in reference 7). The AP-2 complex has been shown to be involved in endocytosis of cell surface proteins and is the only AP complex known to be localized to the inner leaf of the plasma membrane (7). The other AP complexes function in different aspects of membrane protein targeting and are localized to the late-Golgi/trans-Golgi network, endosomes, and lysosomes (7). Since RID acts at the plasma membrane to internalize cell surface receptors, RIDβ may interact with μ2. Experiments are under way to test this hypothesis.
The idea that RIDβ contains a functional Yxxφ sorting motif is supported by studies that examined the sequences required for sorting signal function and binding to μ subunits. These studies show that the tyrosine residue is important for function but that in some cases activity could be retained by substitution with phenylalanine (10, 15, 50, 55, 56). Notably, alanine cannot substitute for tyrosine (10, 15, 50, 55, 56). In accordance with these studies, Y124F RIDβ was functional in all of the assays, whereas Y124A RIDβ was not functional in any assay in which it was tested (Fig. 5 to 8). Taken together, these observations suggest that tyrosine 124 forms part of the core of a functional Yxxφ sorting motif.
In argument against the conclusion stated above, however, were the results with L127A RIDβ. This mutant protein was functional (Fig. 5 to 8) despite evidence from previous studies of other proteins that contain a Yxxφ motif, which showed that when alanine occupies the φ position of the Yxxφ motif the protein is not efficiently internalized (15, 20, 24). In addition, studies with combinatorial peptide libraries, yeast two-hybrid interaction assays, or glutathione _S_-transferase fusion proteins that examined the sequence requirements for Yxxφ motif binding to the μ subunit of the AP complex have shown that when alanine is in the φ position the peptide or fusion protein does not bind efficiently (10, 55, 56). These studies indicate that leucine is preferred in the φ position, but that isoleucine, phenylalanine, methionine and, to a lesser extent, valine are acceptable (10, 55, 56). It is possible that some aspect of RIDα function could compensate for a partially defective Yxxφ motif in RIDβ.
The ability of a viral protein to use sorting motifs to subvert the host trafficking machinery has been demonstrated previously for the Nef protein of human and simian immunodeficiency viruses (HIV and SIV, respectively). Nef protein contains tyrosine-based (SIV) (59) and dileucine-based (HIV and SIV) (1, 12, 64) sorting signals that are required for cell surface down-regulation of the CD4 receptor. In addition, Le Gall et al. (42) proposed that the HIV Nef protein unveils a cryptic Yxxφ motif present in HLA-A and -B molecules that is required for Nef-mediated down-regulation of MHC I expression. Interestingly, the consensus sequences of HLA-A and -B indicate that the cryptic Yxxφ motif should be nonfunctional, since the sequence is YSQA. Those authors suggested that Nef functions by directly or indirectly mediating interaction of MHC I molecules with AP complexes, thereby overcoming what would normally be a nonfunctional Yxxφ motif (42). As described for Nef, RIDα might compensate for the mutation present in L127A RIDβ. Together, these data show that Nef acts as a bridge connecting certain cell surface receptors with the sorting machinery (27, 49). Furthermore, these data demonstrate that a viral protein can coerce cellular proteins into interacting with the sorting machinery in a manner that is not normally appropriate.
RID may similarly act as a bridge that connects cell surface receptors to the sorting machinery. RID might act on its target receptors by enhancing recognition of their Yxxφ and/or dileucine sorting motifs. Alternatively, cryptic sorting signals may be revealed within the receptors that RID down-regulates. Whereas Nef has been shown to bind directly to CD4 (28), RID binding to any of its target receptors has yet to be conclusively demonstrated, thus raising the question of how RID attains specificity. Specificity may be achieved by RID binding to another cellular protein(s), which in turn binds to specific receptors.
