Characterization of a highly variable eutherian microRNA gene (original) (raw)
Abstract
Mouse microRNAs (miRNAs) miR-290–miR295 are encoded by a cluster of partially homologous pre-miRNA hairpins and are likely to be functionally important in embryonic stem (ES) cells and preimplantation embryos. We present evidence that a spliced, capped, and polyadenylated primary transcript spans this entire Early Embryonic microRNA Cluster (EEmiRC). Partial Drosha processing yields additional large nuclear RNA intermediates. A conserved promoter element containing a TATA-box directs EEmiRC transcription. Sequence analysis shows that the EEmiRC transcription unit is remarkably variable and can only be identified bioinformatically in placental (eutherian) mammals. Consistent with eutherian-specific function, EEmiRC is expressed in trophoblastic stem (TS) cells. When analyzing evolutionary and functional relationships, the organization of the entire miRNA loci should be considered in addition to the mature miRNA sequences. Application of this concept suggests that EEmiRC is a recently acquired rapidly evolving gene important for eutherian development.
Keywords: miRNA, ES cells, TS cells, EEmiRC, transcription unit, primary transcript
INTRODUCTION
microRNAs (miRNAs) are a family of single-stranded 21–25-nt-long RNA species that target cognate mRNAs for degradation or translational repression (For review, see Bartel 2004). The founding members, lin-4 and let-7, were identified genetically in Caenorhabditis elegans and are important developmental regulators (Lee et al. 1993; Reinhart et al. 2000). Numerous miRNAs were subsequently discovered in worms, flies, and human cells (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Presently the miRNA database contains several hundred miRNAs cloned from a variety of sources (Ambros et al. 2003; Griffiths-Jones 2004). Bioinformatic analyses suggest that the majority of animal miRNAs have been discovered (Lim et al. 2003a,b) and that miRNAs regulate a wide array of biological phenomena that are probably not restricted to developmental transitions (Lewis et al. 2003, 2005).
Mature miRNAs are embedded in characteristic hairpin folds within their primary transcript (pri-miRNA) (Lee et al. 1993, 2002 Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). This feature distinguishes miRNAs from the related short interfering RNAs (siRNAs), which are double-stranded 21–25-nt species with 2-nt 3′ overhangs produced from long double-stranded RNA precursors (Hamilton and Baulcombe 1999; Zamore et al. 2000; Elbashir et al. 2001).
Some miRNAs are present in the sense orientation within introns and may, therefore, be cotranscribed with protein coding mRNAs. However, since most miRNAs are located in intergenic regions or within introns but in the antisense orientation, dedicated miRNA transcription units must exist (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Aravin et al. 2003). Indeed, capped and polyadenylated pri-miRNAs were recently characterized and dedicated miRNA promoter regions were identified, suggesting that most if not all miRNAs are transcribed by RNA polymerase II (Bracht et al. 2004; Cai et al. 2004; Lee et al. 2004).
Drosha, a member of the RNAse III family of nucleases, excises the hairpin stem-loop from the nuclear pri-miRNA and generates one end of the mature miRNA (Lee et al. 2003). This event depends on additional protein cofactors that, together with Drosha, form the so-called Microprocessor complex (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). The ~65-nt pre-miRNA hairpin is subsequently exported to the cytoplasm by exportin-5 and Ran/GTP (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). A double-stranded break within the pre-miRNA catalyzed by Dicer, another RNAse III family member, results in the generation of a transient double-stranded siRNA-like intermediate (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001). While subsequent miRNA processing has not been studied directly, it is thought to be very similar to siRNA processing (Hutvagner and Zamore 2002). A multiprotein complex measures the thermodynamic stability of the siRNA and loads the strand whose 5′ end is less tightly paired to its complement into the effector RNA-induced silencing complex (RISC) (Caudy et al. 2002; Schwarz et al. 2003; Tomari et al. 2004). mRNA target recognition results in cleavage or translational repression depending on the degree of miRNA-mRNA complementarity and the type of Argonaute (PAZ/PIWI) family member present within RISC (Hutvagner and Zamore 2002; Doench et al. 2003; Liu et al. 2004; Song et al. 2004).
Previously, we identified a cluster of six pre-miRNA precursors that have similar sequences and encode a family of miRNAs that appear to be embryonic stem (ES) cell or early embryo specific by four criteria (Houbaviy et al. 2003): (1) their sequences are distinct from those of previously described miRNAs, including miRNAs cloned from adult mouse organs; (2) they cannot be detected in adult mouse organs by Northern analyses; (3) they are repressed during ES cell differentiation in vitro; and (4) all ESTs that map within the cluster are derived from ES cells or preimplantation embryos. This Early Embryonic microRNA Cluster (EEmiRC) may have a role in the maintenance of the pluripotent cell state and in the regulation of early mammalian development.
Here, we investigate the initial events that lead to EEmiRC expression. Bioinformatic analysis and ectopic overexpression identify a TATA box containing minimal promoter and a transcription unit that displays considerable variation between different mammalian orders. Northern analyses detect several large RNAs originating from EEmiRC. These RNAs are down-regulated upon ES cell differentiation, suggesting that EEmiRC expression is transcriptionally regulated. The EEmiRC pre-miRNA hairpins are transcribed within a common capped and polyadenylated primary transcript. The remaining large EEmiRC RNAs result from partial Drosha processing.
The RNA species from which the nuclease Drosha excises the pre-miRNA hairpin precursors has been designated pri-miRNA (from primary microRNA) because it was thought to be identical to the primary transcript in which the miRNAs are initially incorporated. In this study, we show that the EEmiRC primary transcript is spliced and, therefore, it need not be the direct Drosha substrate. Thus, we use the term pri-miRNA to designate a Drosha substrate regardless of its splicing status.
RESULTS
Comparative bioinformatics of EEmiRC
Our previous in silico analysis suggested the presence of an EEmiRC counterpart in the human genome (Houbaviy et al. 2003). Recent cloning of the corresponding microRNA homologs from human ES cells provided experimental support for this prediction (Suh et al. 2004).
While individual mouse and human EEmiRC miRNAs are sufficiently different from each other to warrant different numerical designations (Ambros et al. 2003), multiple sequence alignment reveals that they have related sequences both within each individual cluster and across species. Furthermore, sequence conservation extends beyond the mature miRNAs to the entire pre-miRNA hairpin sequences. Thus, an EEmiRC homolog is a locus that contains one or more pre-miRNA hairpins that match the multiple sequence alignment of experimentally identified (at this time mouse and human) EEmiRC pre-miRNA hairpins.
BLAST searches with the entire mouse and human EEmiRC pre-miRNAs identified at least one homolog in the chimpanzee, rat, dog, and cow genomes. Scanning the flanking genomic sequences with the program HMMER (Eddy et al. 1995) for matches to the human/mouse EEmiRC pre-miRNA consensus identified additional homologous sequences that could be folded into pre-miRNA hairpins (Fig. 1). Systematic folding did not identify additional hairpins. Interestingly, HMMER found a seventh hairpin in the mouse locus (mm-X), which we had not identified in our original analysis.
