Multiple links between transcription and splicing (original) (raw)
Abstract
Transcription and pre-mRNA splicing are extremely complex multimolecular processes that involve protein–DNA, protein–RNA, and protein–protein interactions. Splicing occurs in the close vicinity of genes and is frequently cotranscriptional. This is consistent with evidence that both processes are coordinated and, in some cases, functionally coupled. This review focuses on the roles of _cis_- and _trans_-acting factors that regulate transcription, on constitutive and alternative splicing. We also discuss possible functions in splicing of the C-terminal domain (CTD) of the RNA polymerase II (pol II) largest subunit, whose participation in other key pre-mRNA processing reactions (capping and cleavage/polyadenylation) is well documented. Recent evidence indicates that transcriptional elongation and splicing can be influenced reciprocally: Elongation rates control alternative splicing and splicing factors can, in turn, modulate pol II elongation. The presence of transcription factors in the spliceosome and the existence of proteins, such as the coactivator PGC-1, with dual activities in splicing and transcription can explain the links between both processes and add a new level of complexity to the regulation of gene expression in eukaryotes.
Keywords: splicing, alternative splicing, RNA polymerase II, transcription, coupling
INTRODUCTION
The largest human gene (2400 kb) encodes dystrophin. This gene would require ~16 h to be transcribed, and it was demonstrated that its pre-mRNA is spliced cotranscriptionally (Tennyson et al. 1995). Cotranscriptional splicing appears here as a very intuitive concept. In fact, it would be very difficult to conceive that the splicing of the dozens of dystrophin introns would “wait” until the synthesis of a huge 2400-kb pre-mRNA substrate molecule were completed. In agreement with this observation, direct visualization of nascent transcripts of early Drosophila embryo genes by electron microscopy clearly demonstrated that splicing occurs cotranscriptionally with a reasonable frequency and that splice site selection precedes polyadenylation (Beyer and Osheim 1988). Nevertheless most biology (and even molecular biology) textbooks keep showing drawings in which a fully transcribed primary transcript, with all its introns, appears as the substrate for splicing (Fig. 1 ▶). Indeed, for years gene transcription and pre-mRNA processing were thought to be independent events until a series of biochemical, cytological, and functional experiments demonstrated that all three processing reactions (capping, splicing, and cleavage/polyadenylation) can be tightly coupled to RNA polymerase II (pol II) transcription (excellent reviews have been recently published by Bentley 2002; Howe 2002; Maniatis and Reed 2002; Neugebauer 2002; Proudfoot et al. 2002; Proudfoot 2003).
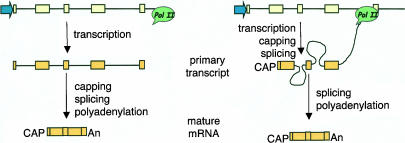
FIGURE 1.
(Left) Classical textbook picture in which all pre-mRNA processing reactions are depicted as posttranscriptional (cf. Alberts et al. 2002, Figs. 6–21). (Right) pre-mRNA processing is cotranscriptional. In the depicted pre-mRNA molecule, splicing of intron 1 has already occurred, introns 2 and 3 are being processed, and exon 4 has not been transcribed yet.
In the case of splicing, cotranscriptionality seems to be a reasonable prerequisite for coupling, but the existence of cotranscriptionality per se does not necessarily imply that transcription and pre-mRNA splicing are coupled. It is worth noting that cotranscriptional splicing is not obligatory. As Karla Neugebauer pointed out clearly in her recent review, “…the time that it takes for pol II to synthesize each intron defines a minimal time and distance along the gene in which splicing factors can be recruited and spliceosomes formed. The time that it takes for pol II to reach the end of the transcription unit defines the maximal time in which splicing could occur cotranscriptionally…” (Neugebauer 2002). In a long gene, for example, some introns could be spliced out cotranscriptionally, whereas others could be processed well after transcription has been completed. In most cases, we do not know which introns follow each pattern. We do not even know if a particular intron always follows the same pattern of processing. In certain cases, the position of the intron along the gene seems to be relevant to the excision pattern. For example, in the Balbiani ring 1 (BR1) gene, intron 3, located 3 kb from the 5′-end of the 40-kb pre-mRNA, is excised cotranscriptionally. However, intron 4, located 0.6 kb from the poly(A) site, is excised cotranscriptionally in ~10% of the molecules, but posttranscriptionally in the remaining molecules (Bauren and Wieslander 1994). Further studies on another Balbiani ring gene, BR3, allowed the investigators to propose that spliceosomes assemble rapidly as introns appear in the pre-mRNA, but intron-specific constraints result in cotranscriptional excision of some introns, preferentially those located in the 5′-part of the primary transcript, and posttranscriptional excision of other introns, preferentially those located in the 3′-part. (Wetterberg et al. 1996). It is worth noting that if splicing were strictly cotranscriptional, that is, if the elimination of one intron would necessarily be completed before the transcription of the following intron has begun, widely distributed mechanisms such as exon definition (Robberson et al. 1990) or alternative splicing by exon skipping would simply not exist.
This review focuses on discussing the evidence supporting the existence of functional links between transcription and splicing. Both processes are extremely complex, involving thousands of protein factors, RNA molecules, and DNA sequences. The added complexity of both processes probably hinders any attempt at generalization and simplification. The reader should bear in mind that certain molecular interactions or kinetic constraints might be relevant for a particular gene or set of genes but not for others. Part of the evidence discussed, although robust, is rather indirect. Nevertheless, because the regulation of transcription and splice site selection are paramount events in eukaryotic cell regulation and differentiation, such indirect evidence sets the necessary framework for more direct investigation in a dynamic emerging field.