Serine 116 was identified as the site at which RIDβ is phosphorylated, since mutant proteins that substituted alanine for serine at position 116 were clearly no longer phosphorylated (Fig. 3). However, lack of phosphorylation did not result in any gross defect in the ability of these mutant proteins to internalize and degrade receptors and to protect cells from death receptor-mediated apoptosis (Fig. 5 to 8). These studies did not rule out more subtle effects on RID function, such as altered kinetics of receptor clearance. Alternatively, the S116A mutant proteins could have differential effects on the clearance of TRAIL receptor 1 or TRAIL receptor 2 that may not have been detected in the experiment that was performed (Fig. 8). In addition, the other known function of RID, namely inhibition of cPLA2 translocation to membranes, was not investigated and, thus, may be affected in the phosphorylation-defective mutant proteins. Likewise, the lack of any observed defect for the three mutant proteins in which alanine was substituted for proline (P88A, P112A, and P114A), the acidic domain mutant protein (D131A/D132A), and the Y124F and Y127A mutant proteins could also be explained as described above. Further experiments assaying the kinetics of receptor clearance, internalization and degradation of additional receptors, assessment of TRAIL receptor 1 versus TRAIL receptor 2 clearance, and the translocation of cPLA2 should be performed to address the lack of any observed defect for these mutants.
Acknowledgments
We thank Chris Wells for technical assistance and Dawn Schwartz for help in preparing the manuscript.
This research was supported by grant CA58538 from the National Institutes of Health.
REFERENCES
- 1.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76**:**853-864. [DOI] [PubMed] [Google Scholar]
- 2.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Immunol. Today 21**:**447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, M., A. McMichael, and P. A. Peterson. 1987. Reduced allorecognition of adenovirus-2 infected cells. J. Immunol. 138**:**3960-3966. [PubMed] [Google Scholar]
- 4.Andersson, M., S. Pääbo, T. Nilsson, and P. A. Peterson. 1985. Impaired intracellular transport of class I MHC antigens as a possible means for adenoviruses to evade immune surveillance. Cell 43**:**215-222. [DOI] [PubMed] [Google Scholar]
- 5.Benedict, C., P. Norris, T. Prigozy, J. L. Bodmer, J. A. Mahr, C. Garnett, F. Martinon, J. Tschopp, L. R. Gooding, and C. F. Ware. 2001. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J. Biol. Chem. 276**:**3270-3278. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, E. M., J. R. Bennink, J. W. Yewdell, and F. M. Brodsky. 1999. Cutting edge: adenovirus E19 has two mechanisms for affecting class I MHC expression. J. Immunol. 162**:**5049-5052. [PubMed] [Google Scholar]
- 7.Boehm, M., and J. S. Bonifacino. 2002. Genetic analyses of adaptin function from yeast to mammals. Gene 286**:**175-186. [DOI] [PubMed] [Google Scholar]
- 8.Boldin, M. P., T. M. Goncharov, Y. V. Goltsev, and D. Wallach. 1996. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85**:**803-815. [DOI] [PubMed] [Google Scholar]
- 9.Boldin, M. P., E. E. Varfolomeev, Z. Pancer, I. L. Mett, J. H. Camonis, and D. Wallach. 1995. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J. Biol. Chem. 270**:**7795-7798. [DOI] [PubMed] [Google Scholar]
- 10.Boll, W., H. Ohno, Z. Songyang, I. Rapoport, L. C. Cantley, J. S. Bonifacino, and T. Kirchhausen. 1996. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15**:**5789-5795. [PMC free article] [PubMed] [Google Scholar]
- 11.Bradshaw, J. D., P. Lu, G. Leytze, J. Rodgers, G. L. Schieven, K. L. Bennett, P. S. Linsley, and S. E. Kurtz. 1997. Interaction of the cytoplasmic tail of CTLA-4 (CD152) with a clathrin-associated protein is negatively regulated by tyrosine phosphorylation. Biochemistry 36**:**15975-15982. [DOI] [PubMed] [Google Scholar]
