siRNA function in RNAi: A chemical modification analysis (original) (raw)
Abstract
Various chemical modifications were created in short-interfering RNAs (siRNAs) to determine the biochemical properties required for RNA interference (RNAi). Remarkably, modifications at the 2′-position of pentose sugars in siRNAs showed the 2′-OHs were not required for RNAi, indicating that RNAi machinery does not require the 2′-OH for recognition of siRNAs and catalytic ribonuclease activity of RNA-induced silencing complexes (RISCs) does not involve the 2′-OH of guide antisense RNA. In addition, 2′ modifications predicted to stabilize siRNA increased the persistence of RNAi as compared with wild-type siRNAs. RNAi was also induced with chemical modifications that stabilized interactions between A–U base pairs, demonstrating that these types of modifications may enhance mRNA targeting efficiency in allele-specific RNAi. Modifications altering the structure of the A-form major groove of antisense siRNA–mRNA duplexes abolished RNAi, suggesting that the major groove of these duplexes was required for recognition by activated RISC*. Comparative analysis of the stability and RNAi activities of chemically modified single-stranded antisense RNA and duplex siRNA suggested that some catalytic mechanism(s) other than siRNA stability were linked to RNAi efficiency. Modified or mismatched ribonucleotides incorporated at internal positions in the 5′ or 3′ half of the siRNA duplex, as defined by the antisense strand, indicated that the integrity of the 5′ and not the 3′ half of the siRNA structure was important for RNAi, highlighting the asymmetric nature of siRNA recognition for initiation of unwinding. Collectively, this study defines the mechanisms of RNAi in human cells and provides new rules for designing effective and stable siRNAs for RNAi-mediated gene-silencing applications.
Keywords: RNAi, siRNA, human, nucleotide modification, GFP
INTRODUCTION
The evolutionarily conserved phenomenon RNA interference (RNAi), the process by which specific mRNAs are targeted for degradation by complementary short-interfering RNAs (siRNAs), has increasingly become a powerful tool for genetic analysis and is likely to become a potent therapeutic approach for gene silencing (for review, see Hammond et al. 2001; McManus and Sharp 2002). Consequently, understanding the mechanism of RNAi has become critical for developing the most effective RNAi methodologies for both laboratory and clinical applications. The general mechanism of RNAi involves the cleavage of double-stranded RNA (dsRNA) to short 21–23-nt siRNAs. This processing event is catalyzed by Dicer, a highly conserved, dsRNA-specific endonuclease that is a member of the RNase III family (Hammond et al. 2000; Zamore et al. 2000; Bernstein et al. 2001; Hamilton et al. 2002; Provost et al. 2002; Zhang et al. 2002). Processing by Dicer results in siRNA duplexes that have 5′-phosphate and 3′-hydroxyl termini, and subsequently, these siRNAs are recognized by the RNA-induced silencing complex (RISC; Hammond et al. 2000). Active RISC complexes (RISC*) promote the unwinding of the siRNA through an ATP-dependent process, and the unwound antisense strand guides RISC* to the complementary mRNA (Nykanen et al. 2001). The targeted mRNA is then cleaved by RISC* at a single site that is defined with regard to where the 5′-end of the antisense strand is bound to the mRNA target sequence (Hammond et al. 2000; Elbashir et al. 2001b). For RNAi-mediated mRNA cleavage and degradation to be successful, 5′-phosphorylation of the antisense strand must occur, and the double helix of the antisense-target mRNA duplex must be in the A form (Chiu and Rana 2002).
One highlighted difference between mammalian RNAi and RNAi in other eukaryotes is the lack of an amplification system for long-term persistence of RNAi in mammalian cells. For example, in Drosophila, ~35 molecules of dsRNA can silence ~1000 copies of the targeted mRNA per cell and can persist over the course of many generations (Kennerdell and Carthew 1998; Zamore 2001). In mammalian cells, RNAi only persists effectively for an average of ~66 h before the siRNA is likely diluted out over the course of several cell divisions (Chiu and Rana 2002). The amplification that is seen in flies and other lower eukaryotes can potentially be attributed to three factors. One is that the conversion of long trigger dsRNA to smaller 21–23-nt siRNAs by Dicer adds a degree of RNAi amplification, whereas in mammalian cells long trigger dsRNA invokes the interferon response that activates the protein kinase PKR (Stark et al. 1998). This suggests that only siRNA transfections successfully trigger RNAi in mammalian cells without other side effects, and thus, no amplification would take place through the processing of longer RNAs. A second factor in amplification is the presence of RNA-dependent RNA polymerase (RdRP), which has been found in plants, worms, fungi, and flies (Cogoni and Macino 1999; Dalmay et al. 2000; Sijen et al. 2001). RdRP has been postulated to amplify target mRNA, through a random, degradative PCR model (Lipardi et al. 2001; Nishikura 2001; Sijen et al. 2001), into dsRNA, which can be targeted by Dicer. However, no RdRP homologs have been found in mammalian cells, and the 3′-OH that is required for RdRP-dependent degradative PCR is not required for RNAi in mammalian cells (Chiu and Rana 2002; Schwarz et al. 2002; Stein et al. 2003), indicating that PCR-based amplification likely does not occur in mammals. A third factor in amplification may be the high enzymatic turnover rate of RISC* during the targeting and cleavage of mRNA (Hutvagner and Zamore 2002), which may add a degree of amplification to RNAi induction in all eukaryotes, including mammals. However, as the persistence of RNAi occurs for only a short period of time, finding methods for increasing the longevity of siRNAs in human cells will be fundamental for applying RNAi to laboratory and therapeutic applications.
To address this issue of siRNA stability for prolonging the duration of dsRNA-mediated gene silencing and to further dissect the mechanism of RNAi in human cells, various chemically modified nucleotides predicted to affect siRNA stability were incorporated into siRNAs to study whether specific modifications increased or decreased the efficacy and persistence of RNAi in vivo. The most important of these modifications was to the 2′-OH of the ribonucleotide that distinguishes RNA from DNA and is required for the nucleophilic attack occurring during the hydrolysis of the RNA backbone, the reaction catalyzed by degradative RNases. Our results showed that the 2′-OH was not required for RNAi, indicating that structural rather than chemical properties of siRNA–mRNA duplexes were the key to inducing RNAi and that RISC* did not require the 2′-OH for ribonuclease activity. 2′-modified siRNAs also increased the persistence of RNAi in human cells. Modifications that stabilized base-pairing interactions were also incorporated into the antisense strand of siRNAs and were able to initiate RNAi, signifying that this class of chemical modifications could be used to increase the targeting efficiency of siRNAs for mRNA target sequences and for allele-specific inhibition of gene expression.
Other chemical modifications affected the formation of the major groove of the A-form helix of the antisense-siRNA–target-mRNA duplex, and potentially disrupted H-bonds or sterically hindered protein contacts, most probably preventing the RISC* complex from stably interacting with the dsRNA duplex. These modifications completely abolished RNAi, demonstrating that an intact major groove in the A-form helix and stable RNA–protein interactions were required for RNAi in human cells. Finally, previous observations of psorelan cross-linked siRNAs implied that unwinding of siRNA occurred from the 5′-end of the antisense strand and that complete unwinding may not be necessary for effective RNAi (Chiu and Rana 2002). By using mismatched or chemically modified nucleotides on either the 3′ or 5′ half of the antisense strand within the siRNA duplex, we have shown here that RNAi depended on the integrity of the 5′, and not the 3′, half of the siRNA duplex, as defined by the antisense strand. Altogether, these results gave insight into the essential biochemical properties of functional siRNAs and how specific changes in the siRNA structure can affect the efficacy of RNAi. Furthermore, these studies present new methodologies for improving the stability and utility of siRNAs for future RNAi applications.