POL II CTD AND COUPLING
Coupling of transcription and pre-mRNA processing may be in part due to the ability of pol II to bind and “piggyback” some of the processing factors in a complex known as the “mRNA factory” (Bentley 2002). Because this review is focused on splicing, we do not discuss in detail the roles of RNA polymerase II in the coupling of the other pre-mRNA processing reactions and transcription. The C-terminal domain (CTD) of pol II plays a central role in the coupling process: truncation of the CTD causes defects in capping, cleavage/polyadenylation, and splicing (McCracken et al. 1997). The human CTD comprises 52 heptad repeats with the consensus sequence YSPTSPS. Fong and Bentley (2001) found that the CTD C terminus including heptads 27–52 and a unique 10-amino-acid sequence (ISPDDSDEEN) located at the C terminus of heptad 52 supported all three processing reactions but the N terminus supported only capping, concluding that different CTD regions can display different functions in pre-mRNA processing. The heptad repeats alone are not sufficient to support pre-mRNA processing, but the 10-amino-acid C-terminal motif was reported to confer efficient processing capability to pol II with only heptads 1–25 or with 27 consensus heptads (Fong et al. 2003). It was recently reported that different pre-mRNAs might have different dependencies on the number of the CTD repeats as well as the need for the nonrepeated C-terminal sequence for efficient processing (Rosonina and Blencowe 2004). In any case, the C terminus or the repeats might act by binding factors that participate in transcriptions and/or processing directly, in the control of pol II elongation or affecting pol II subnuclear localization. Assignment of a specific role in pre-mRNA processing to the C-terminal sequence seems to be in contradiction with recent findings that this segment might simply act by conferring stability to the CTD. Chapman et al. (2004) found that variants lacking the C-terminal motif suffer proteolytic degradation of the whole CTD in vivo, giving rise to the previously known IIb isoform of the RNA polymerase II large subunit. The abundance and ability to transcribe of this CTD-less isoform vary among the different cell types, which indicates that further investigation is needed to elucidate the apparent contradiction.
Purified phosphorylated RNA pol II is able to activate splicing in vitro (Hirose et al. 1999). Isolated CTD fragments cannot duplicate this effect unless the precursor RNA is recognized via exon definition, that is, it offers to the splicing machinery at least one complete internal exon with its 3′- and 5′-splice sites. CTD does not activate splicing of precursors in which pairs of splice sites are in intronic polarity (Zeng and Berget 2000). These findings support a direct role for the CTD in exon recognition and led to the speculation that the CTD would not only be a landing path for splicing factors but also for bringing closer consecutive exons, which would facilitate spliceosomal assembly (Fig. 2 ▶).
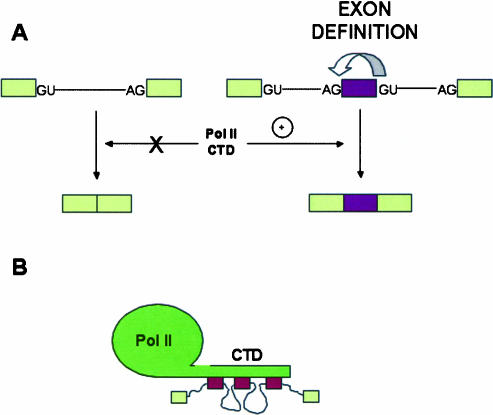
FIGURE 2.
(A) The C-terminal domain (CTD) of the RNA polymerase II largest subunit stimulates splicing of pre-mRNAs with exons governed by an exon definition mechanism (right), but has no effect on the splicing of precursors with an intronic configuration of splice sites (left). (B) Putative model according to Zeng and Berget (2000), in which the CTD brings together distant exons governed by exon definition and helps splicing.
Dynamic changes in CTD phosphorylation seem to play significant roles in RNA processing. Consistently, the peptidyl-prolyl isomerase Pin 1, which stimulates CTD phosphorylation by cdc2/cyclin B by affecting CTD structure, was shown to inhibit Pol II-dependent splicing in vitro (Xu et al. 2003).
In addition, the CTD seems to play a role in the nuclear distribution of components of the transcription and splicing machineries. In fact, transcriptional activation of pol II genes increases association of splicing factors to sites of transcription, but this relocalization does not occur if pol II has a truncated C-terminal domain (Misteli and Spector 1999). This is consistent with previous findings that over-expression of CTD-containing large subunits of pol II in mammalian cells induces selective nuclear reorganization of splicing factors (Du and Warren 1997).
CROSS-TALK PROTEINS
A comprehensive proteomic analysis of the human spliceosome (Zhou et al. 2002; for review, see Jurica and Moore 2003) reveals that at least 30 out of the 145 spliceosomal proteins are either known or candidate participants in the coupling between splicing and other gene expression steps. For instance, the transcription cofactor TAT-SF1 (see below) and the transcription factors CA150, XAB2, and SKIP are present in the spliceosome. On the other hand, the promoter itself could be responsible for recruiting splicing factors, such as SR proteins, to the site of transcription, possibly through transcription factors that bind the promoter or the transcriptional enhancers. Some proteins display a dual function; acting in both processes as is the case of a transcriptional activator of the human papilloma virus (Lai et al. 1999), or the thermogenic coactivator PGC-1. Interestingly, PGC-1 can affect alternative splicing, but only when it is recruited to complexes that interact with gene promoters (Monsalve et al. 2000). The product of the WT-1 gene, which is essential for normal kidney development, could also be involved in both transcription and splicing. Although generally considered a transcription factor, WT1 isoforms that include three amino acids, KTS, interact with the essential splicing factor U2AF65 in vitro (Davies et al. 1998). Another example of dual function is the transcription/splicing factor p54nrb, which associates with the 5′-splice site within large complexes in HeLa cells together with the hyperphosphorylated form of RNA pol II and U1 or U1 and U2 snRNPs. These macromolecular complexes also contain other transcription/splicing factors, such as PSF and TLS, as well as factors known to control elongation, such as P-TEFb, TAT-SF1, and TFIIF (Kameoka et al. 2004). Other proteins, such as SAF-B, which mediate chromatin attachment to the nuclear matrix, have been implicated in the coupling of transcription and pre-mRNA splicing (Nayler et al. 1998). The RNA polymerase itself could be responsible for recruiting these proteins, perhaps through its CTD. Three proteins carrying WW and/or FF domains, and whose activities might be related to the coupling between transcription and splicing, were found to bind specifically to phosphorylated CTD: (1) the yeast splicing factor Prp40 (Morris and Greenleaf 2000); (2) Ess1, a yeast peptidyl prolyl isomerase, proposed to act in cis/trans protein isomerizations that could play a crucial role in the recognition of CTD by other proteins (Myers et al. 2001); and (3) CA150, a human nuclear factor implicated in transcriptional elongation (Carty et al. 2000). Other candidates to function in the coupling of splicing and transcription are a group of proteins known as SCAFs (SR-like CTD associated factors). These are CTD-interacting proteins that, similarly to SR proteins, contain an RS domain and an RNA-binding domain (Yuryev et al. 1996). The fact that SR-like proteins interact with the CTD might not be related to splicing. Indeed, SR and SR-like proteins have been implicated in other coupling events. For instance, the yeast poly(A)+ RNA-binding proteins Gbp2 and Hrb1, which resemble members of the SR family, specifically bind to the TREX (transcription/export) complex, which couples transcription elongation to the nuclear export of mRNAs. TREX-bound Gbp2 and Hrb1 could be then transferred from the TREX complex to the nascent pre-mRNA during transcription (Hurt et al. 2004). It is claimed that cotranscriptional recruitment of these mRNA-binding proteins might increase export efficiency to ensure their later function as part of the mRNP in the cytoplasm.