- 12.Bresnahan, P. A., W. Yonemoto, and W. C. Greene. 1999. Cutting edge: SIV Nef protein utilizes both leucine- and tyrosine-based protein sorting pathways for down-regulation of CD4. J. Immunol. 163**:**2977-2981. [PubMed] [Google Scholar]
- 13.Burgert, H.-G., and S. Kvist. 1985. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 41**:**987-997. [DOI] [PubMed] [Google Scholar]
- 14.Burgert, H.-G., J. L. Maryanski, and S. Kvist. 1987. “E3/19K” protein of adenovirus type 2 inhibits lysis of cytolytic T lymphocytes by blocking cell-surface expression of histocompatibility class I antigens. Proc. Natl. Acad. Sci. USA 84**:**1356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canfield, W. M., K. F. Johnson, R. D. Ye, W. Gregory, and S. Kornfeld. 1991. Localization of the signal for rapid internalization of the bovine cation-independent mannose 6-phosphate/insulin-like growth factor-II receptor to amino acids 24-29 of the cytoplasmic tail. J. Biol. Chem. 266**:**5682-5688. [PubMed] [Google Scholar]
- 16.Carlin, C. R., A. E. Tollefson, H. A. Brady, B. L. Hoffman, and W. S. M. Wold. 1989. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell 57**:**135-144. [DOI] [PubMed] [Google Scholar]
- 17.Cesareni, G., S. Panni, G. Nardelli, and L. Castagnoli. 2002. Can we infer peptide recognition specificity mediated by SH3 domains? FEBS Lett. 513**:**38-44. [DOI] [PubMed] [Google Scholar]
- 18.Chen, P., J. Tian, I. Kovesdi, and J. B. Bruder. 1998. Interaction of the adenovirus 14.7K protein with FLICE inhibits Fas ligand-induced apoptosis. J. Biol. Chem. 273**:**5815-5820. [DOI] [PubMed] [Google Scholar]
- 19.Chinnaiyan, A. M., K. O'Rourke, M. Tewari, and V. M. Dixit. 1995. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81**:**505-512. [DOI] [PubMed] [Google Scholar]
- 20.Collawn, J. F., M. Stangel, L. A. Kuhn, V. Esekogwu, S. Q. Jing, I. S. Trowbridge, and J. A. Tainer. 1990. Transferrin receptor internalization sequence YXRF implicates a tight turn as the structural recognition motif for endocytosis. Cell 63**:**1061-1072. [DOI] [PubMed] [Google Scholar]
- 21.Dimitrov, T., P. Krajcsi, T. W. Hermiston, A. E. Tollefson, M. Hannink, and W. S. M. Wold. 1997. Adenovirus E3-10.4K/14.5K protein complex inhibits tumor necrosis factor-induced translocation of cytosolic phospholipase A2 to membranes. J. Virol. 71**:**2830-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elsing, A., and H.-G. Burgert. 1998. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. USA 95**:**10072-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman, J. M., and M. S. Horwitz. 2002. Inhibition of tumor necrosis factor alpha-induced NF-κB activation by the adenovirus E3-10.4/14.5K complex. J. Virol. 76:5515-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girones, N., E. Alverez, A. Seth, I. M. Lin, D. A. Latour, and R. J. Davis. 1991. Mutational analysis of the cytoplasmic tail of the human transferrin receptor. Identification of a sub-domain that is required for rapid endocytosis. J. Biol. Chem. 266**:**19006-19012. [PubMed] [Google Scholar]
- 25.Gooding, L. R., L. W. Elmore, A. E. Tollefson, H. A. Brady, and W. S. M. Wold. 1988. A 14,700 MW protein from the E3 region of adenovirus inhibits cytolysis by tumor necrosis factor. Cell 53**:**341-346. [DOI] [PubMed] [Google Scholar]
- 26.Gooding, L. R., I. O. Sofola, A. E. Tollefson, P. Duerksen-Hughes, and W. S. M. Wold. 1990. The adenovirus E3-14.7K protein is a general inhibitor of tumor necrosis factor-mediated cytolysis. J. Immunol. 145**:**3080-3086. [PubMed] [Google Scholar]
- 27.Greenberg, M. E., S. Bronson, M. Lock, M. Neumann, G. N. Pavlakis, and J. Skowronski. 1997. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J. 16**:**6964-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grzesiek, S., S. J. Stahl, P. T. Wingfield, and A. Bax. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 35**:**10256-10261. [DOI] [PubMed] [Google Scholar]
