Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition (original) (raw)
Abstract
The anaphase-promoting complex/cyclosome (APC/C) is a multisubunit E3 ligase required for ubiquitin-dependent proteolysis of cell-cycle-regulatory proteins, including mitotic cyclins and securin/Pds1. Regulation of APC/C activity and substrate recognition, mediated by the coactivators Cdc20 and Cdh1, is fundamental to cell-cycle control. However, the precise mechanism by which coactivators stimulate APC/C ubiquitylation activity and the nature of the substrate-binding sites on the activated APC/C are not understood. Here, we show that the optimal interaction of substrate with APC/C is dependent specifically on the simultaneous association of coactivator. This is consistent with a model whereby both core APC/C subunits and coactivators contribute recognition sites for substrates, accounting for the bipartite nature (D and KEN boxes) of most APC/C degradation signals. A direct and stoichiometric function for the coactivators could explain how specific substrates are recognized by APC/C in a cell-cycle-specific manner, and how coactivator stimulates APC/C ubiquitylation activity.
Keywords: APC/C, cell cycle, E3 ligase, substrate recognition, ubiquitin
Introduction
Progression through the cell cycle is controlled by means of an interplay of phosphorylation by cyclin-dependent kinases (CDKs), and ubiquitin-mediated proteolysis, triggered by the anaphase-promoting complex/cyclosome (APC/C) and SCF (Skp1–Cul1–F-box protein; Reed, 2003). The proteolytic events triggered by APC/C are required to release sister chromatid cohesion during anaphase, specify the exit from mitosis and prevent premature entry into S phase.
Budding yeast APC/C is composed of 13 core subunits, most of which are conserved in all eukaryotes (Harper et al, 2002; Peters, 2002; Yoon et al, 2002; Hall et al, 2003; Passmore et al, 2003). Regulatory phosphorylations and coactivator proteins (Cdc20/fizzy or Cdh1/Hct1/fizzy-related) activate APC/C to ubiquitylate substrates containing destruction (D) and/or KEN boxes in a cell-cycle-dependent manner (Schwab et al, 1997; Visintin et al, 1997; Fang et al, 1998; Kramer et al, 1998; Zachariae et al, 1998; Jaspersen et al, 1999; Harper et al, 2002; Passmore et al, 2003). Cdc20 activates APC during mitosis, whereas Cdh1 promotes APC activity during G1. Coactivators are themselves regulated during the cell cycle by transcription, proteolysis and phosphorylation. Functional and biochemical studies of APC/C in vitro have shown that coactivators are required for APC/C E3 ligase activity (Passmore et al, 2003). APC/C activation was correlated with the ability of coactivator to promote APC/C–substrate interactions, indicating that at least one role of the coactivators is to generate a high-affinity substrate recognition site on APC/C. However, the mechanism of coactivator-induced APC/C–substrate recognition remains unclear, partly because the nature of the substrate-binding sites in the activated APC/C is unknown. Both specific APC/C subunits (Meyn et al, 2002; Passmore et al, 2003; Nourry et al, 2004; Yamano et al, 2004) and coactivators (Pfleger & Kirschner, 2000; Burton & Solomon, 2001; Hilioti et al, 2001; Pfleger et al, 2001; Passmore et al, 2003) have been implicated in substrate recognition, and this has led to diverse and conflicting proposals for coactivator functions (Fig 1). For example, the coactivators could act as substrate (co-)receptors (Fig 1, scheme Ia), functioning in a similar manner to F-box proteins in the SCF E3 ligase. In support of this, several studies have reported a direct association between purified coactivators and substrates (Burton & Solomon, 2001; Hilioti et al, 2001; Pfleger et al, 2001; Schwab et al, 2001). Nevertheless, coactivator is not sufficient for substrate binding, as APC/C–coactivator complexes lacking the Doc1/Apc10 subunit cannot bind to the substrate, a defect that can be fully restored by addition of purified Doc1 (Passmore et al, 2003). A second potential role for the coactivators would be to mediate a conformational change in APC/C, creating a substrate-binding site coincident with stable coactivator association (Fig 1, scheme Ib). In a contrasting model, prompted by the finding that mitotic Xenopus APC/C interacts with the substrate D box in the absence of coactivator, Yamano et al (2004) proposed that coactivator would act in a substoichiometric manner to convert APC/C into a form capable of binding to the substrate. In this model (Fig 1, schema IIa,b), coactivator functions catalytically and transiently, either as a (co-)chaperone to alter APC/C conformation or by recruiting enzymes (such as kinases or phosphatases) to post-translationally modify APC/C.
