Kaposi's Sarcoma-Associated Herpesvirus K3 Utilizes the Ubiquitin-Proteasome System in Routing Class I Major Histocompatibility Complexes to Late Endocytic Compartments (original) (raw)
Abstract
Human herpesvirus 8 (HHV8) downregulates major histocompatibility complex (MHC) class I complexes from the plasma membrane via two of its genes, K3 and K5. The N termini of K3 and K5 contain a plant homeodomain (PHD) predicted to be structurally similar to RING domains found in E3 ubiquitin ligases. In view of the importance of the ubiquitin-proteasome system in sorting within the endocytic pathway, we analyzed its role in downregulation of MHC class I complexes in cells expressing K3. Proteasome inhibitors as well as cysteine and aspartyl protease inhibitors stabilize MHC class I complexes in cells expressing K3. However, proteasome inhibitors differentially affect sorting of MHC class I complexes within the endocytic pathway and prevent their delivery to a dense endosomal compartment. In this compartment, the cytoplasmic tail of MHC class I complexes is cleaved by cysteine proteases. The complex is then cleaved within the plane of the membrane by an aspartyl protease, resulting in a soluble MHC class I fragment composed of the lumenal domain of the heavy chain, β2-microglobulin (β2m), and peptide. We conclude that K3 not only directs internalization, but also targets MHC class I complexes to a dense endocytic compartment on the way to lysosomes in a ubiquitin-proteasome-dependent manner.
Antigen presentation via the major histocompatibility complex (MHC) class I pathway is targeted for interference at different stages by several viruses (26, 46). Virtually every herpesvirus studied encodes at least one gene product that negatively affects MHC class I expression in the infected cell. Human herpesvirus 8 (HHV8), a gamma-2 herpesvirus, also known as Kaposi's sarcoma-associated herpesvirus (KSHV), downregulates expression of MHC class I complexes at the cell surface via two gene products, K3 and K5 (7, 22). Both genes independently induce internalization of MHC class I complexes from the plasma membrane, which ultimately results in lysosomal degradation of MHC class I complexes. The specificity of K3 and K5 differs for different MHC class I alleles and depends on the transmembrane domain of the class I molecule. K3 induces internalization of HLA-A, -B, -C, and -E, while K5 induces internalization of HLA-A and -B and only a small fraction of HLA-E. In addition, K5 downregulates B7-2 and ICAM-1 (8, 21).
K3 shares 40% amino acid identity with K5 (30). Both genes contain in their N terminus a variant of the C4HC3 zinc finger domain class, termed the plant homeodomain (PHD)/leukemia-associated protein (LAP) motif. It has been shown recently that an intact PHD is required for MHC class I downregulation by K3 (28). While PHDs (for review, see reference 1) are similar to RING fingers and LIM (Lin11/Isl-1/Mec3) domains, their function remains unknown. Structural analysis by nuclear magnetic resonance of the PHD in the transcriptional corepressor KAP-1 shows its structural similarity with the prototype C3HC4 pattern RING fingers (6). Nicholas et al. (39) have categorized the PHDs of K3 and K5 as the BKS (bovine herpesvirus 4, KSHV, swinepox virus) subclass of the prototypical PHD/LAP motif present in many cellular proteins, such as human AF10 and MLLa.
RING finger-containing, E3 ubiquitin protein ligases participate in transfer of ubiquitin to other proteins and to themselves (24). E3 ubiquitin protein ligases facilitate direct transfer of activated ubiquitin from E2, the ubiquitin-conjugating enzymes, to the substrate and confer specificity for this reaction (16). Ubiquitin serves a widespread role in the regulation of the fate of cellular proteins (5, 17, 33). There are at least two major functions for ubiquitin modification. The best known role is in targeting proteins for proteasomal degradation. It is now clear that the functions of ubiquitin modification extend to many other processes, notably the control of internalization of surface receptors and the determination of their subsequent fate (5, 18). KSHV K3 and K5 may utilize a pathway that exploits the latter aspect of ubiquitin function.
The ubiquitin-proteasome pathway helps regulate internalization and sorting into the endolysosomal compartment. The connection between the ubiquitin machinery and regulated receptor internalization from the plasma membrane is illustrated by Ste2p, a G protein-coupled receptor specific for α-factor (19). Upon ligand binding, this receptor is monoubiquitinated at the plasma membrane and rapidly internalized and degraded by vacuolar hydrolases, a process in which Ent1 plays a critical role. Ent1 binds ubiquitin directly and interacts with clathrin, thus linking ubiquitin with the endocytosis machinery (for review, see references 33 and 51). Moreover, the involvement of the proteasome can be direct, as in the case of the c-Met receptor (23), or indirect, as in the case of the growth hormone receptor (GHR) (48). The role of the proteasome in endocytosis is not understood in any mechanistic detail.
Furthermore, the ubiquitin-proteasome system is involved in sorting of proteins within compartments of the endocytic pathway (12, 17). Sorting of the internalized receptor tyrosine kinase ErbB1 into late endocytic compartments instead of recycling endosomes is dependent on the RING-containing E3 ubiquitin-protein ligase, c-cbl (13, 50). Ubiquitination is also involved in the distribution of proteins within the endocytic compartment known as the multivesicular body. The internal vesicles that characterize a multivesicular body derive from invagination of its limiting membrane in a process that is in some cases dependent on ubiquitin (25, 35).
Here we report results that indicate sorting of MHC class I complexes within the endocytic compartment of cells expressing KSHV K3 to be dependent on the ubiquitin-proteasome system. Furthermore, MHC class I complexes are sorted to a dense endocytic compartment, where the MHC class I heavy chain (HC) is cleaved in its cytoplasmic tail by a cysteine protease and within its transmembrane domain by an aspartyl protease.
MATERIALS AND METHODS
Cell lines and cell culture.
BJAB is an Epstein-Barr virus-negative Burkitt's lymphoma cell line. BJAB cells stably transfected with pEF (BJAB) and pEF-K3 (K3-BJAB), as previously described (22), were grown in HEPES-buffered RPMI, supplemented with 5% fetal calf serum, 5% calf serum, 50 U of penicillin per ml, 50 μg of streptomycin sulfate per ml, 2 mM l-glutamine, and 0.5 mg of Geneticin per ml (GIBCO, Fredrick, Md.).
Plasmids and DNA constructs.
The vectors pEF and pEF-K3 were previously described (22). HLA-A 0201 (HLA-A2312) (amino acids [aa] 1 to 312) was generated as described previously (43); HLA-A2322 (aa 1 to 322) and HLA-A2271 (aa 1 to 271) were generated with similar PCR primers to introduce a stop codon at positions 323 and 272, respectively. HLA-A2275 has been described previously (14).
Materials and inhibitors.
The proteasome inhibitor carboxylbenzyl-leucyl-leucyl-leucine vinyl sulfone (ZL3VS) was synthesized as described previously (3). Epoxomicin and lactacystin were purchased from Affiniti Research Products, Exeter, United Kingdom. Leupeptin and pepstatin A were purchased from Sigma Chemical Co. (St. Louis, Mo.) and concanamycin B from Ajinimoto Co. (Kanagawa, Japan).
Antibodies.
