Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p (original) (raw)
Abstract
The Ski complex (composed of Ski3p, Ski8p, and the DEVH ATPase Ski2p) is a central component of the 3′–5′ cytoplasmic mRNA degradation pathway in yeast. Although the proteins of the complex interact with each other as well as with Ski7p to mediate degradation by exosome, a 3′-exonuclease complex, the nature of these interactions is not well understood. Here we explore interactions within the Ski complex and between the Ski complex and Ski7p using a directed two-hybrid approach combined with coimmunoprecipitation experiments. We also test the functional significance of these interactions in vivo. Our results suggest that within the Ski complex, Ski3p serves as a scaffold protein with its C terminus interacting with Ski8p, and the sub-C terminus interacting with Ski2p, while no direct interaction between Ski2p and Ski8p was found. Ski7p interacts with the Ski complex via its interaction with Ski8p and Ski3p. In addition, inactivating the Ski complex by mutating conserved residues in the DEVH helicase motif of Ski2 did not abrogate its interaction with Ski7p, indicating that Ski2p function is not necessary for this interaction.
Keywords: mRNA degradation, SKI2, RNA helicase, yeast
INTRODUCTION
mRNA degradation is an important mechanism for regulating gene expression in eukaryotic cells. Two general cytoplasmic mRNA degradation pathways have been described so far in yeast (Decker and Parker 1994; Beelman and Parker 1995; Caponigro and Parker 1996; Jacobs Anderson and Parker 1996; Mitchell and Tollervey 2000a). The primary pathway is initiated by deadenylation followed by decapping and subsequent 5′-to-3′ degradation by Xrn1p. A second but less robust deadenylation-dependent pathway uses the 3′-exonuclease activity of the exosome, as well as the DEVH box ATPase Ski2p in complex with Ski3p and Ski8p (the Ski2/3/8 complex). Although the relative contributions of these two pathways to mRNA decay in higher eukaryotes are not well characterized, in vitro and in vivo analyses suggest that 3′-decay is the predominant pathway in human cells (Wang and Kiledjian 2001; Mukherjee et al. 2002). This pathway also requires the GTPase Ski7p (Benard et al. 1999; van Hoof et al. 2000; Araki et al. 2001). We have shown previously that Ski2p, Ski3p, and Ski8p exist as a stable complex in the cytoplasm (Brown et al. 2000). Ski7p is a multidomain protein and appears to bridge the interaction between the exosome and the Ski2/3/8 complex (Araki et al. 2001), although a stable complex containing both the Ski2/3/8 complex and the exosome has not been observed (Brown et al. 2000; Araki et al. 2001).
The SKI (superkiller) genes were first identified from mutations that led to the increased expression of the endogenous double-strand RNA “killer” virus in yeast (Tohe et al. 1978; Ridley et al. 1984). Thus, the SKI genes can act as an antiviral system. However, even in the absence of dsRNA “killer” virus, mutations in the SKI genes are synthetic lethal with loss of XRN1, encoding a cytoplasmic exoribonuclease (Hsu and Stevens 1993), suggesting that the SKI genes have a general function in regulating gene expression (Johnson and Kolodner 1995). Indeed, Ski4p and Ski6p are components of the exosome that, along with Ski2p, Ski3p, Ski7p, and Ski8p, are required for 3′-RNA degradation in the cytoplasm (Jacobs Anderson and Parker 1998). Defects in the SKI genes lead to increased translation of poly(A) minus mRNAs (Benard et al. 1998, 1999). Recently, our lab has shown that the effect of ski mutants to enhance translation of poly(A) minus mRNA (Widner and Wickner 1993) can be phenocopied for a given mRNA by the inclusion of RNA elements in cis within the 3′-UTR of the transcript that inhibit 3′-decay (Brown and Johnson 2001). Thus, the effect of ski mutations on poly(A) minus mRNA is likely due to defects in 3′-decay rather than a direct effect on the translation machinery itself (Widner and Wickner 1993; Benard et al. 1998, 1999).
More recently, it has been shown that the 3′-decay pathway also acts on “nonstop transcripts” lacking a stop codon (Frischmeyer et al. 2002; van Hoof et al. 2002). It has been proposed that the EF1 α-like GTPase domain of Ski7p recognizes ribosomes stalled at the end of a transcript without a stop codon (van Hoof et al. 2002). Ski7p would then recruit the exosome and the Ski complex to degrade the transcript. Furthermore, the Ski complex also acts on transcripts containing premature termination codons (Mitchell and Tollervey 2003; Takahashi et al. 2003). In this case as well, recruitment of the 3′-decay factors appears to be mediated by Ski7p, although here it is through interaction with the nonsense decay factor Upf1p (Takahashi et al. 2003).
We previously showed that Ski2/3/8 exists as a stable cytoplasmic complex but how these proteins interact with each other and with other components of the 3′-decay pathway is poorly understood. In this study, we have used a directed two-hybrid approach together with coimmunoprecipitation of proteins expressed in vivo to investigate the domain interactions within Ski2/3/8 complex, and between the Ski2/3/8 complex and Ski7p.
RESULTS
Domain interactions within ski2/3/8 identified by directed two hybrid
We showed previously that efficient Ski2p interaction with Ski8p depends on the presence of Ski3p; however, Ski3p and Ski8p interacted in the absence of Ski2p (Brown et al. 2000). These results suggested that Ski3p provides the scaffold for the interaction of Ski8p and Ski2p. To further examine the protein interactions within the Ski2/3/8 complex, we used a directed two-hybrid approach to identify the protein interaction domains of Ski2p and Ski3p that are important for complex formation.
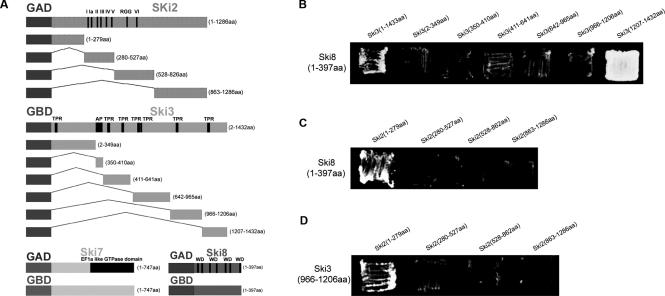
Ski2p is a putative ATPase of the DEVH family within helicase superfamily 2 (Widner and Wickner 1993). It contains seven motifs which are conserved among the DEVH helicase family, as well as an arginine–glycine-rich (RGG) box that may be important for RNA binding. Ski2p is a 146-kDa cytoplasmic protein in yeast, and homologs are found in mammalian cells, suggesting a conserved function in mammalian cells as well. In yeast, Mtr4p is a closely related nuclear protein thought to act as the adapter for the nuclear exosome (Jacobs Anderson and Parker 1996; Mitchell and Tollervey 2000b). Ski3p is a 164-kDa protein that contains seven TPR (Tetratricopeptide Repeat) motifs (Rhee et al. 1989), commonly found in proteins within large complexes and believed to be important for protein–protein interaction (Goebl and Yanagida 1991). Ski8p is a 44-kDa protein whose structure reveals seven WD repeats that are commonly involved in protein–protein interactions as well (Matsumoto et al. 1993; Cheng et al. 2004; Madrona and Wilson 2004). Ski7p is an 85-kDa protein that contains an elongation factor 1 α-like GTPase domain at its C terminus (Benard et al. 1999). A cartoon of the primary structures of Ski2p, Ski3p, Ski8p, and Ski7p is shown in Figure 1A.