RESULTS
2′-OH is not required for siRNA to enter the RNAi pathway
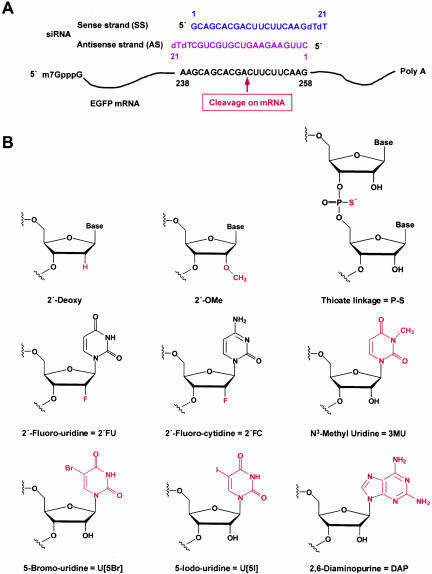
Previous results showed that RNAi effects typically peaked between 42 and 54 h posttransfection, and targeted gene expression started to be restored by 66 h posttransfection (Chiu and Rana 2002). To determine if the duration of RNAi could be prolonged by increasing the half-life of siRNAs, various chemical modifications were made to nucleotides that affected siRNA stability. These modified siRNAs were then tested in an improved dual fluorescence reporter assay based on the original assay developed previously (Chiu and Rana 2002). Briefly, GFP and RFP were constitutively expressed from pEGFP-C1 and pDsRed2-N1, respectively. EGFP mRNA was targeted for RNAi using a 21-nt siRNA targeted to nucleotides 238–258 of the EGFP mRNA (Fig. 1A ▶). The fluorescence intensity ratio of target (GFP) to control (RFP) fluorophores was determined in the presence of siRNA duplexes and normalized to that observed in the mock-treated cells. The sequence of EGFP siRNA and EGFP mRNA, the specific mRNA cleavage site, plus the structures of the chemically modified nucleotides are diagrammed in Figure 1 ▶. As outlined previously, the cleavage site was defined precisely 11 nt downstream of the target position complementary to the 3′-most nucleotide of the antisense guide siRNA (Elbashir et al. 2001a). The specific chemical modifications, the particular siRNA strand(s) where modifications were made, and the effect of the chemically modified siRNA on RNAi activity are summarized in Table 1 ▶. The RNAi activity of siRNAs was evaluated with eight different siRNA concentrations (ranging from 1 to 200 nM). Each experiment was completed in duplicate and repeated twice.
FIGURE 1.
Structures of EGFP siRNA and chemical modifications. (A) Graphical representation of dsRNAs used for targeting EGFP mRNA. EGFP was encoded by the pEGFP-C1 reporter plasmid. siRNAs were synthesized with 2-nt deoxythymidine overhangs at the 3′-end. The position of the first nucleotide of the mRNA target site is indicated relative to the start codon of EGFP mRNA. The sequence of the antisense strand of siRNA is exactly complementary to the mRNA target site. (B) Structure and nomenclature of chemical modifications.
TABLE 1.
RNA interference mediated by chemically modified siRNAs
| Row no. | EGFP siRNA | Sense strand | Antisense strand | RNAi activity (%) | RNAi activity (+ or −) |
|---|---|---|---|---|---|
| 1 | DS (WT) | Unmodified | Unmodified | 93 ± 0.70 | ++++ |
| 2 | SS/AS-2′-FU, FC | Unmodified | 2′-FU, FC | 83 ± 3.48 | ++++ |
| 3 | SS-2′-FU, FC/AS | 2′-FU, FC | Unmodified | 92 ± 0.98 | ++++ |
| 4 | DS-2′-FU, FC | 2′-FU, FC | 2′-FU, FC | 83 ± 0.01 | ++++ |
| 5 | SS/AS-2′-FU, FC + (9, 10, 13) dA, dG | Unmodified | 2′-FU, FC + (9, 10, 13) dA, dG | 85 ± 2.10 | ++++ |
| 6 | SS-2′-FU, FC/AS-2′-FU, FC + (9, 10, 13) dA, dG | 2′-FU, FC | 2′-FU, FC + (9, 10, 13) dA, dG | 64 ± 2.89 | +++ |
| 7 | SS/AS-2′-FU, FC + dA, dG | Unmodified | 2′-FU, FC + dA, dG | 42 ± 1.66 | ++ |
| 8 | SS-2′-FU, FC/AS-2′-FU, FC + dA, dG | 2′-FU, FC | 2′-FU, FC + dA, dG | 44 ± 0.60 | ++ |
| 9 | SS/AS-Deoxy | Unmodified | Deoxy | 0 ± 5.97 | − |
| 10 | SS-Deoxy/AS | Deoxy | Unmodified | 38 ± 2.95 | + |
| 11 | DS-Deoxy | Deoxy | Deoxy | 0 ± 0.01 | − |
| 12 | SS/AS-2′-OMe | Unmodified | 2′-OMe | 16 ± 4.41 | − |
| 13 | SS-2′-OMe/AS | 2′-OMe | Unmodified | 25 ± 1.75 | + |
| 14 | DS-2′-OMe | 2′-OMe | 2′-OMe | 0 ± 0.01 | − |
| 15 | SS/AS-P-S | Unmodified | P-S | 42 ± 6.03 | ++ |
| 16 | SS-P-S/AS | P-S | Unmodified | 62 ± 0.07 | +++ |
| 17 | DS-P-S | P-S | P-S | 47 ± 0.03 | ++ |
| 18 | SS/AS-2′-FU, FC + P-S | Unmodified | 2′-FU, FC + P-S | 22 ± 0.03 | + |
| 19 | SS/AS-U[5Br] | Unmodified | U[5Br] | 70 ± 1.88 | +++ |
| 20 | SS/AS-U[5I] | Unmodified | U[5I] | 59 ± 11.2 | +++ |
| 21 | SS/AS-DAP | Unmodified | DAP | 51 ± 0.57 | ++ |
| 22 | SS-2′-FU, FC/AS-U[5Br] | 2′-FU, FC | U[5Br] | 31 ± 1.88 | + |
| 23 | SS-2′-FU, FC, FC/AS-U[5I] | 2′-FU, FC | U[5I] | 42 ± 5.02 | ++ |
| 24 | SS-2′-FU, FC/AS-DAP | 2′-FU, FC | DAP | 35 ± 7.69 | + |
| 25 | SS/AS-3MU | Unmodified | 3MU | 0 ± 6.65 | − |
| 26 | SS/AS-(11) 3MU | Unmodified | (11) 3MU | 0 ± 1.71 | − |
| 27 | SS/AS-(1, 2) mm | Unmodified | (1, 2) mm | 35 ± 5.69 | + |
| 28 | SS/AS-(18, 19) mm | Unmodified | (18, 19) mm | 77 ± 2.00 | +++ |
| 29 | SS/AS-2′-FU, FC + (1-13) dA, dG | Unmodified | 2′-FU, FC + (1-13) dA, dG | 43 ± 0.09 | ++ |
| 30 | SS-2′-FU, FC/AS-2′-FU, FC + (1-13) dA, dG | 2′-FU, FC | 2′-FU, FC + (1-13) dA, dG | 45 ± 2.23 | ++ |
| 31 | SS/AS-2′-FU, FC + (9-19) dA, dG | Unmodified | 2′-FU, FC + (9-19) dA, dG | 91 ± 0.36 | ++++ |
| 32 | SS-2′-FU, FC/AS-2′-FU, FC + (9-19) dA, dG | 2′-FU, FC | 2′-FU, FC + (9-19) dA, dG | 64 ± 0.42 | +++ |
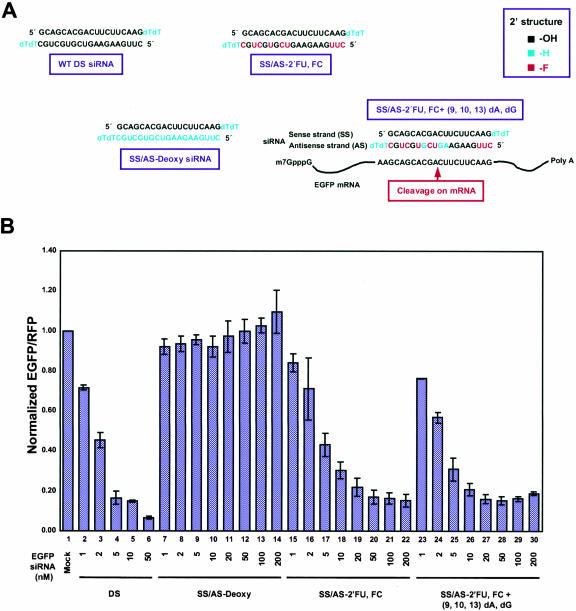
The effects of modifying the 2′-OH of nucleotides on RNAi were studied by replacing uridine and cytidine in the antisense strand of siRNA with 2′-fluoro-uridine (2′-FU) and 2′-fluoro-cytidine (2′-FC), respectively, which have a fluoro group at the 2′-position in place of the 2′-OH (Fig. 1B ▶). Where these modified 2′-FU, 2′-FC nucleotides reside in the siRNA sequence are highlighted in red in Figure 2A ▶. Addition of a 2′-fluoro group should increase the stability of the siRNA by making the siRNAs less recognizable to RNases, thereby providing siRNAs protection from degradation (see below). When measured in the dual fluorescence assay, 2′-FU, 2′-FC siRNAs, modified only in the sense strand, only in the antisense strand, or in both strands, all showed decreased EGFP fluorescence when normalized to nontargeted RFP fluorescence that was comparable to the normalized decrease seen with wild-type siRNAs (Fig. 2 ▶; Table 1 ▶, rows 1–4). These results indicated that the 2′-OH was not required for RNAi and that nucleotides modified with 2′-fluoro groups could be used in siRNA constructs to successfully induce RNAi-mediated gene silencing.