A summary of protein factors that are candidates for linking transcription and splicing is presented in Table 1 ▶. As a general remark, although evidence of CTD recruitment of processing factors explains satisfactorily the coupling of transcription with capping and cleavage/polyadenylation, evidence for a link between recruitment and splicing is still circumstantial and needs further investigation.
TABLE 1.
Candidate proteins linking transcription and pre-mRNA splicing
| Protein | Link to transcription | Link to splicing | Reference |
|---|---|---|---|
| Mammalian cells | |||
| SCAFs | Bind CTD | SR-like proteins | Yuryev et al. 1996 |
| WT-1 | Transcription factor | Binds U2AF65 | Davies et al. 1998 |
| PGC-1 | Coactivator | Regulates alternative splicing | Monsalve et al. 2000 |
| CA150 | Elongation factor | Binds SF1 | Goldstrohm et al. 2001 |
| Present in spliceosome | Zhou et al. 2002 | ||
| XAB2 | Transcription factor | Present in spliceosome | Zhou et al. 2002 |
| SKIP | Transcription factor | Present in spliceosome | Zhou et al. 2002 |
| TAT-SF1 | Elongation factor | Binds UsnRNPs | Fong and Zhou 2002 |
| p54nrb | Transcription factor | In complexes with UsnRNPs | Kameoka et al. 2004 |
| PSF | Transcription factor | In complexes with UsnRNPs | Kameoka et al. 2004 |
| TLS | Transcription factor | In complexes with UsnRNPs | Kameoka et al. 2004 |
| CoAA | Coactivator | hnRNP-like protein | Auboeuf et al. 2004b |
| Regulates alternative splicing | |||
| Yeast | |||
| PrP40 | Binds phosphoCTD | Splicing factor | Morris and Greenleaf 2000 |
| Spt5 | Elongation factor | Activates splicing | Lindstrom et al. 2003 |
ALTERNATIVE SPLICING REVEALS STRONG LINKS BETWEEN TRANSCRIPTION AND SPLICING
Alternative splicing appears as a widespread means for producing polypeptide diversity from a single gene (Black 2003). In human fibronectin (FN), for example, up to 20 different polypeptide variants arise from alternative splicing in three regions of a single gene (Gutman and Kornblihtt 1987). However, this figure remains modest when compared with that of the Drosophila dscam gene, where an extremely complex array of alternative exons could potentially give rise to 38,016 DSCAM proteins (Schmucker et al. 2000). In spite of the estimation that 60% of human genes are expressed through alternative splicing and the sophisticated functional, cell-type, and developmental specificities documented in many cases, the mechanisms of alternative splicing regulation are poorly understood. A key role in splice site choice regulation is played by members of the SR (Ser/Argrich) family of proteins. These proteins participate both in constitutive and alternative splicing. By binding to splicing enhancers they can stimulate or repress spliceosome assembly at adjacent splice sites. It is conceivable that alternative splicing in different cell types or different points in time is regulated by variation in the relative abundance of SR proteins. However, although relative proportions of SR proteins and their antagonistic splicing factors (namely, heterogeneous nuclear ribonucleoprotein) vary naturally in several rat tissues and cell lines in culture (Hanamura et al. 1998), SR proteins do not seem to have a highly specific tissue distribution, which suggests the existence of more complex regulatory mechanisms.
The demonstration that differences in promoter structure lead to differences in alternative splicing of the transcript (Cramer et al. 1997) supports the concept that splicing and transcription are coupled and that this coupling may offer an additional level of regulation of alternative splicing. The system analyzed in our laboratory involved transient transfection of mammalian cells with minigenes carrying the EDI exon, which encodes a facultative repeat of FN. EDI contains an exonic splicing enhancer (ESE), which is targeted by the SR proteins SF2/ASF and 9G8. Overexpression of SF2/ASF and 9G8 markedly stimulates EDI inclusion, but the effect of these proteins is modulated by the promoter (Cramer et al. 1999). These effects are not the trivial consequence of different mRNA levels produced by each promoter (promoter strength) but depend on some qualitative properties conferred by promoters to the transcription/RNA processing machinery. The promoter effect is also observed in cell lines stably transfected with the same minigenes used as episomal templates, indicating that a physiological chromatin assembly of the integrated minigenes is compatible with the promoter mechanism (Kadener et al. 2001).
The promoter effect is not restricted to the FN EDI exon. Similar effects have been found independently in other genes. Reporter minigenes whose products are subject to alternative splicing decisions in the CD44 and the calcitonin gene related product (CGRP) genes were put under the control of steroid-sensitive promoters (mouse mammary tumor virus and synthetic promoters containing either the progesterone or the estrogen response elements) or promoters that do not respond to steroid hormones (CMV and thymidine kinase). Steroid hormones affected splice site selection only of pre-mRNAs produced by the first type of promoters. As in the case of FN EDI, promoter-dependent hormonal effects on splicing were not a consequence of an increase in transcription rate or of a saturation of the splicing machinery (Auboeuf et al. 2002). Promoter-dependent alternative splicing patterns have been also found when reporter minigenes for the cystic fibrosis transmembrane regulator (CFTR) exon 9 (Pagani et al. 2003) or for the fibroblast growth factor receptor 2 (Robson-Dixon and García-Blanco 2004) were expressed in mammalian cells.