FIGURE 1.
Domain interactions within Ski2/3/8 complex identified by a directed two-hybrid approach. (A) Primary structures of Ski2p, Ski3p, Ski8p, and Ski7p as well as all constructs made for directed two-hybrid analysis. Lengths of the proteins are given to the right of each diagram. The motifs in Ski2p characteristic of the DEVH family of ATPase/RNA helicases are indicated by Roman numerals. I, II (DEVH box) are ATPase motifs; III, helicase; and VI, RNA binding. (RGG) Arginine–glycine-rich RNA-binding motif; (TPR) tetratricopeptide repeat; (WD) β-propeller repeat; and (AP) acidic patch. Pairwise interaction was determined by transforming yeast strain PJ69–4A with different GAD (activation domain) and GBD (binding domain) plasmids. (B) pAJ964 (GAD-SKI8) together with different GBD plasmids containing different fragments of SKI3. (C) pAJ965 (GBD-SKI8) with different GAD plasmids containing different fragments of SKI2. (D) pAJ951 (GAD-SKI2) together with different GBD plasmids containing different fragments of SKI3. Transformants were patched onto trp+ leu+ his+ triple-dropout plates supplemented with 6 mM 3-AT and incubated at 30°C.
A series of SKI3 “bait” plasmids was made by fusing the _GAL4_-binding domain to six regions of SKI3 spanning the entire protein. The boundaries of these regions were selected based on apparent boundaries of domains identified from secondary structure prediction and hydrophilicity plots. Among the six Ski3p fragments used as bait proteins, all contained at least one TPR domain with the exception of the fusion spanning amino acids 350 to 410, which encompassed a highly acidic patch. A set of SKI2 fusions was made in a similar fashion. The N-terminal (amino acids 1–279) and C-terminal (amino acids 863–1286) fragments of Ski2p correspond to the N-terminal and C-terminal extensions, beyond the central ATPase and helicase domains, that are suggested to be important for protein–protein interaction and/or subcellular localization (de la Cruz et al. 1999). The second Ski2p fusion (amino acids 280–527) encompassed motifs I through V, including the ATPase motifs (I, II) and helicase motif (III) that are responsible for enzymatic activity. The third Ski2p fusion (amino acids 528–862) contained the RGG-box and the RNA-binding domain VI, both of which are likely important for substrate recognition. Fusions of intact Ski2p, Ski3p, Ski7p, and Ski8p were made as well (see Fig. 1A for a summary of the constructs). All “bait” and “prey” constructs were tested for pairwise interactions in a strain developed for yeast two hybrid. Figure 1, B–D, shows a subset of the interactions tested. All pairwise combinations not shown did not reveal interactions.
Strong interaction was observed between the C terminus (amino acids 1206~1432) of Ski3p and Ski8p (Fig. 1B). Weak interactions were observed between the N terminus of Ski2p (amino acids 1–279) and Ski8p (Fig. 1C), and the N terminus of Ski2p (amino acids 1–279) and the sub-C terminus (amino acids 966–1206) of Ski3p (Fig. 1D).
We did not observe two-hybrid interactions between Ski7p and the individual Ski2p, Ski3p, or Ski8p proteins, which may indicate that Ski7p interacts with the proteins of the Ski2/3/8 complex only when they form a complex. Alternatively, tagging Ski7p with GAD or GBD at its N terminus may inactivate the protein or reduce its interaction with the Ski2/3/8 complex. Consistent with this idea, the N terminus of Ski7p (amino acids 1–80) has been shown to be important for Ski2/3/8 complex interaction (Takahashi et al. 2003). We also did not observe interactions with full-length SKI2, possibly because of low expression or low nuclear levels of the fusion protein.
Confirmation of two-hybrid interactions within Ski2/3/8 by immunoprecipitation
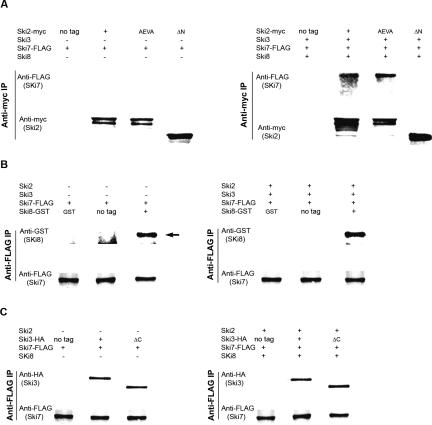
To verify the protein–protein interaction information obtained from two hybrid, we carried out immunoprecipitation (IP) experiments. We made functional epitope-tagged Ski2, Ski3, Ski7, and Ski8 proteins. We also made partial deletions of SKI2 (ski2 Δ_N_) by deleting the N-terminal putative Ski3p interaction domain (amino acids 1~279) and SKI3 (ski3- Δ_C_) that removed the C terminus (amino acids 1206~1432) of Ski3p, which, according to the two-hybrid result, is responsible for Ski8p binding. These mutant proteins were expressed pairwise in wild-type cells with epitope-tagged wild-type copies of each Ski protein. All proteins were assayed for interaction by immunoprecipitation of the bait protein and by Western blotting for the presence of the second protein.
As the two-hybrid analysis was carried out in a strain wild type for the SKI genes, interactions that were identified could be direct or bridged by the endogenous wild-type proteins. Consequently, coimmunoprecipitations were also done in a yeast strain containing simultaneous deletions of SKI2, SKI3, SKI7, and SKI8, in which bridging interactions between Ski proteins would be eliminated. Because these genes all act in the same nonessential cytoplasmic pathway, the quadruple mutant is viable and has no noticeable growth defect. For domain–domain interactions within the Ski2/3/8 complex, pairwise IPs with or without the presence of the third protein in this complex were carried out in this quadruple deletion strain.
The N terminus of Ski2p (amino acids 1–279) is required and sufficient for Ski3p interaction
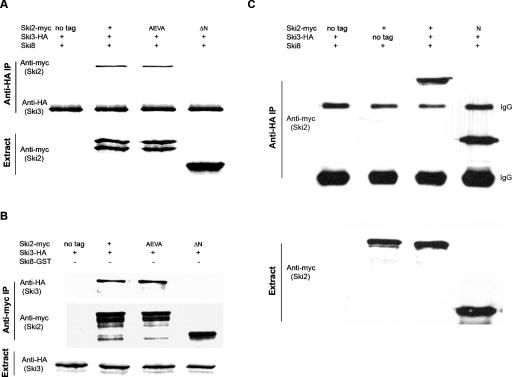
We first examined the interaction between Ski2p and Ski3p to follow up the two-hybrid interaction between the N terminus of Ski2p (amino acids 1–279) and the sub-C terminus of Ski3p (amino acids 966–1206).