FIGURE 2.
siRNA 2′-OH is not required to guide mRNA cleavage. (A) Sequence and structure of siRNA duplexes with modification at the 2′-position of the sugar unit. Nucleotides with 2′-hydroxyl groups (-OH) are black. Nucleotides with 2′-deoxy groups (-H) are cyan. Nucleotides with 2′-fluoro groups (-F) are red. The cleavage site on the target mRNA is also shown (red arrow). (B) Ratios of normalized GFP to RFP fluorescence intensity in lysates from modified siRNA-treated HeLa cells. The fluorescence intensity ratio of target (GFP) to control (RFP) fluorophores was determined in the presence of EGFP siRNA duplexes with modifications at the 2′-position of the sugar unit. Normalized ratios at <1.0 indicate specific RNA interference effects. For comparison, results from unmodified duplex siRNA-treated cells are included.
To support the conclusion that the 2′-OH was not required for RNAi, adenine and guanine deoxynucleotides that inherently have 2′-H in place of the 2′-OH (Fig. 1B ▶) were incorporated into the sense, antisense, or both strands of 2′-FU, 2′-FC-modified EGFP siRNAs to determine their effect on RNAi (Fig. 2A ▶; green nucleotides). When 2′-FU, 2′-FC nucleotides were incorporated into the EGFP siRNA antisense strand with guanine and adenine deoxynucleotides at positions 9, 10, and 13, which base pair with nucleotides lining the cleavage site (Fig. 2A ▶), EGFP RNAi effects were almost indistinguishable from wild-type levels (Fig. 2B ▶; Table 1 ▶, row 5). This same antisense construct base-paired to 2′-FU, 2′-FC-modified sense strands also showed considerable EGFP silencing at ~64% (Table 1 ▶, row 6). In addition, siRNAs that had the entire antisense strand replaced with 2′-FU, 2′-FC, dATP, and dGTP nucleotides still showed moderate levels of RNAi activity at ~42%, or ~44% if the sense strand was also modified with 2′-FU, 2′-FC (Table 1 ▶, rows 7,8). All together, these results demonstrated that a 2′-OH group was not required for RNAi-mediated degradation and, even more specifically, was not required for nucleotides base-paired with nucleotides lining the mRNA cleavage site. There was, however, a limit on the extent to which deoxynucleotides could substitute for ribonucleotides because replacing the entire siRNA sense strand with deoxynucleotides decreased EGFP gene silencing to ~38% inhibition, and replacing either the antisense strand or both strands entirely with deoxynucleotides completely abolished EGFP RNAi (Fig. 2B ▶; Table 1 ▶, rows 9–11). Nonetheless, these results collectively showed that nucleotides with either 2′-F or 2′-H groups can selectively replace ribonucleotides within the siRNA sequence to effectively induce RNAi.
An interesting result was seen by modifying the 2′-OH to a bulky methyl group to create 2′-OMe nucleotides that were incorporated into sense, antisense, or both strands of EGFP siRNAs (Fig. 1B ▶). This modification was hypothesized to improve RNAi efficacy because 2′-OMe groups are thought to increase RNA stability by inducing an altered RNA conformation that is more resistant to nucleases (Cummins et al. 1995). This modification is also thought to increase RNA affinity for RNA targets and improve hybridization kinetics (Majlessi et al. 1998). Despite these potential benefits, 2′-OMe nucleotides incorporated into either the sense or antisense strand greatly diminished EGFP gene silencing to ~25% or ~16%, respectively, whereas double-stranded 2′-OMe-modified siRNAs completely abolished RNAi (Table 1 ▶, rows 12–14). These results indicated that the methyl group, as a bulky group, may severely limit the interactions between siRNAs, target mRNAs, and the RNAi machinery required for successfully mediating RNAi. It is worth noting that because the bulkiness of the methyl group would likely be the cause of decreased RNAi activity rather than the actual lack of the 2′-OH specifically, these studies still supported the conclusion that the 2′-OH was not required for RNAi.
In a final analysis of modifications that may potentially increase siRNA stability without disrupting RNAi potency, a thioate linkage (P–S) was integrated into the backbone of the EGFP siRNA strand(s). P–S linkages were previously used in antisense methodology for increasing resistance to ribonucleases (for review, see Stein 1996) and, therefore, were postulated to enhance the stability of siRNAs. Incorporating the P–S linkages into the double-stranded siRNA sense strand led to moderate levels of RNAi activity (62% inhibition), whereas P–S linkages in either the antisense or both strands of the siRNAs led to just less than ~50% RNAi-induced inhibition (Table 1 ▶, rows 15–17). These results implied that the P–S modifications did not prohibit RNAi-mediated degradation and only moderately affected the efficiency of RNAi. Interestingly, incorporating 2′-FU, 2′-FC modifications into the antisense strand in addition to the added P–S linkages showed lower levels of EGFP gene silencing (Table 1 ▶, row 18), indicating that there was a synergistic effect that decreased but did not inhibit RNAi-mediated degradation when both the 2′-F groups and the P–S linkages were incorporated into siRNAs.
Stability of modified siRNAs and the persistence of their RNAi activity in vitro and in vivo
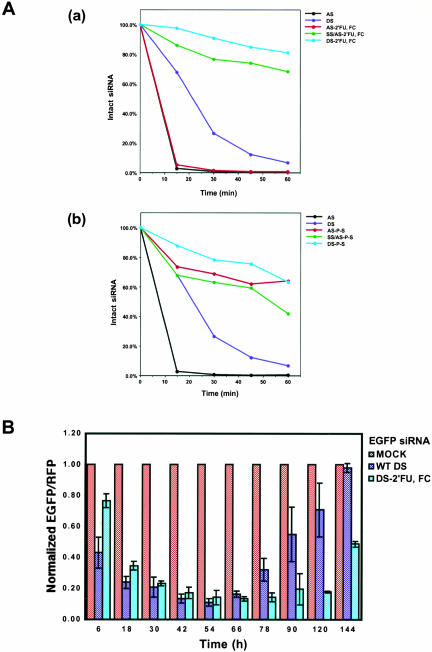
As the above experiments showed that siRNAs modified with stabilizing 2′-FU, 2′-FC groups could effectively mediate RNAi to levels comparable to wild type, it was necessary to show that these modifications did in fact enhance siRNA stability. To measure the stability of siRNA in cell extracts, unmodified or 2′-FU, 2′-FC-modified EGFP antisense strand siRNAs 5′-labeled with [γ-32P]ATP were annealed with sense strand siRNAs to form duplex siRNAs, which were then incubated in HeLa cell extracts. At various time points, siRNAs were extracted, analyzed on a 20% polyacrylamide gel containing 7 M urea, and visualized by phosphorimager analysis. Smaller siRNA degradation products were visualized in this analysis (data not shown), indicating that the lost of intact siRNA observed during these experiments was not caused by dephosphorylation of siRNAs. The top panel (a) of Figure 3A ▶ shows the stability of the various 2′-FU, 2′-FC-modified siRNAs as compared with wild-type siRNAs over time. Wild-type double-stranded siRNAs showed a steady loss of intact siRNAs over the course of the experiment, with only ~7% of the original concentration of intact siRNAs remaining after 1 h in extract (Fig. 3A ▶[a], dark blue line). Intact modified or unmodified single-stranded antisense siRNAs were quickly lost over the time course and were virtually undetectable by 30 min in extract (Fig. 3A ▶[a], black and red lines). This result showed that single-stranded modified siRNA was as susceptible to degradation as wild-type siRNA, indicating that single-stranded siRNAs, modified or unmodified, are inherently less stable than duplex siRNA. Double-stranded siRNAs with 2′-FU, 2′-FC modifications in either the antisense strand or both strands remained predominantly intact over the course of the experiment with ~68% or ~81%, respectively, of the original siRNA population remaining intact throughout the duration of the experiment (Fig. 3A ▶[a], green and light blue lines). These results indicated that the 2′-FU, 2′-FC modifications did, indeed, increase the stability of the siRNAs upon exposure to extract and that having these modifications in both strands provided the siRNAs with the most stability.
FIGURE 3.