THE ROLES OF TRANSCRIPTIONAL ACTIVATORS IN ALTERNATIVE SPLICING
The finding that promoter structure is important for alternative splicing predicts that factors that regulate alternative splicing could be acting through promoters and that cell-specific alternative splicing may not simply result from the differential abundance of ubiquitous SR proteins, but from a more complex process involving cell-specific promoter occupation. However, promoters are not swapped in nature, and because most genes have a single promoter, the only conceivable way by which promoter architecture could control alternative splicing in vivo should be the differential occupation of promoters by transcription factors of different natures and/or mechanistic properties. Accordingly, it has been found that transcriptional activators affect alternative splicing. Class I activators (Blau et al. 1996), such as SW6, Sp1 and CTF/NF1, which only stimulate transcriptional initiation, have little effect on EDI splicing. On the contrary, class IIB activators such as VP16, which stimulate both initiation and elongation, provoke EDI exon skipping. HIV-1 Tat, a member of class IIA activators, which has little effect on transcription in the absence of other activators, has no effect on EDI splicing. However, Tat synergizes with SW6, Sp1, and CTF, but not with VP16, in promoting transcriptional elongation and therefore in provoking EDI exclusion (Nogués et al. 2002).
Promoters and enhancers are _cis_-acting elements that control gene transcription via complex networks of protein–DNA and protein–protein interactions. Whereas promoters deal with putting in place the RNA polymerase, both enhancers and promoters can control transcriptional initiation and elongation. The presence of the SV40 transcriptional enhancer near a promoter stimulates pol II elongation (Yankulov et al. 1994). Consistently, deletion of the SV40 enhancer provokes a 3–10-fold reduction in exon skipping, independently of the promoter used (Kadener et al. 2002).
Transcriptional coregulators have also been implicated in the control of alternative splicing. Steroid hormones affect the processing of pre-mRNA synthesized from steroid-sensitive promoters, but not from steroid-unresponsive promoters, in a steroid-receptor-dependent and receptor-selective manner. Several coregulators of these nuclear receptors showed differential effects on alternative splicing (Auboeuf et al. 2002, 2004a). Some coregulators act by recruiting coactivators. It was recently shown that the coactivator CoAA (coactivator activator), an hnRNP-like protein that interacts with the transcriptional coregulator TRBP, which is in turn recruited to promoters through interactions with activated nuclear receptors, regulates alternative splicing in a promoter-dependent manner. CoAA similarly enhanced transcriptional activities fired by the MMTV or CMV promoters, but only affected alternative splicing of transcripts synthesized from the progesterone-activated MMTV promoter (Auboeuf et al. 2004b). It was recently shown that transcriptional activators not only modulate alternative but also constitutive splicing in a pol II CTD-dependent manner (Rosonina et al. 2003).
POL II ELONGATION
An alternative, but not exclusive, model suggests that transcription might control splicing via the regulation of pol II elongation rate or processivity. Low pol II elongation rate or internal pauses for elongation would favor the inclusion of alternative exons governed by an exon skipping mechanism, whereas a highly elongating pol II, or the absence of internal pauses, would favor exclusion of these kinds of exons. The mechanism by which the elongation rate would affect EDI splicing is a consequence of EDI pre-mRNA sequence. EDI exon skipping occurs because the 3′-splice site of the upstream intron is suboptimal compared with the 3′-splice site of the downstream intron. If the polymerase pauses anywhere between these two sites, only elimination of the upstream intron can take place. Once the pause is passed or the polymerase proceeds, there is no option for the splicing machinery but to eliminate the downstream intron, which leads to exon inclusion. A highly processive elongating pol II, or the absence of internal pauses, would favor the simultaneous presentation of both introns to the splicing machinery, a situation in which the stronger 3′-splice site of the downstream intron outcompetes the weaker 3′-splice site of the upstream intron, resulting in exon skipping. Figure 3 ▶ shows how when a weak 3′-splice site is followed by a strong one, as in many alternative splicing examples, pol II elongation rates affect the relative amounts of splicing isoforms. On the contrary, when two consecutive strong 3′-splice sites occur, as in constitutive splicing, pol II elongation rates are irrelevant.
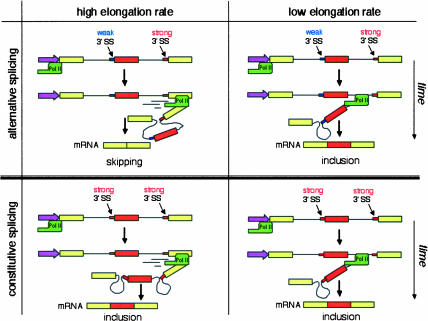
FIGURE 3.
Influence of RNA polymerase II elongation rate on alternative splicing by “exon skipping.” Alternative splicing (top): when the 3′-splice site (SS) by the alternative exon is weaker than the 3′-SS of the downstream intron, low transcriptional elongation rates (right) favor exon inclusion, whereas high elongation rates (left) favor skipping. Constitutive splicing (bottom): when both 3′-SSs are strong, the exon is included constitutively independently of the elongation rate.
A kinetic role for transcription on splicing was originally suggested by Eperon et al. (1988), who found that the rate of RNA synthesis may affect its secondary structure, which may, in turn, affect splicing. A similar mechanism involving a kinetic link between transcription and splicing was suggested from experiments in which RNA pol II pause sites affect alternative splicing by delaying the transcription of an essential splicing inhibitory element (DRE) required for regulation of tropomyosin exon 3 (Roberts et al. 1998).