Figure 1.
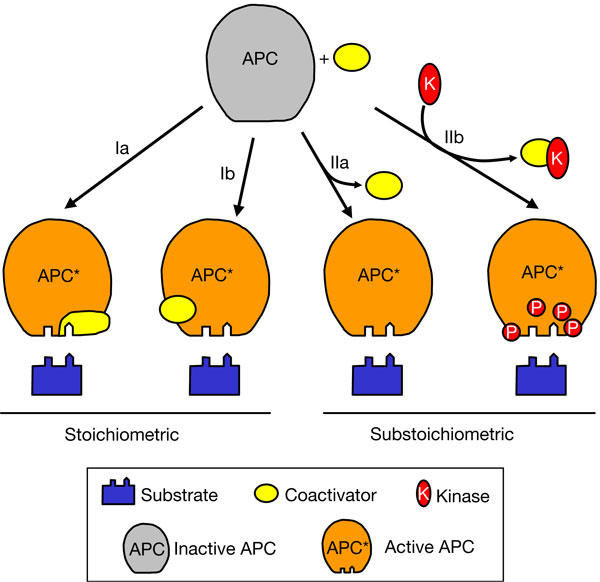
Possible roles for coactivators in activating the anaphase-promoting complex/cyclosome. Substrates contain two known motifs (D and KEN boxes) required for anaphase-promoting complex/cyclosome (APC/C) binding. See text for details.
Each of these contrasting modes of coactivator action has different implications for understanding how APC/C achieves substrate specificity and how checkpoint proteins (e.g. Mad1, Mad2 and Bub1) inhibit APC/C and/or coactivators. A possible explanation for the opposing models of coactivator function is that most studies investigate the substrate-binding properties of coactivator or APC/C in isolation, but have not considered how an APC/C–coactivator binary complex recognizes substrate. Therefore, we have examined how coactivators influence the ability of APC/C to recognize substrate in a defined in vitro system. To investigate the composition of APC/C–substrate complexes and the mechanism of coactivator-induced APC/C–substrate recognition, we developed a unique native gel binding assay that can detect simultaneous substrate and coactivator interactions with APC/C. Here, we show that substrate association with APC/C is dependent on the formation of a stoichiometric complex with APC/C and coactivator.
Results
Visualization of APC/C complexes on native gels
To visualize APC/C–coactivator complexes, 35S-labelled Cdh1 was produced using an in vitro transcription/translation (IVT) system, incubated with purified APC/C and run on a native/non-denaturing gel. 35S-labelled free Cdh1 and Cdh1–APC/C complexes can be resolved and visualized by autoradiography (Fig 2A). APC/CCdh1 migrates as two distinct species, possibly corresponding to monomer and dimer, and both of these complexes undergo band shifts after the addition of antibodies to the APC/C subunits Cdc16 and Cdc27 (Passmore et al, 2003). Cdh1 also interacts with the chaperonin CCT in the reticulocyte lysate (Passmore et al, 2003), consistent with a requirement for CCT to facilitate coactivator function in vivo (Camasses et al, 2003).
Figure 2.