Monoclonal antibody W6/32 recognizes properly folded class I molecules (31). Rabbit anti-class I HC serum (HC70) was generated against the bacterially expressed lumenal fragment of HLA-A2 and HLA-B27 (47). Rabbit polyclonal p8 was generated against the cytoplasmic tail of HLA-A2 (exon 8), anti-protein disulfide isomerase (PDI) rabbit polyclonal antibody was generated against bacterially expressed PDI, and antihuman transferrin receptor antibody was a gift from I. S. Trowbridge (The Salk Institute, La Jolla, Calif.). Monoclonal antibody HC10, produced against free HLA-B HCs, was used for detection of HCs in immunoblots (41). Monoclonal antibody against Rab9 was purchased from Affinity BioReagents (Golden, Colo.). Monoclonal anti-γ-adaptin AP-1 (clone 100/3; Sigma Chemical Co., St. Louis, Mo.) and rabbit polyclonal anti-C9 (α-3 subunit of 20S proteasome; gift from J. J. Monaco, University of Cincinnati, Cincinnati, Ohio) antibodies were used as described above.
Pulse-chase analysis.
Prior to the pulse, 106 cells per ml were incubated at 37°C for 1 h in starvation medium (cysteine- and methionine-free Dulbecco's modified Eagle's medium [DMEM] supplemented with 5% fetal calf serum, 5% calf serum) and either the specified inhibitor or the same volume of carrier alone (DMSO or acetonitrile). Cells were pulse-labeled at 106 cells per 50 μl of starvation medium with 500 μCi of [35S]methionine-cysteine (1,200 Ci/mmol; NEN-Dupont, Boston, Mass.)/ml for the specified time at 37°C. Following the pulse, cells were chased at 106 cells per ml for the indicated times in starvation medium containing 2.5 mM methionine and 0.5 mM cysteine. The indicated inhibitors were present throughout the starvation, pulse, and chase. From 1 × 106 to 2 × 106 cells were taken at each time point and lysed on ice in 1 ml of NP-40 lysis buffer (10 mM Tris [pH 7.8], 150 mM NaCl, 5 mM MgCl2, 0.5% NP-40) supplemented with 1.5 μg of aprotinin per ml, 1 μM leupeptin, and 2 mM phenylmethylsulfonyl fluoride (PMSF), followed by immunoprecipitation (described below). Sodium dodecyl sulfate (SDS) lysis was carried out in 100 μl of a mixture of 1% SDS, 50 mM Tris (pH 7.4), and 1 mM dithiothreitol (DTT). The samples were vortexed vigorously to reduce viscosity and then heated. Prior to immunoprecipitation, the SDS concentration was adjusted to 0.1% by addition of NP-40 lysis buffer.
Immunoprecipitations and endo H and _N_-glycanase digestion.
NP-40 lysates were centrifuged for 10 min at 10,000 × g to remove cell debris. Nonspecifically binding proteins were removed from the cell lysates by incubation with 4 μl of normal mouse or rabbit serum per ml for 20 min followed by a 40-min incubation with formalin-fixed heat-killed Staphylococcus aureus cells at 4°C with gentle agitation. After centrifugation at 8,000 × g for 1 min, the supernatant was transferred to a new tube. Immunoprecipitations were carried out by incubation at 4°C with antiserum for 20 min followed by the addition of S. aureus for 40 min. The pelleted immunoprecipitates were washed three times in washing buffer (0.5% NP-40, 50 mM Tris [pH 7.4], 150 mM NaCl, 5 mM EDTA). For lysates containing 0.1% SDS, SDS was added to the wash buffer to a final concentration of 0.1%. The immunoprecipitates were released from the S. aureus cells by addition of reducing sample buffer (4% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.025% bromophenol blue in 62.5 mM Tris [pH 6.8]) and subjected to SDS-polyacrylamide gel electrophoresis (PAGE [12.5% polyacrylamide]) as described previously (34). For reimmunoprecipitations, immunoprecipitates were released in 100 μl of a mixture of 1% SDS, 50 mM Tris (pH 7.4), and 1 mM DTT at 95°C for 1 min. The released material was immunoprecipitated as described above after the addition of 1 ml of NP-40 lysis buffer. Treatment with endoglycosidase H (endo H) (New England Biolabs, Beverly, Mass.) and _N_-glycanase (Roche/Boehringer Mannheim, Germany) was performed as suggested by the manufacturers.
Na2CO3 extraction.
A total of 10 × 106 BJAB or K3-BJAB cells were pulsed for 30 min and chased for 2 h as described above. Cell pellets were resuspended in 1.5 ml of homogenization buffer (0.25 mM sucrose, 1 mM EDTA, 2 mM PMSF, 1.5 μg of aprotinin per ml, 1 μM leupeptin) and homogenized with four strokes in a ball-bearing homogenizer (1-μm gap). Homogenates were incubated on ice with 0.1 M Na2CO3 or buffer alone for 30 min, followed by centrifugation at 150,000 × g for 1.5 h. The supernatant and pellet fractions were lysed in SDS, followed by immunoprecipitation as described above.
In vitro transcription and translations.
In vitro transcription was performed as described previously (20). In vitro translations were performed for 1 h 30 min at 30°C in a reaction mixture containing 17.5 μl of Flexi rabbit reticulocyte lysate (Promega), 0.8 μl of KCl (2.5 M; Promega), 0.5 μl of amino acid mixture minus methionine (1 mM; Promega), 2.5 μl of [35S]methionine (10 mCi/ml; translation grade; NEN), 0.2 to 1 μl of the indicated mRNA, and 0.5 μl of canine pancreas microsomal membranes, prepared as described previously (49). After translation, the reaction mixture was centrifuged at 10,000 × g for 10 min. The microsomal pellet was resuspended in 1× reducing sample buffer and subjected to SDS-PAGE (12.5% polyacrylamide).
Percoll gradient fractionation.
Cells were pulsed for 30 min and chased for 2 h as described above in the presence of the specified inhibitors. Two-step Percoll gradient fractionations were performed as described previously (10). Aprotinin was included in the homogenization buffer at 1.5 μg/ml.
Characterization of subcellular fractions.
Markers for various cellular compartments were used to determine their distribution in both gradients. Fractions collected from the two-step Percoll gradients were analyzed by colorimetric assay for β-hexosaminidase activity (37) and by immunoblotting for marker localization. PDI (endoplasmic reticulum [ER]), AP-1 (Golgi), transferrin receptor (early endosome and plasma membrane), and rab9 (late endosome) and the α-3 20S proteasome subunit were used as markers for immunoblotting. Biotin-JPM, an active site covalent cysteine protease inhibitor, containing a biotin moiety (JPM-biotin, synthesized in the laboratory as described previously [15]), was used as a marker for lysosomes. For JPM-biotin labeling, fractions were lysed at pH 5.0.
Immunoblotting.
One hundred microliters of each fraction collected from the Percoll gradients was lysed by addition of 4× NP-40 lysis buffer and centrifuged at 100,000 × g for 30 min to sediment the Percoll. The supernatant was boiled in reducing sample buffer and subjected to SDS-PAGE (12.5% polyacrylamide). After electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and blocked in 5% milk in phosphate-buffered saline containing 0.2% Tween. Primary antibody incubations were performed with the specified antiserum. Secondary antibodies (horseradish peroxidase [HRP] conjugated; Southern Biotechnologies, Birmingham, Ala.) were used at a dilution of 1/10,000. JPM-biotin was detected with HRP-conjugated streptavidin (Amersham, Life Science). Enhanced chemiluminescence (NEN) was used for visualization. The signal obtained was quantitated with ALPHAIMAGER software by Alpha Innotech (San Leandro, Calif.).