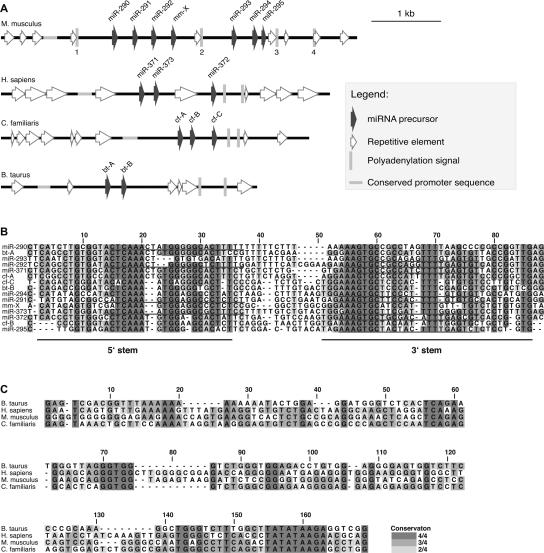
FIGURE 1.
Bioinformatic analysis of EEmiRC. (A) Schematic representation of the murine, human, canine, and bovine EEmiRC loci. Sequence features are given according to the legend. (B) Multiple sequence alignment of the murine (experimentally identified miR-290–295 and predicted mm-X), human (experimentally identified miR-371–373), canine (predicted cf-A–C), and bovine (predicted bt-A,B) pre-miRNA hairpins. The positions of cloned mature miRNAs and the two strands of the hairpin stem are underlined. (C) Multiple sequence alignment of the four conserved EEmiRC core promoter sequences.
Scanning the entire zebra fish, puffer fish, and chick genomes with HMMER failed to identify strong matches to the EEmiRC pre-miRNA consensus. HMMER also failed to identify EEmiRC paralogs within the mouse genome. Preliminary data suggest that EEmiRC is also absent in marsupials. Thus, EEmiRC is likely to be a uniquely eutherian locus present at one copy per haploid genome.
Having identified six mammalian EEmiRC loci we tried to find conservation beyond the pre-miRNA hairpins. The chimpanzee and rat EEmiRC loci are so similar to their respective human and mouse counterparts that they were excluded from further consideration so as not to bias the analysis toward the primate and rodent lineages. The remaining loci are distributed among four distinct mammalian orders (Rodentia, Primates, Carnivora, and Artiodactyla). Smith–Waterman local alignments revealed a statistically significant region of homology, ~150 bp long and positioned <1 kb upstream of the first pre-miRNA in each cluster (Fig. 1A,C).
The most obvious feature of this conserved sequence element is a TATATAAGA motif, which contains a canonical TATA box, centered at alignment position 159, that is conserved in all four species. Other highly conserved regions are a TCANAN(N)G motif at position 59, a GGTGG motif at position 72, and a TGNG motif at position 143, all of unknown function. The presence of a conserved TATA box, taken together with the recent evidence that RNA polymerase II is responsible for miRNA transcription (Lee et al. 2004), strongly suggests that the conserved region described above constitutes the core promoter responsible for the transcription of EEmiRC. This putative promoter element is the only sequence motif conserved between the four EEmiRC loci, apart from the pre-miRNAs and some homologous repetitive elements.
Consistent with transcription by RNA polymerase II, there are four polyadenylation motifs in the murine EEmiRC (Fig. 1A, vertical bars labeled 1–4). Of the seven ESTs that map within the mouse EEmiRC (Houbaviy et al. 2003) the 3′ ends of four align adjacent to polyadenylation signal 3 immediately downstream of miR-295, strongly suggesting that this is the bona fide polyadenylation site. Thus, bioinformatic analysis predicts that the mouse EEmiRC primary transcript is ~3.1 kb long. Polyadenylation signals were also found in the remaining mammalian EEmiRC loci (Fig. 1A).
The seed sequence, consisting of nucleotides 2–7 at the miRNA 5′ end, is the major determinant of miRNA target recognition (Lewis et al. 2003, 2005). Five of the experimentally identified EEmiRC miRNAs (miR-291-as, miR-294, miR-295, miR-372, and miR-373-as) share seeds with miR-302a–d and miR-93, whereas the remaining seven (miR-292-as, miR-293, and miR-371, the 5′ ends of which are shifted, and miR-290, miR-291-s, miR-292-s, and miR-373-s, which are excised from the opposite strand of the hairpin stem) have unique seeds. Together with pre-miR-367, pre-miR-302a–d form a cluster, which, like EEmiRC is expressed in ES cells (Suh et al. 2004). Neither miR-93, which is not ES cell specific, nor the cluster consisting of miR-302a–d and miR-367, can be considered EEmiRC paralogs. miR-93 is excised from the 5′ stem of its pre-miRNA hairpin and the sequence of pre-miR-93 does not match the EEmiRC pre-miRNA consensus sequence. While multiple sequence alignment reveals similarities between the EEmiRC pre-miRNAs and the individual hairpins of pre-miRNAs 302a–302d, the miR-302a–302d–367 cluster is not an EEmiRC paralog because it does not contain sequences corresponding to the EEmiRC promoter and because pre-miR-367 is not homologous to any sequences within EEmiRC.
Large RNAs from the mouse EEmiRC and identification of the pri-miRNA
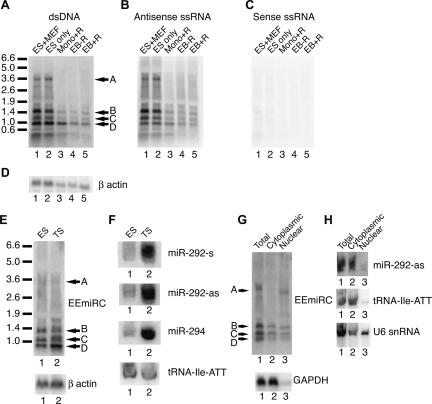
To obtain experimental support for our bioinformatic predictions, we looked for large RNAs originating from EEmiRC. Northern analyses of ES cell total RNA readily detected four major large RNA species, all of which had the same polarity as the mature miRNAs (Fig. 2A–C, bands A–D). All RNAs were down-regulated upon differentiation of ES cells into embryoid bodies or upon induction of differentiation with retinoic acid in monolayer (Fig. 2A–C, cf. lanes 1 and 2 with lanes 3–5). This is consistent with the previously determined expression pattern of the mature EEmiRC miRNAs (Houbaviy et al. 2003). Band A has an apparent length consistent with that predicted for the EEmiRC pri-miRNA. The remaining bands may be EEmiRC pri-miRNAs corresponding to alternative initiation and/or termination events or they may be processing products of band A.
FIGURE 2.
Large RNAs from the EEmiRC locus. (A_–_C) Northern analysis of total RNA from ES cells grown on feeders (lane 1), ES cells grown without feeders (lane 2), ES cells differentiated with retinoic acid in monolayer (lane 3), and ES cells differentiated into embryoid bodies without (lane 4) and with (lane 5) retinoic acid. Hybridization was performed with a random primed DNA probe (A) or with single-stranded RNA probes antisense (B) or sense (C) to the mature microRNAs. The β-actin loading control is given in D. (E) Northern analysis of the large EEmiRC RNA intermediates in ES (lane 1) and TS (lane 2) cells is shown in the top panel. The β-actin mRNA (bottom panel) serves as a loading control. The corresponding analyses of the mature miR-292-s, miR-292-as, miR-294, and the tRNA-Ile-ATT loading control are shown in F. (G) Subcellular localization of the large EEmiRC RNAs (top panel) or the GAPDH mRNA control (bottom panel). Northern analysis of total (lane 1), cytoplasmic (lane 2), and nuclear (lane 3) RNA was performed as in A. The apparent change in the mobility of band A RNA in lanes 1 and 3 is due to differences in the total RNA amounts loaded. (H) Short RNA Northern analysis of the subcellular localization of mature miR-292-as (top), tRNA-Ile-ATT (middle), and U6 snRNA (bottom). Total, cytoplasmic, and nuclear RNAs are analyzed in lanes 1,2,3, respectively.