A strain with SKI3 genomically tagged with the hemagglutinin (HA) epitope was transformed individually with nontagged SKI2, myc-tagged SKI2 (SKI2-myc), or _ski2-Δ_N-myc. We also introduced a SKI2-myc ATPase mutant (DEVH → AEVA), in which the canonical DEVH has been changed to AEVA. Cell extracts were prepared and IPs were performed with anti-HA antibody, followed by SDS PAGE and Western blotting using anti-myc antibody. As shown in Figure 2A, Ski2-myc was specifically associated with Ski3-HA, while no signal was observed when immunoprecipitations were performed with untagged Ski2p or with the N-terminal deletion of Ski2p (deleted of amino acids 1–279). These data suggest that the N terminus of Ski2p is necessary for interacting with Ski3p, which is consistent with the direct two-hybrid result. Interestingly, the DEVH → AEVA mutation of Ski2p, which inactivates the protein in vivo (see below) and destroys the ATPase activity of Ski2p in vitro (L. Wang and A. Johnson, unpubl.), immunoprecipitated Ski3p as efficiently as did wild-type Ski2p. This result indicates that the ATPase activity of Ski2p is not required for its association with Ski3p. Similar results were also observed when using anti-myc antibody for IP and anti-HA for Western blotting (data not shown).
FIGURE 2.
The N terminus of Ski2p (amino acids 1–279) is required and sufficient for Ski3p interaction. (A) Extracts from strain AJY245 (SKI3–3xHA) carrying pAJ39 (SKI2), pAJ159 (SKI2–3xmyc), pAJ819 [SKI2 (DEVH → _AEVA)-3xmyc_], or pAJ832 (ski2_Δ_N-3xmyc), respectively, were immunoprecipitated with anti-HA antibody followed by Western blotting with anti-myc antibody. (B) Immunoprecipitations were performed on extracts from AJY2074 (quadruple mutant) carrying pAJ703 (GAL-SKI2), pAJ705 (GAL-SKI2–3xmyc), pAJ820 [GAL-SKI2 (DEVH → _AEVA)-3xmyc_], or pAJ832 together with pAJ709 (GAL-SKI3–3xHA) with anti-myc antibody. Western blots were done using anti-HA antibody. (C) Anti-HA IPs were performed on extracts from AJY2052 transformed with different combinations of SKI2 (pAJ703, pAJ705, or pAJ1241) and SKI3 (pAJ270 or pAJ709), followed by Western blotting against myc.
We next asked whether the Ski2p and Ski3p interaction that we observed through two hybrid and the above IP was direct or was bridged by Ski8p. To answer this, we carried out the IP experiments in the quadruple-mutant strain. Nontagged SKI2, SKI2-myc, ski2 (DEVH → AEVA)-myc, or ski2_Δ_N-myc was transformed individually into the quadruple-mutant strain together with SKI3-HA. Cell extracts were prepared and IPs were carried out using anti-myc antibody; Western blots were performed using anti-HA antibody. Without the presence of Ski8p, both full-length Ski2p and the AEVA mutant of Ski2p could pull down Ski3p, while no Ski3p was detected with non-tagged Ski2p and the N-terminal deletion of Ski2p (Fig. 2B). Similar results were obtained when using anti-HA antibody for IP and anti-myc for Western blotting (data not shown).
Next, we tested if the N terminus of Ski2p by itself was sufficient for interaction with Ski3p. Different combinations of SKI2 together with SKI3 were transformed into a ski2Δski3Δ double-mutant strain, and anti-HA IP was performed on the extracts. In Figure 2C, the N terminus of ski2p by itself was sufficient to coimmunoprecipitate with Ski3p, which is consistent with our two-hybrid data and strongly suggests that Ski2p interacts with Ski3p via its N terminus (amino acids 1–279).
Taken together, these data argue that Ski2p and Ski3p interact via the N terminus (amino acids 1–279) of Ski2p independent of Ski8p and that the N terminus of Ski2p is sufficient for interaction with Ski3p. In addition, active Ski2p is not required for this interaction.
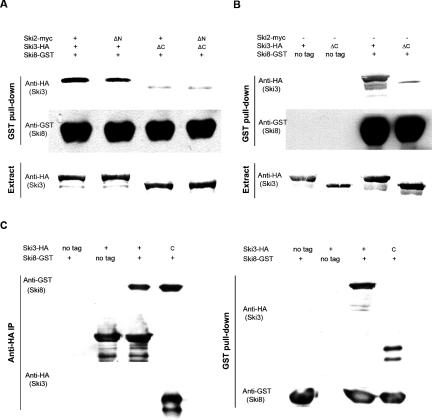
The C terminus of Ski3p (amino acids 1206–1432) is important and sufficient for efficient Ski8p interaction
The strong two-hybrid interaction between the C terminus (amino acids 1206–1432) of Ski3p and Ski8p was tested by immunoprecipitation. Different combinations of SKI2 and SKI3 together with GST-SKI8 and a wild-type SKI7 were transformed into the quadruple-mutant strain. Glutathione agarose beads were added to the cell extracts to pull down GST-Ski8, and anti-HA Western blotting was carried out to check the copurification of Ski3p. GST-Ski8 was specifically associated with full-length Ski3-HA, no matter whether full-length Ski2p or the N-terminal deletion of Ski2p was present (Fig. 3A). The C-terminal deletion of Ski3p did not efficiently interact with GST-Ski8 as the Ski3ΔC-HA signal was significantly reduced. These data support the two-hybrid observation that the C terminus of Ski3p is responsible for Ski8p interaction. Moreover, full-length Ski3p can interact with Ski8p in the presence of Ski2ΔN (which has presumably lost interaction with Ski3p and Ski8p), suggesting that the Ski3p and Ski8p interaction is direct and independent of Ski2p.
FIGURE 3.
The C terminus of Ski3p (amino acids 1206–1432) is important and sufficient for Ski8p interaction. (A) Extracts from strain AJY2074 carrying different combinations of SKI2 (pAJ705 or pAJ832), SKI3 [pAJ709 or pAJ833 (ski3_Δ_C-3xHA)] together with pAJ706 (GST-SKI8) were prepared. Glutathione agarose beads were added to the cell extracts to pull down GST-Ski8, and anti-HA Western blotting was carried out to monitor the co-sedimentation of Ski3p. (B) GST pull-downs were performed on extracts from AJY2074 transformed with different combinations of SKI8 [pAJ267 (SKI8) or pAJ706] and SKI3 (pAJ709 or pAJ833), followed by Western blotting against HA. (C) Anti-HA IPs (left) or GST pull-downs (right) were performed on extracts from AJY2045 transformed with different combinations of SKI8 (pAJ267 or pAJ706) and SKI3 (pAJ709 or pAJ1238) plasmids. Western blotting was done using anti-GST or anti-HA.
To further verify that the interaction between the C terminus of Ski3p and Ski8p was direct, we repeated the co-IPs in the quadruple-mutant strain in the absence of Ski2p. As seen in Figure 3B, without Ski2p, GST-Ski8 was specifically immunoprecipitated with full-length Ski3-HA. This interaction was lost with the C-terminal truncation of Ski3p.
