Comparative analysis detects dependencies among the 5′ splice-site positions (original) (raw)
Abstract
Human–mouse comparative genomics is an informative tool to assess sequence functionality as inferred from its conservation level. We used this approach to examine dependency among different positions of the 5′ splice site. We compiled a data set of 50,493 homologous human–mouse internal exons and analyzed the frequency of changes among different positions of homologous human–mouse 5′ splice-site pairs. We found mutual relationships between positions +4 and +5, +5 and +6, −2 and +5, and −1 and +5. We also demonstrated the association between the exonic and the intronic positions of the 5′ splice site, in which a stronger interaction of U1 snRNA and the intronic portion of the 5′ splice site compensates for weak interaction of U1 snRNA and the exonic portion of the 5′ splice site, and vice versa. By using an ex vivo system that mimics the effect of mutation in the 5′ splice site leading to familial dysautonomia, we demonstrated that U1 snRNA base-pairing with positions +6 and −1 is the only functional requirement for mRNA splicing of this 5′ splice site. Our findings indicate the importance of U1 snRNA base-pairing to the exonic portion of the 5′ splice site.
Keywords: mRNA splicing, 5′ splice site, U1 snRNA, mutual relationship, familial dysautonomia, comparative genomics
INTRODUCTION
In mRNA splicing, the 5′ splice site (5′ ss) is recognized by three small nuclear ribonuclear particles (snRNPs)—a complex of small nuclear RNA (snRNA) and proteins. First, U1 snRNP binds to the 5′ ss via base-pairing of U1 snRNA. Before the first splicing catalytic step, U1 is replaced by U5 and U6 snRNPs, for which the snRNA binds to the exonic and intronic portion of the 5′ ss, respectively (Brow 2002). The 5′ end of U1 snRNA is complementary to the consensus sequence of the 5′ splice site, and, indeed, U1 snRNA base pairs with the 5′ ss as a prerequisite step for splicing (Lerner et al. 1980; Rogers and Wall 1980; Zhuang and Weiner 1986; Wassarman and Steitz 1992; Alvarez and Wise 2001). This model still holds, in view of other studies suggesting that the base-pairing of U1 snRNA with the 5′ ss is nonessential for splicing (Bruzik and Steitz 1990; Crispino et al. 1994; Hwang and Cohen 1996; Du and Rosbash 2001 Du and Rosbash 2002; Lund and Kjems 2002), which are likely to be the exception (Maroney et al. 2000).
In mammals, five positions in the intronic portion of the 5′ ss (positions +1 to +3, +5, and +6; for clarity, positions “+” and “−” indicate nucleotides downstream from and upstream of the exon/intron boundary, respectively) were shown to base pair with U1 snRNA by compensatory mutations (Zhuang and Weiner 1986; Aebi et al. 1987; Weber and Aebi 1988; Hitomi et al. 1998). Only one indication for base-pairing to the exonic portion of the 5′ ss was recorded, using cross-linking with psoralen (position −2 of the 5′ ss with position +10 of U1; Malca et al. 2003). In yeast, Saccharomyces cerevisiae or Schizosaccharomyces pombe, there is evidence for 5′ ss:U1 snRNA base-pairings with positions +1 and +3 to +6 (Seraphin et al. 1988; Siliciano and Guthrie 1988; Seraphin and Rosbash 1989; Nandabalan et al. 1993; Alvarez and Wise 2001). The base-pairing of U1 with the exonic portion of the 5′ ss at positions −1 and −2 was recorded in S. cerevisiae (Seraphin and Kandels-Lewis 1993), although in this organism, the exonic portion of the 5′ ss is not conserved well (Fig. 1 ▶, lower part; Spingola et al. 1999). Additionally, in Drosophila melanogaster, U1 snRNA was shown to base pair with positions −1 and +6 (Lo et al. 1994).
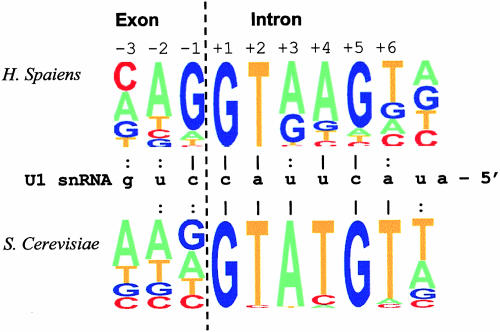
FIGURE 1.
Potential base pairs of U1 snRNA and 5′ ss in human and yeast (S. cerevisiae). We used 49,778 human U2-dependent 5′ ss sequences with the canonical GT nucleotides in the invariant positions taken from the homologous human–mouse exon database compiled by us (see text) and 253 yeast 5′ ss sequences from the Yeast Intron Database (YIDB; Lopez and Seraphin 2000). The 5′ ss consensus sequences of human and yeast are represented graphically (Burge http://genes.mit.edu/pictogram.html; upper part and lower part, respectively). Potential base pairs with the appropriate nucleotide of U1 snRNA are marked either by a vertical bar for highly frequent base pairs (>0.7) or by a colon for less abundant ones (<0.7). The consensus sequence of the constitutive and alternative 5′ ss subsets in human was not significantly different.
The 5′ ss in metazoans provides nine potential positions for U1 snRNA:5′ ss base-pairing. However, mutation analysis of the factor IX gene suggests that U1 snRNA:5′ ss base-pairing requires a minimal number of 5–6 Watson–Crick base pairs with U1 snRNA for mRNA splicing (Ketterling et al. 1999). The idea of a minimal number of base pairs was also derived from another study (Zhuang and Weiner 1986). Additionally, when the number of U1 snRNA:5′ ss base pairs is relatively high (>7), the reaction efficiency is reduced under low RNA concentrations in humans (Lund and Kjems 2002). In S. cerevisiae, hyperstabilization of the U1 snRNA:5′ ss base-pairing with 10 bp was detrimental (Staley and Guthrie 1999). Therefore, it is inferred that an average 5′ ss prefers less than seven potential Watson–Crick base pairs to U1 snRNA. This also suggests that the different positions of the 5′ ss might have a mutual relationship: A mispair between U1 snRNA and one position of the 5′ ss is compensated by a base pair of U1 snRNA to another position(s), thus maintaining the base pairs’ number above the minimum. Indeed, evidence for such a mutual relationship among the 5′ ss positions were found: The existence of G at position +3 depends on the concordance that the residues at positions +4 to +6 have with U1 snRNA (Ohno et al. 1999); base-pairing of U1 snRNA with positions +3 and +4 compensates for a mispair of U1 snRNA with position +5 (Nelson and Green 1990); and the dependence of positions +5 with position +3 (Burge and Karlin 1997; Clark and Thanaraj 2002) and with positions +3 and +4 was demonstrated (Lund and Kjems 2002). Additionally, a linkage between the exonic and intronic portions of the 5′ ss was indicated in a few studies (Burge and Karlin 1997; Rogozin and Milanesi 1997; Thanaraj and Robinson 2000). In S. cerevisiae, it was recorded that the nonconserved positions +7 and +8 can compensate for a mismatch at position +3 of the 5′ ss (Nandabalan et al. 1993), and a mispair at position +5 compensated for base pairing of the slightly conserved last two exonic positions (Seraphin and Kandels-Lewis 1993).
In contrast to the intronic 5′ ss, the base pairing of U1 snRNA with the exonic 5′ ss is supported by few genetic evidences, and the conservation of the exonic positions was mainly attributed to base pairing with the invariant loop of U5 snRNA (Newman and Norman 1992; Cortes et al. 1993). However, this base pairing is dispensable for mRNA splicing in humans (Segault et al. 1999). In addition, several other splicing factors, such as U1(C) and U5(p220), and U1(Sm) proteins were shown to bind to the exonic portion of the 5′ ss (Wyatt et al. 1992; Rossi et al. 1996; Zhang et al. 2001; Du and Rosbash 2002). We, therefore, set out to examine whether the conservation of the exonic portion of the 5′ ss can be attributed to base pairing with U1 snRNA in mammals. Using a large data set of ~50,000 5′ ss, we found a linkage between the exonic and intronic 5′ ss, with respect to base pairing with U1 snRNA, that further supports the importance of the base pairing of U1 snRNA and the exonic portion of the 5′ ss. This linkage was also demonstrated experimentally in an ex vivo system that mimics the mutation leading to familial dysautonomia (FD). In addition, we used comparative genomics to detect a mutual relationship among the 5′ ss positions. We compiled a data set of homologous human–mouse internal exons and analyzed the frequency of changes among different positions of homologous human–mouse 5′ ss pairs. We found mutual relationships between positions +4 and +5, +5 and +6, −2 and +5, and −1 and +5.