Quantitation of immunoprecipitations.
Quantitation of the immunoprecipitations from Percoll gradient fractions was performed with a Storm PhosphorImager (Molecular Dynamics) and analyzed with ImageQuant software.
RESULTS
MHC class I HC stability in K3-expressing BJAB cells.
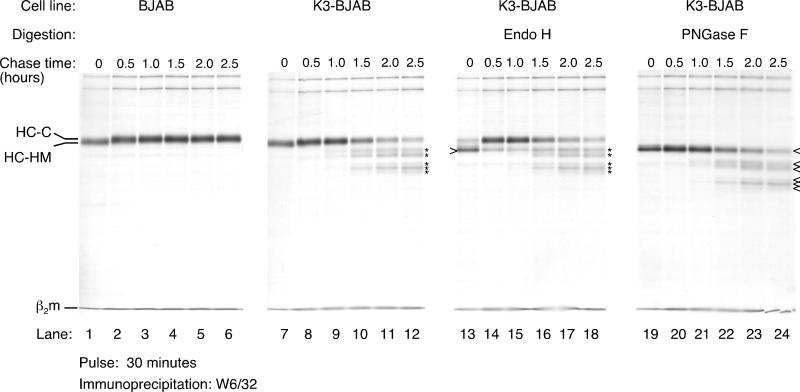
We analyzed, by pulse-chase analysis, the stability of MHC class I HC in cells that stably express K3 (K3-BJAB). K3-BJAB cells were pulsed with [35S]methionine for 30 min and chased for the indicated times (Fig. 1). Detergent lysates were subjected to immunoprecipitation with the W6/32 antibody, a pan-class I antibody that recognizes the trimeric MHC class I complex (HC, β2-microglobulin [β2m], and peptide), and the immunoprecipitates were analyzed by SDS-PAGE. The level of HC reactive with W6/32 increases from the 0- to the 30-min chase point as the trimeric complexes assemble from their subunits. The conversion of the single N-linked high-mannose glycan in HC (HC-HM) to the complex oligosaccharide-containing form (HC-C) occurs at approximately 30 min of chase (Fig. 1, compare lanes 1 and 2 and 7 and 8) and results in resistance to endo H digestion (data not shown). From this point on, HCs are stable in BJAB cells transfected with a control plasmid (Fig. 1, lanes 1 to 6). Maturation of HC appears unaffected in K3-BJAB cells, as judged by acquisition of endo H resistance (compare lanes 13 and 14). Most of the W6/32 reactive material in BJAB (data not shown) and in K3-BJAB is endo H resistant by 30 min of chase (Fig. 1, lanes 14 to 18). However, recovery of full-length mature HC decreases over time in K3-BJAB cells (Fig. 1, lanes 7 to 12). In addition, several HC intermediates (described below) are present at one 1 h of chase (Fig. 1, lane 9) in K3-BJAB cells. Five distinct HC intermediates of progressively lower apparent molecular weight are observed at the 2-h chase point (Fig. 1, lane 11). All intermediates are sensitive to endo F (PNGase F) digestion, showing that they have retained their N-linked glycan (Fig. 1; compare lanes 7 to 12 with lanes 19 to 24). The intermediates are all endo H resistant (Fig. 1; compare lanes 9 to 12 with lanes 15 to 18), indicating that they are generated in a post-Golgi compartment.
FIG. 1.
MHC class I HC maturation and stability in K3-BJAB cells. Properly folded MHC class I complexes were recovered by immunoprecipitation with the W6/32 antibody from NP-40 lysates of BJAB (lanes 1 to 6) and K3-BJAB (lanes 7 to 12) cells at the indicated chase points after a 30-min pulse with [35S]methionine. Indicated are the HC-HM and HC-C forms of the MHC class I HC as well as the light chain, β2m. The asterisks denote MHC class I HC fragments seen only in K3-BJAB cells. In lanes 13 to 18, the W6/32 immunoprecipitates were digested with endo H, and in lanes 19 to 24, they were digested with _N_-glycanase (PNGase F). The open arrowhead indicates deglycosylated MHC class I HC. Note that the MHC class I HC fragments marked by the asterisks are endo H resistant and PNGase F sensitive.
The ubiquitin-proteasome pathway is necessary for downregulation of MHC class I by KSHV K3.
An intact ubiquitination machinery and proteasomal activity are required for downregulation of some receptors from the plasma membrane, as well as for sorting of proteins within the endocytic pathway. In addition, RING domain-containing E3-ubiquitin protein ligases are involved in sorting of vesicles within the endocytic pathway. Since K3 contains a zinc finger domain in its N terminus predicted to be structurally similar to RING-H2 domains, we examined the effect of the covalent proteasome inhibitor ZL3VS on downregulation of MHC class I by K3.
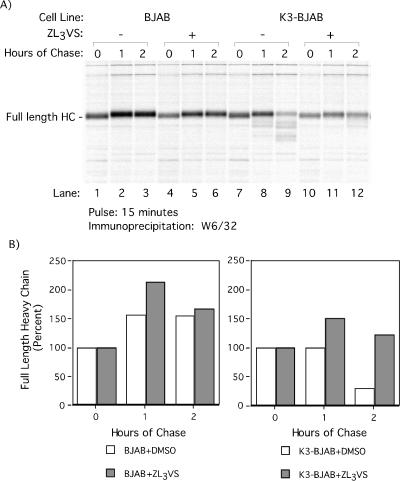
Stability of HC in BJAB and K3-BJAB cells was analyzed by pulse-chase analysis in the absence or presence of 50 μM ZL3VS. The inclusion of 50 μM ZL3VS results in increased stability of full-length HC in K3-BJAB cells compared to that in controls (Fig. 2A, compare lanes 9 and 12). In addition, 50 μM ZL3VS prevents generation of the lower-molecular-weight HC species found in DMSO-treated or untreated K3-BJAB cells at 2 h of chase (Fig. 2A; compare lanes 9 and 12).
FIG. 2.
Proteasome inhibitor ZL3VS stabilizes MHC class I HC in K3-BJAB cells. (A) BJAB and K3-BJAB cells in the presence of DMSO (lanes 1 to 3 and 7 to 9) or 50 μM ZL3VS (lanes 4 to 6 and 10 to 12) were pulsed for 15 min with [35S]methionine and chased for the indicated times. Properly folded MHC class I HCs were recovered from NP-40 lysates with the W6/32 antibody and subjected to SDS-PAGE (12.5% polyacrylamide). (B) Quantitation of the autoradiogram in panel A, depicted as the percentage of full-length MHC class I HC recovered at each time point. Open bars, DMSO treated; gray bars, treated with 50 μM ZL3VS.
Quantitation of full-length HC in control and K3-BJAB cells illustrates the increased stability of full-length HC in K3-BJAB cells treated with 50 μM ZL3VS (Fig. 2B, gray bars) compared to the DMSO control (Fig. 2B, open bars). Treatment with 50 μM ZL3VS has no effect on the levels of HC recovered from BJAB up to 2 h of chase. However, in K3-BJAB cells, 50 μM ZL3VS restores the recovery of full-length HC (from 30% to 122%) to a level similar to that in BJAB (167%) at 2 h of chase. This suggests that the ubiquitin-proteasome pathway is involved in K3-mediated downregulation of HC. Of note, at no time did we detect the occurrence of intermediates of higher molecular weight, such as would be expected for ubiquitin-conjugated forms of HC.