The EEmiRC miRNAs were initially discovered in ES cells, suggesting a function in the blastocyst inner cell mass (ICM) (Houbaviy et al. 2003). To address a potential function of EEmiRC in the other blastocyst lineage, the trophoblast, we looked for EEmiRC expression in trophoblastic stem (TS) cells (Tanaka et al. 1998). Northern analyses detected both the large EEmiRC RNA intermediates and the mature EEmiRC miRNAs in TS cells. (Fig. 2E,F). The latter were ~10 times more abundant in TS cells than ES cells. Thus, EEmiRC is probably expressed in both the epiblast and the trophoblast and may be involved in the development of the placenta.
Initial processing of pri-miRNAs into pre-miRNAs by the nuclease Drosha takes place in the nucleus (Lee et al. 2002, 2003). Thus, we sought to determine the subcellular distribution of the EEmiRC RNA species. Northern analysis of total, nuclear, and cytoplasmic ES cell RNA preparations revealed that band A was predominantly nuclear whereas bands B, C, and D were approximately evenly distributed between the nucleus and the cytoplasm (Fig. 2G, top panel). As expected, the mature miRNAs were predominantly cytoplasmic as were the tRNA and GAPDH controls (Fig. 2G,H). A significant fraction of the U6 snRNA was detected in the cytoplasm, suggesting leakage of nuclear components (Fig. 2H, bottom panel). Thus, the presence of bands B–D in the cytoplasm could be an artifact of the nuclear isolation procedure.
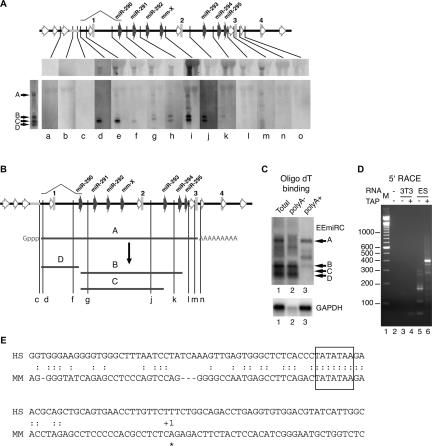
To unravel the relationship between EEmiRC RNA bands A–D we performed Northern blot hybridizations of ES cell RNA with oligonucleotide probes spanning the EEmiRC locus (Fig. 3A; Table 1). As a negative control, total RNA from NIH/3T3 cells was used. Due to the much lower specific activity of radiolabeled oligonucleotides as compared to the random primed probes, gels had to be overloaded in order to obtain robust detection of band A (Fig. 3A, top panels). This band was detected by probes d–m but not by probes a–c or n and o. Thus, the 5′ end of the RNA corresponding to band A is between the positions of probes c and d or immediately downstream of the conserved putative core promoter region, whereas the 3′ end of band A is between probes m and n, which flank polyadenylation signal 3. Therefore, band A most likely corresponds to the theoretically predicted EEmiRC pri-miRNA.
FIGURE 3.
Mapping of the large EEmiRC RNAs. (A) Mapping of the EEmiRC RNAs with oligonucleotide probes. Northern hybridizations with oligonucleotide probes (Table 1) corresponding to the indicated positions within the EEmiRC locus are shown. In each individual panel, the left lane contains total RNA from ES cells and the right lane contains total RNA from NIH/3T3 cells. Hybridization with a random primed probe and the positions of the four major RNA species are given on the left. Robust detection of band A can only be achieved after overloading the membranes (top panels). The EEmiRC locus representation is according to Figure 1A and includes a schematic of the intron preceding pre-miR-290. (B) Schematic representation of the RNA species deduced from A. (C) Polyadenylation of the EEmiRC transcripts. Northern analysis of total ES cell RNA (lane 1), ES cell RNA not bound to oligo(dT) cellulose (lane 2), and RNA bound to oligo(dT) cellulose (lane 3). Analysis of the EEmiRC transcripts and the polyadenylated GAPDH mRNA control are shown in the top and bottom panels, respectively. (D) Mapping of the 5′ end of the EEmiRC pri-miRNA. Final PCR amplification of the 5′ RACE products from NIH/3T3 (lanes 3,4) or ES (lanes 5,6) cell total RNA without (lanes 3,5) or with (lanes 4,6) TAP treatment. No 5′ RACE template was added to the PCR reaction shown in lane 2. The molecular weight marker is shown in lane 1. (E) The transcription start site (asterisk, +1) determined from the experiment shown in D is superimposed onto the alignment of the putative mouse (MM) and human (HS) EEmiRC promoter regions. The box highlights the conserved TATA element.
TABLE 1.
Hybridization and RNAse protection probes
| Probe sequencea | Positionb | |
|---|---|---|
| a | AAAGGAAGAATGTGGGAGGTTCAGTTTATTCTTTCGTTTG | −396 −357 |
| b | TTACTCTACCACCCTGCTTCCTCTGAGCTGAGTTTCCCTG | −140 −101 |
| c | AGGCTCTAGGTTCTTATATAGTCTGAAGGCTCA | −47 −15 |
| d | CAGCACGCCGGAGGTATCCCTGAAGACCGCAGAGACCAGC | +27 +66 |
| e | ATCCTGGATCTACTTTCGCCTCTCTAAACCAGGTAGATTA | +133 +171 |
| f | GTTAGTACATCGGTCTAACTCAAGGTATAGTGAAACTCAA | +655 +694 |
| g | CCATCCCTAGCCTTTCGCTATACTCAGTCTCATTCCTTTC | +954 +993 |
| h | CTCTTTAGGATAGGACCCATTAAACTCCAAGCCTAAACCC | +1225 +1264 |
| i | AAGCAGGTAAGCGATTCCAGGTTGCAGCTGCAGCCGGCTT | +1740 +1779 |
| j | GGTATTATGGGTATTATCTACCCGGCTGTCCACCAAGCTT | +2231 +2270 |
| k | AATGCAGCTATATGTAATACAGTACCTGTCAAATCTGGGT | +2699 +2738 |
| l | AAAGCAGCCGACCTGTGAATGGTGCCCACAGGAGAGACTC | +2997 +3035 |
| m | ATTCATGTTTGGAGGCTGAGGGCACTGGTTGCTCCCATAG | +3120 +3159 |
| n | CCCAGTGATTAAAAGGGTGCTTGCATGCTTGCAAGCTTTT | +3238 +3277 |
| o | TGCCCTATTACTGTTGAAAACACTTCCAGCTATGTGTTGG | +3345 +3384 |
| rpA | CAGAGGAAGCAGGGTGGTAG / GCAGGAAGATCTCTCTTCTGGA | −125 +195 |
| rpB | TTCATTAACTACGTGGTTTCATTGT / GTTACCGTCTACTGGGCAGG | +623 +1017 |
The relationships between EEmiRC RNAs A–D, revealed by the above analysis, are illustrated in Figure 3B. The mapping data suggest that RNAs B–D are processing intermediates of the RNA corresponding to band A, which is the only EEmiRC primary transcript. Band C RNA could be a product of band B RNA and it appears, at this level of resolution, that a single event produces the 3′ end of band D RNA and the 5′ ends of bands B and C RNAs.