Both Burge (Burge and Karlin 1997) and Thanaraj and Robinson (Thanaraj and Robinson 2000) surveyed the dependencies among the 5′ ss positions. The work presented here extends their work by using a very large data set, providing an experimental validation for the linkage between the exonic and the intronic portions of the 5′ ss, average number of U1 snRNA:5′ ss base-pairing, and detecting dependencies using comparative genomics.
RESULTS
The human–mouse ortholog 5′ ss database
Approximately 75–130 million years have passed since the human and mouse common ancestor was speciated into two separate lineages (Yang and Yoder 1999; Waterston et al. 2002). Most of the genes (99%) are orthologs, and the majority of these genes (86%) share the same intron/exon arrangement. In homologous exons sequences, a high degree of conservation (88%) is observed, whereas only 40% of the introns are alignable, with poor sequence identity (69%) (Waterston et al. 2002). We thus assumed that the level of conservation of different 5′ ss positions of human–mouse homologous 5′ ss would be an indicative of their functional importance.
Therefore, we assembled a database of 50,493 homologous human–mouse internal exons (see Materials and Methods). From this database, we extracted 49,778 5′ ss pairs of exons flanked by AG-GT splice sites, in which the flanking introns are U2 dependent (termed U2-dependent 5′ ss motifs pairs). The human–mouse homologous 5′ ss pairs included 45,519 and 4259 pairs of constitutive- and alternative-spliced exons, respectively.
The 5′ ss weight matrix of all the 5′ ss motifs was calculated (Fig. 1 ▶, upper part) and was not significantly different from the 5′ ss consensus of its alternative and constitutive subsets (χ2, p > 0.97). Neither set (alternative or constitutive) was significantly different, either from the other (χ2, p > 0.97) or from the weight matrix of Shapiro and Senapathy (1987; χ2, _P_-values ranging between 0.4 and 0.99).
All the bioinformatic analyses further described here were preformed on each of the alternative and constitutive 5′ ss homologous pairs. The results were similar in all of the cases, excluding one case, indicated below, in which the alternative set of exons was too small to deduce valuable conclusions. We thus showed the results of the analysis performed on the 5′ ss motifs of the constitutive exons.
Mutual relationship among the different positions of the 5′ ss
Using the human–mouse comparison, we examined dependency among the 5′ ss positions. We analyzed the substitution events per position between human–mouse homologous 5′ ss. We then examined whether we could identify mutual relationships among various 5′ ss positions, namely, whether a change in one of the 5′ ss positions reduces or increases the occurrence of substitution events in other 5′ ss positions.
From our data set, we used 45,519 constitutive-spliced U2-dependent 5′ ss motifs pairs. We divided these pairs into sets, with respect to the number of 5′ ss positions that differs between human and mouse (Fig. 2A ▶). We ruled out the possibility that the difference between ortholog 5′ ss pairs represents an intron sliding event, because each exons pair shares the same length and open reading frame. Next, we calculated the distribution per position of the 5′ ss sets of pairs that have 1 nt difference between human and mouse homologous 5′ ss. Positions +3 and +6 are the most variable positions (Fig. 2B ▶), which is in agreement with the consensus sequence (Fig. 1 ▶, upper part) and other comparative analyses of the 5′ ss (Waterston et al. 2002; Sorek and Ast 2003). Positions −3 to −1 are relatively conserved, supposedly due to coding constraints (further examined below).
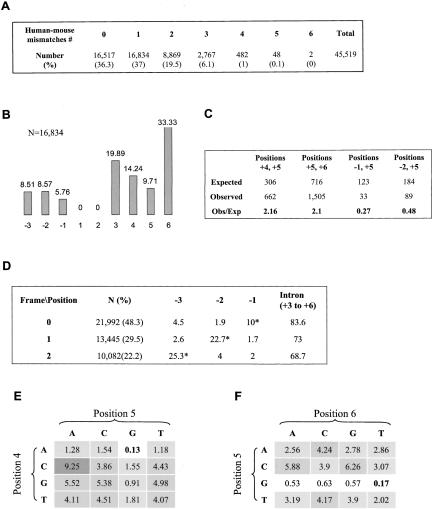
FIGURE 2.
Comparative analysis of human–mouse homologous 5′ ss. (A) A data set of 45,519 homologous human–mouse U2-dependent 5′ ss sequences with GT at the invariant positions of introns for which their upstream exon is constitutively spliced and for which they share the same reading frame was sorted by the number of different bases between each homologous 5′ ss sequence. The size and percentage (in parentheses) of either set are shown. (B) The distribution by positions of the 5′ ss homologous sequences that contain one base difference of the 5′ ss pairs. The percentage and the position are indicated above and below each column, respectively. (C) Assuming independence of a single substitution event, we calculated the expected number of human–mouse homologous 5′ ss sequences with two base differences at each two positions of the 5′ ss (see text). The expectation was then compared with the observed numbers. A considerable gap between our expectation and the observed values was found at positions +4 and +5, positions +5 and +6, positions −1 and +5, and at positions −2 and +5. The exact values regarding these positions are shown. Detailed analysis is described in Figure 1S in the supplementary materials (http://www.tau.ac.il/~gilast/sup_mat.htm). (D) The 5′ ss pairs were sorted into three groups, according to their reading frame, and the same analysis demonstrated in panel B was performed on each of the groups. The size and the percentage of each group are indicated in the column titled by N(%). Detailed frequency of the exonic positions and the frequency of substitution events in the intronic positions are shown. The percentage of changes in positions −1 to −3 is shown when an asterisk marks the wobble position in each frame. (E,F) The variability degree (see text) of base combinations at positions +4 and +5 (E) and at positions +5 and +6 (F). Bold numbers correspond with the consensual base combination. The intensity of the gray level in the background of each value corresponds with the degree of variability.
Based on the assumption that substitution events in each position of the 5′ ss are independent, we examined the occurrence of mutual relationship between each two 5′ ss positions. We used the frequency of changes in each position (Fig. 2B ▶) to calculate the expected number of 5′ ss pairs that are distinct in each two positions of the 5′ ss and compared this value with the observed number. In general, a notable deviation between the expected and the observed numbers were found (χ2, p < 10−5). The prominent deviations were at positions +4 and +5, positions +5 and +6, positions −2 and +5, and finally at positions −1 and +5 (Fig. 2C ▶). These deviations suggest that the independency assumption regarding substitution events at these positions were invalid, and a mutual relationship between these positions is inferred. This detailed analysis is described at Figure 1S in the Supplemental Materials (see http://www.tau.ac.il/~gilast/sup_mat.htm).
The observed number of 5′ ss pairs with two substitution events at positions −1 and +5 and at positions −2 and +5 were smaller, with respect to the expectation (Fig. 2C ▶, 0.27-fold and 0.48-fold, respectively). This observation suggests that, in these positions, a substitution event in one position of the pair reduces the occurrence of a substitution event at the other position. At positions +4 and +5 and at positions +5 and +6, the number of observed 5′ ss changes is higher, with respect to the expectation (2.16- and 2.1-fold, respectively). This might indicate that a substitution event in one position encourages the substitution of the other. These results are in agreement with previous works that describe dependencies between the very same positions (Burge and Karlin 1997; Thanaraj and Robinson 2000). The small data set of alternatively spliced exons (4259) makes it impossible to repeat this analysis on those exons and to deduce valuable conclusions.