MHC class I HC fragments in K3-BJAB cells.
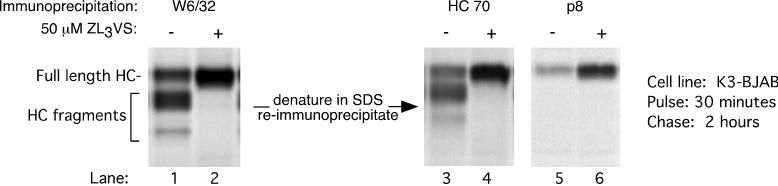
The difference in size between the observed class I HC intermediates and full-length HC cannot be due to their glycosylation state (Fig. 1) and therefore must be attributed to proteolytic cleavage, because no other posttranslational modification produces this type of change in mass. To characterize these intermediates, we first recovered all W6/32 reactive complexes from the 2-h chase point, which include the proteolytic intermediates, and then fully denatured this material. The resulting free HCs were then immunoprecipitated with antibodies that recognize either the lumenal domain of unfolded HC (HC70) or the cytoplasmic tail of HC (p8) (Fig. 3). Both HC70 and p8 recover full-length HC in this assay (Fig. 3, lanes 4 and 6, respectively). All five of the W6/32 reactive HC intermediates seen at 2 h of chase are recognized by the HC70 antiserum, while none of them are recovered by p8 (Fig. 3, compare lane1 with lanes 3 and 5). The HC intermediates must therefore be fragments of HC that lack the cytoplasmic tail. Their reactivity with W6/32 suggests that they contain an intact N terminus that is part of a properly folded complex, since the entire lumenal domain is required for association with β2m and generation of the W6/32 epitope.
FIG. 3.
MHC class I HC fragments in K3-BJAB cells lack the cytoplasmic tail. MHC class I complexes were immunoprecipitated from K3-BJAB lysates after a 30-min pulse and 2 h of chase in the absence (lane 1) or presence (lane 2) of 50 μM ZL3VS. The immunoprecipitates were boiled in SDS, and the released material was then immunoprecipitated with HC70 (lanes 3 and 4) or p8 (lanes 5 and 6). HC70 recognizes the lumenal fragment of unfolded MHC class I HC; p8 recognizes the cytoplasmic tail of MHC class I HC.
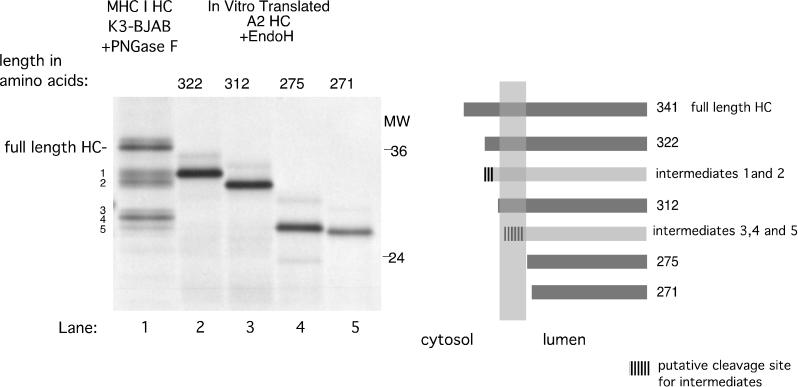
In order to establish the location of the cleavage sites that generate these HC fragments, we compared PNGase F-treated HC fragments obtained from K3-expressing cells to endo H-digested, in vitro-translated C-terminal truncations of HLA-A2 (Fig. 4). The C-terminal truncation sites are depicted diagrammatically in Fig. 4. K3-mediated HC fragments 1 and 2 (Fig. 4, lane 1) are approximately equal in size to HLA-A2 HC truncated at aa 322 (Fig. 4, lane 2) and 312 (Fig. 4, lane 3), respectively. Both HLA-A2322 and HLA-A2312 contain the entire lumenal domain and transmembrane domain of HLA-A2 HC, while HLA-A2322 has 14 cytoplasmic tail residues and HLA-A2312 has only 4. HC fragments 3, 4, and 5 (Fig. 4, lane 1) appear to have a higher apparent molecular weight than HLA-A2 truncated at aa 275 (Fig. 4, lane 4) and 271 (Fig. 4, lane 5), both of which lack the transmembrane domain. This suggests that HC fragments 3, 4, and 5 contain the lumenal domain and only part of the transmembrane domain of HC.
FIG. 4.
MHC class I HC fragments in K3-BJAB cells could be generated by cleavages within its cytoplasmic tail and transmembrane segment. PNGase F-treated W6/32 immunoprecipitates from K3-BJAB cells at the 2-h chase point (lane 1, same as lane 23 in Fig. 1) were compared in SDS-PAGE (12.5% polyacrylamide) with endo H-treated, in vitro-translated HLA-A2 HC truncated at aa 322, 312, 275, and 271. The panel on the right side is a schematic representation of the HLA-A2 truncations (dark gray bars) and the estimated cleavage region (hatched area) of HC intermediates (light gray bars).
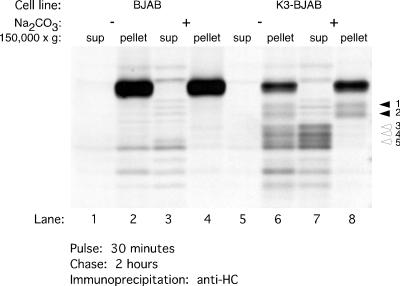
We reasoned that if HC fragments 3, 4, and 5 contain only a small segment of the transmembrane anchor, they should be either soluble or loosely associated with the membrane. Thus, we examined whether fragments 3, 4, and 5 are extractable by treatment with sodium carbonate (Fig. 5). After a 30-min pulse and 2 h of chase, BJAB and K3-BJAB cells were homogenized in the absence of detergent. The homogenates were treated with 0.1 M Na2CO3 and centrifuged at 150, 000 × g to sediment all particulate matter. HC was immunoprecipitated from the supernatant and pellet fractions after their extraction with detergent. As expected, full-length HCs in BJAB and K3-BJAB cells remain associated with the pellet fraction in both the presence (Fig. 5, lanes 4 and 8) and absence (Fig. 5, lanes 2 and 6) of 0.1 M Na2CO3. HC fragments 1 and 2 are also found in the pellet fraction, regardless of 0.1 M Na2CO3 treatment (Fig. 5, lanes 6 and 8). However, HC fragments 3, 4, and 5 are found in the pellet fraction in the untreated homogenates (Fig. 5, lane 6), but are released into the supernatant after treatment with 0.1 M Na2CO3 (Fig. 5, lane 7). Thus, fragments 3, 4, and 5 are either soluble or only loosely associated with the membrane.
FIG. 5.
Some MHC class I HC intermediates in K3-BJAB cells are soluble or loosely associated with membranes. BJAB or K3-BJAB cells pulsed for 30 min and chased for 2 h were homogenized as described for the Percoll gradient fractionations. The postnuclear supernatants were treated on ice with 0.1 M Na2CO3 (lanes 3, 4, 7, and 8) or buffer alone (lanes 1, 2, 5, and 6), followed by centrifugation at 150,000 × g for 1.5 h. The supernatant (sup) and pellet fractions were lysed in SDS, and the free MHC class I HC was immunoprecipitated with HC70. Solid arrowheads indicate MHC class I HC fragments 1 and 2, which remain associated with the pellet fraction after treatment with 0.1 M Na2CO3. Open arrowheads indicate MHC class I HC fragments 3, 4, and 5, which are released into the supernatant upon treatment with 0.1 M Na2CO3.