As we observed a very strong two-hybrid interaction between the C terminus of Ski3p and Ski8p, we tested if the C terminus of Ski3 by itself was sufficient for interaction with Ski8p. Different combinations of SKI3 together with GST-SKI8 were transformed into a ski3Δski8Δ double-mutant strain, and a GST-pull-down or anti-HA IP was performed on the extracts. We found that the C terminus of Ski3p coimmunoprecipitated with Ski8p, and that Ski8p can also pull down the C terminus of Ski3p (Fig. 3C). These data indicate that Ski3p can interact with Ski8p via its C terminus independently of Ski2p. The C terminus of Ski3p is sufficient for interaction with Ski8p. Subsequent subcloning of the C terminus of SKI3 indicates that the extreme 123 amino acids are sufficient for strong two-hybrid interaction with Ski8p (data not shown).
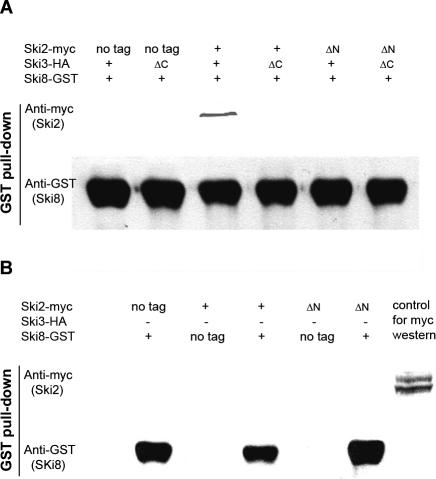
The N terminus of Ski2p (amino acids 1–279) is required for interaction with Ski8p, and this interaction is mediated by Ski3p
We next examined the Ski2p and Ski8p interaction via IP as the N terminus of Ski2p (amino acids 1–279) and Ski8p showed weak interaction through the directed two-hybrid approach. The quadruple-mutant strain was transformed with different combinations of SKI2 and SKI3 constructs and together with GST-SKI8 expression vector individually (Fig. 4A). Full-length Ski2-myc associated specifically with GST-Ski8 only in the presence of full-length Ski3p, while no significant Ski2ΔN-myc signal was observed even in the presence of full-length Ski3p (Fig. 4A). Surprisingly, Ski2p and Ski8p interaction was disrupted when the C terminus of Ski3p was removed. As the C terminus of Ski3p interacts with Ski8p, these data suggest that the Ski2p and Ski8p interaction observed by two hybrid and by IP was bridged by Ski3p.
FIGURE 4.
The interaction of the N terminus of Ski2p (amino acids 1–279) with Ski8p is mediated by Ski3p. (A) Extracts from strain AJY2074 carrying different combinations of SKI2 (pAJ703, pAJ705, or pAJ832), SKI3 (pAJ709 or pAJ833) together with pAJ706 were prepared. GST pull-downs were performed, followed by Western blotting against myc. (B) GST pull-downs were performed on extracts from AJY2074 transformed with different combinations of SKI8 (pAJ267 or pAJ706) and Ski2p (pAJ703, pAJ705, or pAJ832), followed by Western blotting against myc.
To test the requirement for Ski3p, IPs were carried out in the quadruple mutant but in the absence of Ski3p. As expected, Ski2p and Ski8p interaction was totally abolished in the absence of Ski3p (Fig. 4B). Together, these results suggest that Ski2p and Ski8p interaction is bridged by Ski3p, and that the N terminus (amino acids 1–279) of Ski2p is responsible for Ski3p interaction and thus required for Ski2p and Ski8p interaction.
Interactions between Ski2/3/8 and Ski7p
By a directed two-hybrid approach we did not identify any interactions between the individual components of the Ski2/ 3/8 complex and Ski7p. This would be explained if stable Ski7p interaction requires Ski2/3/8 complex formation and would be consistent with work by Araki et al. (2001) showing that Ski2p interacted with Ski7p in the presence of wild-type Ski3p and Ski8p, but not in the absence of Ski8p. Thus, the interaction of Ski7p with Ski2/3/8 complex may require complex formation or it could be mediated by Ski8p or Ski3p. To discriminate between these possibilities, we carried out immunoprecipitation experiments to test the interaction of Ski7p with Ski2p, Ski3p, or Ski8p individually in the presence or the absence of the other two proteins. If Ski7p interaction requires Ski2/3/8 complex formation, then we would not see interaction of Ski7p with any component of the Ski2/3/8 complex without the presence of the other two Ski proteins of this complex. If Ski8p is mediating the interaction of Ski7p with Ski2p, then we would expect to see direct interaction between Ski7p and Ski8p even in the absence of Ski2p and Ski3p.
Ski2p and Ski7p interact in the presence of Ski3p and Ski8p
Nontagged SKI2, SKI2-myc, ski2 (DEVH → AEVA)-myc, or ski2_Δ_N-myc was each cotransformed with Flag-SKI7 into the quadruple deletion mutant or a _ski2_Δ_ski7_Δ double-mutant strain. Immunoprecipitations using anti-myc antibody were performed, followed by Western blotting using anti-Flag antibody to detect Ski7p.
No specific Flag-Ski7 signal was observed when immunoprecipitations were performed in the absence of Ski3p and Ski8p (Fig. 5A, left panel). Full-length Ski2-myc coimmunoprecipitated with Flag-Ski7 in the _ski2_Δ_ski7_Δ double-mutant strain (expressing Ski3p and Ski8p), while the N-terminal deletion of Ski2p (which has lost the ability to interact with Ski3p and thus Ski8p) could not (Fig. 5A, right panel). Interestingly, the ski2 (DEVH → AEVA) mutant protein also coimmunoprecipitated Ski7p, indicating that Ski2p activity is not required for its interaction with Ski7p. These data suggest that Ski2p and Ski7p interact indirectly, probably mediated by either Ski3p or Ski8p or both via the N terminus of Ski2p. To identify which protein is mediating Ski2p and Ski7p interaction, we carried out similar IPs for Ski3p, Ski7p and also Ski7p, Ski8p.
FIGURE 5.
Interactions between Ski2/3/8 and Ski7p identified by immunoprecipitation. (A) Ski2p and Ski7p coimmunoprecipitation requires the N terminus of Ski2p (amino acids 1–279) and both Ski3p and Ski8p. Extracts from strain AJY2074 carrying pAJ703, pAJ705, pAJ820, or pAJ832 together with pAJ1206 (Flag-SKI7) were immunoprecipitated with anti-myc antibody, followed by Western blotting with anti-Flag antibody (left). The same immunoprecipitations were carried out in AJY2062 (right). (B) Ski8p and Ski7p can interact in the absence of Ski2p and Ski3p. Extracts from strain AJY2074 carrying pAJ689, pAJ267, or pAJ706 together with pAJ1206 were immunoprecipitated with anti-Flag antibody, followed by Western blotting with anti-GST antibody (left). GST-Ski8p is indicated by an arrow. The same immunoprecipitations were carried out in AJY2055 (right). (C) Ski3p and Ski7p can interact in the absence of Ski2p and Ski8p. Extracts from strain AJY2074 carrying pAJ270 (SKI3), pAJ709, or pAJ833 together with pAJ1206 were immunoprecipitated with anti-Flag antibody, followed by Western blotting with anti-HA antibody (left). The same immunoprecipitations were carried out in AJY2057 (right).