We sorted our data set according to the reading frame of the exonic 5′ ss and repeated the analysis described above (Fig. 2D ▶). The data set of 45,519 human–mouse constitutives AG-GT ortholog exons pairs that have U2-dependent 5′ ss with the same reading frames was divided as follows: 21,992 (48.3%) homologous 5′ ss pairs have the wobble position in the last nucleotide of the exon (and, hence, followed by a phase zero introns); in 13,445 (29.5%) and 10,082 (22.2%) 5′ ss pairs, the wobble position is in positions −2 and −3 upstream of the 5′ ss, respectively (Fig. 2D ▶; followed by phase 2 and 1 introns, respectively). This distribution was in agreement with previous work (Long et al. 1995). In all the 5′ ss reading frames, the distribution by position of the 5′ ss pairs that differs in one position between human and mouse showed a picture similar to that described in Figure 2B ▶, with regard to the intronic positions (data not shown). As for the exonic positions, a variation in the conservation level between the different reading frames is notable (Fig. 2D ▶). In general, the wobble position is always the most variable position in the exonic 5′ ss. However, the wobble position in the last nucleotide of the exon (position −1) showed relatively small variability, compared with positions −2 and −3 (10% with respect to 22.7% and 25.3%, respectively). Apparently, the regulatory constraint on position −1 is stronger, as implied from the consensus sequence.
According to the analysis presented in Figure 2C ▶, we also examined the mutual relationship between each two positions of the 5′ ss of the 5′ ss groups that were sorted by frames. We detected the same dependencies among the positions shown in Figure 2C ▶ of all the frames together (see Supplemental Table 2S, at http://www.tau.ac.il/~gilast/sup_mat.htm; compare rows 1, 2, and 3 with the last row). However, the degree of the linkage (the observed/expected values) between the 5′ ss positions is different in each frame, and an additional mutual relationship that was unique to each frame was also observed (see Supplemental Table 1S at http://www.tau.ac.il/~gilast/sup_mat.htm). Interestingly, in each reading frame, we found an additional mutual relationship that encourages substitution events between the two exonic positions that were not in the wobble position (see Supplemental Table 1S at http://www.tau.ac.il/~gilast/sup_mat.htm).
We further tested whether there is a preference for a G nucleotide presence at positions −1 and +5. Using the human 5′ ss of the data set above, we calculated the percentage of G at these positions (80% at position −1 and 78.1% at position +5) and the expected number of 5′ ss with and without G at either of the positions. The expectation deviated from the observed numbers (χ2, p < 0.001); we expected that 1997 (4.4%) of the 5′ ss would be without G at these positions. However, only 204 5′ ss (0.4%) actually do not contain G in either of the positions. This indicates a bias of having at least one G at positions −1 or +5 of the 5′ ss and further supports the repressive mutual relationship shown in Figure 2C ▶, in which a substitution in position −1 reduces the occurrence of substitutions in position +5, and vice versa. A similar bias was attained in positions −2 and +5 (p < 0.001); we expected that 3550 (7.8%) of the 5′ ss would be without A at position −2 and G at position +5, but found only 1151 (2.5%) such events.
Next, we checked whether there is a characteristic event of substitution at the positions that encourage substitution events: pairs +4 and +5, and +5 and +6 (Fig. 2C ▶). We looked for prevalent transitions between one base combination and the other at these positions. The CA ↔ TG transition was found at 14.8% (98 of 662) of the 5′ ss pairs that differ at positions +4 and +5 and in 7.4% of the cases (111 of 1505) at positions +5 and +6. This transition may originate as a result of a single change at the hypermutable CpG sequence of the ancestor 5′ ss that substitutes to either TpG or CpA (Cooper and Krawczak 1990). However, given the evolutionary time scale, multiple changes may also be considered.
Assuming that the transition TG ↔ CA is promoted by the CpG mechanism, we checked whether the CG → TG and CG → CA transitions provide more immovability to the 5′ ss motif. Thus, we conducted a test calculating the variability degree (see Materials and Methods) of all the possible base combinations at positions +4 and +5, and positions +5 and +6 (Fig. 2 ▶, E and F, respectively). In general, positions +4 and +5 are less tolerant to base combinations that differ from the consensus sequence, as compared with positions +5 and +6, namely, the values of variability degree at these positions are much higher (compare the gray intensity at Fig. 2E,F ▶). Corresponding with the 5′ ss consensus, the presence of G at position +5 and A at position +4 notably decreased the variability degree of either sequence that includes those nucleotides. Interestingly, the presence of T at position +6 does not decrease the variability degree (particularly without G at position +5). At positions +4 and +5, the sequence CG is less variable than either CA or TG; at positions +5 and +6, CG is more variable than TG and as variable as CA. Thus, if the transitions CG → CA and CG → TG originate from hypermutable CpG, this transition works against the stability of the 5′ ss sequence at positions +4 and +5 and +5 and +6, and only the transition CG → TG at positions +5 and +6 reduces the variability of the sequence.
A linkage between the exonic and the intronic sequence of the 5′ ss motif
We then set out to establish the importance of the base-pairing of U1 snRNA to the exonic portion of the 5′ ss in metazoans. We examined possible correlations between the exonic and the intronic portion of the 5′ ss by analyzing the effect of base-pairing of U1 snRNA and the exonic portion of the 5′ ss on the base-pairing of U1 snRNA and the intronic portion of the 5′ ss and the other way around. Thus, we extracted 45,519 U2-dependent 5′ ss of human constitutive GT-AG internal exons. For each 5′ ss, we predicted the free energy value (ΔG) created by base-pairing of U1 sn-RNA and the intronic (positions +1 to +6) and the exonic (positions −3 to +2) 5′ ss and examined a possible correlation between these values. Positions +1 and +2 were added to the exonic 5′ ss sequences to “fasten” the exonic 5′ ss to U1 (positions 7 to 11 of U1 snRNA) in the right alignment (Fig. 1 ▶, upper part). The predicted additional energetic contribution of positions +1 and +2 is constant and, hence, does not bias the correlation, because all the 5′ ss contain GT by definition. This also ensures that the staking energy between position −1 and +1 is calculated. A significant inverse correlation (Spearman, _r_2 = 0.185, p < 0.001) was found between the exonic and the intronic portion of the 5′ ss, with respect to the predicted ΔG values of base-pairing with U1 snRNA. As expected, this correlation was also reflected by correlation in the number of hydrogen bonds (_r_2 = 0.127, p < 0.001) and the number of base pairs created by base-pairing of U1 snRNA and either portion of the 5′ ss, exonic and intronic (_r_2 = 0.123, p < 0.001 and _r_2 = 0.188, p < 0.001 for calculation that counts or ignores G:U as a base pair, respectively). This intronic/exonic linkage was termed “seesaw” linkage and was in agreement with a previous studies based on a smaller data set (Burge and Karlin 1997; Rogozin and Milanesi 1997).
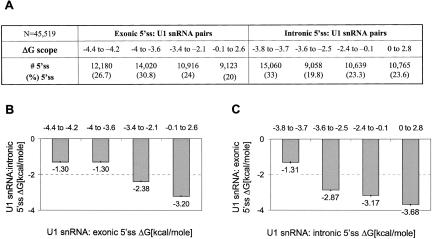
To further demonstrate this linkage, we divided the 5′ ss sequences into four groups, with a similar number of members in each group, according to scopes of the predicted ΔG created by base-pairing with U1 snRNA and the exonic portion of the 5′ ss (Fig. 3A ▶). For each set, we calculated the average ΔG of base-pairing of U1 snRNA and the intronic portion of these 5′ ss (Fig. 3B ▶). We found a notable linkage in which an increase in the ΔG from a range of (−4 to −3.6) to (−3.4 to 2.6) kcal/mole of U1 snRNA base-pairing with the exonic portion of the 5′ ss led to a decrease of more than 2 kcal/mole in the ΔG value created by base-pairing of U1 snRNA and the intronic portion of the 5′ ss (Fig. 3B ▶).
FIGURE 3.