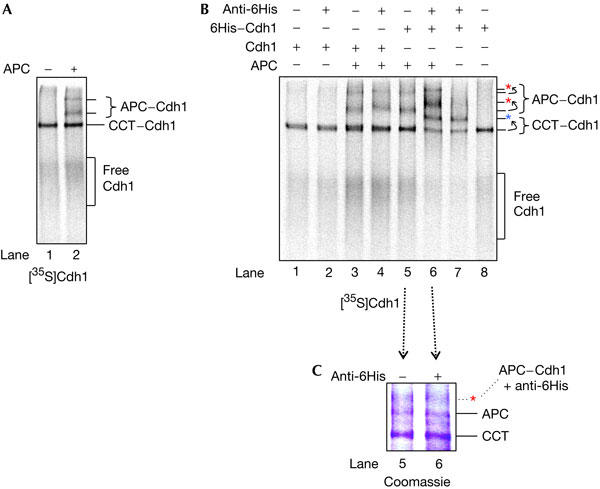
Native gel assays showing an interaction between Cdh1 and the anaphase-promoting complex (APC). (A) Purified APC/cyclosome (APC/C) was added to in vitro transcription/translation-produced [35S]Cdh1, run on a native (non-denaturing) gel and visualized by autoradiography. APC/CCdh1 complexes migrate as two species on a native gel. The positions of free [35S]Cdh1 and [35S]Cdh1 in complex with APC/C or CCT are marked. (B) [35S]Cdh1 (lanes 1–4) and [35S]6His–Cdh1 (lanes 5–8) were run on a native gel in the presence or absence of purified APC/C and a 6His antibody. CCT6His–Cdh1 and APC/C6His–Cdh1 complexes undergo large band shifts following the addition of the antibody (dotted arrows). The shifted complexes are indicated by red (APC/C6His–Cdh1) and blue (CCT6His–Cdh1) asterisks. A small shift in the migration of APC/CCdh1 is observed after the addition of the 6His antibody (lane 4). This change in migration is small compared with the supershift of APC/C6His–Cdh1 caused by the specific binding of the 6His antibody (compare lanes 4 and 6), allowing APC/CCdh1 and APC/C6His–Cdh1 to be clearly distinguished using the 6His antibody. (C) Coomassie blue stain of a native gel containing 6His–Cdh1 and APC/C, in the absence and presence of a 6His antibody (this is the Coomassie blue stain of lanes 5 and 6 of the gel in (B)). The upper APC/C6His–Cdh1 band is not clearly visible by Coomassie blue staining. The red asterisk marks the expected position of bandshifted APC/CCdh1, on the basis of its position on the autoradiograph of (B). Unlabelled bands represent unknown proteins present in the rabbit reticulocyte lysate.
We tested whether an antibody directed towards an amino-terminal 6His tag on Cdh1 would induce a band shift of Cdh1-containing complexes. Fig 2B shows that the 6His antibody binds to [35S]6His–Cdh1 in the native gel, retarding the mobilities of both free 6His–Cdh1 and 6His–Cdh1 complexes. In particular, the migrations of the two APC/C6His–Cdh1 complexes (compare lanes 5 and 6) and CCT6His–Cdh1 (compare lanes 7 and 8) are retarded following the addition of the 6His antibody. An untagged version of Cdh1 does not bind to the 6His antibody (lanes 1–4).
Fig 2A,B shows that the position of APC/CCdh1 on native gels can be visualized by autoradiography owing to the 35S-labelled Cdh1. An antibody to 6His–Cdh1 quantitatively shifts all the APC/C6His–Cdh1 complexes. Although apo-APC/C cannot be detected using autoradiography, it was important to determine the relative proportions of apo-APC/C and APC/CCdh1 in the IVT reactions expressing Cdh1. APC/C can be visualized on a native gel by Coomassie blue staining, and apo-APC/C and labelled APC/CCdh1 have the same mobilities (Passmore et al, 2003). Fig 2C shows that the migration position of APC/C (lane 5) is not significantly altered by addition of a 6His antibody (lane 6). Because the unshifted APC/C band in lane 6 corresponds to apo-APC/C, and we are unable to detect the shifted APC/C6His–Cdh1 complex by Coomassie blue staining, apo-APC/C must be present in a large excess of the APC/C6His–Cdh1 complex.