Extending the half-life of siRNA duplexes prolongs the persistence of RNA interference in vivo. (A) Comparing the stability of unmodified siRNAs with siRNAs containing 2′-fluoro-uridine and 2′-fluoro-cytidine (2′-FU, 2′-FC) modifications (a) and thioate linkage (P–S) modifications (b). Unmodified or modified EGFP antisense strand siRNAs (AS) were 5′-labeled with [γ-32P]ATP by T4 polynucleotide kinases. Duplex siRNAs were formed by annealing equal molar ratios of sense-strand (SS) siRNAs with the 5′-32P-labeled antisense strand. To analyze siRNA stability in HeLa cell extract, 50 pmole of siRNA was incubated with 500 μg of HeLa cell extract in 50 μL of reaction mixture containing 20 mM HEPES (pH 7.9), 100 mM KCl, 10 mM NaCl, 2 mM MgCl2, and 10% glycerol. At various time points, siRNAs were extracted and analyzed on 20% polyacrylamide gels containing 7 M urea followed by phosphorimage analysis (Fugi). (B) Kinetics of RNAi effects of duplex siRNA with 2′-fluoro-uridine and 2′-fluoro-cytidine modification in HeLa cells over a 144-h time course. The fluorescence intensity ratio of target (GFP) to control (RFP) protein was determined in the presence of unmodified dsRNA (blue bars) and duplex siRNA with 2′-fluoro-uridine and -cytidine modifications (DS-2′-FU, 2′-FC, cyan bar) and normalized to the ratio observed in the presence of mock-treated cells (red bars). Normalized ratios at <1.0 indicated specific RNA interference.
In a similar experiment, the stability of P–S-modified EGFP siRNAs was evaluated. Unmodified, double-stranded antisense siRNAs showed about the same rate of siRNA loss as described in the above experiment (Fig. 3A ▶[b], dark blue lines). However, P–S-modified single-stranded antisense siRNAs demonstrated a markedly increased rate of stability over the course of the experiment, showing ~63% of the original siRNAs remaining intact after 1 h in extract as compared with 0% intact for single-stranded unmodified antisense siRNAs (Fig. 3A ▶[b], black and red lines). The stability of double-stranded siRNAs with P–S modifications in both strands was comparable to the stability seen with the modified single-stranded antisense strand, with ~63% of the original siRNA population remaining intact after 1 h (Fig. 3A ▶[b], light blue lines). Double-stranded siRNAs with P–S modifications in only the antisense strand showed weaker but still significant stability with ~42% of the original siRNA population remaining intact through 1 h in extract (Fig. 3A ▶[b], green lines). These results showed that the P–S modifications increased the stability of the siRNAs and, most notably, increased the stability of both single- and double-stranded siRNAs.
These in vitro results indicated that siRNA stability is prolonged by these different modifications; however, it is important to note that these experiments address the general stability of siRNA in the context of endonucleases present in whole-cell extracts. Therefore, these experiments cannot distinguish whether the endonucleases affecting siRNAs in the in vitro assay would necessarily affect the stability of these various siRNAs in vivo. To address whether increased stability seen with modified siRNAs prolonged the duration of RNAi in vivo, RNAi, induced by unmodified and 2′-FU, 2′-FC-modified double-stranded EGFP siRNAs, was assayed in the dual fluorescence reporter assay over a period of 144 h. To visualize RNAi effects over an even longer period of time, HeLa cells were transfected with modified or unmodified siRNA and, 36 h later, transfected with dual fluorescence reporter plasmids; RNAi activity persisted but was tapering by 168 h (data not shown). Also, growth of cells containing modified siRNAs was comparable to cells containing wild-type siRNA, indicating that modified siRNAs were not affecting cell division (data not shown). Although 2′-FU, 2′-FC-modified EGFP siRNAs were slower to show RNAi effects by 6–18 h, maximal RNAi effects occurred by 42 h posttransfection for both modified and unmodified siRNAs. The maximal activity for both siRNAs was also in the same range, with both showing ~85%–90% inhibition of GFP expression. However, the RNAi effects observed over the period of 66–120 h revealed that the effect of modified siRNAs was much more persistent than that of unmodified siRNAs. By 120 h posttransfection, the effect of modified siRNAs still remained at ~80% inhibition of GFP expression but the effect of unmodified siRNAs had dropped to less than ~40% inhibition. Similarly, prolonged RNAi activity was observed with 2′-FU, 2′-FC-modified siRNAs targeting endogenous human Cyclin T1 mRNA when compared with wild-type siRNAs targeting Cyclin T1 (see Discussion; Y.L. Chiu and T.M. Rana, unpubl.). Altogether, these results strongly indicated that there was a direct link between the duration of the RNAi effects and siRNA stability in human cells. Furthermore, these results showed conclusively that siRNAs stabilized by chemical modifications, like the 2′-FU, 2′-FC modifications, can be used to effectively induce and significantly prolong RNAi-mediated gene silencing in vivo.
Modified siRNAs that stabilize A–U base-pair interactions can induce RNAi
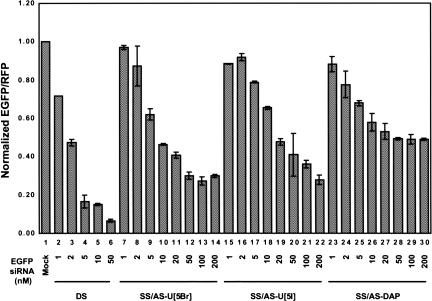
In addition to incorporating modifications that affected the stability of siRNAs, nucleotides chemically modified to strengthen the base-pair interactions between two complementary bases were analyzed. In theory, increasing the stability of base-pair interactions may increase the targeting efficiency of siRNAs to target mRNA sequences. Increasing targeting efficiency may then induce more robust RNAi effects with siRNAs that are weaker at binding to their target sequence or have mismatched sequences, and thus, are not showing a high degree of RNAi. This type of approach may also be used to significantly inhibit expression of one allele over another when both alleles are present in the same cell. To bolster base-pairing interactions, 5-bromo-uridine (U[5Br]), 5-iodo-uridine (U[5I]), or 2,6-diaminopurine (DAP; Fig. 1B ▶), which are modified nucleotides known to increase the association constant between A–U base pairs (Saenger 1984), were incorporated into siRNAs and tested in the dual fluorescence report assay. Double-stranded siRNAs having U[5Br], U[5I], or DAP modifications incorporated into the antisense strand were capable of inducing RNAi activity at levels of ~70% for U[5Br], ~59% for U[5I], and ~51% for DAP (Fig. 4 ▶; Table 1 ▶, rows 19–21). Interestingly, when 2′-FU, 2′-FC stabilizing modifications in the sense strand were combined with these modifications in the antisense strand, gene silencing was not as efficient as wild type in inducing RNAi. EGFP gene silencing was 31% for the 2′-FU, 2′-FC-modified sense siRNA base-paired with U[5Br]-modified, ~42% for U[5I]-modified, or ~35% for DAP-modified antisense siRNAs (Fig. 4 ▶; Table 1 ▶, rows 22–24). These results indicated that enhancing the interactions between base pairs through these siRNA modifications was a viable option for increasing mRNA targeting efficiency, but that there was a limit to how stable the base-pairing interactions can be made before they interfere with siRNA unwinding (see Discussion).
FIGURE 4.
RNA interference mediated by siRNAs harboring modifications that stabilize base-pairing interactions. 5-Bromo-uridine (U[5Br]) or 5-iodo-uridine (U[5I]) replaced uridine or 2,6-diaminopurine (DAP) replaced adenine in siRNAs to stabilize base-pairing interactions. The activity of siRNAs with base modifications was quantified by the dual fluorescence assay. For comparison, results from unmodified duplex siRNA (DS, lanes 2_–_6)-treated cells are included.
The major groove of the A-form helix is required for RNAi
Previously, the A-form helix was shown to be required for the mechanism of RNAi, as 2-nt bulges that distort A-form helices between antisense siRNAs and target mRNAs abolished RNAi (Chiu and Rana 2002). To test whether the major groove of the A-form helix was required for RNAi, siRNAs were modified with _N_3-methyl uridine (3MU) nucleotides that remove an H-bond donor at _N_3-H. Structurally, the bulky _N_3-methyl group would jut into the major groove of the A-form helix, potentially introducing a steric clash between base pairs. In addition, the presence of 3MU in the major groove may also introduce a steric clash between RNA and RNA-interacting proteins (Saenger 1984). Therefore, both steric hindrance and the loss of an H-bond donor by the addition of the _N_3-methyl group should destabilize RNA–protein interactions in the major groove. 3MU-modified EGFP siRNAs introduced into HeLa cells completely abolished RNAi (Table 1 ▶, row 25). RNAi was also abolished if only one 3MU modification was introduced specifically at U11 of the antisense strand, which is one of the nucleotides that base pairs with A248 of the target EGFP mRNA cleavage site (Fig. 1A ▶; Table 1 ▶, row 26). These results indicated that disrupting the functional groups of the major groove of the A-form helix formed by the antisense strand and its target mRNA specifically at the cleavage site inhibited RNAi. These data also indicated that the major groove was required for mediating RNAi and for RNA–RISC interactions that subsequently lead to mRNA cleavage.