- 29.Hattula, K., and J. Peranen. 2000. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 10**:**1603-1606. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman, P., M. B. Yaffe, B. L. Hoffman, S. Yei, W. S. M. Wold, and C. Carlin. 1992. Characterization of the adenovirus E3 protein that down-regulates the epidermal growth factor receptor. Evidence for intermolecular disulfide bonding and plasma membrane localization. J. Biol. Chem. 267**:**13480-13487. [PubMed] [Google Scholar]
- 31.Horton, T. M., T. S. Ranheim, L. Aquino, D. I. Kusher, S. K. Saha, C. F. Ware, W. S. M. Wold, and L. R. Gooding. 1991. Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis. J. Virol. 65**:**2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horwitz, M. 2001. Adenovirus immunoregulatory genes and their cellular targets. Virology 279**:**1-8. [DOI] [PubMed] [Google Scholar]
- 33.Hsu, H., J. Huang, H.-B. Shu, V. Baichwal, and D. V. Goeddel. 1996. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 4**:**387-396. [DOI] [PubMed] [Google Scholar]
- 34.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1993. Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 121**:**317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchhausen, T. 1999. Adaptors for clathrin-mediated traffic. Annu. Rev. Cell Dev. Biol. 15**:**705-732. [DOI] [PubMed] [Google Scholar]
- 36.Krajcsi, P., T. Dimitrov, T. W. Hermiston, A. E. Tollefson, T. S. Ranheim, S. B. Vande Pol, A. H. Stephenson, and W. S. M. Wold. 1996. The adenovirus E3-14.7K protein and the E3-10.4K/14.5K complex of proteins, which independently inhibit tumor necrosis factor (TNF)-induced apoptosis, also independently inhibit TNF-induced release of arachidonic acid. J. Virol. 70**:**4904-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krajcsi, P., A. E. Tollefson, C. W. Anderson, A. R. Stewart, C. R. Carlin, and W. S. M. Wold. 1992. The E3-10.4K protein of adenovirus is an integral membrane protein that is partially cleaved between Ala22 and Ala23 and has a Ccyt orientation. Virology 187**:**131-144. [DOI] [PubMed] [Google Scholar]
- 38.Krajcsi, P., A. E. Tollefson, C. W. Anderson, and W. S. M. Wold. 1992. The adenovirus E3 14.5-kilodalton protein, which is required for down-regulation of the epidermal growth factor receptor and prevention of tumor necrosis factor cytolysis, is an integral membrane protein oriented with its C terminus in the cytoplasm. J. Virol. 66**:**1665-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krajcsi, P., A. E. Tollefson, and W. S. M. Wold. 1992. The E3-14.5K integral membrane protein of adenovirus that is required for down-regulation of the EGF receptor and for prevention of TNF cytolysis is O-glycosylated but not N-glycosylated. Virology 188**:**570-579. [DOI] [PubMed] [Google Scholar]
- 40.Krajcsi, P., and W. S. M. Wold. 1992. The adenovirus E3-14.5K protein which is required for prevention of TNF cytolysis and for down-regulation of the EGF receptor contains phosphoserine. Virology 187**:**492-498. [DOI] [PubMed] [Google Scholar]
- 41.Laster, S. M., W. S. M. Wold, and L. R. Gooding. 1994. Adenovirus proteins that regulate susceptibility to TNF also regulate the activity of PLA2. Semin. Virol. 5**:**431-442. [Google Scholar]
- 42.Le Gall, S., L. Erdtmann, S. Benichou, C. Berlioz-Torrent, L. Liu, R. Benarous, J. M. Heard, and O. Schwartz. 1998. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8**:**483-495. [DOI] [PubMed] [Google Scholar]
- 43.Li, Y., J. Kang, J. Friedman, L. Tarassishin, J. Ye, A. Kovalenko, D. Wallach, and M. S. Horwitz. 1999. Identification of a cell protein (FIP-3) as a modulator of NF-κB activity and as a target of an adenovirus inhibitor of tumor necrosis factor α-induced apoptosis. Proc. Natl. Acad. Sci. USA 96**:**1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, Y., J. Kang, and M. S. Horwitz. 1997. Interaction of an adenovirus 14.7-kilodalton protein inhibitor of tumor necrosis factor alpha cytolysis with a new member of the GTPase superfamily of signal transducers. J. Virol. 71**:**1576-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, Y., J. Kang, and M. S. Horwitz. 1998. Interaction of an adenovirus E3 14.7-kilodalton protein with a novel tumor necrosis factor alpha-inducible cellular protein containing leucine zipper domains. Mol. Cell. Biol. 18**:**1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104**:**487-501. [DOI] [PubMed] [Google Scholar]