The elongation factor P-TEFb converts the polymerase from a nonprocessive to a processive form, which is consistent with the fact that inhibitors of this kinase such as DRB (dichlororibofuranosylbenzimidazole) or flavopiridol inhibit pol II elongation (Price 2000; Ni et al. 2004). Cells transfected with EDI splicing reporters and treated with DRB displayed a threefold increase in EDI inclusion into mature mRNA compared with untreated cells (Nogués et al. 2002).
Changes in chromatin structure provoked by histone acetylation also affect splicing. In fact, trichostatin A, a potent inhibitor of histone deacetylation, inhibits EDI inclusion (Nogués et al. 2002). This supports the hypothesis that acetylation of the core histones would facilitate the passage of the transcribing polymerase, which is, in turn, consistent with the proposal of chromatin opening mediated by DNA tracking by a transcribing pol II complex piggybacking a histone acetyltransferase activity (Travers 1999).
The above mentioned weakness of the 3′-splice of the upstream intron of EDI is caused by its suboptimal polypyrimidine tract. By mutating EDI’s polypyrimidine tract and therefore generating pre-mRNAs with increasing degrees of exon recognition, it was shown that responsiveness of exon skipping to elongation is inversely proportional to the 3′-splice site strength, which means that the better the alternative exon is recognized by the splicing machinery, the less its degree of inclusion is affected by transcriptional elongation (Nogués et al. 2003).
SLOW POLYMERASES AND ALTERNATIVE SPLICING
A more direct proof for the elongation mechanism in the transcriptional control of alternative splicing in human cells was provided by the use of a mutant form of pol II (called C4) with a lower elongation rate (Chen et al. 1996). Human cells were transfected with reporter minigenes for alternative splicing together with vectors expressing an α-amanitin-resistant human pol II large subunit carrying the C4 mutation. After treatment with α-amanitin, the endogenous pol II becomes inhibited and the reporter minigenes are transcribed by the recombinant, elongation-defective mutant. The slow polymerase was shown to stimulate the inclusion of fibronectin EDI exon by threefold, confirming the hypothesis of inverse correlation between elongation rate and inclusion of this alternative exon. The C4 mutation also affected the splicing of Adenovirus E1a, by favoring the use of the most upstream of the three alternative 5′-splice sites that compete for a common 3′-splice site. The explanation for this effect is that a reduction in elongation rate would allow more time to assemble splicing complexes at the upstream 5′-splice site. However, inclusions of other alternative exons such as exon 7B of the heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1-E7B) gene were not affected by the slow polymerase (de la Mata et al. 2003).
Similar effects of pol II elongation rates on splicing were found in yeast. Alternative splicing is a very rare event in yeast. By mutating the branchpoint upstream of the con stitutive internal exon of the DYN2 gene, Howe et al. (2003) created an artificial cassette exon that becomes alternatively spliced. Skipping of this exon can be partially prevented when expressed in a yeast mutant carrying a slow pol II or in the presence of elongation inhibitors. This supports the hypothesis that what is important to the balance between exon skipping and exon inclusion is the relative rates of spliceosome formation and pol II processivity (Fig. 4 ▶).
FIGURE 4.
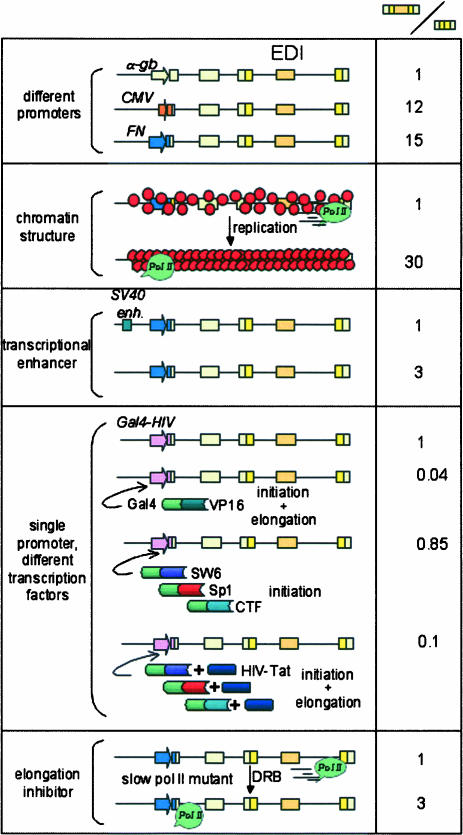
Effects of several _cis_- and _trans_-acting factors that affect pol II elongation on the alternative splicing of the fibronectin EDI (extra domain I) exon. Promoters, enhancers, and chromatin structure changes caused by template replication act in cis. Transcription factors and drugs such as DRB (dichlororibofuranosylbenzimidazole) act in trans. The right column displays the ratios of the amounts of mRNA isoforms containing versus lacking the EDI exon. Ratio standardizations are valid only within each condition analyzed.
PAUSES TO POL II ELONGATION AFFECT SPLICING
Even if the elongation rates of pol II were not regulatable, experiments with slow polymerases in mammalian cells and yeast put into the scene the importance of the presence of pauses for pol II transcription on splicing. Pauses for pol II have been found in the 3′-flanking regions of genes regulating termination and polyadenylation (Enriquez-Harris et al. 1991), between closely spaced genes (Ashfield et al. 1991), and at several internal sites in the c and N-myc genes (Keene et al. 1999). Artificial arrests (ARTAR) to pol II elongation have been created and shown to be effective in pausing pol II at positions far downstream from the promoter (Kulish and Struhl 2001). The effect of a MAZ-type pol II pause on the alternative splicing of the tropomyosin gene has been commented on above (Roberts et al. 1998). Insertions of MAZ pauses at certain positions of the fibro-blast growth factor receptor 2 gene deeply affect alternative splicing of its mutually exclusive exons IIIb and IIIc when minigenes are transfected into cells in culture. However, the effects on splicing are not observed when in vitro transcribed pre-RNAs, containing similar pauses, are directly transfected into the cells, strongly demonstrating the co-transcriptional nature of the MAZ effects (Robson-Dixon and García-Blanco 2004). Most importantly, a natural pol II pause was shown to contribute to the regulation of the alternative processing of immunoglobulin μ RNA, where a combination of alternative polyadenylation and splicing determine the switch from membrane to soluble immunoglobulin synthesis. The pause site, located between a poly(A) and a splicing site enhances the use of the upstream poly(A) site, which leads to the soluble form of IgM. Interestingly, nuclear run-ons demonstrated a stalling of RNA polymerase just downstream from the soluble μ poly(A) site (Peterson et al. 2002).