A linkage between the exonic and intronic sequences of the 5′ ss. The values of free energy (ΔG) created by base-pairing of U1 snRNA with either the exonic portion (positions −3 to +2) or the intronic portion (positions +1 to +6) of the 5′ ss were predicted. All the analyzed 5′ ss sequences to “fasten” the exonic 5′ ss base-pairing to U1 snRNA in the right alignment (base-pairing with positions 11 to 7 of U1 snRNA), and also to include the stacking energy of positions +1 and −1 in the prediction. All of the 5′ ss contain GT; thus, by definition, the additional predicted energetic contribution of the base-pairing of positions +1 and +2 is constant. (A) The 5′ ss sequences were sorted into four groups of similar sizes, which define scopes of the predicted free-energy values of base-pairing of U1 snRNA and the exonic and intronic portions of the 5′ ss. The size (#5′ ss) and percentage of each subset is shown. (B) The average predicted ΔG value created by base-pairing of U1 snRNA and positions +1 to +6 of the 5′ ss (U1snRNA:intronic 5′ ss ΔG) was examined, with respect to the scopes of ΔG created by base-pairing of U1 snRNA and positions −3 to +2 of the 5′ ss (U1 snRNA:exonic 5′ ss ΔG). The exact values are indicated below each column; the error bars are shown as ±1 standard error. (C) Similar to panel B, but in the inverse direction, the predicted ΔG from base-pairing of U1 snRNA to positions −3 to +2 of the 5′ ss (U1 snRNA:exonic 5′ ss ΔG), with respect to the ΔG created by base-pairing of U1 snRNA and positions +1 to +6 of the 5′ ss (U1 snRNA:intronic 5′ ss ΔG).
This observation was further supported when we performed the same analysis in the inverse direction (Fig. 3C ▶): A reduction of up to 2 kcal/mole in the ΔG value of base-pairing of U1 snRNA and the exonic portion of the 5′ ss compensates for an increase in the ΔG level between −3.8 to −3.7 and −3.6 to −2.5 kcal/mole of U1 snRNA base-pairing with the intronic portion of the 5′ ss, on average. Besides the first two sets of intronic 5′ ss (Fig. 3B ▶), the average ΔG of U1 snRNA and the other portion of the 5′ ss of either set described in Figure 3A ▶ was found to be significantly distinct (Mann–Whitney, p < 0.001).
This seesaw linkage between the exonic and the intronic portions of the 5′ ss, in which coding and noncoding sequences interact, further highlights the importance of the U1 snRNA:exonic 5′ ss base-pairing in mRNA splicing.
The contribution of each position of the exonic or intronic portion of the 5′ ss to the predicted ΔG value of the base-pairing of U1 snRNA with the other portion
We next examined the contribution of each position of the exonic (see Supplemental Fig. 2S, at http://www.tau.ac.il/~gilast/sup_mat.htm) or intronic portion (see Supplemental Fig. 3S, at http://www.tau.ac.il/~gilast/sup_mat.htm) of the 5′ ss on the intronic/exonic linkage presented in Figure 3 ▶. We examined the effect of each mispair between U1 snRNA and a specific position in one portion of the 5′ ss on the predicted ΔG value created by base-pairing of U1 snRNA and the other portion. Additionally, we checked 5′ ss sequences (N = 45,519) were taken from human constitutive internal exons flanked by GT-AG dinucleotides of U2-dependent introns. Positions +1 and +2 were added to the exonic whether there is a dependency among different 5′ ss positions: Does the content of the neighboring positions influence the effect created by a mispair between U1 snRNA and a specific 5′ ss position? This analysis is described in the supplementary material in detail.
From this analysis, we drew the following conclusions: In the exonic portion of the 5′ ss, the level of the intronic compensation for a mispair at position −2 is dependent upon the base-pairing at position −1. Position −3 slightly affects the ΔG level of intronic 5′ ss:U1 snRNA base-pairing. In the intronic portion of the 5′ ss, an increase in the exonic compensation for a mispair at positions +3, +4, and +6 was detected when position +5 base pairs with U1 snRNA. Finally, a mispair in a location adjacent to a paired position has a larger compensation effect than a mispair at a position for which its adjacent position mismatches (Supplemental Figs. 2S, 3S, at http://www.tau.ac.il/~gilast/sup_mat.htm), which corresponds with the base-stacking effect. These results are also in agreement with previous observations (Burge and Karlin 1997; Thanaraj and Robinson 2000).
Analysis of the 5′ ss according to the potential number of base pairs with U1 snRNA
Our results indicate that positions −1 and +5 are the major contributors to the exonic/intronic 5′ ss linkage. We thus examined a possible hierarchy at the 5′ ss positions. The data set of 45,519 U2-dependent 5′ ss from human constitutive exons that are flanked by AG-GT was sorted to sets, according to the number of potential base pairs with U1 snRNA (includes G:U pairing). Figure 4A ▶ shows each set size from three to nine potential base pairs of U1 snRNA with the 5′ ss (including base-pairing with positions +1 and +2). The average number of potential base pairs of U1 snRNA and the 5′ ss is 7.05 ± 0.004 (6.38 ± 0.004 excludes G:U base pairs). Around this figure, we observed a Gaussian-like distribution of the 5′ ss motifs (Fig. 4A ▶; Kolmogorov–Smirnov, p = 0.605). The average number of putative hydrogen bonds and free energy was also calculated (17.04 ± 0.1, −6.53 ± 0.1 kcal/mole, respectively).
FIGURE 4.
Analysis of the 5′ ss according to the potential number of base pairs with U1 snRNA. (A) 45,519 human 5′ ss sequences from constitutive exons were sorted into sets, according to the total number of base pairs with U1 snRNA (includes G:U). The number of motifs in each set is indicated above each point. (B) The consensus sequences of each set are represented graphically (Burgehttp://genes.mit.edu/pictogram.html). The number of base pairs with U1 snRNA and the size of each set are indicated on the left of each diagram. The same analysis that takes into account only Watson–Crick base pairs is demonstrated in Figure 4S in the Supplementary Materials (see http://www.tau.ac.il/~gilast/sup_mat.htm).
The 5′ ss consensus of each set, from three to nine potential base pairs with U1 snRNA, was calculated (Fig. 4B, ▶top to bottom). In Figure 4B ▶, 4 to 9 bp, the following phenomena are notable. First, 5′ ss that have 3 and 4 bp with U1 snRNA base pair mostly at positions −1 to +3, whereas positions −3, +4, and +6 hardly base pair at all. Second, a dependency between position +3 and positions +4 to +6 is shown; an increase in the base-pairing frequency at positions +3 is associated with a decrease of the base-pairing level of positions +4 to +6, and the other way around (Fig. 4B ▶, cf. 5 and 6 bp with 8 and 9). This dependency was described in previous studies (Burge and Karlin 1997; Ohno et al. 1999; Thanaraj and Robinson 2000). Third, G at position −1 was selected before G at position +5 (Fig. 4B ▶, cf. 4 bp and 5 bp), suggesting that G at position −1 is more dominant than G at position +5 in 5′ ss with minimal base-pairing with U1. Finally, 5′ ss with 3 to 6 bp have a surprising preference for A at position −3, which might correspond to a possible base-pairing with U5 snRNA (Malca et al. 2003), rather than with U1 snRNA.
An ex vivo validation of the importance of U1 snRNA base-pairing with position −1
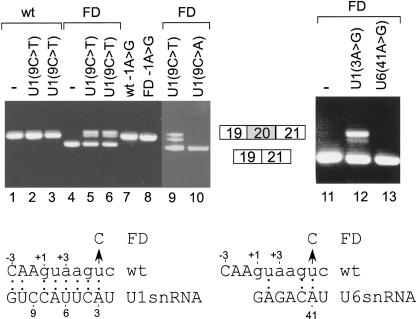
We next checked, experimentally, if an aberrant splicing caused by a mispair between U1 snRNA and an intronic position can be restored by an additional base-pairing of the exonic portion of the 5′ ss with U1 snRNA. We selected the IkappaB kinase complex-associated protein (IKBKAP) gene, in which mutation in position +6 of intron 20 [IVS20(+6T → C)] induces aberrant mRNA splicing, which leads to familial dysautonomia (FD). FD is an autosomal recessive congenital neuropathy that occurs almost exclusively in the Ashkenazi Jewish population, and IVS20(+6T → C) is the major mutation (99.5%) causing FD (Fini and Slaugenhaupt 2002). We cloned a mini-gene of IKBKAP containing exons 19, 20, and 21 (and the introns in between), without mutation (wt) or with the IVS20(+6T → C) mutation (FD; Fig. 5 ▶). The indicated plasmids were introduced into 293T cells by transfection, total cytoplasmic RNA was extracted, and splicing products were separated in 2% agarose gel following RT-PCR. The 5′ ss motif of exon 20 has 8 potential bp with U1 snRNA and is constitutively spliced (Fig. 5 ▶, lane 1; the lower left part shows the mutation, positions, and potential base-pairings of U1 with that 5′ ss). However, the IVS20 (+6T → C) mutation led to the almost constitutive skipping of exon 20 (Fig. 5 ▶, lane 4).