It was recently shown that many pri-miRNAs are capped and polyadenylated RNA polymerase II transcripts (Lee et al. 2004). Because the EEmiRC miRNAs are flanked by a TATA-box and a polyadenylation signal we expected its pri-miRNA to be capped and polyadenylated as well. Thus, we looked for the presence of a poly(A) tail and a cap structure in the large EEmiRC RNAs. Fractionation of total ES cell RNA on an oligo(dT) column showed that the putative pri-miRNA corresponding to band A is polyadenylated whereas RNAs corresponding to bands B, C, and D are not (Fig. 3C).
To test for the presence of a cap structure at the 5′ end of the EEmiRC pri-miRNA and to map the transcription start site at single nucleotide resolution, we used a modified 5′-RACE protocol that specifically amplifies capped RNAs. A similar procedure was used to characterize a capped plant pri-miRNA (Aukerman and Sakai 2003). To amplify capped RNAs by 5′ RACE the cap structure must be converted to a 5′ phosphate. This is achieved by treating the RNA with tobacco acid pyrophosphatase (TAP). When 5′ RACE was performed with dephosphorylated and then TAP treated ES cell RNA a strong band migrating near the 400-bp marker was observed (Fig. 3D, lane 6). This band constituted at least 90% of the final PCR product and was not amplified from dephosphorylated TAP treated NIH/3T3 RNA or ES cell RNA that was dephosphorylated but not treated with TAP (Fig. 3D, cf. lane 6 with lanes 4 and 5, respectively). The 5′ RACE adaptor adds 38 bp to the final PCR products. Therefore, the 3′ PCR primer maps ~360 bp downstream of the transcription start site. Sequencing of the major 5′ RACE product from TAP treated ES cell RNA positioned the transcription start site at an A residue 35 bp downstream of the first T residue of the conserved TATA box (Fig. 3E). The 3′ PCR primer spans positions +341 to +359 relative to the transcription start site, consistent with the size of the 5′ RACE product. Note that 35 bp is the canonical distance between the TATA box and the transcription start site.
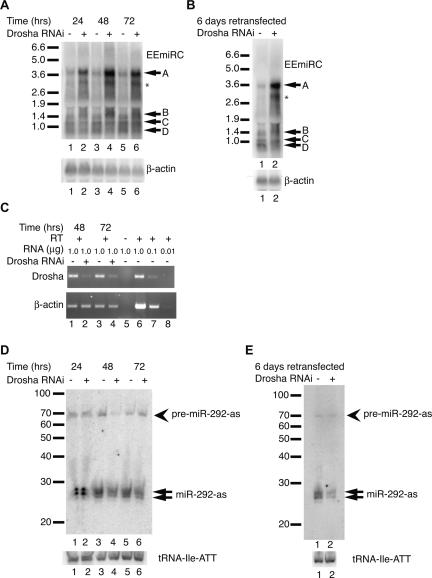
To consider the observed EEmiRC RNA intermediates in the context of the pri-miRNA processing pathway, we performed RNAi mediated Drosha knockdowns in ES cells. Transfection of a Drosha siRNA resulted in the dramatic up-regulation of a band migrating similar to band A, a modest increase of band B, and up-regulation of an additional minor band and a smear of material migrating between bands A and B (Fig. 4A). A GFP control siRNA did not change the levels of the EEmiRC RNAs (Fig. 4A, cf. the odd with the even lanes). The effect of the Drosha knockdown was evident as early as 24-h post-transfection, reached a maximum at 48 h, and began to decrease by 72 h. Retransfecting the cells sustained the increase in EEmiRC RNA levels over a period of 6 d. No significant changes were observed in the levels of bands C and D at any time point.
FIGURE 4.
Effects of Drosha knockdown on EEmiRC RNA expression. (A) ES cells were transfected with a Drosha siRNA (even lanes) or an EGFP control siRNA (odd lanes). Large EEmiRC RNAs (top panel) were monitored by Northern analysis at 24, 48, and 72 h post-transfection. The positions of bands A–D are indicated. The position of an additional RNA that was up-regulated in response to Drosha knockdown is shown by an asterisk. The β-actin loading control is given in the bottom panel. (B) ES cells were transfected twice with an EGFP siRNA (lane 1) or a Drosha siRNA (lane 2) and the large EEmiRC RNAs were monitored by Northern analysis at 6 d after the first transfection (top panel). The β-actin loading control is shown in the bottom panel. (C) Drosha mRNA levels (top panel) in cells transfected with an EGFP siRNA (lanes 1,3) or a Drosha siRNA (lanes 2,4) monitored by RT-PCR at 48 or 72 h post-transfection. Amplification of the β-actin mRNA is shown in the bottom panel. No reverse transcriptase was added to the reactions analyzed in lanes 5. Lanes 1_–_5 contained 1 μg total RNA. The titration of 1, 0.1, and 0.01 μg total RNA in lanes 6_–_8 shows that the assay is in the linear range. (D,E) Short RNA Northern analyses of miR-292-as and pre-miR-292-as (top panels) or tRNA-Ile-ATT (bottom panels) in the experiments shown in panels A and B, respectively.
The above experiments imply that RNAs migrating as bands A and B are Drosha substrates. Importantly, these observations reinforce our conclusion that RNA migrating as band A is an EEmiRC pri-miRNA whereas RNAs corresponding to bands B–D are processing intermediates. However, while it is not surprising that the level of band D RNA, which does not contain any pre-miRNA hairpins, did not depend on Drosha, it was surprising that the level of band C did not change since it contains at least three pre-miRNAs (Fig. 3A,B) and is, therefore, expected to be a Drosha substrate.
The changes in EEmiRC RNA levels correlated inversely with the levels of Drosha mRNA (Fig. 4C). Approximately 10% of the initial Drosha mRNA remained in cells subjected to Drosha RNAi, suggesting that some residual Drosha activity was still present. Changes in the levels of the mature miRNAs and pre-miRNAs were consistent with a role for Drosha in their production—mature miR-292-as decreased in cells transfected with Drosha siRNA and reached a minimum level in retransfected cells (Fig. 4D,E). Interestingly, pre-miR-292-as showed only a transient decrease at 48 h post-transfection as well as in retransfected cells. Its levels remained constant at all other time points (Fig. 4D,E). This behavior is consistent with reestablishment of the normal steady-state rate of pre-miRNA production at a decreased concentration of Drosha compensated by an increased concentration of Drosha substrate. Thus, the apparent insensitivity of the levels of band C RNA to the Drosha knockdown could be explained by similar kinetic effects. For example, if band C RNA is produced by Drosha cleavage of band A RNA, then a decrease in Drosha accompanied by an increase in band A RNA levels could, by saturating the available Drosha, possibly generate a constant level of band C RNA. A similar argument may also explain why changes in the intensity of band B were much less pronounced than the increase in RNAs migrating as band A.