Substrates associate with APC/C–coactivator complexes
Together, the above results validate our assay system, showing that the use of a 6His antibody specifically alters the migration of APC/C6His–Cdh1, distinguishing it from apo-APC/C. A 6His-tagged protein ligand of APC/C (in this instance, 6His–Cdh1) can be used to detect the resultant APC/C complex both by virtue of its 35S label and by its ability to be supershifted by a 6His antibody. Therefore, we could use this assay to determine whether two ligands associate with APC/C simultaneously, by labelling one ligand with 35S and the other with a 6His tag. Thus, we investigated whether the substrate Clb2 can bind to APC/C in a ternary complex with Cdh1, or whether Clb2 and Cdh1 can interact with APC/C separately.
[35S]Clb2 binds to APC/C in native gels, but only in the presence of coactivator (Passmore et al, 2003; Fig 3A, lane 3). After addition of an antibody to the 6His tag on Cdh1, the migration of the APC/C–Clb2 complex is retarded in the gel (Fig 3A, lane 6). This band shift is similar to that seen for APC/C6His–Cdh1 (Fig 2B, lane 6). Importantly, as the 6His tag is on Cdh1 and not on [35S]Clb2, the shifted complexes must contain both Cdh1 and Clb2. When untagged Cdh1 is used instead of 6His–Cdh1, no band shift is observed (Fig 3A, compare lanes 4 and 6). This result shows, for the first time, that substrate can bind to APC/C as an APC/C–coactivator–substrate ternary complex. Moreover, in this assay, the substrate exclusively bound to APC/C6His–Cdh1 and not to apo-APC/C because the addition of a 6His antibody shifted all of the 35S-labelled APC/C–Clb2 complexes. If Clb2 were capable of binding to apo-APC/C, an unshifted APC/CClb2 band would have been observed. As Clb2 is polyubiquitylated by APC/C isolated from budding yeast, and its interaction with the complex is dependent on intact D and KEN boxes (Passmore et al, 2003), the interactions that we observe here are likely to be physiologically relevant. Significantly, our finding that APC/C, Cdh1 and Clb2 co-associate correlates with Cdh1-induced activation of APC/C-mediated ubiquitylation of Clb2.
Figure 3.
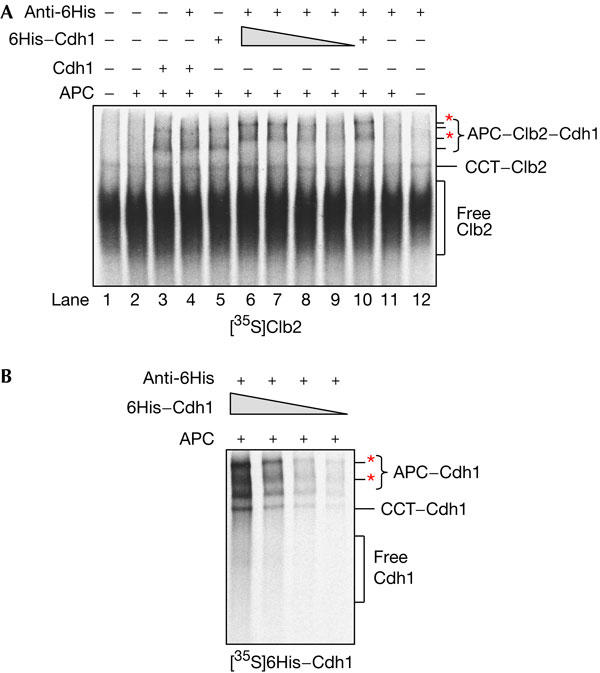