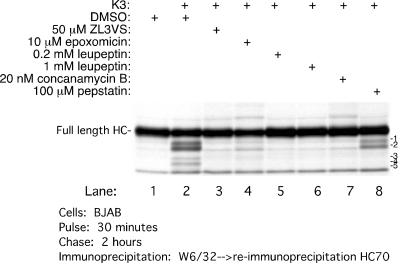
Since generation of these HC fragments is sensitive to ZL3VS, we examined the effect of other inhibitors on the generation of HC fragments (Fig. 6). All cells were pretreated for an hour prior to the pulse with the inhibitors indicated, and HC was then recovered at 2 h of chase. A smaller amount of full-length HC or its fragments is recovered from K3-BJAB cells treated with DMSO than was recovered from BJAB cells (Fig. 6, lanes 1 and 2). Treatment with 50 μM ZL3VS or 10 μM epoxomicin, a structurally unrelated proteasome inhibitor, suppresses the generation of HC fragments (Fig. 6, compare lanes 3 and 4 to lane 2), and both agents yield an increase in recovery of full-length HC. Similar results were obtained with 5 μM lactacystin (data not shown). The cysteine protease inhibitor leupeptin, at concentrations of 0.2 and 1 mM (Fig. 6, lanes 5 and 6), and an inhibitor of the vacuolar H+ ATPase, concanamycin B at 10 nM (Fig. 6, lane 7), stabilize HC and prevent generation of the fragments in K3-BJAB cells. The only compound that we identified that selectively affects the generation of some HC fragments is the aspartyl protease inhibitor pepstatin. It inhibits generation of fragments 3, 4, and 5 only (Fig. 6, lane 8). Based on our site comparisons with in vitro-translated HC (Fig. 4), we suggest that the cleavage(s) executed by aspartyl protease(s) must be within the plane of the hydrophobic transmembrane segment, in the manner of a γ-secretase-like cleavage.
FIG. 6.
Effect of various proteasome and protease inhibitors on MHC class I HC stability in K3-BJAB cells. MHC class I HCs were recovered after a 30-min pulse and 2 h of chase, in the presence of the indicated compound, from BJAB or K3-BJAB cells. The MHC class I HC intermediates are labeled 1 to 5.
Subcellular distribution of MHC class I in K3-expressing cells as determined by subcellular fractionation.
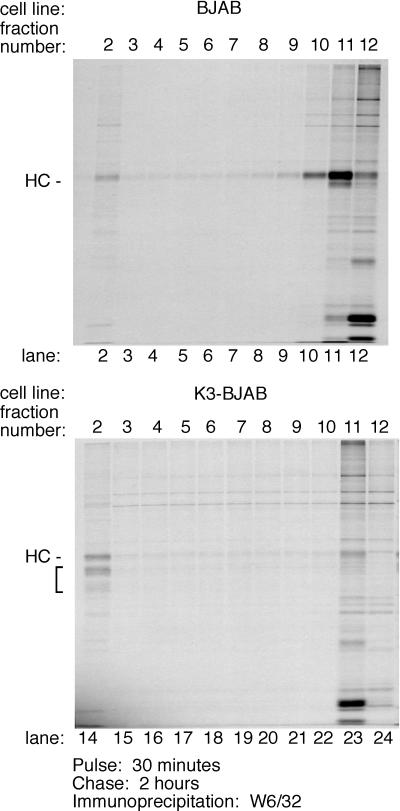
To determine the subcellular distribution of MHC class I HC and its intermediates 2 h 30 min after synthesis, BJAB and K3-BJAB cells were pulsed for 30 min with [35S]methionine and chased for 2 h with cold methionine, and cell homogenates were fractionated with a two-step Percoll gradient (Fig. 7). Markers for various cellular compartments were used to characterize their distribution in the gradients at steady state (see Materials and Methods) (data not shown). In brief, equal levels of β-hexosaminidase activity are found in the top (lighter) and bottom (denser) fractions of the 10% gradients for all cell lines. Markers analyzed by immunoblotting include PDI (ER), AP-1 (Golgi), α-3 20S proteasome subunit, transferrin receptor (early endosome and plasma membrane), MHC class I HC (plasma membrane), and rab9 (late endosome). At steady state, these markers are found in the top fractions of the 10% gradient for all cell lines examined. As expected at steady state, HC is mostly found in the top fractions from BJAB cells (fractions 10 to 12 [data not shown]), while a small amount of HC is found in the bottom fractions of all the gradients from K3-BJAB cells (fraction 2, data not shown). Biotin-JPM, an active site covalent cysteine protease inhibitor containing a biotin moiety, was used as a marker for lysosomes. Most JPM-biotin is detected in the top fractions, while a small amount is seen in fraction 2 in both BJAB and K3-BJAB cells (data not shown). Thus, lighter organelles, such as ER, Golgi, plasma membrane, and early endosomes, are found at the top of the gradient, while denser organelles, such as lysosomes are found at the bottom of the gradient. The patterns for HC in K3-BJAB cells and JPM-biotin in K3-BJAB and BJAB cells are very similar, suggesting that in K3-BJAB cells, HC is delivered to lysosomes or a prelysosomal, dense compartment.
FIG. 7.
Subcellular distribution of MHC class I HC within the endocytic compartment in K3-BJAB cells. BJAB and K3-BJAB cells were pulsed for 30 min and chased for 2 h, followed by homogenization in the absence of detergent. Cell homogenates were loaded on a 27% Percoll gradient, and 1-ml fractions were collected. The lighter fractions with high β-hexosaminidase activity were loaded onto the 10% Percoll gradient, and 1-ml fractions were collected. MHC class I complexes were recovered by immunoprecipitation with the W6/32 antibody from each 10% Percoll gradient fraction (lanes 2 to 12 for BJAB, lanes 14 to 24 for K3-BJAB). The bracket indicates the MHC class I HC fragments seen in K3-BJAB cells. Lanes 2 and 14 correspond to the bottom of the gradient, and lanes 12 and 24 correspond to the top of the gradient.
MHC class I complexes were immunoprecipitated from all fractions from the 10% Percoll gradient with the W6/32 antibody and subjected to SDS-PAGE, and radiolabled HC was detected by autoradiogram (Fig. 7). In BJAB cells, the majority of HC is found at the top of the gradient (Fig. 7, lanes 10, 11, and 12), with only trace amounts found at the bottom of the gradient (Fig. 7, lane 2). In K3-BJAB cells, full-length HC is distributed equally between the top and bottom of the gradient (Fig. 7, lanes 23 and 14, respectively), while K3-dependent HC fragments are found exclusively at the bottom of the gradient (Fig. 7, lane14). This suggests that the K3-dependent HC fragments are generated at a location corresponding to the denser organelles in the endocytic pathway.