Structural integrity of the 5′ half of siRNA duplex, as defined by the antisense strand, is important for mediating RNAi
Previous data using psoralen photochemistry suggested that complete unwinding of the siRNA duplex was not required for RNAi in vivo because psoralen cross-linked siRNAs did not completely abolish gene silencing (Chiu and Rana 2002). In describing these results, it was proposed that a single cross-linking event occurring near the 3′-end of the antisense strand still allowed for the initial unwinding of duplex siRNAs from the 5′-end, freeing enough of the nucleotides in the antisense strand to hybridize to the target mRNA and induce RNAi, even if unwinding was not complete. If this were the case, then unwinding of siRNAs must start from the 5′-end of the antisense strand, a conclusion supported by the fact that blocking either the 3′-end of the antisense siRNA strand or the 5′-end of the sense siRNA strand had no significant effect on RNAi activity (Chiu and Rana 2002).
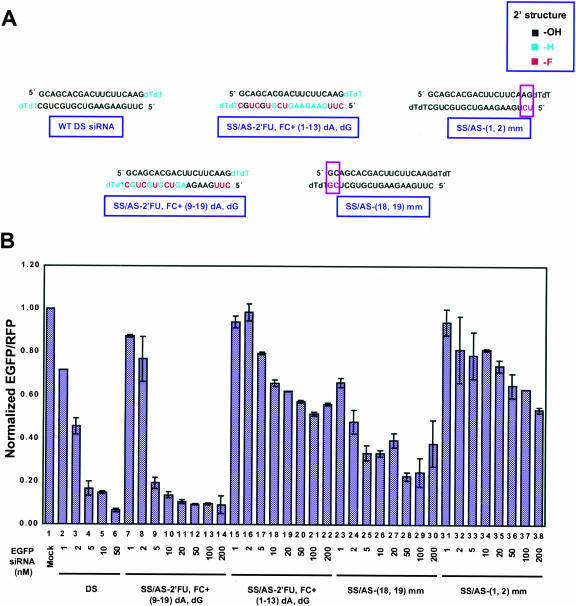
If this 5′-to-3′ unwinding model was correct, sequences within the 3′ half of the siRNA duplex as defined by the antisense siRNA strand should be changeable without significantly interfering with RNAi. To test this hypothesis, EGFP siRNAs with mismatched base pairs at either the internal 5′ (nt 1, 2) or 3′ (nt 18, 19) positions within the duplex were introduced into the antisense strand (Fig. 5A ▶, purple box, purple nucleotides). siRNAs with mismatches near the 5′ half of the duplex structure showed only ~35% inhibition in the dual fluorescence reporter assay, whereas mismatches at the 3′ half retained a significant level of gene silencing at ~77% (Fig. 5B ▶; Table 1 ▶, rows 27–28). These results strongly indicated that the integrity at the 5′ half of the duplex, as defined by the antisense strand, was functionally more important than that at the 3′ half.
FIGURE 5.
Structural integrity of the 5′ half of siRNA duplexes, as defined by the antisense strand, was functionally more important than at the 3′ half. (A) Graphical description of asymmetric requirement of duplex siRNA structure. The structure of unmodified (WT DS) siRNA duplexes, siRNAs with 2′-fluoro-uridine and 2′-fluoro-cytidine, 2′-deoxy modification at the 3′ half (SS/AS-2′-FU, 2′-FC + [9–19] dA, dG) or 5′ half (SS/AS-2′-FU, 2′-FC + [1–13] dA, dG) of the antisense strand, and siRNA duplexes with mismatches within the antisense 3′ half (SS/AS-[18,19] mm) or 5′ half (SS/AS-[1,2] mm) of the siRNA duplex are shown here. (B) Results from cells treated with duplex siRNAs with asymmetrically modified siRNA duplexes. For comparison, results from unmodified duplex siRNAs (DS, lanes 2_–_6)-treated cells are included.
To explore this idea even further, 2′-FU, 2′-FC plus dATPs, dGTPs were incorporated within internal positions within the antisense-strand siRNAs predominantly at the 5′ (nt 1–13) or predominantly within the 3′ half (nt 9–19; Fig. 5A ▶, red and cyan nucleotides). In the dual fluorescence reporter assay, predominantly 5′-modified antisense (AS-2′-FU, 2′-FC + [1–13] dA, dG) EGFP siRNAs were only moderately effective, inducing RNAi at ~43%, or at 45% if the sense strand was also modified to 2′-FU, 2′-FC (Fig. 5B ▶; Table 1 ▶, rows 29,30). However, predominantly 3′-modified and 5′-unmodified antisense (AS-2′-FU, 2′-FC + [9–19] dA, dG) siRNAs significantly induced RNAi activity at ~91%, or at 64% if the sense strand was also modified to 2′-FU, 2′-FC (Fig. 5B ▶; Table 1 ▶, rows 31,32). These contrasting results indicated that sequence structure within the 5′ region of the antisense strand was more sensitive to modification than in the 3′region. All together, these data indicated that recognition of siRNA duplexes by an as-yet-unidentified RNA helicase occurs asymmetrically with the structure of the antisense 5′-end of the duplex preferentially distinguished from the 3′-end during the initiation of unwinding.
Modified siRNAs enter into the RNAi pathway in vitro
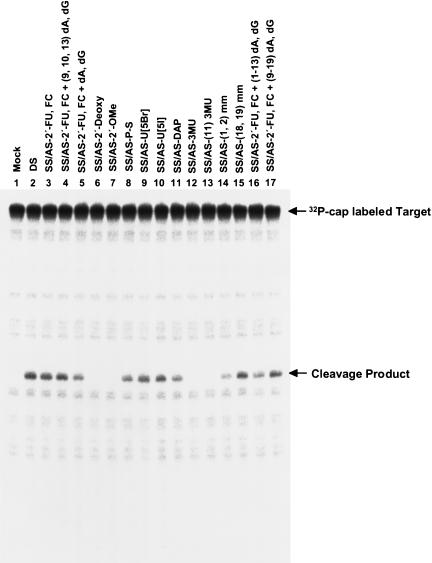
Although the dual fluorescence reporter assay did detect changes in EGFP gene expression with the modified siRNAs created herein, it was possible that gene silencing was being induced by a mechanism other than RNAi-mediated degradative pathways. To test whether the targeted mRNA was, indeed, being cleaved upon exposure to modified siRNAs, an in vitro RNAi assay was performed to measure the cleavage of a 32P-cap-labeled mRNA target upon incubation with modified siRNAs and HeLa cytoplasmic extract. Cleavage products were resolved on an 8% polyacrylamide-7 M urea gel. Mock-treated mRNAs did not show an observable cleavage product (Fig. 6 ▶, lane 1), but wild-type and all modified siRNAs that displayed gene silencing effects in vivo showed clearly visible cleavage products in vitro (Fig. 6 ▶, lanes 2,8–11,14–17). Furthermore, modified siRNAs that did not show any marked gene-silencing effects in vivo did not show any distinct cleavage products in the in vitro assay (Fig. 6 ▶, lanes 1,6,7,12,13), implying that the cleavage events observed were specifically dependent on functional siRNAs. These in vitro results provided a strong correlation between the in vivo gene silencing observed with the modified siRNAs and target mRNA degradation, indicating that the modified siRNAs were distinctly targeting mRNAs for cleavage and subsequent degradation through the in vivo RNAi pathway.
FIGURE 6.
For analysis of siRNA-mediated target mRNA cleavage in vitro, 10 nM cap-labeled target EGFP mRNA was incubated with 100 nM siRNA and HeLa cytoplasmic extracts, as described in Materials and Methods. Reaction products were resolved on an 8% polyacrylamide-7 M urea gel. Arrows indicate the capped target EGFP mRNA and the 5′ cleavage product, which were expected to be 124 nt and 55 nt, respectively. The identity of the cleavage product was assigned according to RNase T1 partial digestion and a molecular weight marker of RNA (data not shown). The 3′ fragment is unlabeled, and therefore, invisible.
DISCUSSION
RNAi has moved toward the forefront of reverse genetic analysis in human cells for characterizing loss of gene function phenotypes and establishing connections between gene structure and function. In light of its versatility, understanding the detailed mechanism behind the RNAi phenomenon and developing methods to extend the limits of its current capabilities is crucial for implementing this methodology even further into the laboratory and therapeutic realms. By introducing various chemical modifications into siRNAs and measuring their effects on RNAi, this study revealed new insights into the mechanism of RNAi and outlined new approaches for increasing the efficacy of RNAi in vivo in human cells for use in future applications.