Ectopic expression of EEmiRC
The above results strongly suggest that the conserved sequence upstream of the EEmiRC miRNAs constitutes a core promoter. To obtain further experimental evidence for this conclusion, we investigated the activity of this DNA element in transient transfections.
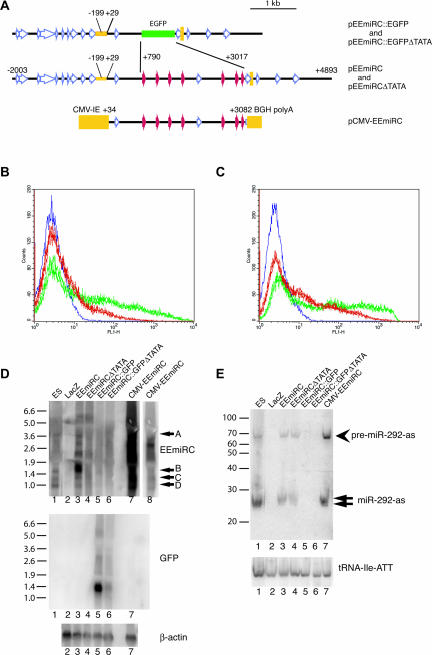
The genomic fragment spanning positions −2003 to +4893 relative to the EEmiRC pri-miRNA transcription start site was subcloned into pBluescript, resulting in plasmid pEEmiRC (Fig. 5A). Next, the region from +790 to +3017 of pEEmiRC was replaced with a DNA fragment encoding EGFP. In the resulting plasmid, designated pEEmiRC::EGFP, all pre-miRNA hairpins are eliminated, but the putative promoter and polyadenylation sites are intact. Transfection of pEEmiRC::EGFP into ES cells resulted in reproducible expression of EGFP as measured by FACS (Fig. 5B, green traces). Deleting DNA between positions −199 and +29 (plas-mid pEEmiRC::EGFPΔTATA; for reference the conserved mouse sequence element shown in Fig. 1C spans positions −181 to −20) resulted in a significant decrease in EGFP expression but did not eliminate it completely (Fig. 5B, red traces; compare with the blue trace, which corresponds to a transfection of a LacZ expressing construct). Similar results were obtained when the plasmids were transfected in HEK-293 cells, although deletion of the putative core promoter had a less pronounced effect on EGFP expression (Fig. 5C).
FIGURE 5.
Expression of EEmiRC in transient transfections. (A) Maps of the transfected constructs. pEEmiRC contains a genomic fragment spanning positions −2003 to +4893 relative to the EEmiRC transcription start site. In pEEmiRC::EGFP positions +790 to +3017 are replaced by the EGFP coding sequence. In plasmids pEEmiRC(ΔTATA) and pEEmiR-C::EGFP(ΔTATA) positions −199 to +29 have been deleted. In pCMV-EEmiRC a fragment spanning the region from +34 to +3082 is placed between the cytomegalovirus immediate early promoter and the bovine growth hormone polyadenylation signal. (B) EGFP expression in ES cells monitored by FACS. Cells were transfected with pEEmiRC::EGFP (green traces, superposition of three independent transfections) or pEEmiRC::EGFP(ΔTATA) (red traces, superposition of three independent transfections). The blue trace shows the background fluorescence of cells transfected with a LacZ expressing construct. (C) Transfection of HEK-293 cells as in B. (D) Northern analysis of the expression of large EEmiRC RNAs (top panel) or EGFP mRNA (middle panel) in HEK-293 cells transfected with a LacZ expressing construct, pEEmiRC, pEE-miRC(ΔTATA), pEEmiRC::EGFP, pEEmiRC::EGFP(ΔTATA), or pCMV-EEmiRC (lanes 2_–_7, respectively). RNA from untransfected ES cells is shown in lane 1. Lane 8 is a shorter exposure of lane 7. The β-actin loading control is given in the bottom panel. (E) The levels of miR-292-as and pre-miR-292-as (top panel) and the tRNA-Ile-ATT loading control (bottom panel) in the samples from D are monitored by short RNA Northern analysis.
Is the low level of transcription observed from pEEmiRC::EGFPΔTATA physiologically relevant? To address this question, we assayed the EGFP mRNA levels in transfected HEK-293 cells by Northern analysis. A random primed EGFP probe detected a single RNA species in cells transfected with pEEmiRC::EGFP but not in cells transfected with pEEmiRC::EGFPΔTATA or untransfected cells (Fig. 5D, middle panel, cf. lanes 5 and 6). This RNA migrated slightly faster than expected for the full-length EGFP mRNA (apparent size ~1400 nt; predicted size 1678 nt). An identical result was obtained in ES cells (data not shown). Failure to detect defined transcripts in cells transfected with pEEmiRC::EGFPΔTATA argues that the low level of EGFP expression may be an artifact due to nonspecific transcription initiation and termination at sites not normally used in vivo. We note that several repetitive elements are present in pEEmiRC::EGFPΔTATA. Some of them are LINEs and, therefore, potentially contain RNA polymerase II promoters. The detection of EEmiRC promoter activity in transiently transfected HEK-293 cells suggests that ES cell specific factors are not required for basal transcription of EEmiRC and that silencing of EEmiRC expression in somatic tissue may occur via heterochromatinization of the locus.
The endogenous EEmiRC locus is not expressed in HEK-293 cells (Fig. 5D, top panel, lane 2, and Fig. 5E, lane 2). Transfection of pEEmiRC into HEK-293 cells resulted in the production of all four large EEmiRC RNAs (bands A–D) similar to those observed in ES cells plus additional RNA species (Fig. 5D, top panel, cf. lane 3 to lane 1). Interestingly, the abundance of band B relative to bands A, C, and D in transfected HEK-293 cells was much higher than in untransfected ES cells. Deletion of the putative promoter element (pEEmiRCDTATA, deletion identical to pEEmiRC::EGFPΔTATA) abolished expression of all large EEmiRC RNAs (Fig. 5, top panel, lane 4).
Consistent with expression of the large EEmiRC RNAs, both miR-292-as and pre-miR-292-as were detected by short RNA Northern analysis in HEK-293 cells transfected with pEEmiRC (Fig. 5E). Deletion of the putative promoter element, however, had almost no effect on the levels of miR-292-as and pre-miR-292-as. Again, we attribute this to nonspecific transcription that generates heterogeneous transcripts that direct the production of mature miRNAs. Ectopic expression from pEEmiRC in HEK-293 cells resulted in miRNA and pre-miRNA levels similar to the endogenous levels in ES cells. Furthermore, EEmiRC expression from the strong CMV promoter (plasmid pCMV-EEmiRC, Fig. 5A) caused only a modest increase in miRNA and pre-miRNA production (Fig. 5E, cf. lanes 7 and 3) even though the levels of some large EEmiRC RNAs increased by at least one order of magnitude (Fig. 5D, top panel, lane 7). Thus, in transiently transfected HEK-293 cells, the EEmiRC miRNA synthesis pathway is probably at or close to saturation, regardless of promoter strength. Even the low-level nonspecific transcription from pEEmiRCDTATA may be enough to saturate the miRNA synthesis machinery, which would explain why deletion of the putative promoter had little effect on the miRNA and pre-miRNA levels.