Given that the ubiquitin-proteasome pathway is involved in sorting in the endocytic pathway and that various proteasome inhibitors block MHC class I downregulation by K3, we examined the effect of ZL3VS on the distribution of HC in K3-BJAB cells. To test whether proteasome inhibitor blocked internalization of MHC class I from the plasma membrane, we used cytofluorimetry to determine the surface level of MHC class I in K3-BJAB cells treated with ZL3VS. K3-BJAB cells, regardless of drug treatment, display an equivalent low level of MHC class I at the surface, as do untreated K3-BJAB cells (data not shown). The proteasome must therefore be involved in a step subsequent to internalization from the plasma membrane.
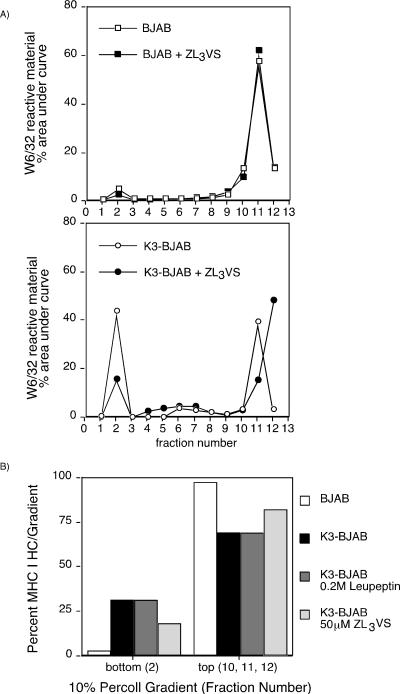
To determine where the proteasome inhibitor exerts its effect within the endocytic pathway, we analyzed the distribution of HCs in K3-BJAB cells treated with ZL3VS by using a two-step Percoll gradient (Fig. 8A) as described above. The distribution of W6/32 reactive HC is not affected by ZL3VS in BJAB cells. However, in K3-BJAB cells, the amount of total HC found in the dense portion of the gradient decreases when cells are treated with ZL3VS (15%) compared to the amount found in DMSO-treated K3-BJAB cells (43%). In contrast, leupeptin, while stabilizing the level of HC in K3-BJAB cells, has no effect on the distribution of HC in K3-BJAB cells in these subcellular fractionations (Fig. 8B). The total amount of HC found in the dense fraction from K3-BJAB cells, which consists of full-length HC and the HC fragments (31%), equals the total amount of HC found in the dense fraction from K3-BJAB cells treated with leupeptin (31%), which consists of only full-length HC. However, the total amount of HC found in the dense fraction from K3-BJAB cells treated with ZL3VS (18%; only full-length HC) is smaller than that seen in untreated K3-BJAB cells. Thus, we conclude that the ubiquitin-proteasome system is involved in the sorting of HC within the endocytic pathway in the presence of K3. Moreover, once HC has been sorted to the correct compartment within the endocytic pathway, cysteine proteases are responsible for initiating the generation of HC fragments.
FIG. 8.
ZL3VS and leupeptin differentially affect the subcellular distribution of MHC class I HC in K3-BJAB cells. (A) Quantitation of total MHC class I HC recovered in a W6/32 immunoprecipitation from the 10% Percoll gradient fractions from BJAB (squares) or K3-BJAB (circles) cells pulsed and chased in the absence (open symbols) or presence (closed symbols) of 50 μM ZL3VS. Levels of MHC class I HC are plotted for each fraction as the percent area under the curve within each gradient. (B) Quantitation of total MHC class I HC at the top and bottom of a 10% Percoll gradient of BJAB and K3-BJAB cells after a 30-min pulse and 2-h chase in the presence of the inhibitors indicated.
DISCUSSION
The ubiquitin-proteasome system plays a critical role in regulation within the endocytic pathway. The structural similarity of PHDs and RING-H2 domains found in E3 ubiquitin-protein ligases suggests a connection between KSHV K3 and the ubiquitin-proteasome system in downregulation of MHC class I complexes. Our results indicate that active proteasomes are required for degradation of MHC class I molecules by K3, as inferred from its sensitivity to proteasome inhibitors. More specifically, we show that an active proteasome is necessary in sorting of MHC I complexes to a dense endocytic compartment in cells that express K3. So far, we have been unable to observe accumulation of ubiquitin-conjugated HC. While this lack of obvious ubiquitin-conjugated intermediates might be due to technical difficulties in detecting them, it is also possible that some other protein is the prime target for ubiquitin conjugation.
Direct involvement of the proteasome in downregulation of proteins from the plasma membrane is seen for the tyrosine kinase receptor c-Met, which is ubiquitinated at the plasma membrane and subsequently degraded by the proteasome (23). However, the proteolytic intermediates of MHC class I HC that we observe in K3 cells are generated not by the proteasome, but rather by cysteine and aspartyl proteases. Three structurally different proteasome inhibitors stabilize MHC class I HC in K3-BJAB cells, ruling out as a possible explanation the inhibition of cysteine proteases by these inhibitors. And while both proteasome inhibitors and cysteine protease inhibitors result in a comparable level of stabilization of MHC class I HC in pulse-chase experiments, they differentially affect the distribution of MHC class I HC within endocytic compartments. These observations effectively rule out inhibition of cysteine proteases by proteasome inhibitors as an explanation for our results.
Indirect involvement of the proteasome has been invoked in downregulation of other surface receptors, such as the GHR. A _trans_-acting protein appears to be the target of the ubiquitin-proteasome system in ligand-induced downregulation of GHR, while the lysine residues in the cytoplasmic tail of GHR, the possible targets for ubiquitination, are not required for ligand-induced downregulation (48). Likewise, rsp5 ubiquitin ligase, a Nedd4 homolog in yeast, is required for downregulation of a nonubiquitinated yeast chimeric receptor (11). Of note, Nedd4 is also involved in retrovirus particle release, which may be considered a form of reverse vesicle budding (44). K3 may remove HC from the plasma membrane in a set of reactions that utilize this common mechanism of regulated vesicle budding. The ubiquitin-proteasome system might then target a negative regulator of MHC class I endocytosis in K3-expressing cells. While our inability to detect any mono- or polyubiquitination of MHC class I HC would fit this model, proteasomal inhibition by proteasome inhibitors does not increase the level of MHC class I at the cell surface in K3-BJAB cells, as determined by fluorescence-activated cell sorter analysis (data not shown). It is possible that a similar mechanism is utilized in regulation of vesicle budding later in the endocytic pathway.
The epidermal growth factor receptor tyrosine kinase ErbB-1 exploits a different ubiquitin-proteasome-dependent sorting system (see review in reference 17). Sorting of internalized ErbB1 into late endocytic compartments instead of recycling endosomes is dependent on the RING-containing E3 ubiquitin protein ligase, c-cbl. K3 might serve a function in MHC class I sorting similar to that of c-cbl for ErbB-1, particularly if the PHDs are functionally and structurally homologous to RING-H2 domains.
The murine gammaherpesvirus 68-encoded K3 (MK3) downregulates MHC class I complexes before they traverse the secretory pathway (42). MK3 is homologous to K3 and K5 in its PHD and overall structure. Thus, despite their different sites of action, MK3 and KSHV K3 might share a general mechanism. As in K3 cells, MHC class I complexes are stabilized by proteasome inhibitors in cells expressing MK3 (4). However, the MHC class I HC appears to be ubiquitinated in MK3 cells. Removal of the cytoplasmic tail of MHC class I HC or the three lysines in its cytoplasmic tail results in stabilization of HC, while the apparent ubiquitination is not affected. It has been difficult to establish the biochemistry of this ubiquitination reaction, and extremely long exposure times were required for the autoradiographic detection of ubiquitin-conjugated MHC class I HC.