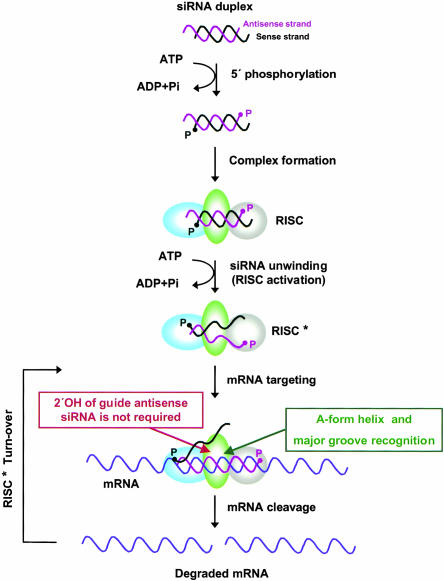
From these data, a more complete picture of the stepwise process of RNAi can be envisioned and is depicted in Figure 7 ▶. In the first step of RNAi induction, the 5′-ends of the siRNA duplex are phosphorylated, resulting in the formation of an siRNA–RISC complex. The data presented here showing the asymmetric nature of unwinding then indicate an ATP-dependent event during which siRNA is unwound from the 5′-end of the antisense strand and RISC is activated. Following RISC activation, the antisense strand of the unwound siRNA guides the siRNA–RISC* complex to the target mRNA. The guide antisense strand base pairs with the target mRNA, forming an A-form helix, and the RISC* protein complex recognizes the major groove of the A-form helix, an event that occurs independently of the RNA 2′-OH of the guide antisense siRNA. In the final step of this process, the target mRNA is cleaved by RISC*, which is another event that occurs independently of the 2′-OH of the guide antisense siRNA. RISC* is then recycled to catalyze another cleavage event.
FIGURE 7.
Model for RNAi in human cells highlighting the requirement of the A-form helix and major groove for mRNA cleavage with the steps not requiring the RNA 2′-OH of the guide antisense siRNA. See text for details.
The requirement for the A-form helix supercedes the requirement for the 2′-OH in RNAi
Several important mechanistic findings were presented here that not only more clearly defined the mechanism of the RNAi pathway, but will also increase the utility of RNAi in various applications. Our results showing that the 2′-OH was not essential for RNAi have several important implications for the structural and catalytic elements required for the RNAi pathway. Remarkable functional implications were that the RNAi machinery does not require the 2′-OH for recognition of siRNAs, and the catalytic ribonuclease activity of RISC does not involve 2′-OH groups of the guide antisense RNA. Another consequence of this discovery was that a variety of chemical groups, including fluoro or deoxy groups, could substitute for the 2′-OH in siRNAs, indicating that no distinguishing chemical specificity was required for RNAi at the 2′-position. These findings would imply that other properties of the siRNA–mRNA duplexes, such as core structural elements, were essential for siRNA. If helical structure was the key to RNAi induction, then the A-form helix that forms between siRNAs and the target mRNA would, indeed, be required for RNAi, as was previously shown (Chiu and Rana 2002). Furthermore, the 2′-fluoro or combined 2′-fluoro, deoxy-modified antisense siRNAs lacking the 2′-OH would have to competently form an A-form helix to induce RNAi as shown here. This will likely turn out to be the case because 2′-fluoro-modified RNA–RNA hybrids were previously reported to exhibit an A-form helical conformation (Cummins et al. 1995; Luy and Marino 2001), lending significant merit to the idea that helical structure strongly influences RNAi efficiency. Still another implication of these particular results was that alternate chemical groups at the 2′-position that allow the A-form helix to be retained but help siRNAs evade recognition by RNases can increase siRNA stability and prolong RNAi effects induced in vivo.
It was previously shown in Caenorhabditis elegans and Drosophila extracts that completely substituting one or both siRNA strands with deoxynucleotides abolished RNAi (Parrish et al. 2000; Elbashir et al. 2001b), and those observations were consistent with the data presented here. The failure of true DNA–RNA hybrids to induce RNAi most plausibly relates to the argument that structure, and thus the A-form helix, was an essential determinant for RNAi induction. Based on circular dichroism spectra, DNA–RNA hybrids displayed characteristics that were intermediate between A- and B-form helices (Cummins et al. 1995). Following the contention that the A-form helix was an absolute requirement for RNAi induction, 2′-deoxy siRNA–mRNA target duplexes would not be recognized by the RNAi machinery because they would not be forming the proper A-form helical structure. Therefore, RNAi would not be induced by DNA–RNA hybrids, as has been observed. It is also worth mentioning that microRNAs (miRNAs) induce posttranscriptional gene silencing (PTGS) through the same pathway as RNAi but, ultimately, only inhibit translation machinery instead of inducing RNA degradation, the event that defines RNAi. The only observable difference between the two mechanisms is that RNAi requires the A-form helix, but miRNA-induced PTGS does not, as miRNAs often mismatch with their target mRNAs, forming a bulge that would distort the helical structure. This would indicate that the differences between the miRNA-induced silencing mechanism and siRNA-mediated RNAi may solely be attributable to differences in RNA–RNA helical structure, and further support a model in which helical structure was the sole determinant for whether RNAi was induced.
It was also previously reported that replacement of uridine with 2′-FU, corresponding to one-fourth of the bases of long dsRNAs, elicited RNAi effects in C. elegans, whereas deoxycytodine incorporated into long dsRNAs diminished RNAi effects (Parrish et al. 2000). However, exactly where these modified nucleotides fell within the sequence structure of RNAi-inducing siRNAs and whether these modified nucleotides in the longer RNAs corresponded to the mRNA cleavage site or major groove after being processed to siRNAs was not clear. It has also been reported that siRNAs in which 3′ overhangs and two of the 3′-end ribonucleotides were replaced with deoxyribonucleotides retained RNAi activity upon exposure to Drosophila extracts (Elbashir et al. 2001b). Presumably, replacing two of the 3′-end base-paired nucleotides with deoxynucleotides would not disrupt the overall A-form structure of the siRNA–mRNA duplex required for RNAi and would thereby allow RNAi induction.
Neither analyses in C. elegans nor in Drosophila extracts ascertained whether there was a distinct requirement for the 2′-OH for cleavage-site recognition and the cleavage event itself during RNAi induction. The results presented here demonstrated that exclusively using 2′-FU, 2′-FC modifications in siRNAs and selectively substituting in deoxyribonucleotides for nucleotides base-paired with the nucleotides lining the mRNA cleavage site, or even replacing the entire sequence of siRNA with a combination of 2′-fluoro- and 2′-deoxynucleotides, elicited RNAi induction. Therefore, it has now been definitively established that recognition of the mRNA-target cleavage site and subsequent cleavage did not require the 2′-OH of the antisense siRNA to induce RNAi. As a final point, the inhibitory RNAi effects seen with the bulky 2′-OMe modification, which was also shown previously with Drosophila (Elbashir et al. 2001b), did demonstrate that there were steric constraints on the types of 2′ modifications that would be amenable for inducing RNAi. As 2′-OMe modifications probably did not disrupt the A-form helix of the siRNA–mRNA duplex (Cummins et al. 1995), the methyl group may be sterically interfering with protein–RNA interactions, thereby preventing RNAi. Nevertheless, steric constraints notwithstanding, this analysis conclusively showed that the nonessential nature of the 2′-position could very much be exploited for improving the efficacy of RNAi in a variety of applications.
Improving the efficacy of RNAi using chemical modifications
Modifications like the 2′-fluoro and P–S linkages both increased the half-life of siRNAs upon exposure to cytoplasmic extracts, and in vivo studies with 2′-FU, 2′-FC siRNAs showed that increasing the half-life of siRNAs did, in fact, prolong the effects of RNAi. In addition, we have observed similar prolonged RNAi activity with silencing lasting over 90 h using these same modifications in siRNAs targeting the endogenous target, human Cyclin T1 (Y.L. Chiu and T.M. Rana, unpubl.). These observations demonstrated that the siRNA modifications studied here can be used to effectively silence endogenous human genes over prolonged periods of time. Our results also indicated that short-lived RNAi effects usually observed in human cells were caused at least in part by the degradation of siRNAs. Our findings that the stabilizing siRNA modifications still allowed for a substantial level of RNAi induction showed that these modifications will be invaluable for studying the phenotypic effects of prolonged gene silencing in cell culture or in increasing the long-term in vivo effects of siRNAs in clinical applications. Interestingly, the P–S-modified, single-stranded antisense strand did not show increased RNAi effects in the dual fluorescence reporter assay used here (data not shown), despite showing significantly increased stability (Fig. 3A ▶[a]). Similarly, single-stranded antisense siRNAs modified with 2′-OMe or 2′-FU, 2′-FC did not cause RNAi efficiently (data not shown). This strongly indicated that siRNA stability was not the main reason that single-stranded antisense RNAi was not as effective in inducing RNAi as dsRNA. Nonetheless, creating P–S modifications in the siRNA backbone showed that stabilizing the siRNA backbone did not inhibit RNAi and signified that using chemical modifications that stabilized phosphate linkages was a viable option for prolonging RNAi effects.