Ski8p and Ski7p can interact directly
GST alone, nontagged SKI8, or GST-SKI8 was transformed together with Flag-SKI7 into the quadruple-mutant strain or a _ski8_Δ_ski7_Δ double-mutant strain, and IPs were carried out using anti-Flag antibody. GST-Ski8 bound Flag-Ski7 with or without the presence of Ski2p and Ski3p (Fig. 5B), indicating that Ski8p and Ski7p can interact with each other directly. Similar results were also observed using glutathione beads to bind Ski8p (data not shown).
Ski3p and Ski7p can interact directly
We next looked at Ski3p and Ski7p interaction in the presence or absence of Ski2p and Ski8p. Nontagged SKI3, SKI3-HA, or ski3_Δ_C-HA was transformed together with Flag-SKI7 into the quadruple-mutant strain or a _ski3_Δ_ski7_Δ double-mutant strain. IPs using anti-Flag antibody were performed, followed by Western blotting to detect Ski3-HA. Both full-length Ski3-HA and Ski3ΔC-HA associated with Flag-Ski7 with or without Ski2p and Ski8p (Fig. 5C, right and left panels), indicating that Ski3p and Ski7p can interact with each other directly. The C terminus (amino acids 1206~1432) of Ski3p is not important for interaction with Ski7p. Similar results were also observed using anti-HA antibody to pull down Ski3-HA followed by Western blotting to detect Flag-Ski7 (data not shown).
Together, these observations suggest that both Ski3p and Ski8p interact directly with Ski7p, but that Ski7p interaction with Ski2p requires both Ski3p and Ski8p. Furthermore, active Ski2p is not required for Ski7p interaction with the Ski2/3/8 complex.
Defining domain(s) in Ski3 that are responsible for Ski7p interaction
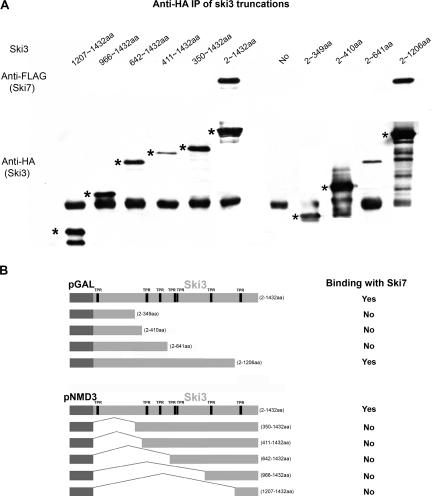
We have shown that different domains of Ski3p are responsible for interacting with Ski2p and Ski8p separately. We were also interested in finding the domains in Ski3p that are responsible for Ski7p interaction. Two series of Ski3p truncations starting from the N terminus or the C terminus were made (Fig. 6B) under control of the GAL10 or NMD3 promoters. The interaction between each truncation with Ski7p was tested in a ski3Δ ski7Δ double-deletion strain.
FIGURE 6.
Domains in Ski3p that are responsible for Ski7p interaction. (A) Extracts from strain AJY2057 carrying pAJ1201 together with different fragments of Ski3 [pAJ1224 (amino acids 1207–1432), pAJ1225 (amino acids 966–1432), pAJ1226 (amino acids 642–1432), pAJ1227 (amino acids 411–1432), pAJ1228 (amino acids 350–1432), pAJ709 (amino acids 2–1432), pAJ1233 (amino acids 2–349), pAJ1234 (amino acids 2–410), pAJ1235 (amino acids 2–641), pAJ833 (amino acids 2–1206), or an empty vector] were immunoprecipitated with anti-HA antibody, followed by Western blotting with anti-Flag antibody. Asterisks denote the Ski3p fragments. (B) Diagram of the different Ski3p constructs and their ability to interact with Ski7p.
In Figure 6A, only full length but not the N-terminal truncations of Ski3p was able to coimmunoprecipitate Ski7p, which suggests that the N terminus of Ski3p (amino acids 2–349) is required for Ski7p interaction. In addition, the region of Ski3p from amino acids 641 to 1206 of Ski3p is also important for interaction with Ski7p, as a Ski3p truncation containing 2–1206 amino acids was able to pull down Ski7p, while further truncations were not. Taken together, multiple domains in Ski3p appear to be responsible for Ski7p interaction, including the N terminus of Ski3p (amino acids 2–349) and amino acids 641–1206. This is consistent with our directed two-hybrid data, as we did not observe interactions between Ski7p and the individual regions of Ski3p.
Ski2-ΔN, ski2 (DEVH fi AEVA) and Ski3-ΔC are nonfunctional
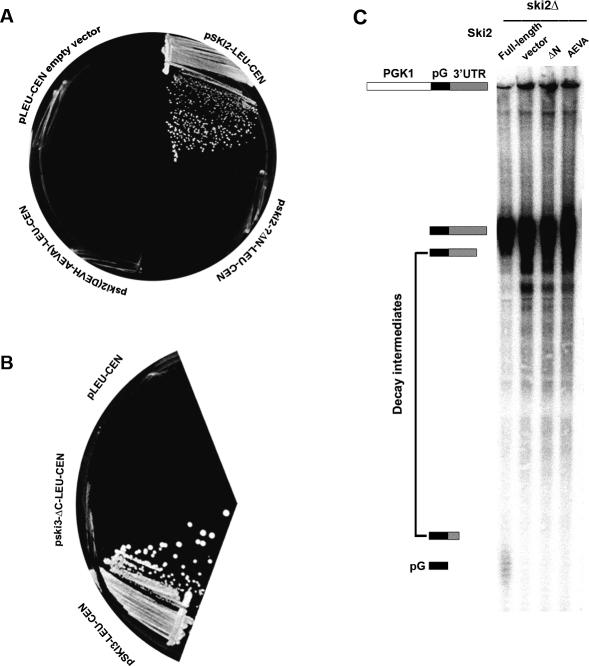
As the interactions we identified from the directed two-hybrid approach were confirmed by Co-IPs, we next tested the functional consequence of deleting these interacting domains. Plasmid-shuffling experiments were performed to determine whether SKI2 and SKI3 deletion mutants can functionally substitute for the corresponding wild-type proteins in vivo. Deletion of SKI2, SKI3, or SKI8 by themselves blocks 3′-decay but confers no obvious growth defect on yeast. However, these deletions are lethal when combined with a deletion of XRN1, encoding the 5′-exoribonuclease responsible for most cytoplasmic RNA degradation. Consequently, the function of a given mutation can be assessed by its ability to support growth of a double mutant with _xrn1_Δ. Yeast strain RDKY2055 (_ski2_Δ _xrn1_Δ), containing XRN1 on a centromeric URA3 plasmid, was transformed individually with pAJ832 (_ski2_-Δ_N_-LEU2-CEN), pAJ819 [ski2 (DEVH → _AEVA)-LEU2-CEN_], pAJ159 (SKI2-LEU2-CEN), or pAJ24 (LEU2-CEN). Leu+ transformants were selected on appropriate plates and then restreaked on a 5-FOA plate to select for the loss of the XRN1-URA3 plasmid.