- 47.Lukashok, S., L. Tarassishin, Y. Li, and M. Horwitz. 2000. An adenovirus inhibitor of tumor necrosis factor alpha-induced apoptosis complexes with dynein and a small GTPase. J. Virol. 74**:**4705-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahr, J. A., and L. R. Gooding. 1999. Immune evasion by adenoviruses. Immunol. Rev. 168**:**121-130. [DOI] [PubMed] [Google Scholar]
- 49.Mangasarian, A., M. Foti, C. Aiken, D. Chin, J. L. Carpentier, and D. Trono. 1997. The HIV-1 Nef protein acts as a connector with sorting pathways in the Golgi and at the plasma membrane. Immunity 6**:**67-77. [DOI] [PubMed] [Google Scholar]
- 50.McGraw, T. E., and F. R. Maxfield. 1990. Human transferrin receptor internalization is partially dependent upon an aromatic amino acid on the cytoplasmic domain. Cell Regul. 1**:**369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moise, A. R., J. R. Grant, T. Z. Vitalis, and W. A. Jefferies. 2002. Adenovirus E3-6.7K maintains calcium homeostasis and prevents apoptosis and arachidonic acid release. J. Virol. 76**:**1578-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muzio, M., A. M. Chinnaiyan, and F. C. Kischkel. 1996. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85**:**817-827. [DOI] [PubMed] [Google Scholar]
- 53.Muzio, M., B. R. Stockwell, H. R. Stennicke, G. S. Salvesen, and V. M. Dixit. 1998. An induced proximity model for caspase-8 activation. J. Biol. Chem. 273**:**2926-2930. [DOI] [PubMed] [Google Scholar]
- 54.Nilsson, T., M. Jackson, and P. A. Peterson. 1989. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell 58**:**707-718. [DOI] [PubMed] [Google Scholar]
- 55.Ohno, H., M. C. Fournier, G. Poy, and J. S. Bonifacino. 1996. Structural determinants of interaction of tyrosine-based sorting signals with the adaptor medium chains. J. Biol. Chem. 271**:**29009-29015. [DOI] [PubMed] [Google Scholar]
- 56.Ohno, H., J. Stewart, M. C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, and J. S. Bonifacino. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269**:**1872-1875. [DOI] [PubMed] [Google Scholar]
- 57.Owen, D. J., and P. R. Evans. 1998. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science 282**:**1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pääbo, S., B. M. Bhat, W. S. M. Wold, and P. A. Peterson. 1987. A short sequence in the COOH-terminus makes an adenovirus membrane glycoprotein a resident of the endoplasmic reticulum. Cell 50**:**311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piguet, V., Y. L. Chen, A. Mangasarian, M. Foti, J. L. Carpentier, and D. Trono. 1998. Mechanism of Nef-induced CD4 endocytosis: Nef connects CD4 with the mu chain of adaptor complexes. EMBO J. 17**:**2472-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280**:**248-253. [DOI] [PubMed] [Google Scholar]
- 61.Prill, V., L. Lehmann, K. von Figura, and C. Peters. 1993. The cytoplasmic tail of lysosomal acid phosphatase contains overlapping but distinct signals for basolateral sorting and rapid internalization in polarized MDCK cells. EMBO J. 12**:**2181-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rawle, F. C., A. E. Tollefson, W. S. M. Wold, and L. R. Gooding. 1989. Mouse anti-adenovirus cytotoxic T lymphocytes. Inhibition of lysis by E3 gp19K but not E3 14.7K. J. Immunol. 143**:**2031-2037. [PubMed] [Google Scholar]
- 63.Rudoll, T., K. Phillips, S. W. Lee, S. Hull, O. Gaspar, N. Sucgang, E. Gilboa, and C. Smith. 1996. High-efficiency retroviral vector mediated gene transfer into human peripheral blood CD4+ T lymphocytes. Gene Ther. 3**:**695-705. [PubMed] [Google Scholar]