The anaphase-promoting complex/cyclosome (APC/C) binds to the substrate Clb2 as an APC/C–coactivator–substrate ternary complex. (A) In vitro transcription/translation (IVT)-produced [35S]Clb2 was analysed by native gels and autoradiography. When purified APC/C and Cdh1 or 6His–Cdh1 is incubated with [35S]Clb2, APC/C–[35S]Clb2 complexes can be observed. The addition of a 6His antibody causes band shifts (red asterisks) in the APC/C6His–Cdh1–[35S]Clb2 complexes (lane 6) but not the APC/CCdh1–[35S]Clb2 complexes (lane 4). In lanes 6–9, varying amounts of 6His–Cdh1 (2, 1, 0.5 or 0.25 μl IVT reaction) were used. (B) The native gel assay was used to analyse varying amounts of [35S]6His–Cdh1 (2, 1, 0.5 or 0.25 μl IVT reaction) mixed with purified APC/C and a 6His antibody. Red asterisks indicate APC/C6His–Cdh1 complexes retarded by the 6His antibody. In all other native gels, 2 μl Cdh1 IVT reaction was used.
Coactivator and substrate bind APC/C stoichiometrically
The Coomassie gel in Fig 2C suggests that APC/C is present in a large excess of Cdh1. However, it was important to confirm that apo-APC/C is available, which could in principle interact with substrates, and that the amount of Cdh1 was substantially less than that of APC/C. Thus, to detect the presence of possible apo-APC/C–substrate complexes, we titrated 6His–Cdh1 in the native gel assay. Fig 3A (lanes 6–9) shows that even with approximately eightfold less 6His–Cdh1, [35S]Clb2 binds to APC/C6His–Cdh1 and not to apo-APC/C. In addition, in the presence of lower amounts of 6His–Cdh1, the quantity of [35S]Clb2 that binds to APC/C is reduced, suggesting that Cdh1 is limiting and that formation of an APC/C–substrate complex is dependent on the availability of Cdh1. Fig 3B shows that the amount of the APC/C6His–Cdh1 complex also decreases following a reduction in the quantity of [35S]6His–Cdh1, supporting the idea that Cdh1 is limiting and that the concentration of apo-APC/C greatly exceeds that of APC/CCdh1 complexes. These results confirm the stoichiometric model in which substrate binds to APC/C in a ternary complex with coactivator.
Although our results show that substrate binding to APC/C is dependent on Cdh1, it was important to establish whether all APC/CCdh1 is competent to bind to substrate. Thus, we performed a reciprocal native gel binding experiment in which we used 6His-tagged Clb2 and untagged [35S]Cdh1. [35S]Cdh1 binds to APC/C and its migration in the native gel is not affected significantly by the addition of an antibody to a 6His tag or by the addition of 6His–Clb2 (Fig 4A, lanes 3,4,6). However, when APC/C, 6His–Clb2 and the 6His antibody are added together with [35S]Cdh1, all of the APC/CCdh1 complex undergoes a band shift (Fig 4A, lane 5). This indicates that all APC/CCdh1 is competent to bind to substrate and confirms the existence of a ternary complex between APC/C, coactivator and substrate.
Figure 4.