FROM TRANSFECTED MINIGENES TO ENDOGENOUS GENES
Several laboratories, including ours, use minigenes transfected into mammalian cells to study splicing regulation, which proved to be extremely useful to look at the mechanisms by which transcription controls alternative splicing. However, transfection experiments may be of poor physiological relevance because minigenes are chimeric constructs in which alternatively spliced regions are positioned at incorrect distances with respect to promoters. Furthermore, transfected minigenes act in a different gene environment and at high copy numbers. A more physiological approach to the coupling between transcription and splicing should necessarily study an endogenous gene in its natural environment. A first step in this direction is an observation obtained in Drosophila: flies carrying the C4 mutation show changes in the alternative splicing profile of the large ultrabithorax (Ubx) gene (de la Mata et al. 2003). The observed changes are consistent with a kinetic mechanism that allows more time for early splicing events. Most interestingly, Drosophila with the C4 allele in heterozygosis but wild type for both Ubx alleles show a mutant phenotype called “Ubx effect” that resembles the one seen in flies haploinsufficient for the Ubx protein (Greenleaf et al. 1980). It will be important to investigate to what extent this alteration in splicing isoform proportions is causative for the display of the _Ubx_-like phenotype.
RECIPROCAL COUPLING: HOW SPLICING AFFECTS TRANSCRIPTION
Fong and Zhou (2002) have found that spliceosomal U small nuclear ribonucleoproteins (UsnRNPs) interact with the human transcription elongation factor TAT-SF1 and strongly stimulate pol II elongation, probably via the binding of TAT-SF1 to the elongation factor P-TEFb. Because the TAT-SF1–UsnRNP complex also stimulates splicing in vitro, these results not only reveal that splicing factors function directly to promote transcriptional elongation but that reciprocal interactions exist in the coupling process. Consistently, the presence of an intron or simply a 5′-splice site immediately downstream from a promoter greatly enhances transcription, both in mammalian and yeast genes (Furger et al. 2002), which indicates that factors controlling intron removal are important for normal levels of transcription. This confirms old observations of inefficient expression of recombinant cDNAs transfected into mammalian cells, compared with the corresponding intron-containing constructs. Although the underlying mechanism for the positive influence of splicing on transcription is still unknown, these results provide further support that both processes are tightly coupled. The authors favor an RNA-mediated process in which the nascent transcript, associated with splicing factors that, in turn, associate with components of the transcription machinery, promotes transcriptional elongation and perhaps regulates initiation. Such a mechanism is supported by the unexpected finding that U1 snRNA associates with the general transcription factor TFIIH, functioning in regulating transcription by pol II in addition to its role in splicing (Kwek et al. 2002). In addition, U1 snRNP is recruited cotranscriptionally in vivo to intron-containing genes in yeast (Kotovic et al. 2003). High levels of U1 snRNP were detected in intronic regions of actively transcribing genes, but not in promoter regions or along the length of intron-less genes. This kind of cotranscriptional recruitment is clearly different from the one demonstrated for capping enzymes, which bind directly to the CTD of pol II. This opens the question whether the coupling between transcription and splicing is based on the recruitment of splicing factors to the CTD or to intron-containing nascent transcripts, as shown for U1 snRNP. The latter seems to be the case at least in yeast, where it was demonstrated that the CTD is dispensable for efficient splicing.
CONCLUDING REMARKS
Findings that splicing factors increase transcriptional elongation (Fong and Zhou 2002) and that introns are necessary for efficient pol II transcription (Furger et al. 2002) suggest that the strong connection between transcription and splicing might be the consequence of a combination of both recruitment of factors to the sites of transcription and elongation control by RNA pol II. Although evidence for the elongation mechanism is stronger in terms of the variety of molecular approaches that support it, certain data allow us to speculate that recruitment and elongation might be interconnected. For instance, the CTD is preferentially phosphorylated at Ser 5 when pol II is recruited at promoter sites but becomes phosphorylated at Ser 2 when located at the coding region (Cho et al. 2001). This change in phosphorylation quality might be relevant for the recruitment of splicing factors. Simultaneously, it would be important to determine whether a pausing pol II has the same phosphorylation status and recruitment properties of a fast-elongating pol II. Chromatin immunoprecipitation experiments with antibodies to different kinds of phosphopol II and characterization of protein complexes throughout different segments of transcribed regions should help to test the combined hypothesis.
Studies on the yeast Spt5 factor also suggest a combined mechanism. This factor has been proposed to regulate pol II elongation through nucleosomes. General elongation factors such as TFIIF and TFIIS coimmunopurify with Spt5, which, in turn, is able to interact with capping enzymes. Lindstrom et al. (2003) found that spt5 mutations lead to accumulation of unspliced pre-mRNAs. Such an inhibition of splicing may occur because splicing factors fail to interact with the transcription machinery in the absence of Spt5.
A complex panorama emerges when trying to summarize the factors involved in the regulation of splice site selection. On the “_cis_” side, we should take into account not only the specific sequences acting at the RNA level (splice sites, splicing enhancers and silencers, and determinants of pre-mRNA secondary structures) but also those acting at the DNA level such as promoters, transcriptional enhancers, and the pol II pausing architecture of a gene. On the “_trans_” side, the abundance, cell localization, and phosphorylation state of SR and hnRNP proteins should be complemented with those of transcription factors, coactivators, chromatin factors, CTD kinases, transcriptional elongation factors, and factors with dual activities in both transcription and splicing.
Alternative splicing has been associated with increased evolutionary change in vertebrates. Comparative genomic analysis has shown that whereas constitutive exons are strongly conserved in the mouse and human genomes, alternative exons are mostly not conserved and are the product of recent exon creation or loss events (Modrek and Lee 2003). Selection of the _cis_- and _trans_-acting regulatory factors of splicing, involving multiple links with the transcription machinery, must have occurred concomitantly in a short evolutionary time, contributing to the high adaptive benefit of alternative splicing.