Neither band D RNA nor the EGFP mRNA could be detected by an EEmiRC probe in cells transfected with pEEmiRC::EGFP even though the primary EGFP transcript should contain sequences complementary to the probe (Fig. 5D, top panel, lane 5). Thus, consistent with the detection of a single mRNA band by the EGFP probe (Fig. 5D, middle panel), the proposed single cleavage that generates the 3′ end of band D and the 5′ ends of bands B and C in the native EEmiRC pri-miRNA does not occur when the pre-miRNA hairpins are replaced by EGFP. Furthermore, the 1.4-kb EGFP mRNA expressed from pEEmiRC::EGFP is probably not colinear with the plasmid sequence from the initiation to the polyadenylation site.
Splicing of the EEmiRC pri-miRNA and Drosha processing at pre-miR-290
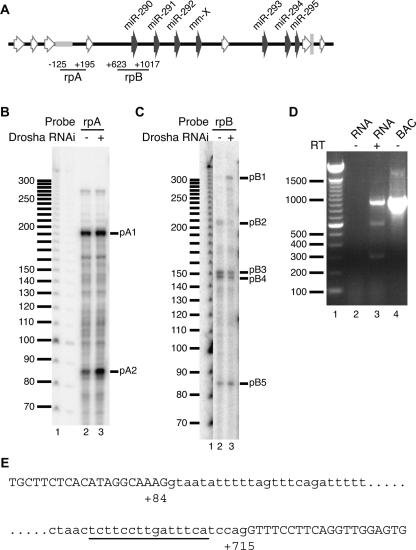
The simplest explanation for the above discrepancy is the presence of an intron spanning most of the primary transcript sequence between the transcription start site and pre-miR-290. To obtain evidence for splicing of the native EEmiRC pri-miRNA, we performed RNAse protection assays with RNA probes that spanned the expected splice junctions (Fig. 6 and Table 1, probes rpA and prB, respectively).
FIGURE 6.
Splicing of the EEmiRC primary transcript and Drosha processing at pre-miR-290. (A) The positions of the RNAse protection probes (rpA and rpB) superimposed onto the schematic representation of the EEmiRC locus as in Figure 1. (B,C) Total RNA from ES cells transfected with an EGFP control siRNA (lane 2) or a Drosha siRNA (lane 3) was hybridized with probe rpA (B) or rpB (C) and subjected to RNAse protection analysis. The molecular weight marker is shown in lane 1. (D) The RT-PCR products obtained from total ES cell RNA with probes flanking the predicted intron are shown in lane 3. The reaction analyzed in lane 2 did not contain reverse transcriptase. The molecular weight marker and PCR of the genomic EEmiRC BAC are shown in lanes 1 and 4, respectively. (E) Positions of the splice sites deduced from D. Exon sequences are given in capital letters; the intron is in lowercase. The polypyrimidine tract is underlined.
Two RNAse resistant fragments, designated pA1 and pA2, were obtained when a probe spanning the transcription start site (rpA) was hybridized with total ES cell RNA. (Fig. 6B). No protected bands were seen when NIH/3T3 cell RNA was used in the assay, confirming that both protected fragments were EEmiRC specific (data not shown). Probe rpA extends from −125 to +195 relative to the transcription start site. Thus, pA1, which is ~190 nt long, likely corresponds to the 5′ end of the primary transcript. Similarly, pA2 (~82 nt) probably corresponds to an exon extending from +1 to ~+82 relative to the transcription start site. Interestingly, Drosha knockdown by RNAi caused a significant increase in pA2 but only a modest increase in pA1 (Fig. 6B cf. lanes 2 and 3). Thus, it appears that the spliced pri-miRNA is up-regulated upon Drosha depletion, and this is probably the band A RNA that accumulated in Figure 4A,B after treatment with Drosha siRNA.
Probe prB, which spans the region between positions +623 and +1017 relative to the transcription start site, yielded four protected fragments (pB2-pB5) when hybridized to ES cell total RNA (Fig. 6C, lane 2). Drosha knockdown resulted in a decrease of protected fragments pB2–pB4 and in the appearance of an additional fragment designated pB1 (~310 nt; Fig. 6C, cf. lanes 2 and 3). The level of fragment pB5 (~85 nt) remained unchanged. Thus, it is likely that pB2–pB4 are generated by Drosha cleavages. Indeed, the sizes of pB3 (~155 nt) and pB4 (~150 nt) are consistent with RNA species whose 5′ ends are within the 3′ stem of the pre-miR-290 hairpin, and extend from approximately position +867 to the 5′ end of probe rpB at position +1017. Likewise, the length of pB2 (~210 nt) implies that it has a 3′ end resulting from cleavage within the 5′ stem of the pre-miR-290 hairpin. This fragment would then correspond to a protected RNA species between positions +623 and +828. Thus, the apparent single event that was postulated to produce bands B, C, and D from band A (Fig. 3A,B) is, in fact, Drosha processing of the pre-miR-290 hairpin. This conclusion is further supported by the fact that band D was not detected in HEK-293 cells transfected with pEEmiRC::EGFP, which lacks all pre-miRNA hairpins (cf. lanes 5 and 6 in Fig. 5, top and middle panels).
Consistent with the up-regulation of protected fragment pA2, fragment pB1, which is also upregulated upon Drosha knockdown, likely corresponds to a 5′ exon junction around position +707 and extends to the 5′ end of the probe at position +1014. The nature of fragment pB5 is presently unclear. Since its length and the length of fragment pB1 add up approximately to the size of probe rpB (310 + 85 = 395 nt vs. 356 nt), it may represent the predicted intron, consistent with the constant levels of pB5 upon knockdown of Drosha.
The above data strongly suggest the presence of an intron between positions +82 and +707 of the EEmiRC primary transcript. Thus, we performed RT-PCR with primers flanking these positions (Fig. 6D). Three DNA species with apparent lengths of ~300 bp, 600 bp, and 950 bp were amplified from total ES cell RNA in a complete RT-PCR reaction but not when reverse transcriptase was omitted (Fig. 6, cf. lanes 2 and 3). Only the 950-bp band was amplified in a PCR reaction that used the genomic DNA as a template (Fig. 6, lane 4). This is consistent with the 950-bp cDNA product representing the unspliced primary EEmiRC transcript. Both the 950-bp and 300-bp RT-PCR products could be reamplified by PCR whereas reamplification of the 600-bp band yielded a mixture of the 950-bp, 600-bp, and 300-bp products. Thus, the 300-bp band likely corresponds to the spliced EEmiRC pri-miRNA whereas the 600-bp band is probably a heteroduplex of spliced and unspliced cDNA. Sequencing of the 300-bp cDNA product and comparison with the genomic EEmiRC sequence revealed a splice junction of positions +84 and +715 relative to the transcription start site (Fig. 6E). This is consistent, within experimental error, with the predictions of the RNAse protection mapping.