Cells transformed with control wild-type SKI2 grew on 5-FOA as expected (Fig. 7A). Cells transformed with _SKI2-Δ_N-LEU-CEN, SKI2 (DEVH → AEVA)-LEU-CEN, or an empty LEU2 vector (pRS315) as a negative control, failed to grow on 5-FOA. The functional defect of the mutant Ski2 proteins was not due to reduced expression of the mutant protein (data not shown). Similarly, plasmid shuffle was carried out to check the function of SKI3 mutants in a _ski3_Δ _xrn1_Δ double-mutant strain. Cells transformed with wild-type SKI3 grew on 5-FOA, while cells transformed with _ski3-Δ_C and empty vector failed to grow (Fig. 7B). As Ski2-ΔN is deficient for interaction with Ski3p and thus Ski8p, and Ski3-ΔC has lost interaction with Ski8p, these results suggest that the assembly of Sk2p, Ski3p, and Ski8p into a complex is required for their function.
FIGURE 7.
Ski2-ΔN and Ski3-ΔC are nonfunctional. (A) Yeast strain RDKY2055 (ski2_Δ xrn1_Δ/pXRN1-URA3) was transformed individually with pAJ832 (ski2_Δ_N), pAJ819 (ski2 AEVA), pAJ159 (ski2), or pRS315 (vector). Leu+ transformants were selected and restreaked on a 5′-FOA. (B) Yeast strain RDKY2067 (ski3_Δ xrn1_Δ/pXRN1-URA3) was transformed individually with pAJ840 (_ski3-Δ_C), pAJ263 (SKI3), or pRS315 (vector). Leu+ transformants were streaked onto 5′-FOA to select against the XRN1-URA3 plasmid. (C) Yeast strain AJY1478 was transformed with pAJ832, pAJ159, pAJ819, or pRS315 together with pRP469, expressing PGK1 with an oligo(G) tract in its 3′-UTR. Transformants were grown at 30°C to mid-log phase (OD600 = 0.5) in galactose-containing medium. RNA was extracted and subjected to Northern blotting using oligonucleotide oRP140, specific for the oligo(G) insert.
Ski2-ΔN, ski2(DEVH fi AEVA) and Ski3-ΔC mutants are defective in 3′–5′ mRNA degradation
To confirm that the loss of function reflects a defect in 3′-degradation, we examined the effects of these mutants on 3′-decay in vivo using a published procedure (Jacobs Anderson and Parker 1998). This assay examines the fate of a 3′-terminal fragment of a transcript resulting from inhibition of the 5′-decay pathway with a short (18 nt) oligo(G) tract. PGK1, containing oligo(G) in its 3′-untranslated region, was expressed under control of a galactose-inducible and glucose-repressible promoter. Transcription of the reporter is induced by growing cells in the presence of galactose and is abruptly shut off by the addition of glucose. In mutants defective for 3′-decay, degradation intermediates extending from the oligo(G) to the 3′-end of the transcript accumulate and can be detected by Northern blotting. Loss of function of the SKI genes will lead to accumulation of these intermediates.
The ski2-ΔN and ski2(DEVH → AEVA) mutants accumulated a higher level than wild-type cells of the poly(G) → 3′-end fragment from the PGK1pG transcript (Fig. 7C). In addition, several smaller RNA species also accumulated. Notably, the oligo(G) tract, degraded from both the 5′- and 3′-ends, was apparent only in wild-type cells. Similar results were obtained with ski3_-Δ_C (data not shown). These results indicate that Ski2/3/8 complex formation is required for efficient 3′-to-5′ degradation of mRNAs.
DISCUSSION
In this study, we have investigated the domain–domain interactions within the Ski2/3/8 complex and the interaction between the Ski complex and Ski7p. A model summarizing these interactions is shown in Figure 8. In this model, the N terminus (amino acids 1–279) of Ski2p is responsible for interaction with the sub-C terminus (amino acids 966–1206) of Ski3p, and the C terminus of Ski3p (amino acids 1207–1433) interacts with Ski8p. Ski2p and Ski8p do not interact directly, but, rather, their interaction is bridged by Ski3p. Ski7p interacts with both Ski3p and Ski8p, and the interaction between Ski2p and Ski7p is mediated by Ski3p and Ski8p. Although these experiments were carried out in cell extracts, it is likely that these interactions among Ski2p, Ski3p, and Ski8p are direct because co-overexpression of these proteins allows for the purification of a complex that sediments at ~370–380 kDa on glycerol gradients (see Supplemental Material at (http://www.utexas.edu/research/rnadecay/Supplement.pdf) and does not contain other proteins at stoichiometric levels (L. Wang and A. Johnson, unpubl.). This interaction model is consistent with our previous work showing that Ski3p and Ski8p can interact independently of Ski2p (Brown et al. 2000) and with that of Araki et al. (2001) showing that Ski2p interaction with Ski7p requires either Ski3p or Ski8p. However, this arrangement of proteins is different from and not compatible with that in a recent prediction of the oligomerization of the proteins within the Ski complex (Aloy et al. 2004). In that work, Ski2p was suggested to bridge the interaction of Ski3p and Ski8p, with no obvious direct contacts between Ski3p and Ski8p. Because the Ski complex was not available for electron microscopy, this modeling was based entirely on predicted protein structures and pairwise interactions of apparently related proteins with known structures. A cryoEM structure combined with the protein contact information presented here should be useful for improved modeling of this complex. More recently, the crystal structure of yeast Ski8p has been solved by two groups (Cheng et al. 2004; Madrona and Wilson 2004). Mutational analysis suggests that a hydrophobic patch on the top surface of the β-propeller of Ski8p is involved in binding Ski3p (Cheng et al. 2004). Based on our deletion analysis, a corresponding hydrophobic loop or surface should be present in the C terminus of Ski3p (amino acids 1207–1432).
FIGURE 8.
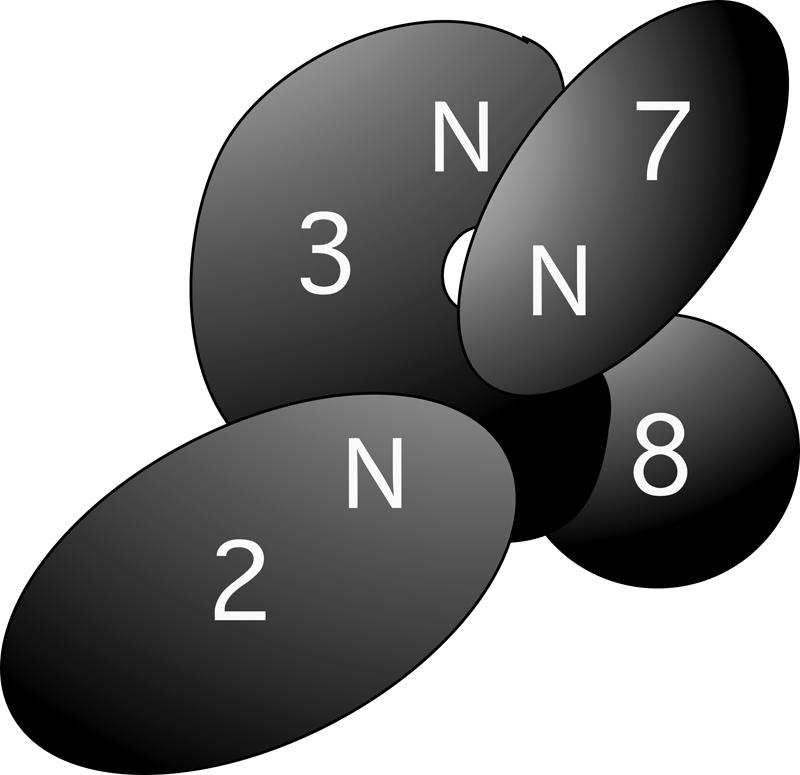
Cartoon of the domain interactions within the Ski2/3/8 complex and the interaction between Ski2/3/8 and Ski7p. See text for description of the model.