Acknowledgments
This work was supported by grants from the Howard Hughes Medical Institute, the Fundación Antorchas, the Agencia Nacional de Promoción de Ciencia y Tecnología of Argentina, and the Consejo Nacional de Investigaciones Científicas y Técnicas of Argentina (CONICET). M.d.l.M., J.P.F., and M.J.M. are recipients of fellowships from the CONICET. A.R.K is a career investigator of the CONICET and a Howard Hughes Medical Institute international research scholar.
REFERENCES
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. 2002. Molecular biology of the cell, 4th ed. Garland Science, New York.
- Ashfield, R., Enriquez-Harris, P., and Proudfoot, N.J. 1991. Transcriptional termination between the closely linked human complement genes C2 and factor B: Common termination factor for C2 and c-myc? EMBO J. 10**:** 4197–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf, D., Hönig, A., Berget, S.M., and O’Malley, B.W. 2002. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298**:** 416–419. [DOI] [PubMed] [Google Scholar]
- Auboeuf, D., Dowhan, D.H., Kang, Y.K., Larkin, K., Lee, J.W., Berget, S.M., and O’Malley, B.W. 2004a. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc. Natl. Acad. Sci. 101**:** 2270–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf, D., Dowhan, D.H., Li, X., Larkin, K., Ko, L., Berget, S.M., and O’Malley, B.W. 2004b. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 24**:** 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren, G. and Wieslander, L. 1994. Splicing of Balbiani ring 1 pre-mRNA occurs simultaneously with transcription. Cell 76**:** 183–192. [DOI] [PubMed] [Google Scholar]
- Bentley, D. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14**:** 336–342. [DOI] [PubMed] [Google Scholar]
- Beyer, A.L. and Osheim, Y.N. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 2**:** 754–765. [DOI] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72**:** 291–336. [DOI] [PubMed] [Google Scholar]
- Blau, J., Xiao, H., McCracken, S., O’Hare, P., Greenblatt, J., and Bentley, D. 1996. Three functional classes of transcriptional activation domains. Mol. Cell. Biol. 16**:** 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty, S.M., Goldstrohm, A.C., Sune, C., Garcia-Blanco, M.A., and Greenleaf, A.L. 2000. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc. Natl. Acad. Sci. 97**:** 9015–9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, R.D., Pancalade, B., Lang, A., Bensaude, O., and Eick, D. 2004. The last CTD repeat of the mammalian RNA polymerase II large subunit is important for its stability. Nucleic Acids Res. 32**:** 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Chafin, D., Price, D.H., and Greenleaf, A.L. 1996. Drosophila RNA polymerase II mutant that affects transcription elongation. J. Biol. Chem. 271**:** 5993–5999. [PubMed] [Google Scholar]
- Cho, E.J., Kobor, M.S., Kim, M., Greenblatt, J., and Buratowski, S. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 15**:** 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, P., Pesce, C.G., Baralle, F.E., and Kornblihtt, A.R. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. 94**:** 11456–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, P., Cáceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E., and Kornblihtt, A.R. 1999. Coupling of transcription with alternative splicing, RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4**:** 251–258. [DOI] [PubMed] [Google Scholar]
- Davies, R.C., Calvio, C., Bratt, E., Larsson, S.H., Lamond, A.I., and Hastie, N.D. 1998. WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into spliceosomes. Genes & Dev. 12**:** 3217–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata, M., Alonso, C.R., Kadener, S., Fededa, J.P., Blaustein, M., Pelisch, F., Cramer, P., Bentley, D., and Kornblihtt, A.R. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12**:** 525–532. [DOI] [PubMed] [Google Scholar]
- Du, L. and Warren, S.L. 1997. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J. Cell Biol. 136**:** 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris, P., Levitt, N., Briggs, D., and Proudfoot, N.J. 1991. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 10**:** 1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon, L.P., Graham, I.R., Griffiths, A.D., and Eperon, I.C. 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: Is folding limited to a region behind the transcribing RNA polymerase? Cell 54**:** 393–401. [DOI] [PubMed] [Google Scholar]
- Fong, N. and Bentley, D.L. 2001. Capping, splicing and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 15**:** 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, Y.W. and Zhou, Q. 2002. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414**:** 929–933. [DOI] [PubMed] [Google Scholar]
- Fong, N., Bird, G., Vigneron, M., and Bentley, D.L. 2003. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 22**:** 4274–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger, A., O’Sullivan, J.M., Binnie, A., Lee, B.A., and Proudfoot, N.J. 2002. Promoter proximal splice sites enhance transcription. Genes & Dev. 16**:** 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm, A.C., Albrecht, T.R., Suñé, C., Bedford, M., and García-Blanco, M.A. 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21**:** 7617–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf, A.L., Weeks, J.R., Voelker, R.A., Ohnishi, S., and Dickson, B. 1980. Genetic and biochemical characterization of mutants of an RNA polymerase II locus in D. melanogaster. Cell 21**:** 785–792. [DOI] [PubMed] [Google Scholar]
- Gutman, A. and Kornblihtt, A.R. 1987. Identification of a third region of cell-specific alternative splicing in human fibronectin mRNA. Proc. Natl. Acad. Sci. 84**:** 7179–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura, A., Cáceres, J.F., Mayeda, A., Franza Jr., B.R., and Krainer, A.R. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4**:** 430–444. [PMC free article] [PubMed] [Google Scholar]
- Hirose, Y., Tacke, R., and Manley, J.L. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes & Dev. 13**:** 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, K.J. 2002. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta 1577**:** 308–324. [DOI] [PubMed] [Google Scholar]
- Howe, K.J., Kane, C.M., and Ares Jr., M. 2003. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA 9**:** 993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt, E., Ming-juan, L., Reed, R., and Sträßer, K. 2004. Cotranscriptional recruitment of the serine–arginine-rich (SR)-like protein Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. 101**:** 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S. and Moore, M.J. 2003. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12**:** 5–14. [DOI] [PubMed] [Google Scholar]