FIGURE 5.
The importance of U1 base-pairing with positions −1 and +6 of the 5′ ss. The indicated plasmids were introduced into 293T cells by transfection, total cytoplasmic RNA was extracted, and splicing products were separated in 2% agarose gel after reverse transcriptase polymerase chain reaction (RT-PCR). (Lower part) A schematic drawing of U1 and U6 snRNA potential base-pairing with the 5′ ss of exon 20 of IKBKAP gene is shown; a base pair is marked by a colon. The positions of the 5′ ss, U1, and U6 snRNA are marked above and underneath, respectively. The U-to-C mutation in position +6, leading to FD, is shown by an arrow. (Upper part, lanes 1_–_3) Splicing products of IKBKAP mini-gene (wt); lane 1 wt only; lanes 2,3, cotransfection with U1 mini-gene containing mutation of C to T at position 9. It is important to state that exogenous and endogenous U1 compete for 5′ ss selection (Zhuang and Weiner 1986). (Lanes 4_–_6) Splicing products of FD mini-gene, which is the IKBKAP mini-gene containing mutation of IVS20(+6T → C); lane 4, FD mutant only, lanes 5,6 cotransfection with U1(9C → T). (Lane 7) IKBKAP mini-gene with A-to-G mutation in the last nucleotide of exon 20. (Lane 8) FD mini-gene with A-to-G mutation in the last nucleotide of exon 20. (Lane 9) Similar to lanes 5,6. (Lane 10) similar to lane 9, except that the U1 cotransfected gene contains a C-to-A mutation in position 9. (Lanes 11_–_13) FD mini-gene; lane 11, FD mini-gene alone; lane 12, cotransfection with U1 gene containing mutation of A to G in position 3; lane 13, cotransfection with U6 gene containing mutation of A to G in position 41. In lanes 5,6,9 and, to some extent, also in lane 12, the upper RT-PCR product contains two closely joined bands (20 nt difference). Their sequence was found to be identical (from one end of the PCR products to the other) to the joint of exons 19–20–21 (no alternative selection of 5′ or 3′ ss; see also Fig. 5S in Supplementary Materials, at http://www.tau.ac.il/~gilast/sup_mat.htm). Compensatory mutations in three different positions of the 5′ end of U1 revealed the same phenomena, and it was detected in two different mini-genes (IKBKAP and ADAR2). re-PCR of the upper band of this doublet led it to migrate in a similar position to that of the wt products in an agarose gel. We have no interpretation for this separation pattern. The procedures of these experiments are identical to those described by Lev-Maor et al. (2003).
During the splicing reaction, the 5′ ss motifs base pair with U1 and U6 snRNA (Wassarman and Steitz 1992). We then examined whether the effect of the mutation in position +6 is related to U1 or U6 base-pairing with position +6. The FD mutant was cotransfected with either U6 or U1 snRNA that can base pair with C at position +6 of the 5′ ss. We found that base-pairing of this position to U1, but not U6 snRNA, partially restored normal splicing of exon 20 (Fig. 5 ▶, cf. lanes 12 and 13, respectively). Thus, the mismatch between the 5′ ss of the FD mutant and U1 snRNA causes the aberrant splicing. However, we cannot rule out the possibility that position 41 of U6 snRNA is required for additional functions besides base-pairing with the 5′ ss, such as U4/U5/U6 assembly.
T-to-C mutation at position +6 on mRNA splicing of exon 20 implies that it weakens the 5′ ss:U1 snRNA interaction. We then examined whether this weakness could be related to the presence of adenosine at position −1, which mispairs with U1 snRNA. Cotransfection of the FD mutation with U1 snRNA, containing compensatory mutation of C to T (Fig. 5 ▶, lanes 5,6,9), but not C to A (Fig. 5 ▶, lane 10), at position +9 of the U1 gene restored the splicing of exon 20 in 24% of the splicing events (which is likely the ratio between the endogenous and exogenous U1). Also, a mutation of A to G at position −1 of exon 20 restored normal splicing of the mRNA derived from the FD mini-gene (Fig. 5 ▶, lane 8). These results both indicate the importance of G at position −1 in 5′ ss selection and provide the first genetic evidence for U1 snRNA base-pairing with position −1 in mammals. It also shows that, in this 5′ ss, the only functional properties of positions +6 and −1 in mRNA splicing are base-pairing with U1, although U5 is known to base pair with the exonic portion of the 5′ ss and U6 with position +6 (Sontheimer and Steitz 1993; Sawa and Shimura 1992). Finally, it shows that increasing the base-pairing of U1 snRNA to the exonic portion of the 5′ ss can compensate for the loss of function due to a mispair with the intronic portion, which is in agreement with the exonic/intronic linkage previously shown.
DISCUSSION
We examined mutual relationships among the various 5′ ss positions that could not be specified by the conventional linear consensus sequence. We compared the 5′ ss of human and mouse and found that position +5 conducts a mutual relationship with positions −2, −1, +6, and +4. This observation is also in agreement with other studies (Burge and Karlin 1997; Thanaraj and Robinson 2000). From different bioinformatics analyses, we concluded that positions −1 and +5 are the most prominent positions in the base-pairing with U1 snRNA, excluding the invariant positions. Thus, mutation at one of these positions is expected to be more harmful than other positions of the 5′ ss (excluding positions +1 and +2). Finally, we provided experimental evidence for the exonic/intronic association; we showed that a base pair of position −1 rescued an aberrant splicing caused by a mispair of position +6 with U1 snRNA. This shows that the exonic portion of the 5′ ss base pairs with U1 snRNA in humans.
U1 snRNA base pairs with the exonic portion of the 5′ ss
The average number of potential base pairs between U1 snRNA and the 5′ ss (positions -3 to +6) is 6.38 (7.05 includes G:U), leaving an average of 2.62 (1.95 with G:U) unpaired positions. The number of positions that base pair with U1 snRNA is likely to be tightly regulated—it must surpass a certain minimum (Zhuang and Weiner 1986; Ketterling et al. 1999), whereas a high number of U1:5′ ss base pairs impairs splicing (Lund and Kjems 2002). This flexibility of the 5′ ss:U1 snRNA base-pairing generates a statistically significant correlation between the exonic and the intronic portions of the 5′ ss, with respect to the predicted free energy level of base-pairing with U1 snRNA (Fig. 3B,C ▶) and the number of hydrogen bonds and base pairs with U1 snRNA.
This linkage, which is in agreement with a previous study (Burge and Karlin 1997), was shown experimentally as well. We demonstrated that in the 5′ ss of exon 20 of the IKBKAP gene, a base pair of U1 snRNA with position −1, was the only requirement needed to restore an aberrant splicing caused by a mispair in position +6 (Fig. 5 ▶). This is also the first genetic evidence for base-pairing of U1 snRNA with position −1 of the 5′ ss in mammals. In mutant rabbit Δ-globin transcripts, 5′ ss selection was also shown to be influenced by base paring of position −1 with the invariant loop of U5 snRNA (Cortes et al. 1993). However, the results presented here, on FD transcript, indicate that 5′ ss selection requires base-pairing of position −1 with U1 snRNA, solely. This discrepancy is likely to reflect different dependencies of subsets of 5′ ss for base-pairing of position −1 with U5, and the same can also be applied to the requirement of U6 base-pairing with position +6. A subset of 5′ ss that may be dependent on U5 base-pairing with the exonic portion of the 5′ ss is that with minimal potential base-pairing with U1 that shows preference to A in position −3 of the 5′ ss (Fig. 4B ▶; 3–5 bp). The A in positions −2 and −3 can form base-pairing with the Us in the invariant loop of U5.