The order of the assembly of Ski proteins and the exosome on transcripts targeted for decay
The order of the assembly of the Ski2/3/8 complex, Ski7p, and the exosome on an mRNA has not been established. Three different scenarios can be envisioned. First, the Ski2/3/8 complex may initiate binding to the transcript through its RGG-box and/or motif VI (see Fig. 1A). The Ski2/3/8 complex could then remodel the mRNP to allow access by the exosome. In this scenario, Ski7p would serve simply as a bridging factor. Alternatively, the exosome could bind a deadenylated transcript first in an inactive form via the predicted RNA-binding domains presented in Rrp4p, Rrp40p, and Csl4p (Mitchell and Tollervey 2000b). Subsequent association with the Ski2/ 3/8 complex via Ski7p would stimulate the exosome by facilitating disassembly of the mRNP or by allosterically activating the exosome (Mitchell and Tollervey 2000b). In the third model, Ski7p is first recruited to a transcript via its interaction with the ribosome or through interaction with ribosome-associated factors. This idea has gained support from recent work suggesting that the EF1α-like GTPase domain of Ski7p could be recruited to the ribosomal A-site on ribosomes stalled on “non-stop” transcripts lacking a stop codon (van Hoof et al. 2002). Ski7p has also been shown to interact directly with the nonsense-mediated decay factor Upf1p (Takahashi et al. 2003), again suggesting initial recruitment of Ski7p. Since Ski7p binds both the Ski2/3/8 complex and the exosome (Araki et al. 2001), it could then actively recruit the degradation machinery.
Active Ski2p is not required for Ski2/3/8 complex formation and Ski7p association
A point mutation in Ski2p that inactivates the ATPase activity in vitro (L. Wang and A. Johnson, unpubl.) and renders the protein inactive in vivo does not prevent Ski2/3/8 complex formation. This is consistent with the notion that Ski2p, Ski3p, and Ski8p form a stable complex in cells (Brown et al. 2000) and that complex assembly is not a dynamic process dependent on substrate. On the other hand, it is somewhat surprising that inactive Ski2/3/8 complex interacts efficiently with Ski7p. This suggests that the interaction of Ski7p with the Ski2/3/8 complex does not depend on molecular signaling from the Ski2/3/8 complex, indicating the presence of an active helicase complex to recruit additional factors in the 3′-decay pathway. Ski7p is suggested to be the limiting factor in 3′-to-5′ mRNA degradation (Araki et al. 2001). Since the binding of mutant Ski2/3/8 complex to Ski7p was as efficient as that of wild-type complex, the mutant complex may compete with the wild-type Ski2/3/8 complex for the limiting Ski7p, thereby inhibiting for 3′-to-5′ mRNA degradation. We are currently testing this possibility.
MATERIALS AND METHODS
Strains and plasmids
The yeast strains used in this study are described in Table 1, and plasmids are listed in Table 2. AJY2045, AJY2052, AJY2055, AJY2057, AJY2062, and AJY2074 were made by crossing the appropriate single-deletion mutants (Research Genetics). All two-hybrid constructs were made by PCR amplification of the corresponding fragment from the respective genomic locus and cloning into the appropriate two-hybrid vector containing the _GAL4_-binding domain or the GAL4 activation domain. pAJ705 was made by replacing XRN1 in pRDK304 (Page et al. 1998): The 5′-end of SKI2 was amplified by PCR using oligonucleo-tides 5′-CTCGCTCGAGAAAAAATGTCTGAGGGATTCAGTA and 5′-CCAGCACCTCTTGAACCTCCA, digested with XhoI and ligated with an XhoI–XbaI fragment from pAJ159 (Brown et al. 2000) containing the 3′-end of SKI2 into XhoI–NheI-cut pRDK304. pAJ703 was made from pAJ705 by replacing the myc-tag-containing SphI–NdeI fragment with the wild-type sequence from pAJ39 (Brown et al. 2000). pAJ709 was made by three-part ligation: the 3xHA tag as a PstI–BglII fragment from PCR amplification of genomic SKI3:HA from AJY245 (Brown et al. 2000) using oligonucleotides 5′-CGCGAATT CAGGCCTGATCTTTCTCAAGGTATT and 5′-GCGGATC CAAGCTTTGATTGACTATCTCGA; SKI3 as a KpnI–PstI fragment from pAJ270; and vector as a KpnI–BamHI fragment from pAJ270. pAJ270 is a GAL1:SKI3 derivative of pRS424. pAJ819 and pAJ820 were made from pAJ159 and pAJ705, respectively, by changing three nucleotides in the SKI2 ORF (A1331C C1339G A1340C). pAJ832 was made by three-part ligation: the promoter and 5′-portion of SKI2 as an XmaI–ClaI fragment from PCR amplification of pAJ159 using oligonucleotides 5′-AGCGGATAACAATTTCACACAGGA and 5′-GGCATCGATCATTAATTAAGTATTAGTACAGTAAATTT TG; the 3′-portion of SKI2 as a ClaI–NdeI fragment from pAJ159; and vector as an XmaI–NdeI fragment from pAJ159. pAJ833 was made from pAJ709 by replacing the ClaI–XmaI fragment of SKI3 with the ski3_Δ_C fragment as a ClaI–XmaI fragment from PCR amplification of genomic SKI3 using oligonucleotides 5′-CGCGAATTCAGGCCTAAAGACCATTTGG GCC and 5′-CGCGGATCCCCCGGGAGCGTTTTCAATAGAG AG. pAJ689 was made from pEG(KT) by EcoRI partial digestion and fill-in to disrupt LEU2. pAJ706 was made from three parts: GAL UAS