Coscoy et al. report that the PHD of K5 can mediate ubiquitination in vitro (9). They show that ubiquitinated polypeptides are present in an immunoprecipitation of HC from cells expressing K3 and K5, but whether these in fact correspond to ubiquitinated MHC class I HC or represent associated proteins was not addressed. In addition, while the lysine residues in the tail of MHC class I are important for downregulation of HC by K5, the presence of ubiquitinated material in an immunoprecipitation of this mutant was not explored. Their results suggest that the PHD of K3 and K5 might indeed mediate ubiquitination of HC or an associated protein in cells and support our proposal that the ubiquitin proteasome system is involved in K3- and K5-mediated downregulation of MHC class I. While we have had no difficulty in detecting ubiquitin-conjugated class I HC under other circumstances in which degradation of class I HC occurs (M. Furman and H. Ploegh, unpublished observations), we have so far not detected ubiquitin-conjugated HC in K3-BJAB cells. Differences in the experimental conditions used by Coscoy et al. (9) and the present study may be responsible. It will be important to establish the stoichiometry and sites of ubiquitin attachment to shed further light on the underlying mechanism of K3-mediated downregulation of MHC class I molecules.
Our findings share some similarity with recently published examples of interleukin-2 receptor β (IL-2Rβ) and Ste6p downregulation, where ubiquitin conjugation prevents recycling of the receptors to the plasma membrane and mediates endosomal sorting. Sorting of the a-factor transporter Ste6p to the yeast vacuole is compromised in a Doa4 mutant (27). Doa4 removes ubiquitin from substrates destined for the lumen of the vacuole and thus contributes to recycling of ubiquitin (45). As a result, mutants in Doa4 have low levels of free ubiquitin. The low ubiquitin levels in Doa4 mutant cells affect sorting of Ste6 to the lumen of the vacuole, yet have no effect on the internalization of Ste6 from the plasma membrane, an effect very similar to that seen for proteasome inhibitors in MHC class I downregulation in K3-BJAB cells. Transient treatment of cells with proteasome inhibitors has been reported to result in an acute depletion of the free ubiquitin pool (29), resulting in a situation analogous to that of the Doa4 mutant. Likewise, the ubiquitin-proteasome pathway is involved in sorting of the IL-2R (36) to late endocytic compartments and not in internalization from the cell surface. While both Ste6p and IL-2Rβ are ubiquitinated, it is possible that K3 or another protein is the target for ubiquitination or that our assays are not sensitive enough to detect a ubiquitinated intermediate for the class I HC. Regardless, the stoichiometry of ubiquitin conjugation could be a key parameter to determine, in most cases, where ubiquitin conjugation has been invoked as the signal for receptor sorting.
Once MHC class I complexes are sorted into a dense endocytic compartment in K3 cells, the cytoplasmic tail of HC is proteolytically cleaved by a cysteine protease. The transmembrane domain of MHC class I HC is subsequently cleaved within the plane of the membrane by an aspartyl protease. Similar cleavages have not been previously reported for MHC class I or for any of the downregulation pathways used by a virus. However, the proteolytic cleavages of HC seen in cells expressing K3 are reminiscent of the proteolytic pattern seen for the yeast vacuolar hydrolase carboxypeptidase S during its maturation in multivesicular bodies and the vacuole (25, 40). Carboxypeptidase S is sorted from the secretory pathway to late endosomes via the trans-Golgi network, and as late endosomes mature into multivesicular bodies, carboxypeptidase S-containing structures are invaginated into the lumen of the multivesicular body. Vesicles containing carboxypeptidase S are then delivered to the vacuole, where the cytoplasmic tail and transmembrane domain of carboxypeptidase S are cleaved, yielding a soluble, mature form of carboxypeptidase S. During this maturation process, the ESCRT-1 complex directs carboxypeptidase S to the budding multivesicular body vesicles via monoubiquitination of the cytoplasmic tail of carboxypeptidase S. The ESCRT-1 complex appears to recognize ubiquitinated carboxypeptidase S via the ubiquitin conjugating (ubc)-like domain of one of its components, the class E protein Vps23. Vps23 is a ubiquitin E2 variant and thus resembles an E2, but does not perform classical E2 functions. Interestingly, the human homolog of Vps23, Tsg101, has been implicated in retrovirus budding (32) and in mediating protein delivery to late endosomal compartments (2, 38). Of note, some transmembrane proteins destined for the lumen of the vacuole are sorted in a ubiquitin-independent manner (35).
The multivesicular body also collects proteins derived from the plasma membrane. Thus, even though MHC class I molecules are not usually found in multivesicular bodies, K3 might utilize a preexisting cellular pathway that sorts plasma membrane constituents to the multivesicular body on their way to the lumen of the lysosome. For example, K3 might mediate monoubiquitination of MHC class I complexes directly or indirectly and thus facilitate recognition of MHC class I complexes by ESCRT-1. Alternatively, K3 or another associated protein might be recognized as the cargo signal, directing MHC class I into the vesicles of the multivesicular body. This would bypass a requirement for monoubiquitination of MHC class I molecules, which we have so far been unable to detect. Either way, K3 might be part of ESCRT-1 or an ESCRT-1-like complex. Analysis by immunofluorescence of MHC class I molecules and K3 in cells by others has failed to detect any colocalization (7, 22). However, high-resolution immunoelectron microscopy would be required to determine the extent of colocalization of MHC class I molecules and K3 in multivesicular bodies.
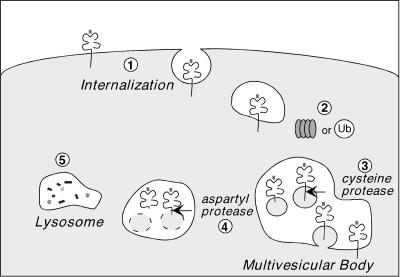
Our findings indicate that KSHV K3 utilizes the ubiquitin-proteasome pathway, after the initial internalization step, in routing MHC class I complexes for destruction in lysosomes (Fig. 9). We show that the HC of properly folded MHC class I complexes is cleaved by cysteine and aspartyl proteases once in this compartment. K3 might be the first viral protein described to utilize ESCRT-1 or a similar complex in sorting molecules within endocytic compartments.
FIG. 9.
Proposed model of the ubiquitin-proteasome pathway in MHC class I downregulation by KSHV K3. (Step 1) MHC class I complexes are internalized from the plasma membrane in K3-expressing cells in a step independent of the ubiquitin-proteasome pathway. (Step 2) Active proteasomes and/or the levels of ubiquitin (Ub) mediate sorting of MHC class I complexes from an early endocytic compartment to a late, dense endocytic compartment, most likely the multivesicular body. (Step 3) Once in this compartment, cysteine proteases remove the cytoplasmic tail of MHC class I HC, while aspartic proteases sequentially cleave MHC class I HC within the plane of the membrane (step 4). (Step 5) The soluble lumenal fragment of MHC class I is then delivered to lysosomes for complete degradation.
Acknowledgments
We thank Domenic Tortorella, Margo Furman, Christoph Driessen, and all members of the Ploegh laboratory for assistance. We also thank Margo Furman for generating HLA-A2322 and HLA-A2271 and Patrick Stern and Brendan Lilly for critical review of the manuscript.