- 64.Salghetti, S., R. Mariani, and J. Skowronski. 1995. Human immunodeficiency virus type 1 Nef and p56lck protein-tyrosine kinase interact with a common element in CD4 cytoplasmic tail. Proc. Natl. Acad. Sci. USA 92**:**349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiratori, T., S. Miyatake, H. Ohno, C. Nakaseko, K. Isono, J. S. Bonifacino, and T. Saito. 1997. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity 6**:**583-589. [DOI] [PubMed] [Google Scholar]
- 66.Shisler, J., C. Yang, B. Walter, C. F. Ware, and L. R. Gooding. 1997. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J. Virol. 71**:**8299-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sorkin, A., and G. Carpenter. 1993. Interaction of activated EGF receptors with coated pit adaptins. Science 261**:**612-615. [DOI] [PubMed] [Google Scholar]
- 68.Stanger, B. Z., P. Leder, T. H. Lee, E. Kim, and B. Seed. 1995. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell 81**:**513-523. [DOI] [PubMed] [Google Scholar]
- 69.Stewart, A. R., A. E. Tollefson, P. Krajcsi, S. P. Yei, and W. S. M. Wold. 1995. The adenovirus E3 10.4K and 14.5K proteins, which function to prevent cytolysis by tumor necrosis factor and to down-regulate the epidermal growth factor receptor, are localized in the plasma membrane. J. Virol. 69**:**172-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ting, A. T., F. X. Pimentel-Muinos, and B. Seed. 1996. RIP mediates tumor necrosis factor receptor 1 activation of NF-κB but not Fas/APO-1-initiated apoptosis. EMBO J. 15**:**6189-6196. [PMC free article] [PubMed] [Google Scholar]
- 71.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. M. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392**:**726-730. [DOI] [PubMed] [Google Scholar]
- 72.Tollefson, A. E., P. Krajcsi, M. H. Pursley, L. R. Gooding, and W. S. M. Wold. 1990. A 14,500 MW protein is coded by region E3 of group C human adenoviruses. Virology 175**:**19-29. [DOI] [PubMed] [Google Scholar]
- 73.Tollefson, A. E., P. Krajcsi, S. P. Yei, C. R. Carlin, and W. S. M. Wold. 1990. A 10,400-molecular-weight membrane protein is coded by region E3 of adenovirus. J. Virol. 64**:**794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tollefson, A. E., A. R. Stewart, S. P. Yei, S. K. Saha, and W. S. M. Wold. 1991. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to down-regulate the epidermal growth factor receptor. J. Virol. 65**:**3095-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tollefson, A. E., K. Toth, K. Doronin, M. Kuppuswamy, O. A. Doronina, D. L. Lichtenstein, T. W. Hermiston, C. A. Smith, and W. S. M. Wold. 2001. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 75**:**8875-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trowbridge, I. S., J. F. Collawn, and C. R. Hopkins. 1993. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 9**:**129-161. [DOI] [PubMed] [Google Scholar]
- 77.Voorhees, P., E. Deignan, E. van Donselaar, J. Humphrey, M. S. Marks, P. J. Peters, and J. S. Bonifacino. 1995. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of _trans_-Golgi network localization and internalization from the cell surface. EMBO J. 14**:**4961-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wold, W. S. M., and G. Chinnadurai. 2000. Adenovirus proteins that regulate apoptosis, p. 200-232. In A. J. Cann (ed.), DNA virus replication: frontiers in molecular biology. Oxford University Press, Oxford, United Kingdom.
- 79.Yaffe, M. B. 2002. Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 3**:**177-186. [DOI] [PubMed] [Google Scholar]
- 80.Ye, J., X. Xie, L. Tarassishin, and M. Horwitz. 2000. Regulation of the NF-κB activation pathway by isolated domains of FIP3/IKKγ, a component of the IκB-α kinase complex. J. Biol. Chem. 275**:**9882-9889. [DOI] [PubMed] [Google Scholar]
- 81.Zilli, D., C. Voelkel-Johnson, T. Skinner, and S. M. Laster. 1992. The adenovirus E3 region 14.7 kDa protein, heat and sodium arsenite inhibit the TNF-induced release of arachidonic acid. Biochem. Biophys. Res. Commun. 188**:**177-183. [DOI] [PubMed] [Google Scholar]