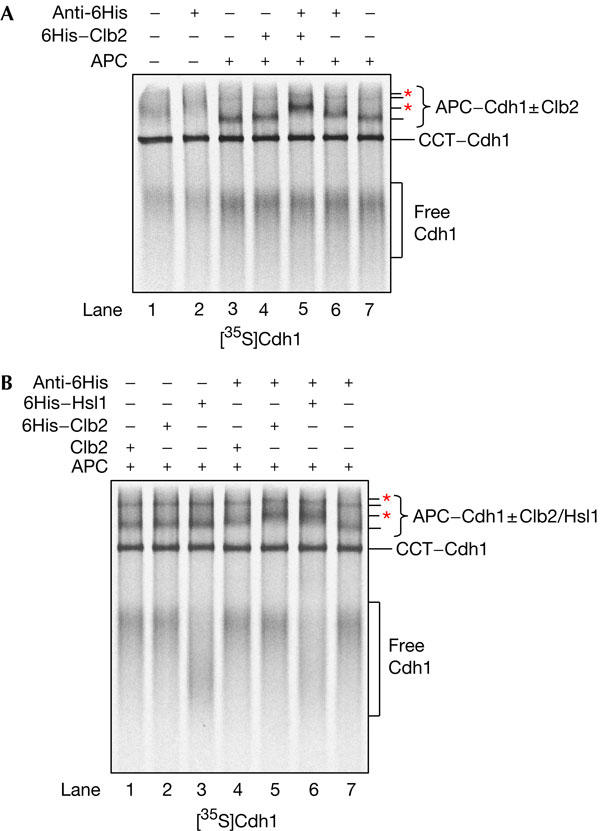
APC/CCdh1 is competent to bind to substrate. In a reciprocal native gel binding assay, [35S]Cdh1 was incubated with purified anaphase-promoting complex/cyclosome (APC/C), a 6His-tagged substrate and an antibody to the 6His tag. The experiment was performed with (A) 6His–Clb2 and with (B) untagged Clb2 (as a negative control), 6His–Clb2 or 6His–Hsl1667−872. APC/C complexes shifted by the antibody are marked with red asterisks.
To extend our findings, we performed a similar experiment using a second APC/C substrate, 6His–Hsl1667−872, a well characterized D-box- and KEN-box-containing fragment of the APC/C substrate Hsl1 (Burton & Solomon, 2001; Passmore et al, 2003). In Fig 4B, the 6His antibody induces a band shift in the APC/CCdh1 complex (formed with [35S]Cdh1) when it is incubated with 6His–Hsl1667−872. Therefore, it is likely that, in general, all substrates interact with APC/C in a ternary complex with coactivator. Intriguingly, 6His–Hsl1667−872 changes the migration pattern of free [35S]Cdh1 (Fig 4B, lanes 3,6), suggesting that free coactivator and substrate may interact in the native gel.
Discussion
Here, we have developed an assay system to distinguish between a stoichiometric and a substoichiometric/catalytic role for coactivator to facilitate APC/C–substrate interactions (Fig 1). Unlike other studies, we have addressed how coactivators influence the ability of the core APC/C to interact with substrates. To our knowledge, this is the first study to analyse APC binding simultaneously to substrate and coactivator. We show unequivocally that only a stoichiometric binary complex of APC/C and coactivator is capable of binding native substrates, and that all APC/CCdh1 is competent to bind to substrates (both Clb2 and Hsl1). Our results exclude the possibility that coactivators increase the affinity of APC/C for substrates by acting substoichiometrically in a catalytic process (Fig 1, schema IIa,b). Instead, our results favour a mechanism whereby the coactivators either contribute directly to the substrate-binding site, and/or cause a conformational change in APC/C to expose a cryptic substrate-binding site (Fig 1, schema Ia,b).
These results allow us to reconcile apparently contradictory reports about the function of coactivator in stimulating APC/C ubiquitylation activity. Our data show that coactivator interacts with APC/C to increase its affinity for substrate. Processive ubiquitylation of substrate is therefore induced because the increased lifetime of the APC/C–substrate complex would be sufficient for the catalytic reaction to proceed. Additionally, the presence of coactivator may optimally position and orientate the substrate at the APC/C catalytic site. Although our study does not exclude the possibility that coactivator functions only to alter the conformation of the core APC/C, most existing data are consistent with the idea that coactivator itself contributes directly to substrate recognition (Burton & Solomon, 2001; Hilioti et al, 2001; Pfleger et al, 2001; Schwab et al, 2001), possibly by acting as a D-box receptor (Kraft et al, 2005). We propose that to generate an optimal high-affinity substrate-binding site, APC/C uses two substrate receptors—one contributed by core APC/C subunits and the other by coactivator.