In S. cerevisiae, a similar phenomena was demonstrated—an aberrant splicing caused by a mispair at position +5 was restored by base-pairing between U1 snRNA and the exonic portion of the 5′ ss (Seraphin and Kandels-Lewis 1993). However, the nonconserved exonic sequence of the 5′ ss of S. cerevisiae (Spingola et al. 1999) suggests that the base-pairing of U1 snRNA with the exonic portion of the 5′ ss is not the typical phenomena in mRNA splicing in S. cerevisiae.
The exonic 5′ ss expansion
The conservation of the exonic 5′ ss sequence is poor in S. cerevisiae (Spingola et al. 1999), relative to metazoans and S. pombe, whereas the conservation level of the intronic sequence is higher in yeast than in metazoans (Burge et al. 1999; see also Fig. 1 ▶, cf. upper and lower parts). This difference might be associated with various percentages of genes that undergo splicing—3.8% in S. cerevisiae, 43% in S. pombe, and most of the protein coding genes in mammals (Lopez and Seraphin 1999; Wood et al. 2002). The difference may be associated with the increase in the level of splicing regulation as well; there are hardly any reported alternative splicing events in yeast, compared with 20%–56% of the human genes (Davis et al. 2000; Barrass and Beggs 2003; Sorek et al. 2004). Thus, the extension of the 5′ ss motif to the exonic portion of the 5′ ss and the reduction in the conservation of the intronic positions of the 5′ ss between S. cerevisiae and metazoan might be concomitant with the evolution of complex splicing-regulation events, such as alternative splicing—namely, correlating with the organism’s complexity.
Positions −1 and +5 play a major role in the exonic/intronic linkage
Although the U1 snRNA:5′ ss base-pairing is not dependent on the complementarity of a specific position, except that of the invariant GT (Aebi et al. 1987; Nelson and Green 1990), it has been speculated that not all the positions contribute to this base-pairing equally (Siliciano and Guthrie 1988; Seraphin and Rosbash 1989). In our results, we found that the positions that contribute the most to the seesaw linkage are the highly conserved positions −1 and +5 (G in 80% and 78.1%, respectively). Further evidence for the importance of positions +5 and −1 are implied from other tests we conducted (Figs. 2 ▶, 4 ▶). Also, an analysis of diseases caused by deleterious mutations in the 5′ ss further supports our findings: 12 of 76 (15.7%) mutations at position −1 and 15 of 76 (19.7%) mutations at +5 caused aberrant splicing, compared with 7 of 76 (9.2%) in all the other positions (excluding the invariant positions; Nakai and Sakamoto 1994). Finally, the importance of position −1 is supported by two experiments (Fig. 5 ▶). First, a mispair of the exonic -1 position in the ADAR2 gene caused an 11-fold increase in exon-skipping events in a 5′ ss that has 6 bp with U1 snRNA (data not shown). Second, a U1 snRNA gene, containing a compensatory mutation that allows A:U base-pairing with position −1 of the 5′ ss at exon 20 of the IKBKAP gene, restored a normal mRNA splicing impaired by C > T mutation in position +6, a mutation that causes FD disease (Fig. 5 ▶). These results indicate that the only functional requirement of the G in position −1 of this 5′ ss is for base-pairing with U1, and not for base-pairing with U5 snRNA or binding to protein factor(s) such as U5(p220) (Wyatt et al. 1992).
A partial explanation for the importance of positions −1 and +5 in base-pairing with U1 snRNA may be that these positions are involved in a G:C base-pairing that forms three hydrogen bonds (Freier et al. 1986). Additionally, G at positions −1 and +1 generates a strong stacking effect between two adjacent purines (Sontheimer and Steitz 1993; Fini and Slaugenhaupt 2002). A mispair at these positions might be more critical for U1 snRNA:5′ ss base-pairing. These positions also pair with other splicing factors (position −1 with U5 snRNA and position +5 with U6 snRNA; Sawa and Shimura 1992; Sontheimer and Steitz 1993), which may contribute to their high degree of conservation and prominence.
MATERIALS AND METHODS
Compiling a database of human–mouse homologous exon pairs
In April 2003, a list of 11,658 pairs of human–mouse homologous mRNA was extracted from the NCBI HomoloGene catalog (Zhang et al. 2000), which joins pairs of homologous mRNA from different organisms. Each sequence was then restricted so that it would appear only once in the pairs list, using a perl script. We successfully retrieved 11,106 pairs from RefSeq. also in April 2003. The mRNA pairs were then masked against repetitive sequences using RepeatMasker (Smit http://ftp.genome.washington.edu/RM/RepeatMasker.html), and each mRNA was aligned against its original genome (April 2003), using blastall (Altschul et al. 1997) with 10−10 expectation cutoff value. The genomic sequence that corresponds to the mRNA molecule of either pair was extracted, and the exon/introns arrangement of the 11,105 pairs was predicted, using sim4 (Florea et al. 1998) configured for high sensitivity (N = 1 W = 6 X = 6 C = 10). Finally, putative homologous exon pairs were extracted with their neighboring exons and their flanking introns using a perl script and gap software from the gcg package (Womble 2000). Each pair of the homologous exons was constrained by the following criteria: (1) they were originated from homologous mRNA pairs, (2) they were internal exons, (3) they shared the same sequence length, (4) the homologous exons revealed 75% of sequence similarity, (5) their original mRNA annotation indicated that they had the same reading frame in human and mouse, and (6) one of their flanking exons (either downstream or upstream) showed 80% similarity in the first 40 nt adjacent to the exon. This constraint enabled the other neighboring exon to undergo alternative splicing. We obtained 50,493 putative homologous exons pairs, of which 197 had at least one U12-dependent 5′ ss (the 5′ ss that contained “[A/G]TATCCT” in their intronic sequence [Dietrich et al. 1997] in either human or mouse), leaving 50,296 with U2-dependent 5′ ss.
Sim4 software attempts to recognize GT-AG (and also CT-AC) splice sites, but may shift the exon/intron boundary when predicting splice sites that does not conform to these signals (Florea et al. 1998). Thus, to avoid possible mistakes, we conducted our analysis on 49,778 of the exons with the U2-dependent 5′ ss that were flanked by the conventional GT-AG splicing consensus, and their 5′ ss was fully sequenced. We therefore omitted from our calculation 515 (1%) of the exon pairs that did not obey the GT-AG rule, mostly containing GC 5′ ss.
Recognition of exons involved in exon-skipping events
For each exon, we constructed a splice-junction probe, joining 25 nt from its upstream and downstream exons. We compared each splice probe against the Expressed Tag Sequences (ESTs) database of the accordant species. An EST that is aligned to a 50-mer, continuously, is an indication for an exon-skipping event. In 46,190 exon pairs, no bridging EST was found; of these, 45,519 obeyed the AG-GT rule and were flanked by U2-dependent 5′ ss. In 4303 of the exon pairs (4259 AG-GT with U2-dependent 5′ ss), we found at least one indication for exon skipping, of which 435 exon pairs were found with bridging EST indication over either human or mouse homologous exons (429 flanked by AG-GT and U2-dependent 5′ ss). It is noteworthy, however, that the given EST data is incomplete; the set of exons that were classified as constitutive may contain a few exon-skipping events that have not yet been documented.
Prediction of free energy (ΔG) created by base-pairing between two short RNA strands
The energy was calculated using the mfold software (Zuker 1989) from the gcg suite of programs (Womble 2000), which predicts the free energy (ΔG) of a single folded RNA strand. To predict the ΔG of two short RNA strands we concatenated these strands into one RNA strand as follows: “NN”–seq1–“NNN”–reversed seq2–“NN”. The software does not hybridize the “N” sequence, because it is considered neutral. Thus, the energy calculations were that of seq2 to seq1 hybrid.