Another option for increasing the efficacy of RNAi was uncovered by the analysis of modifications that should enhance base-pairing interactions between antisense siRNA and targeted mRNA. DAP is a naturally occurring nucleobase that sometimes replaces adenine in phages like the cyanophage S-2L (Kirnos et al. 1977). Incorporation of DAP into RNA strands promotes the formation of three H-bonds between DAP and uridine, increasing the stability of interactions seen between A–U base pairs (Bailly and Waring 1998). U[5Br] and U[5I] have also been shown to have higher association constants when base-paired to A residues than to unmodified uridine (Saenger 1984). When any of these modifications were incorporated into siRNAs, RNAi was still quite efficient, indicating that modifications that stabilize base-pairing interactions can be used in designing siRNAs for various applications. It was also notable that siRNAs with 2′-fluoro modifications introduced into sense strands and base-paired with the DAP, U[Br], or U[5I] antisense strands had decreased RNAi efficiency. 2′-fluoro modifications have been shown to significantly increase the melting temperature between base pairs (Cummins et al. 1995). Consequently, the stabilizing effect on base-pairing interactions when both the 2′-fluoro and DAP, U[Br], or U[5I] modifications were present may have actually hindered the unwinding of the siRNA duplex. These results indicated that the lower rates of siRNA unwinding account for the observed decrease in RNAi activity.
Despite some minor limitations on how much the base-pair interactions can be stabilized, an application for using these types of modifications would be to increase the targeting efficiency of one mRNA sequence over another closely homologous but not identical sequence. Precedent for this type of sequence discrimination was set by Haaima et al. (1997), who showed that DAP improved the ability of an oligomer to discriminate against mismatches. Translated to an RNAi application, these modifications may be useful for specifically targeting a mutant mRNA in a population of both mutant and wild-type mRNAs to recover a recessive wild-type phenotype. These modifications may also be useful in increasing binding affinity between target mRNAs and siRNAs that appear to have weak gene-silencing effects.
Other structural determinants for RNAi induction
Another structural facet of the RNAi mechanism was uncovered using the 3MU modification, which showed that the major groove of the A-form helix was required for RNAi. This finding builds on previous data showing that the A-form helix was required for RNAi (Chiu and Rana 2002). Together, these results indicated that the specific structure of the A-form helical RNA that forms the major groove and contains the mRNA cleavage site was important for recognition by the RNAi machinery. Conceivably, RNA–RISC* contacts depend on the structural integrity of the major groove for precise interactions and, ultimately, to initiate cleavage of the target. By disrupting the major groove, RISC* may no longer be able to interact or only weakly interacts with the siRNA–mRNA target duplex, thereby preventing mRNA cleavage. Alternatively, RISC* might still be able to interact with the destabilized RNA helix but not recognize the cleavage site within the major groove as the catalytic site if the conformation of the RNA helix and more specifically the major groove were altered.
The other structural property of siRNAs defined by these analyses was the asymmetric nature of siRNA unwinding. Initiation of siRNA unwinding from the 5′-end was previously indicated from the ability of single cross-linked siRNAs to still induce RNAi (Chiu and Rana 2002). By using mismatched or modified nucleotides on either the 3′ or 5′ half of the antisense strand within the siRNA duplex, it was shown here that RNAi depended on the integrity of the 5′, and not the 3′, half of the siRNA duplex, as defined by the antisense strand. Previous studies from our laboratory and from other groups have observed that mutations or modifications at the 3′-end overhang of the antisense strands are well-tolerated (Chiu and Rana 2002; Martinez et al. 2002; Amarzguioui et al. 2003; Holen et al. 2003). However, it is important to note that these studies did not address the role of nucleotides and the structure of siRNA at internal sequence positions. Here, we have specifically demonstrated the significance of nucleotides within the RNA duplex and their role in defining the structure of siRNA required for RNAi in vivo. These results indicated that, like RISC*, the RNA helicase, which has not yet been identified, also recognizes structural properties of the siRNA duplex as opposed to specific sequences of the RNA strands. This recognition appears to be asymmetric, with the structure of the antisense 5′ half of siRNA duplexes favored over the 3′ half, and is similar to how restriction enzymes can preferentially cleave the DNA backbone asymmetrically within a palindromic sequence. Further structural analysis of siRNAs to define what properties within the antisense 5′ half of the duplex contribute to the asymmetric nature of the duplex should help elucidate the specific structural elements required for duplex recognition by the RNA helicase for siRNA unwinding.
Our results showing that the modified siRNAs displayed effective RNAi in vivo and in vitro was also significant as it confirmed that the observed gene silencing was mediated by the RNAi pathway. These results also indicated that using chemical modifications that allow for efficient RNAi induction should work in the design of any given siRNA to increase its stability and capacity to specifically induce RNAi in vivo. Using these chemical modifications should take the field of RNAi quite a large step forward beyond the limits presently imposed by unmodified siRNAs with respect to long-term RNAi induction and targeting efficiency. One can imagine numerous applications for all of the chemical modifications used in this analysis, from studying prolonged RNAi effects on multiple genes in human cell cultures to opening up the door for long-term RNAi efficacy in therapeutic realms for curing a variety of genetic diseases.
From the data presented here, the mechanism of RNAi has been further elucidated and the groundwork for incorporating RNAi successfully into therapeutic realms has been laid out. We hope that future studies using the insight garnered here will not only help direct studies for further dissection of the mechanism of RNAi but will lead to new discoveries about gene function and facilitate the introduction of RNAi into vital clinical applications.
MATERIALS AND METHODS
siRNA preparation
The sequences of EGFP target-specific siRNA duplexes were designed as previously described (Chiu and Rana 2002). The 21-nt RNAs were chemically synthesized as 2′-bis(acetoxyethoxy)-methyl ether-protected oligonucleotides by Dharmacon. Synthetic oligonucleotides were deprotected, annealed, and purified; successful duplex formation was confirmed by 20% nondenaturing polyacrylamide gel electrophoresis (Chiu and Rana 2002).
Culture and transfection of cells
HeLa cells were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). Cells were regularly passaged at subconfluence and plated 16 h before transfection at 70% confluency. Lipofectamine (Invitrogen)-mediated transient cotransfections of reporter plasmids and siRNAs were performed in duplicate 6-well plates. A transfection mixture containing 0.16 μg of pEGFP-C1 and 0.33 μg of pDsRed2-N1 reporter plasmids (Clontech), various amounts of siRNA (1.0–200 nM), and 10 μL of lipofectamine in 1 mL of serum-reduced OPTI-MEM (Invitrogen) was added to each well. Cells were incubated in the transfection mixture for 6 h and further cultured in antibiotic-free DMEM. Cells were treated under the same conditions without siRNA for mock experiments. At various time intervals, the transfected cells were washed twice with phosphate-buffered saline (PBS, Invitrogen), flash-frozen in liquid nitrogen, and stored at −80°C for reporter gene assays.
Improved dual fluorescence assay
In an improved dual fluorescence reporter assay, EGFP-C1 encoded enhanced green fluorescence protein (GFP) and DsRed2-N1 encoded red fluorescence protein DsRed2 (RFP), a DsRed variant that has been engineered for faster maturation and lower unspecific aggregation. The extinction coefficient of DsRed2 is 43,800 M−1 cm−1, and the quantum yield is 0.55, a significant quantitative increase when compared with the DsRed1 vector used in the dual fluorescence assay (Chiu and Rana 2002). To quantify RNAi effects, cell lysates were prepared from siRNA duplex-treated cells at 42 h posttransfection, as described previously (Chiu and Rana 2002). Then 240 μg of total cell lysate in 160 μL of reporter lysis buffer was measured by fluorescence spectrophotometry (Photo Technology International). The slit widths were set at 4 nm for both excitation and emission. All experiments were carried out at room temperature. GFP fluorescence in cell lysates was detected by exciting at 488 nm and recording from 498 to 650 nm. The spectrum peak at 507 nm represents the fluorescence intensity of GFP. RFP fluorescence in the same cell lysates was detected by exciting at 568 nm and recording from 588 to 650 nm. The spectrum peak at 583 nm represents the fluorescence intensity of RFP. The fluorescence intensity ratio of target (GFP) to control (RFP) fluorophores was determined in the presence of siRNA duplexes and normalized to that observed in the mock-treated cells. Normalized ratios at <1.0 indicated specific interference.