Evidence that core APC/C subunits contribute to substrate recognition is suggested from several studies. The APC/C subunit, Doc1, is specifically required for substrate recognition, as APC/C lacking Doc1 binds normally to coactivator but fails to bind to substrate and is defective as an E3 ubiquitin ligase (Carroll & Morgan, 2002; Passmore et al, 2003). A recent study has suggested that Doc1 promotes recognition of the substrate D box (Carroll et al, 2005), although this may be an indirect effect, as direct Doc1–substrate interactions have not been observed (data not shown). Moreover, interactions between Xenopus APC/C and D-box substrates have been observed in the absence of coactivator, suggesting the presence of a D-box receptor on APC/C (Yamano et al, 2004). This latter result prompted the idea that coactivator functions substoichiometrically to activate APC/C. However, because ubiquitylation activity of the APC/C–D-box substrate complex was not shown, this study does not formally exclude a stoichiometric role for coactivator in defining increased APC/C–substrate affinity for optimal lifetime and catalytic site positioning necessary for substrate ubiquitylation. It is, therefore, consistent with our new data that coactivators contribute to optimal APC/C–substrate associations.
Recently, Burton et al (2005) published results suggesting that binding of substrate to Cdh1 enhances the formation of APC–Cdh1 complexes. However, our results do not provide support for this hypothesis: in Fig 4, we show that the Cdh1–APC interaction is not enhanced in the presence of substrate, despite the presence of excess free Cdh1.
The role of coactivators in enhancing APC/C–substrate affinity explains their ability to both activate APC/C and define substrate specificity. Dual recognition of the substrate by both core APC/C subunits and coactivator would allow strict regulation of substrate binding and ubiquitylation, therefore providing tight control of cell-cycle transitions. It is also consistent with the presence of bipartite destruction motifs (i.e. a D box and KEN box) in APC/C substrates, a feature that distinguishes APC/C-mediated ubiquitylation from that of the SCF and other E3 ligases.
Methods
Cdc16-TAP APC/C was purified from yeast as described previously (Passmore et al, 2003, 2005). Plasmids containing Clb2, 6His–Clb2, 6His–Hsl1667−872, Cdh1 and 6His–Cdh1 have been described previously (Passmore et al, 2003). Substrates and coactivators were synthesized using the TNT T7 Quick coupled in vitro transcription/translation kit (Promega, Madison, WI, USA) at 30°C for 90 min. Redivue L-[35S]methionine (Amersham, Little Chalfont, Buckinghamshire, UK) was used for labelling proteins.
For binding reactions (Passmore et al, 2005), 2 μl of IVT-produced coactivator, 2 μl of IVT-produced substrate, 2 μl of purified APC/C (∼50 ng), 0.7 μl of 100 mM CaCl2 and 5.3 μl of binding buffer (10 mM Tris pH 8, 150 mM NaCl, 3 mM dithiothreitol, 1 mM magnesium acetate and 2 mM EGTA) were mixed. For control reactions, reticulocyte lysate was substituted for IVT-produced proteins or APC/C storage buffer (Passmore et al, 2003) was substituted for APC/C. Next, 2 μl of 6His monoclonal antibody (Clontech, Mountain View, CA, USA) or 2 μl of antibody buffer (50% glycerol) was added for a total volume of 14 μl. The reactions were incubated at room temperature for 15 min and then 1.5 μl of native gel sample buffer (125 mM Tris pH 8.8, 84% glycerol and bromophenol blue) was added. The reaction was immediately loaded onto a 5.25% non-denaturing polyacrylamide gel. Gels were prepared using the Biorad Mini-PROTEAN system (8 cm × 7.3 cm plates, 1.5 mm spacers, 15 well combs) as follows: resolving gel, 0.37 M Tris pH 8.8, 5.25% 37.5:1 acrylamide:bis-acrylamide; stacking gel, 57 mM Tris pH 8.8, 3.22% 37.5:1 acrylamide:bis-acrylamide; running buffer, 25 mM Tris, 192 mM glycine pH 8.3. Electrophoresis was performed at 110 V and 4°C until 20 min after the dye-front ran off the gel. Gels were fixed, stained with Coomassie blue, dried and exposed to a Kodak BioMax MR-1 film.