Counting the total number of hydrogen bonds between two RNA strands
Using a perl script, we counted the total number of hydrogen bonds between the base pairs of A:T, G:C, and G:T (two, three, and two, respectively). We counted two hydrogen bonds for each G:U base-pairing, following the conventional notion (Ladner et al. 1975; Varani and McClain 2000), although few works indicated that the number of hydrogen bonds of G:U depend on its adjacent neighbors and can be less than two (Chen et al. 2000; Vercoutere et al. 2003).
The variability degree of base composition in two given positions of the 5′ ss motifs
The variability degree is evaluated by

in which: n1, n2 are nucleotides; p1, p2 are positions of the 5′ ss; and PISE stands for Percentage of Involvement in Substitutions Events and is the frequency of the nucleotide combination n1 and n2 at positions p1 and p2 of the 5′ ss, among the 5′ ss homologous pairs that differ at these positions. TF, which stands for Total Frequency, is the frequency of the nucleotide combination n1 and n2 at positions p1 and p2 of all the 5′ ss. For example, PISE(+5, +6, G, T) is the percentage of the base combination GT found at positions +5 and +6 in 1542 constitutive human–mouse homologous 5′ ss motifs pairs, with two base differences between them at these positions (resulting in 9.21%). TF(+5, +6, G, T) is the frequency at which GT is found at positions +5, +6 in all 46,809 constitutive human–mouse 5′ ss pairs in our database (80.88%). The variability degree of (+5, +6, G, T), a base combination that is part of the consensus sequence, is, therefore, 0.11. A value less than 1 indicates that the nucleotides n1 and n2 are involved in substitution events at a frequency lower than their appearance frequency, namely, this combination of nucleotides is considered conserved. Values that are greater than 1 indicate that nucleotides n1 and n2 are a variable combination at the examined positions.
Plasmid constructs
Oligonucleotide primers were designed to amplify (from human genomic DNA) a mini-gene that contains exons 19, 20, and 21 of IKBKAP gene. The sequences of the two primers are: XhoI forward: CCGCTCGAGCATTACAGGCCGGCCTGAGCAGCA, and Pst1 backward: AACTGCAGCTTAGGGTTATGATCATAAATCA GATT.
The PCR product of IKBKAP (1.7 kb) was restriction digested by XhoI and PstI and inserted between those sites in pEGFP-C3 vector (Clontech), at the C terminus of the EGFP. This ensures the same open reading frame of the mini-gene. The human U1 gene inserted in the pUC13 vector is a kind gift of Prof. Alan M. Weiner, University of Washington, Seattle (Zhuang and Weiner 1986).
Site-directed mutagenesis
FD mutation (+6T to C in intron 20) by site-directed mutagenesis was preformed according to Lev-Maor et al. (2003). Briefly, oligonucleotide primers containing the desired mutations were used to amplify a mutation-containing replica of the wild type and the FD mini-gene plasmid. The PCR products were treated with 12U DpnI restriction enzyme (New England Biolabs) for 1 h at 37°C. One to three microliters of the DNA were transformed into Escherichia coli DH5αstrain, followed by colony-picking, mini-prep, and midi-prep extraction (GIBCO/BRL). All plasmids were confirmed by sequencing.
Transfection, RNA isolation and RT-PCR amplification
293T cell lines were cultured in Dulbecco’s Modification of Eagle Medium, supplemented with 4.5 g/mL glucose (Biological Industries), 10% fetal calf serum, 2 mmoles/L L-glutamine and 100 U/mL penicillin–0.1 mg/mL streptomycin–12.5 U/mL nystatin, and cultured in 100-mm dishes under standard conditions at 37°C with 5% CO2. Cells were grown to 50% confluence, and transfection was performed using Metafectene (Biontex) with 10 μg of plasmid DNA or using FuGENE6 (Roche) with 4 μg of plasmid DNA for each transfection. Cotransfection was performed by presenting the mini-gene and the U1 inserted plasmids (4 μg of each using the Fugene 6) to the cells. Cells were harvested after 48 h. Total cytoplasmic RNA was extracted using TriReagent (Sigma), followed by treatment with 1 U RNase-free DNase (Promega). Reverse transcription (RT) was preformed on 2 μg total cytoplasmic RNA for 1 h at 42°C, using a pEGFP-C3-MCS specific reverse primer (5′ -GTTCTATAGATCCGGTGGATG) and 2 U reverse transcriptase of avian myeloblastosis virus (A-AMV, Roche). The spliced cDNA products derived from the expressed mini-genes were detected by PCR, using the pEGFP-C3-specific reverse primer and an exon 19-forward primer for the IKBKAP mini-gene. Amplification was performed for 30 cycles, consisting of 30 sec at 94°C, 45 sec at 62°C, and 1 min at 72°C. The products were resolved on 2% agarose gel and confirmed by sequencing.
URLs
Our homologous exons database is available on http://www.tau.ac.il/~gilast/.
The HomoloGene ortholog list project was downloaded from ftp://ftp.ncbi.nih.gov/pub/homoloGene.
The genomic sequences are from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens (human), ftp://ftp.ncbi.nih.gov/genomes/M_musculus (mouse).
The mRNA sequences were retrieved from ftp://ftp.ncbi.nih.gov/RefSeq (RefSeq), and the EST sequences of human and mouse were downloaded from: ftp://ftp.ncbi.nih.gov/blast/db/.
Acknowledgments
We thank Rotem Sorek, Noam Shomron, and Rani Elkon for comments on the manuscript. This work was supported by a grant from the Israel Science Foundation and by a grant from the FD Hope and MOP India-Israel to G.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Aebi, M., Hornig, H., and Weissmann, C. 1987. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell 50**:** 237–246. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25**:** 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, C.J. and Wise, J.A. 2001. Activation of a cryptic 5′ splice site by U1 snRNA. RNA 7**:** 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrass, J.D. and Beggs, J.D. 2003. Splicing goes global. Trends Genet. 19**:** 295–298. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36**:** 333–360. [DOI] [PubMed] [Google Scholar]
- Bruzik, J.P. and Steitz, J.A. 1990. Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell 62**:** 889–899. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268**:** 78–94. [DOI] [PubMed] [Google Scholar]
- Burge, C.B. http://genes.mit.edu/pictogram.html.