Note that band D is unlikely to correspond to the excised intron RNA, since its production depends on the EEmiRC hairpins (Fig. 5D) and since it can be detected by oligonucleotide probe d, which maps within the first EEmiRC exon (Fig. 3A; Table 1).
DISCUSSION
EEmiRC shows a remarkable degree of evolutionary variation. The only conserved regions within the locus are the pre-miRNA hairpins and the putative minimal promoter. The number and precise sequences of the pre-miRNAs, their distance from the promoter and polyadenylation sites, the regions flanking the hairpins, and the types, positions, and numbers of repetitive element insertions vary in species belonging to different mammalian orders. Such variation is not a general feature of miRNA loci. The pre-miR-302a–d hairpins, which, as pointed out above, are related to the EEmiRC hairpins, show a much greater degree of sequence conservation than the pre-miRNAs within EEmiRC (data not shown). The number and spacing of the individual pre-miR-302 hairpins is conserved, and homology beyond the pre-miRNAs can be easily detected between the chick and mouse loci (data not shown). The microRNA cluster miR-17/91-miR-18-miR-19a-miR-20-miR-19b-1-miR-92 is also very well conserved in mammals (Tanzer and Stadler 2004). Sequences up to 200 nt upstream of most C. elegans pre-miRNA hairpins share significant homology with their C. briggsae counterparts (Ohler et al. 2004).
The common seed shared between some EEmiRC miRNAs, miR-302a–d and miR-93, implies regulation of common targets. Our analysis suggests that these three classes of miRNAs have evolved independently and might have both overlapping and nonoverlapping functions. For example, they may regulate some common targets in different developmental contexts. The variability exhibited by EEmiRC suggests that this gene may have appeared relatively recently in the mammalian lineage and may have mammalian-specific functions. Together with previous evidence on its expression in ES cells and preimplantation embryos, the finding that EEmiRC is expressed in the trophoblast supports the above conclusion.
Several recent studies have reported the characterization of miRNA transcription units (Bracht et al. 2004; Cai et al. 2004; Lee et al. 2004). However, because of their low abundance, the detection and characterization of pri-miRNAs relied extensively on Drosha knockdowns and/or RT-PCR amplification of fractionated RNA species. In contrast, the large EEmiRC RNA precursors reported here are relatively abundant and can be easily detected by standard Northern analyses. Thus, we have analyzed the nucleocytoplasmic distribution, polyadenylation status, and origin of the different EEmiRC RNA species directly. Experimental mapping of the EEmiRC primary transcript agrees with the results of comparative bioinformatic analysis of the homologous EEmiRC loci. The conserved region upstream of the mapped transcription start site contains a canonical TATA-box and likely represents the core EEmiRC promoter since it showed promoter activity that correlated with the presence of the TATA motif.
The bulk of the long RNAs synthesized from the EEmiRC locus consists of cleavage products of the pri-miRNA. Oligonucleotide probe hybridizations suggest that the molar ratio of the pri-miRNA to the shorter RNA species is <5%. The observed EEmiRC pri-miRNA processing products are clearly not an artifact of the RNA isolation procedure. The pri-miRNA is converted into well-defined species that can be resolved on formaldehyde-aga-rose gels and detected by Northern hybridizations. The same pattern of large EEmiRC RNAs is observed in independent Northern analyses of different ES cell RNA preparations as well as in TS cells and can be qualitatively reproduced upon overexpression from transiently transfected constructs in HEK-293 cells.
How do the RNAs corresponding to bands B–D fit within an EEmiRC miRNA processing pathway? The nuclease Drosha is thought to excise the pre-miRNA stem-loop precursor from the pri-miRNA (Lee et al. 2003). The flanking RNA fragments are stable byproducts of this process (Han et al. 2004). Thus, Drosha cleavage at only some of the seven EEmiRC hairpins would result in partial cleavage of the pri-miRNA and the generation of long RNA intermediates such as the band B–D RNAs. Our observations support this scenario. First, the appearance of an additional EEmiRC RNA band, migrating between bands A and B, upon Drosha knockdown is consistent with partial cleavage. Second, RNAse protection analysis of the region surrounding pre-miR-290 detects only Drosha cleavages. Thus, the 3′ end of band D RNA and the 5′ ends of band B and C RNAs are likely due to Drosha processing of the pre-miR-290 hairpin. Similarly, Drosha processing near pre-miR-293 and pre-miR-294 probably generates the 3′ ends of RNA species corresponding to band B and C RNAs respectively. Consistent with its being a Drosha reaction byproduct, band D RNA was no longer generated when the EEmiRC pre-miRNA hairpins were replaced by the EGFP coding sequence.
RT-PCR and RNAse protection assays showed that the EEmiRC primary transcript is spliced between positions +82 and +715. The changes in the RNAse protection patterns upon Drosha knockdown suggest that the spliced pri-miRNA is preferentially cleaved by Drosha. The unspliced primary transcript increased only slightly when Drosha was depleted, whereas RNAs containing the spliced first and second exons increased dramatically. Thus, splicing may be required for, or may enhance, Drosha processing. Indeed, _trans_-splicing of the let-7 pri-miRNA appears to be required for let-7 expression (Bracht et al. 2004). However, splicing is not absolutely necessary for Drosha processing of the EEmiRC pri-miRNA since band D RNA is clearly produced in a Drosha-dependent manner from the unspliced primary transcript. Further studies are needed to elucidate the role of splicing in EEmiRC miRNA expression.
Attempts to assign function to miRNAs have focused mainly on the analysis of the mature miRNA sequences. Here, we show that additional useful information can be obtained by studying the organization of the entire miRNA genes. The remarkable variability of EEmiRC and its presence only in placental mammals reinforce previous conclusions that EEmiRC is likely to have interesting biology (Houbaviy et al. 2003; Suh et al. 2004). Definition of the EEmiRC transcription unit and the characterization of large primary transcript processing intermediates provide a foundation for understanding how this miRNA cluster is regulated during development.
MATERIALS AND METHODS
Bioinformatics
Genomic data were downloaded from the ENSEMBL server (http://www.ensembl.org). Multiple sequence alignments, Smith–Waterman local alignments, HMM searches, and RNA folding were performed with the MacOS X implementations of CLUS-TALW, SSEARCH, HMMER, and MFOLD (Higgins and Sharp 1988; Pearson 1991; Eddy et al. 1995; Zuker et al. 1999).
Cell culture, plasmids, and transfections
Fragments of the EEmiRC locus were PCR amplified from mouse genomic DNA. Detailed information on plasmid construction is available upon request. Synthetic siRNAs against GFP [(NN) GCACCAUCUUCUUCAAGGA(GC)] and Drosha [(NN)CAAC AGUCAUAGAAUAUGA(GC)] were obtained from Dharmacon (option A2) and processed as suggested by the manufacturer. ES cells were differentiated as described previously (Wutz and Jaenisch 2000). Plasmid DNA and siRNAs were transfected into ES cells and HEK-293 cells with Lipofectamine 2000 (Invitrogen) and Fugene 6 (Roche), respectively, as suggested by the manufacturers.