Acknowledgments
We thank C. Fioretto for assistance in preparation of purified APC/C and E.A. McCormack and K.R. Willison for reagents and many useful discussions. This work was supported by Cancer Research UK.
References
- Burton JL, Solomon MJ (2001) D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev 15: 2381–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Tsakraklides V, Solomon MJ (2005) Assembly of an APC–Cdh1–substrate complex is stimulated by engagement of a destruction box. Mol Cell 18: 533–542 [DOI] [PubMed] [Google Scholar]
- Camasses A, Bogdanova A, Shevchenko A, Zachariae W (2003) The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell 12: 87–100 [DOI] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO (2002) The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat Cell Biol 4: 880–887 [DOI] [PubMed] [Google Scholar]
- Carroll CW, Enquist-Newman M, Morgan DO (2005) The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol 15: 11–18 [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell 2: 163–171 [DOI] [PubMed] [Google Scholar]
- Hall MC, Torres MP, Schroeder GK, Borchers CH (2003) Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem 278: 16698–16705 [DOI] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ (2002) The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev 16: 2179–2206 [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Chung YS, Mochizuki Y, Hardy CF, Cohen-Fix O (2001) The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr Biol 11: 1347–1352 [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO (1999) Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol 9: 227–236 [DOI] [PubMed] [Google Scholar]
- Kraft C, Vodermaier HC, Maurerstroh S, Eisenhaber F, Peters JM (2005) The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell 18: 543–553 [DOI] [PubMed] [Google Scholar]
- Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters JM (1998) Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol 8: 1207–1210 [DOI] [PubMed] [Google Scholar]
- Meyn MA III, Melloy PG, Li J, Holloway SL (2002) The destruction box of the cyclin Clb2 binds the anaphase-promoting complex/cyclosome subunit Cdc23. Arch Biochem Biophys 407: 189–195 [DOI] [PubMed] [Google Scholar]
- Nourry C, Maksumova L, Pang M, Liu X, Wang T (2004) Direct interaction between Smad3, APC10, CDH1 and HEF1 in proteasomal degradation of HEF1. BMC Cell Biol 5: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D (2003) Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J 22: 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Barford D, Harper JW (2005) Purification and assay of the budding yeast anaphase-promoting complex. Methods Enzymol 398: 195–219 [DOI] [PubMed] [Google Scholar]
- Peters JM (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell 9: 931–943 [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW (2000) The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev 14: 655–665 [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW (2001) Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev 15: 2396–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI (2003) Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol 4: 855–864 [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90: 683–693 [DOI] [PubMed] [Google Scholar]
- Schwab M, Neutzner M, Mocker D, Seufert W (2001) Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J 20: 5165–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A (1997) CDC20 and CDH1: a family of substratespecific activators of APC-dependent proteolysis. Science 278: 460–463 [DOI] [PubMed] [Google Scholar]
- Yamano H, Gannon J, Mahbubani H, Hunt T (2004) Cell cycle-regulated recognition of the destruction box of cyclin B by the APC/C in Xenopus egg extracts. Mol Cell 13: 137–147 [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Feoktistova A, Wolfe BA, Jennings JL, Link AJ, Gould KL (2002) Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr Biol 12: 2048–2054 [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W (1998) Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282: 1721–1724 [DOI] [PubMed] [Google Scholar]