- Kadener, S., Cramer, P., Nogués, G., Cazalla, D., de la Mata, M., Fededa, J.P., Werbajh, S., Srebrow, A., and Kornblihtt, A.R. 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 20**:** 5759–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener, S., Fededa, J.P., Rosbash, M., and Kornblihtt, A.R. 2002. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. 99**:** 8185–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka, S., Duque, P., and Konarska, M.M. 2004. p54nrb associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23**:** 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, R.G., Mueller, A., Landick, R., and London, L. 1999. Transcriptional pause, arrest and termination sites for RNA polymerase II in mammalian N- and c-myc genes. Nucleic Acids Res. 27**:** 3173–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic, K.M., Lockshon, D., Boric, L., and Neugebauer, K.M. 2003. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 23**:** 5768–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulish, D. and Struhl, K. 2001. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol. Cell. Biol. 21**:** 4162–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek, K.Y., Murphy, S., Furger, A., Thomas, B., O’Gorman, W., Kimura, H., Proudfoot, N.J., and Akoulitchev, A. 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 9**:** 800–805. [DOI] [PubMed] [Google Scholar]
- Lai, M.C., Teh, B.H., and Tarn, W.Y. 1999. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 274**:** 11832–11841. [DOI] [PubMed] [Google Scholar]
- Lindstrom, D.L., Squazzo, S.L., Muster, N., Burckin, T.A., Wachter, K.C., Emigh, C.A., McCleery, J.A., Yates III, J.R., and Hartzog, G.A. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23**:** 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416**:** 499–506. [DOI] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Green-blatt, J., Patterson, S.D., Wickens, M., and Bentley, D.L. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385**:** 357–361. [DOI] [PubMed] [Google Scholar]
- Misteli, T. and Spector, D.L. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3**:** 697–705. [DOI] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C.J. 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34**:** 177–180. [DOI] [PubMed] [Google Scholar]
- Monsalve, M., Wu, Z., Adelmant, G., Puigserver, P., Fan, M., and Spiegelman, B.M., 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6**:** 307–316. [DOI] [PubMed] [Google Scholar]
- Morris, D.P. and Greenleaf, A.L. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275**:** 39935–39943. [DOI] [PubMed] [Google Scholar]
- Myers, J.K., Morris, D.P., Greenleaf, A.L., and Oas, T.G. 2001. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry 40**:** 8479–8486. [DOI] [PubMed] [Google Scholar]
- Nayler, O., Stratling, W., Bourquin, J.P., Stagljar, I., Lindemann, L., Jasper, H., Hartmann, A.M., Fackelmayer, F.O., Ullrich, A., and Stamm, S. 1998. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 26**:** 3542–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer, K.M. 2002. On the importance of being cotranscriptional. J. Cell Sci. 115**:** 3865–3871. [DOI] [PubMed] [Google Scholar]
- Ni, Z., Schwartz, B.E., Werner, J., Suárez, J.R., and Lis, J.T. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13**:** 55–65. [DOI] [PubMed] [Google Scholar]
- Nogués, G., Kadener, S., Cramer, P., Bentley, D., and Kornblihtt, A.R. 2002. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 277**:** 43110–43114. [DOI] [PubMed] [Google Scholar]
- Nogués, G., Muñoz, M.J., and Kornblihtt, A.R. 2003. Influence of polymerase II processivity on alternative splicing depends on splice site strength. J. Biol. Chem. 278**:** 52166–52171. [DOI] [PubMed] [Google Scholar]
- Pagani, F., Stuani, C., Kornblihtt, A.R., and Baralle, F.E. 2003. Promoter architecture modulates CFTR exon 9 skipping. J. Biol. Chem. 278**:** 1511–1517. [DOI] [PubMed] [Google Scholar]
- Peterson, M.L., Bertolino, S., and Davis, F. 2002. An RNA polymerase pause site is associated with the immunoglobulin mus poly(A) site. Mol. Cell. Biol. 22**:** 5606–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, D.H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20**:** 2629–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J. 2003. Dawdling polymerases allow introns time to splice. Nat. Struct. Biol. 10**:** 876–878. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108**:** 501–512. [DOI] [PubMed] [Google Scholar]
- Robberson, B.L., Cote, G.J., and Berget, S.M. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10**:** 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, G.C., Gooding, C., Mak, H.Y., Proudfoot, N.J., and Smith, C.W.J. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26**:** 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson-Dixon, N.D. and García-Blanco, M.A. 2004. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 279**:** 29075–29084. [DOI] [PubMed] [Google Scholar]
- Rosonina, E. and Blencowe, B.J. 2004. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA 10**:** 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina, E., Bakowski, M.A., McCracken, S., and Blencowe, B.J. 2003. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 278**:** 43034–43040. [DOI] [PubMed] [Google Scholar]
- Schmucker, D., Clemens, J.C., Shu, H., Worby, C.A., Xiao, J., Muda, M., Dixon, J.E., and Zipursky, S.L. 2000. Drosophila DSCAM is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101**:** 671–684. [DOI] [PubMed] [Google Scholar]
- Tennyson, C.N., Klamut, H.J., and Worton, R.G. 1995. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 9**:** 184–190. [DOI] [PubMed] [Google Scholar]
- Travers, A. 1999. Chromatin modification by DNA tracking. Proc. Natl. Acad. Sci. 96**:** 13634–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterberg, I., Bauren, G., and Wieslander, L. 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2**:** 641–651. [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.X., Hirose, Y., Zhou, X.Z., Lu, K.P., and Manley, J.L. 2003. Pin 1 modulates the structure and function of human RNA polymerase II. Genes & Dev. 17**:** 2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov, K., Blau, J., Purton, T., Roberts, S., and Bentley, D.L. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77**:** 749–759. [DOI] [PubMed] [Google Scholar]
- Yuryev, A., Patturajan, M., Litingtung, Y., Joshi, R.V., Gentile, C., Gebara, M., and Corden, J.L. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. 93**:** 6975–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, C. and Berget, S. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20**:** 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419**:** 182–185. [DOI] [PubMed] [Google Scholar]