RNA analysis
Total RNA was extracted as described previously (Houbaviy et al. 2003). To prepare nuclear and cytoplasmic RNA, ES cells were lysed in 50 mM HEPES (pH 7.5), 15 mM NaCl, 60 mM KCl, 0.34 M sucrose, 0.5 mM spermidine-HCl, 0.15 mM spermine-HCl, 10 mM DTT, 5 mM EDTA, 0.5 mM EGTA, 1000 u/mL RNAsin, 0.5% Nonidet P40, and the nuclei were separated from the cytoplasm by centrifugation for 5 min at 500_g_. RT-PCR of Drosha mRNA was done with primers 5′-ACTCGGAGGTGTTCGATGTC-3′ and 5′-CATGTTGGCAATCTCCTCCT-3′. The β-actin control primers were 5′-TGTTACCAACTGGGACGACA-3′ and 5′-AAGGAAGGCT GGAAAAGAGC-3′. RT-PCR mapping of the EEmiRC intron was performed with primers 5′-TCTGCGGTCTTCAGGGATAC-3′ and 5′-GTACTCACCACGCTGCAGTT-3′. Random primed EEmiRC DNA hybridization probes were synthesized from a PCR template amplified with primers 5′-TCTGCGGTCTTCAGGGATAC-3′ and 5′-TCCAGGAAACCTTCATCTGG-3′ from genomic DNA. The same primers, but with appended T3 or T7 phage promoter sequences, were used to amplify the in vitro transcription templates from which single-stranded RNA probes were synthesized with the MAXIscript kit (Ambion). Hybridizations with the above probes were performed in the presence of 10 μg/mL mouse Cot-1 DNA (Invitrogen) to suppress repetitive sequences. A random primed DNA probe that did not contain repetitive elements was synthesized from a mixture of PCR products amplified with primer pairs 5′-CGGTTTGGCTGGGTTTACTA-3′/5′-TAGACTCACCACCCCTGG AC-3′ and 5′-GTTGGACTGATGGTTGTGAGTC-3′/5′-GAAAGCA GCCGACCTGTG-3′. Starfire probes (Integrated DNA Technologies) with the sequences shown in Table 1 were used for oligonucleotide Northern hybridization mapping of EEmiRC RNAs. GAPDH and β-actin random primed probes were synthesized from commercially available templates (DECAtemplate, Ambion). The random primed GFP probe was synthesized from a XhoI–NotI fragment of pEGFP-N1 (Clontech). Short RNA Northern analyses were done as described previously (Hamilton and Baulcombe 1999; Houbaviy et al. 2003). The U6 oligonucleotide probe had the sequence 5′-GGGCCATGC TAATCTTCTCTGT-3′. RNAse protection probes were synthesized by in vitro transcription of templates amplified from genomic DNA with the primers shown in Table 1. T7 promoter sequences were appended to the reverse primers. 5′ RACE was performed with the FirstChoice RLM-RACE kit (Ambion). The inner and outer gene-specific PCR primers were 5′-GAGCGAGGAAGGCTGAGTT-3′ and 5′-ACATAGGCTCGTTCCTCCCT-3′, respectively. Oligo-dT RNA fractionation was performed with the MicroPoly(A) purist kit (Ambion).
Acknowledgments
We thank Zhongde Wang for providing TS cells, John Doench for advice on the manuscript, and members of the Sharp and Jaensich laboratories for discussions.
This work was supported by United States Public Health Service MERIT Award R37-GM34277 from the National Institutes of Health, PO1-CA42063 and U19 AI056900 from the National Cancer Institute to PAS, and partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute.
REFERENCES
- Ambros, V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., et al. 2003. A uniform system for microRNA annotation. RNA 9**:** 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5**:** 337–350. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15**:** 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116**:** 281–297. [DOI] [PubMed] [Google Scholar]
- Bohnsack, M.T., Czaplinski, K., and Gorlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10**:** 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht, J., Hunter, S., Eachus, R., Weeks, P., and Pasquinelli, A.E. 2004. _Trans_-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 10**:** 1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, X., Hagedorn, C.H., and Cullen, B.R. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10**:** 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy, A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16**:** 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B.B., Plasterk, R.H., Ketting, R.F., and Hannon, G.J. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432**:** 231–235. [DOI] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17**:** 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, S.R., Mitchison, G., and Durbin, R. 1995. Maximum discrimination hidden Markov models of sequence consensus. J. Comput. Biol. 2**:** 9–23. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15**:** 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, R.I., Yan, K.P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., and Shiekhattar, R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432**:** 235–240. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. 2004. The microRNA registry. Nucleic Acids Res. 32**:** D109–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106**:** 23–34. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J. and Baulcombe, D.C. 1999. A species of small anti-sense RNA in posttranscriptional gene silencing in plants. Science 286**:** 950–952. [DOI] [PubMed] [Google Scholar]
- Han, J., Lee, Y., Yeom, K.H., Kim, Y.K., Jin, H., and Kim, V.N. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Dev. 18**:** 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D.G. and Sharp, P.M. 1988. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73**:** 237–244. [DOI] [PubMed] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5**:** 351–358. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297**:** 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293**:** 834–838. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15**:** 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294**:** 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12**:** 735–739. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294**:** 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294**:** 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75**:** 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21**:** 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425**:** 415–419. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Kim, M., Han, J., Yeom, K.H., Lee, S., Baek, S.H., and Kim, V.N. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23**:** 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115**:** 787–798. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120**:** 15–20. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. 2003a. Vertebrate microRNA genes. Science 299**:** 1540. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003b. The micro-RNAs of Caenorhabditis elegans. Genes & Dev. 17**:** 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305**:** 1437–1441. [DOI] [PubMed] [Google Scholar]
- Lund, E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303**:** 95–98. [DOI] [PubMed] [Google Scholar]
- Ohler, U., Yekta, S., Lim, L.P., Bartel, D.P., and Burge, C.B. 2004. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA 10**:** 1309–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W.R. 1991. Searching protein sequence libraries: Comparison of the sensitivity and selectivity of the Smith–Waterman and FASTA algorithms. Genomics 11**:** 635–650. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403**:** 901–906. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115**:** 199–208. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305**:** 1434–1437. [DOI] [PubMed] [Google Scholar]
- Suh, M.R., Lee, Y., Kim, J.Y., Kim, S.K., Moon, S.H., Lee, J.Y., Cha, K.Y., Chung, H.M., Yoon, H.S., Moon, S.Y., et al. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270**:** 488–498. [DOI] [PubMed] [Google Scholar]
- Tanaka, S., Kunath, T., Hadjantonakis, A.K., Nagy, A., and Rossant, J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282**:** 2072–2075. [DOI] [PubMed] [Google Scholar]
- Tanzer, A. and Stadler, P.F. 2004. Molecular evolution of a microRNA cluster. J. Mol. Biol. 339**:** 327–335. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., and Zamore, P.D. 2004. A protein sensor for siRNA asymmetry. Science 306**:** 1377–1380. [DOI] [PubMed] [Google Scholar]
- Wutz, A. and Jaenisch, R. 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5**:** 695–705. [DOI] [PubMed] [Google Scholar]
- Yi, R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17**:** 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101**:** 25–33. [DOI] [PubMed] [Google Scholar]
- Zuker, M., Mathews, D.H., and Turner, D.H. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In RNA biochemistry and biotechnology (eds. J. Barciszewski and B.F.C. Clark), pp. 11–43. Kluwer Academic, Dordrecht/Boston/London.