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNA by the spliceosome. In The RNA world (eds. R.F. Gesteland et al.), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Chen, X., McDowell, J.A., Kierzek, R., Krugh, T.R., and Turner, D.H. 2000. Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: One hydrogen bond for each G.U pair in r(G GCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2). Biochemistry 39**:** 8970–8982. [PubMed] [Google Scholar]
- Clark, F. and Thanaraj, T.A. 2002. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 11**:** 451–464. [DOI] [PubMed] [Google Scholar]
- Cooper, D.N. and Krawczak, M. 1990. The mutational spectrum of single base-pair substitutions causing human genetic disease: Patterns and predictions. Hum. Genet. 85**:** 55–74. [DOI] [PubMed] [Google Scholar]
- Cortes, J.J., Sontheimer, E.J., Seiwert, S.D., and Steitz, J.A. 1993. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J. 12**:** 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino, J.D., Blencowe, B.J., and Sharp, P.A. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265**:** 1866–1869. [DOI] [PubMed] [Google Scholar]
- Davis, C.A., Grate, L., Spingola, M., and Ares Jr., M. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28**:** 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.C., Incorvaia, R., and Padgett, R.A. 1997. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell 1**:** 151–160. [DOI] [PubMed] [Google Scholar]
- Du, H. and Rosbash, M. 2001. Yeast U1 snRNP–pre-mRNA complex formation without U1snRNA–pre-mRNA base pairing. RNA 7**:** 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419**:** 86–90. [DOI] [PubMed] [Google Scholar]
- Fini, M.E. and Slaugenhaupt, S.A. 2002. Enzymatic mechanisms in corneal ulceration with specific reference to familial dysautonomia: Potential for genetic approaches. Adv. Exp. Med. Biol. 506**:** 629–639. [DOI] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8**:** 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier, S.M., Kierzek, R., Caruthers, M.H., Neilson, T., and Turner, D.H. 1986. Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry 25**:** 3209–3213. [DOI] [PubMed] [Google Scholar]
- Hitomi, Y., Sugiyama, K., and Esumi, H. 1998. Suppression of the 5′ splice site mutation in the Nagase analbuminemic rat with mutated U1snRNA. Biochem. Biophys. Res. Commun. 251**:** 11–16. [DOI] [PubMed] [Google Scholar]
- Hwang, D.Y. and Cohen, J.B. 1996. Base pairing at the 5′ splice site with U1 small nuclear RNA promotes splicing of the upstream intron but may be dispensable for slicing of the downstream intron. Mol. Cell. Biol. 16**:** 3012–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterling, R.P., Drost, J.B., Scaringe, W.A., Liao, D.Z., Liu, J.Z., Kasper, C.K., and Sommer, S.S. 1999. Reported in vivo splice-site mutations in the factor IX gene: Severity of splicing defects and a hypothesis for predicting deleterious splice donor mutations. Hum. Mutat. 13**:** 221–231. [DOI] [PubMed] [Google Scholar]
- Ladner, J.E., Jack, A., Robertus, J.D., Brown, R.S., Rhodes, D., Clark, B.F., and Klug, A. 1975. Structure of yeast phenylalanine transfer RNA at 2.5 Å resolution. Proc. Natl. Acad. Sci. 72**:** 4414–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, M.R., Boyle, J.A., Mount, S.M., Wolin, S.L., and Steitz, J.A. 1980. Are snRNPs involved in splicing? Nature 283**:** 220–224. [DOI] [PubMed] [Google Scholar]
- Lev-Maor, G., Sorek, R., Shomron, N., and Ast, G. 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300**:** 1288–1291. [DOI] [PubMed] [Google Scholar]
- Lo, P.C., Roy, D., and Mount, S.M. 1994. Suppressor U1 snRNAs in Drosophila. Genetics 138**:** 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, M., Rosenberg, C., and Gilbert, W. 1995. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl. Acad. Sci. 92**:** 12495–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, P.J. and Seraphin, B. 1999. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: Implications for splice-site recognition. RNA 5**:** 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. YIDB: the Yeast Intron DataBase. Nucleic Acids Res. 28**:** 85–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M. and Kjems, J. 2002. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA 8**:** 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malca, H., Shomron, N., and Ast, G. 2003. The U1 snRNP base pairs with the 5′ splice site within a penta-snRNP complex. Mol. Cell. Biol. 23**:** 3442–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Romfo, C.M., and Nilsen, T.W. 2000. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 6**:** 317–328. [DOI] [PubMed] [Google Scholar]
- Nakai, K. and Sakamoto, H. 1994. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 141**:** 171–177. [DOI] [PubMed] [Google Scholar]
- Nandabalan, K., Price, L., and Roeder, G.S. 1993. Mutations in U1 snRNA bypass the requirement for a cell type-specific RNA splicing factor. Cell 73**:** 407–415. [DOI] [PubMed] [Google Scholar]
- Nelson, K.K. and Green, M.R. 1990. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc. Natl. Acad. Sci. 87**:** 6253–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A.J. and Norman, C. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68**:** 743–754. [DOI] [PubMed] [Google Scholar]
- Norberg, J. and Nilsson, L. 1995. Stacking free energy profiles for all 16 natural ribodinucleoside monophosphates in aqueous solution. J. Am. Chem. Soc. 117**:** 10832–10840. [Google Scholar]
- Ohno, K., Brengman, J.M., Felice, K.J., Cornblath, D.R., and Engel, A.G. 1999. Congenital end-plate acetylcholinesterase deficiency caused by a nonsense mutation and an A → G splice-donor-site mutation at position +3 of the collagenlike-tail-subunit gene (COLQ): How does G at position +3 result in aberrant splicing? Am. J. Hum. Genet. 65**:** 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J. and Wall, R. 1980. A mechanism for RNA splicing. Proc. Natl. Acad. Sci. 77**:** 1877–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin, I.B. and Milanesi, L. 1997. Analysis of donor splice sites in different eukaryotic organisms. J. Mol. Evol. 45**:** 50–59. [DOI] [PubMed] [Google Scholar]
- Rossi, F., Forne, T., Antoine, E., Tazi, J., Brunel, C., and Cathala, G. 1996. Involvement of U1 small nuclear ribonucleoproteins (snRNP) in 5′ splice site-U1 snRNP interaction. J. Biol. Chem. 271**:** 23985–23991. [DOI] [PubMed] [Google Scholar]
- Sawa, H. and Shimura, Y. 1992. Association of U6 snRNA with the 5′ -splice site region of pre-mRNA in the spliceosome. Genes & Dev. 6**:** 244–254. [DOI] [PubMed] [Google Scholar]
- Segault, V., Will, C.L., Polycarpou-Schwarz, M., Mattaj, I.W., Branlant, C., and Luhrmann, R. 1999. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 19**:** 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin, B. and Rosbash, M. 1989. Mutational analysis of the interactions between U1 small nuclear RNA and pre-mRNA of yeast. Gene 82**:** 145–151. [DOI] [PubMed] [Google Scholar]
- Seraphin, B. and Kandels-Lewis, S. 1993. 3′ splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell 73**:** 803–812. [DOI] [PubMed] [Google Scholar]
- Seraphin, B., Kretzner, L., and Rosbash, M. 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 7**:** 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, M.B. and Senapathy, P. 1987. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 15**:** 7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano, P.G. and Guthrie, C. 1988. 5′ splice site selection in yeast: Genetic alterations in base-pairing with U1 reveal additional requirements. Genes & Dev. 2**:** 1258–1267. [DOI] [PubMed] [Google Scholar]
- Smit, A.G., P. RepeatMasker. http://ftp.genome.washington.edu/RM/RepeatMasker.html.
- Sontheimer, E.J. and Steitz, J.A. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262**:** 1989–1996. [DOI] [PubMed] [Google Scholar]
- Sorek, R. and Ast, G. 2003. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 13**:** 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Shamir, R., and Ast, G. 2004. How prevalent is functional alternative splicing in the human genome? Trends Genet. 20**:** 68–71. [DOI] [PubMed] [Google Scholar]
- Spingola, M., Grate, L., Haussler, D., and Ares Jr., M. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5**:** 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3**:** 55–64. [DOI] [PubMed] [Google Scholar]
- Thanaraj, T.A. and Robinson, A.J. 2000. Prediction of exact boundaries of exons. Brief. Bioinform. 1**:** 343–356. [DOI] [PubMed] [Google Scholar]
- Varani, G. and McClain, W.H. 2000. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1**:** 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoutere, W.A., Winters-Hilt, S., DeGuzman, V.S., Deamer, D., Ridino, S.E., Rodgers, J.T., Olsen, H.E., Marziali, A., and Akeson, M. 2003. Discrimination among individual Watson–Crick base pairs at the termini of single DNA hairpin molecules. Nucleic Acids Res. 31**:** 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman, D.A. and Steitz, J.A. 1992. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 257**:** 1918–1925. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:** 520–562. [DOI] [PubMed] [Google Scholar]
- Weber, S. and Aebi, M. 1988. In vitro splicing of mRNA precursors: 5′ cleavage site can be predicted from the interaction between the 5′ splice region and the 5′ terminus of U1 snRNA. Nucleic Acids Res. 16**:** 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble, D.D. 2000. GCG: The Wisconsin Package of sequence analysis programs. Methods Mol. Biol. 132**:** 3–22. [DOI] [PubMed] [Google Scholar]
- Wood, V., Gwilliam, R., Rajandream, M.A., Lyne, M., Lyne, R., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415**:** 871–880. [DOI] [PubMed] [Google Scholar]
- Wyatt, J.R., Sontheimer, E.J., and Steitz, J.A. 1992. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 6**:** 2542–2553. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Yoder, A.D. 1999. Estimation of the transition/transversion rate bias and species sampling. J. Mol. Evol. 48**:** 274–283. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7**:** 203–214. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Abovich, N., and Rosbash, M. 2001. A biochemical function for the Sm complex. Mol. Cell 7**:** 319–329. [DOI] [PubMed] [Google Scholar]
- Zhuang, Y. and Weiner, A.M. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46**:** 827–835. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244**:** 48–52. [DOI] [PubMed] [Google Scholar]