This work was supported by National Service Research award F31 GM18258 from the National Institute of General Medical Sciences and NIH grants P01 AI42257 and CA91819.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20**:**56-59. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1**:**248-258. [DOI] [PubMed] [Google Scholar]
- 3.Bogyo, M., J. S. McMaster, M. Gaczynska, D. Tortorella, A. L. Goldberg, and H. Ploegh. 1997. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA 94**:**6629-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15**:**627-636. [DOI] [PubMed] [Google Scholar]
- 5.Bonifacino, J. S., and A. M. Weissman. 1998. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 14**:**19-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capili, A. D., D. C. Schultz, I. F. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20**:**165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97**:**8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coscoy, L., and D. Ganem. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Investig. 107**:**1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155**:**1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driessen, C., R. A. Bryant, A. M. Lennon-Dumenil, J. A. Villadangos, P. W. Bryant, G. P. Shi, H. A. Chapman, and H. L. Ploegh. 1999. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J. Cell Biol. 147**:**775-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, R., and L. Hicke. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276**:**25974-25981. [DOI] [PubMed] [Google Scholar]
- 12.Dupre, S., C. Volland, and R. Haguenauer-Tsapis. 2001. Membrane transport: ubiquitylation in endosomal sorting. Curr. Biol. 11**:**R932-R934. [DOI] [PubMed] [Google Scholar]
- 13.Ettenberg, S. A., A. Magnifico, M. Cuello, M. M. Nau, Y. R. Rubinstein, Y. Yarden, A. M. Weissman, and S. Lipkowitz. 2001. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J. Biol. Chem. 276**:**27677-27684. [DOI] [PubMed] [Google Scholar]
- 14.Gewurz, B. E., E. W. Wang, D. Tortorella, D. J. Schust, and H. L. Ploegh. 2001. Human cytomegalovirus US2 endoplasmic reticulum-lumenal domain dictates association with major histocompatibility complex class I in a locus-specific manner. J. Virol. 75**:**5197-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenbaum, D., K. F. Medzihradszky, A. Burlingame, and M. Bogyo. 2000. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 7**:**569-581. [DOI] [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67**:**425-479. [DOI] [PubMed] [Google Scholar]
- 17.Hicke, L. 2001. A new ticket for entry into budding vesicles—ubiquitin. Cell 106**:**527-530. [DOI] [PubMed] [Google Scholar]
- 18.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2**:**195-201. [DOI] [PubMed] [Google Scholar]
- 19.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84**:**277-287. [DOI] [PubMed] [Google Scholar]
- 20.Huppa, J. B., and H. L. Ploegh. 1997. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity 7**:**113-122. [DOI] [PubMed] [Google Scholar]
- 21.Ishido, S., J. K. Choi, B. S. Lee, C. Wang, M. DeMaria, R. P. Johnson, G. B. Cohen, and J. U. Jung. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13**:**365-374. [DOI] [PubMed] [Google Scholar]
- 22.Ishido, S., C. Wang, B.-S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74**:**5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeffers, M., G. A. Taylor, K. M. Weidner, S. Omura, and G. F. Vande Woude. 1997. Degradation of the Met tyrosine kinase receptor by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 17**:**799-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102**:**549-552. [DOI] [PubMed] [Google Scholar]
- 25.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106**:**145-155. [DOI] [PubMed] [Google Scholar]
- 26.Lorenzo, M. E., H. L. Ploegh, and R. S. Tirabassi. 2001. Viral immune evasion strategies and the underlying cell biology. Semin. Immunol. 13**:**1-9. [DOI] [PubMed] [Google Scholar]
- 27.Losko, S., F. Kopp, A. Kranz, and R. Kolling. 2001. Uptake of the ATP-binding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell 12**:**1047-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Means, R. E., S. Ishido, X. Alvarez, and J. U. Jung. 2002. Multiple endocytic trafficking pathways of MHC class I molecules induced by a herpesvirus protein. EMBO J. 21**:**1638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36**:**14418-14429. [DOI] [PubMed] [Google Scholar]
- 30.Nicholas, J., V. Ruvolo, J. Zong, D. Ciufo, H.-G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71**:**1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parham, P., C. J. Barnstable, and W. F. Bodmer. 1979. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C antigens. J. Immunol. 123**:**342-349. [PubMed] [Google Scholar]
- 32.Perez, O. D., and G. P. Nolan. 2001. Resistance is futile. Assimilation of cellular machinery by HIV-1. Immunity 15**:**687-690. [DOI] [PubMed] [Google Scholar]
- 33.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8**:**499-504. [DOI] [PubMed] [Google Scholar]
- 34.Ploegh, H. L. 1995. One-dimensional isoelectric focusing of proteins in slab gels, p. 10.2.1-10.2.8. In J. E. Coligan, B. M. Dunn, H. L. Ploegh, D. W. Speicher, and P. T. Wingfield (ed.), Current protocols in protein science, vol. 1. John Wiley and Sons, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 35.Reggiori, F., and H. R. Pelham. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20**:**5176-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocca, A., C. Lamaze, A. Subtil, and A. Dautry-Varsat. 2001. Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor beta chain to late endocytic compartments. Mol. Biol. Cell 12**:**1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rome, L. H., A. J. Garvin, M. M. Allietta, and E. F. Neufeld. 1979. Two species of lysosomal organelles in cultured human fibroblasts. Cell 17**:**143-153. [DOI] [PubMed] [Google Scholar]
- 38.Ruland, J., C. Sirard, A. Elia, D. MacPherson, A. Wakeham, L. Li, J. L. de la Pompa, S. N. Cohen, and T. W. Mak. 2001. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc. Natl. Acad. Sci. USA 98**:**1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha, V., T. Chaplin, A. Gregorini, P. Ayton, and B. D. Young. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. USA 92**:**9737-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spormann, D. O., J. Heim, and D. H. Wolf. 1992. Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J. Biol. Chem. 267**:**8021-8029. [PubMed] [Google Scholar]
- 41.Stam, N. J., H. Spits, and H. L. Ploegh. 1986. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137**:**2299-2306. [PubMed] [Google Scholar]
- 42.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97**:**8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Story, C. M., M. H. Furman, and H. L. Ploegh. 1999. The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc. Natl. Acad. Sci. USA 96**:**8516-8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97**:**13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swaminathan, S., A. Y. Amerik, and M. Hochstrasser. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10**:**2583-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18**:**861-926. [DOI] [PubMed] [Google Scholar]
- 47.Tortorella, D., C. M. Story, J. B. Huppa, E. J. Wiertz, T. R. Jones, I. Bacik, J. R. Bennink, J. W. Yewdell, and H. L. Ploegh. 1998. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 142**:**365-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Kerkhof, P., R. Govers, C. M. Alves dos Santos, and G. J. Strous. 2000. Endocytosis and degradation of the growth hormone receptor are proteasome-dependent. J. Biol. Chem. 275**:**1575-1580. [DOI] [PubMed] [Google Scholar]
- 49.Walter, P., and G. Blobel. 1983. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96**:**84-93. [DOI] [PubMed] [Google Scholar]
- 50.Waterman, H., and Y. Yarden. 2001. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 490**:**142-152. [DOI] [PubMed] [Google Scholar]
- 51.Wendland, B., K. E. Steece, and S. D. Emr. 1999. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 18**:**4383-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]