Study of duplex siRNA stability in HeLa cell lysate
Unmodified or modified EGFP antisense strand siRNAs were 5′-labeled with [γ-32P]ATP (3000 Ci/mM; ICN) by T4 polynucleotide kinases (New England Biolabs) at 37°C for 1 h and chase-kinased by adding 1 mM ATP at 37°C for 15 min. Free ATP and kinases were removed by the QIAGEN nucleotide removal kit. RNA was then purified by 20% polyacrylamide gel containing 7 M urea. Duplex siRNAs were formed by annealing equal molar ratios of unmodified or modified sense-strand siRNAs with the 5′-32P-labeled antisense strand. Duplex formation was confirmed by 20% PAGE under native condition. We incubated 50 pmole of duplex siRNAs labeled at the 5′-end of the antisense strand with 500 μg of HeLa whole-cell extract in a 50-μL reaction mixture containing 20 mM HEPES (pH 7.9), 100 mM KCl, 10 mM NaCl, 2 mM MgCl2, and 10% glycerol. At various time points, 8-μL aliquots were mixed with 16 μL of loading buffer (0.01% bromophenol blue, 0.01% xylene cyanol, 98% formaldehyde, and 5 mM EDTA). The products were then denatured by heating at 95°C for 10 min and analyzed on 20% polyacrylamide gel containing 7 M urea followed by phosphorimage analysis (Fugi).
Preparation of HeLa cells cytoplasmic extract
Cytoplasm from HeLa cells was prepared following the Dignam protocol for isolation of HeLa cell nuclei (Dignam et al. 1983). The cytoplasmic fraction was dialyzed against cytoplasmic extract buffer (20 mM HEPES at pH 7.9, 100 mM KCl, 200 μM EDTA, 500 μM DTT, 500 μM PMSF, 2 mM MgCl2, 10% glycerol). The extract can be stored frozen at −70°C after quick-freezing in liquid nitrogen. The protein concentration of HeLa cytoplasmic extract varied between 4 and 5 mg/mL as determined by a Biorad protein assay kit.
Preparation of cap-labeled target mRNA
For mapping of the target RNA cleavage, a 124-nt EGFP transcript, corresponding to nucleotides 195–297 relative to the start codon followed by the 21-nt complement of the SP6 promoter sequence, was amplified from template pEGFP-C1 by PCR using the 5′ primer GCCTAATACGACTCACTATAGGACCTACGGCGTGCAGTGC (T7 promoter underlined) and the 3′ primer TTGATTTAGGTGACACTATAGATGGTGCGCTCCTG-GACGT (SP6 promoter underlined). His-tagged mammalian capping enzyme was expressed in Escherichia coli from a plasmid generously provided by Stewart Shuman (Molecular Biology Program, Sloan-Kettering Institute, New York) and purified to homogeneity. Guanylyltransferase labeling was performed by incubating 1 nmole of transcripts with 50 pmole of His-tagged mammalian capping enzyme in the 100-μL capping reaction containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 2.5 mM MgCl2, 1 U/μL RNasin RNase inhibitor (Promega), and [α-32P]GTP at 37°C for 1 h. Reactions were chased for 30 min by supplementing GTP concentration to 100 μM. Cap-labeled target mRNAs were resolved on 10% polyacrylamide-7 M urea gel and purified.
In vitro target mRNA cleavage assay
siRNA-mediated cleavage of target mRNA in human cytoplasmic extract was performed as described (Martinez et al. 2002) with some modifications. siRNA duplexes were preincubated in HeLa cytoplasmic extract at 37°C for 15 min prior to addition of the 124-nt cap-labeled target mRNA generated as described above. After addition of all components, the final concentrations were 500 nM siRNA, 50 nM target mRNA, 1 mM ATP, 0.2 mM GTP, 1 U/μL RNasin, 30 μg/mL creatine kinase, 25 mM creatine phosphate, and 50% S100 extract. Incubation was continued for 1.5 h. Cleavage reactions were stopped by the addition of 8 volumes of proteinase K buffer (200 mM Tris-HCl at pH 7.5, 25 mM EDTA, 300 mM NaCl, and 2% [w/v] SDS). Proteinase K, dissolved in 50 mM Tris-HCl (pH 8.0), 5 mM CaCl2, and 50% glycerol, was added to a final concentration of 0.6 mg/mL. Reaction products were extracted with phenol/chloroform/isoamyl alcohol (25:24:1), chloroform, and precipitated with 3 volumes of ethanol. Samples were separated on 8% polyacrylamide-7 M urea gels.
Acknowledgments
We thank Tamara J. Richman for assistance in editing and critical evaluation of the work. We also thank B. Cullen, C. Mello, and P. Zamore for useful discussions and S. Shuman for kindly providing reagents. This work was supported by grants from the NIH (AI41404, AI45466, and AI43198).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Amarzguioui, M., Holen, T., Babaie, E., and Prydz, H. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31**:** 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, C. and Waring, M.J. 1998. The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA. Nucl. Acids Res. 19**:** 4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409**:** 363–366. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.L. and Rana, T.M. 2002. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell 10**:** 549–561. [DOI] [PubMed] [Google Scholar]
- Cogoni, C. and Macino, G. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399**:** 166–169. [DOI] [PubMed] [Google Scholar]
- Cummins, L.L., Owens, S.R., Risen, L.M., Lesnik, E.A., Freier, S.M., McGee, D., Guinosso, C.J., and Cook, P.D. 1995. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 23**:** 2019–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101**:** 543–553. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11**:** 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001a. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15**:** 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20**:** 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaima, G., Hansen, H.F., Christensen, L., Dahl, O., and Nielsen, P.E. 1997. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucleic Acids Res. 25**:** 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21**:** 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404**:** 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Caudy, A.A., and Hannon, G.J. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2**:** 110–119. [DOI] [PubMed] [Google Scholar]
- Holen, T., Amarzguioui, M., Babaie, E., and Prydz, H. 2003. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 31**:** 2401–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297**:** 2056–2060. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R. and Carthew, R.W. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95**:** 1017–1026. [DOI] [PubMed] [Google Scholar]
- Kirnos, M.D., Khudyakov, I.Y., Alexandrushkina, N.I., and Vanyushin, B.F. 1977. 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature 270**:** 369–370. [DOI] [PubMed] [Google Scholar]
- Lipardi, C., Wei, Q., and Paterson, B.M. 2001. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107**:** 297–307. [DOI] [PubMed] [Google Scholar]
- Luy, B. and Marino, J.P. 2001. Measurement and application of 1H–19F dipolar couplings in the structure determination of 2′-fluorolabeled RNA. J. Biomol. NMR 20**:** 39–47. [DOI] [PubMed] [Google Scholar]
- Majlessi, M., Nelson, N.C., and Becker, M.M. 1998. Advantages of 2′-_O_-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 26**:** 2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110**:** 563–574. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3**:** 737–747. [DOI] [PubMed] [Google Scholar]
- Nishikura, K. 2001. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107**:** 415–418. [DOI] [PubMed] [Google Scholar]
- Nykanen, A., Haley, B., and Zamore, P.D. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107**:** 309–321. [DOI] [PubMed] [Google Scholar]
- Parrish, S., Fleenor, J., Xu, S., Mello, C., and Fire, A. 2000. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol. Cell 6**:** 1077–1087. [DOI] [PubMed] [Google Scholar]
- Provost, P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B., and Radmark, O. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21**:** 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. 1984. Forces stabilizing association between bases: hydrogen bonding and base stacking. In Principles of nucleic acid structure (ed. C.R. Cantor), pp. 116–158. Springer-Verlag, New York.
- Schwarz, D.S., Hutvagner, G., Haley, B., and Zamore, P.D. 2002. Evidence that siRNAs function as guides, not primers, in Drosophila and human RNAi pathways. Mol. Cell 10**:** 537–548. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H., and Fire, A. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107**:** 465–476. [DOI] [PubMed] [Google Scholar]
- Stark, G.R., Kerr, I.M., Williams, B.R., Silverman, R.H., and Schreiber, R.D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67**:** 227–264. [DOI] [PubMed] [Google Scholar]
- Stein, C.A. 1996. Phosphorothioate antisense oligodeoxynucleotides: Questions of specificity. Trends Biotechnol. 14**:** 147–149. [DOI] [PubMed] [Google Scholar]
- Stein, P., Svoboda, P., Anger, M., and Schultz, R.M. 2003. RNAi: Mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 9**:** 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D. 2001. RNA interference: Listening to the sound of silence. Nat. Struct. Biol. 8**:** 746–750. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101**:** 25–33. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F.A., Brondani, V., Billy, E., and Filipowicz, W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21**:** 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]