and GST on an NcoI–NcoI fragment from pAJ261 (Brown et al. 2000); SKI8 on an NcoI–HindIII fragment from pAJ261; and NcoI–HindIII-cut pRS424. pAJ1201 was made by replacing XRN1 in pRDK304 with Flag-SKI7 as an SalI–HindIII fragment from PCR amplification of genomic SKI7 using oligonucleotides 5′-GCGCCC CGGGTCGACATGGACTACAAAGACGATAAATTAATTAAATCG TTATTAGAGCAATTA and 5′-GCGCGAAGCTTGATATGTGTA AAATGG. pAJ1206 was made by moving GAL-Flag-SKI7 as an SpeI–HindIII fragment from pAJ1201 into the same sites of pRS426. pAJ1224 was made by replacing NMD3 in pAJ409 (J.H. Ho and A. Johnson, unpubl.) with ski3-C-terminus-3XHA as an EcoRI–KpnI fragment from PCR amplification of genomic SKI3:HA from AJY245 using oligonucleotides 5′-CGCGAATTCAGGCCTGAT CTTTCTCAAGGTATT (AJO375) and 5′-CCAGATATCTCGA GGTACCTCAGCACTGAGCAGCGTAATC (AJO748).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| AJY245 | MATa ade2 ade3 leu2-3,112 ura3-52 lys2-801SKI3HA:KanR ski8::KanR | Brown et al. 2000 |
| AJY1478 | MATa leu2-3,112 ura3-52 his3-200 ski2::KanR | Research Genetics |
| AJY2045 | MATa trp1-901 leu2-3,112 ura3-52 his3-200 ski3::KanR ski8::KanR | This work |
| AJY2052 | MATa trp1-901 leu2-3,112 ura3-52 his3-200 ski2::KanR ski3::KanR | This work |
| AJY2055 | MATa trp1-901 leu2-3,112 ura3-52 his3-200 ski7::KanR ski8::KanR | This work |
| AJY2057 | MATa trp1-901 leu2-3,112 ura3-52 his3-200 ski3::KanR ski7::KanR | This work |
| AJY2062 | MATa trp1-901 leu2-3,112 ura3-52 his3-200 ski2::KanR ski7::KanR | This work |
| AJY2074 (quadruple) | MAT3 trp1-901 leu2-3,112 ura3-52 his3-200 ski2::KanR ski3::KanR ski7::KanR ski8::KanR | This work |
| PJ69-4A | _MAT_α trp1-901 leu2-3,112 ura3-52 his3-200 gal4_Δ_gal80_Δ_LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. 1996 |
| RDKY2055 | MATa ade2 ade3 leu2 lys2-801 ura3-52 xrn1_Δ_ski2-2-18/pRDK297 | Johnson and Kolodner 1995 |
| RDKY2067 | MATa ade2 ade3 leu2 lys2-801 ura3-52 xrn1_Δ_ski3-3-21/pRDK252 | Johnson and Kolodner 1995 |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pAJ39 | SKI2-2_μ_-LEU2 | Brown et al. 2000 |
| pAJ159 | SKI2-3xmyc-2_μ_-LEU2 | Brown et al. 2000 |
| pAJ263 | SKI3-LEU-CEN | This study |
| pAJ267 | GAL-SKI8-2_μ_-URA3 | Brown et al. 2000 |
| pAJ270 | GAL-SKI3-2_μ_-TRP1 | This study |
| pAJ689 | GAL-GST-2_μ_-URA3 | This study |
| pAJ703 | GAL-SKI2-2_μ_-LEU2 | This study |
| pAJ705 | GAL-SKI2-3xmyc-2_μ_-LEU2 | This study |
| pAJ706 | GAL-GST-SKI8-2_μ_-URA3 | This study |
| pAJ709 | GAL-SKI3-3xHA-2_μ_-TRP1 | This study |
| pAJ819 | SKI2(DEVH_→_AEVA)-3xmyc-2_μ_-LEU2 | This study |
| pAJ820 | GAL-SKI2(DEVH_→_AEVA)-3xmyc-2_μ_-LEU2 | This study |
| pAJ832 | ski2_Δ_N-3xymc-2_μ_-LEU2 | This study |
| pAJ833 | GAL-ski3_Δ_C(aa 2-1206)-3xHA-2_μ_-TRP1 | This study |
| pAJ840 | ski3_Δ_C-LEU-CEN | This study |
| pAJ951 | SKI2 ORF in pGAD-C(1) | This study |
| pAJ952 | ski2(nt 1–840) in pGAD-C(1) | This study |
| pAJ953 | ski2(nt 841–1584) in pGAD-C(1) | This study |
| pAJ954 | ski2(nt 1585–2589) in pGAD-C(1) | This study |
| pAJ955 | ski2(nt 2590–3868) in pGAD-C(1) | This study |
| pAJ956 | SKI3 ORF in pGBD-C(1) | This study |
| pAJ957 | ski3(nt 1–1047) in PGBD-C(1) | This study |
| pAJ958 | ski3(nt 1048–1230) in pGBD-C(1) | This study |
| pAJ959 | ski3(nt 1231–1923) in pGBD-C(1) | This study |
| pAJ960 | ski3(nt 1924–2895) in pGBD-C(1) | This study |
| pAJ961 | ski3(nt 2896–3618) in pGBD-C(1) | This study |
| pAJ962 | ski3(nt 3619–4299) in pGBD-C(1) | This study |
| pAJ963 | SKI7 ORF in pGAD-C(1) | This study |
| pAJ964 | SKI8 ORF in pGAD-C(1) | This study |
| pAJ965 | SKI8 ORF in pGBD-C(1) | This study |
| pAJ966 | SKI7 ORF in pGBD-C(1) | This study |
| pAJ1201 | GAL-FLAG-SKI7-2_μ_-LEU2 | This study |
| pAJ1206 | GAL-FLAG-SKI7-2_μ_-URA3 | This study |
| pAJ1224 | pNMD3-ski3(aa 1207–1432)-3xHA-2_μ_-URA3 | This study |
| pAJ1225 | pNMD3-ski3(aa 966–1432)-3xHA-2_μ_-URA3 | This study |
| pAJ1226 | pNMD3-ski3(aa 642–1432)-3xHA-2_μ_-URA3 | This study |
| pAJ1227 | pNMD3-ski3(aa 411–1432)-3xHA-2_μ_-URA3 | This study |
| pAJ1228 | pNMD3-ski3(aa 350–1432)-3xHA-2_μ_-URA3 | This study |
| pAJ1229 | pNMD3-ski3(aa 2–1432)-3xHA-2_μ_-URA3 | This study |
| pAJ1231 | GAL-3xHA-2_μ_-TRP1 | This study |
| pAJ1233 | GAL-ski3(aa 2–349)-3xHA-2_μ_-TRP1 | This study |
| pAJ1234 | GAL-ski3(aa 2–410)-3xHA-2_μ_-TRP1 | This study |
| pAJ1235 | GAL-ski3(aa 2–641)-3xHA-2_μ_-TRP1 | This study |
| pAJ1238 | GAL-ski3(aa 1207–1432)-3xHA-2_μ_-TRP1 | This study |
| pAJ1241 | GAL-ski2(aa 1–279)-13xmyc-2_μ_-LEU2 | This study |
| pRP469 | GAL-PGK1-pG-2_μ_–URA3 | Muhlrad and Parker 1994 |
pAJ1225, pAJ1226, pAJ1227, pAJ1228, and pAJ1229 were made in the same fashion as pAJ1224 except the 5′-oligonucleotides for PCR amplification were AJO374 (5′-CGC GAATTCAGGCCTCAGTATAGAGATGCTG CT) for pAJ1225, AJO373 (5′-CGCGAA TTCAGGCCTAAAGACCATTTGAGGGCC) for pAJ1226, AJO372 (5′-CGCGAATT CAGGCCTGTCACTGTATTGAGGGAG) for pAJ1227, AJO371 (5′-CGCGAATTCAGGCC TACGGAGAGGAAACAGAT) for pAJ1228, and AJO370 (5′-CGCGAATTCTCGGATAT TAAACAGCTA) for pAJ1229. pAJ1231 was made by replacing the GAL1–10 SKI3-containing KpnI–XmaI fragment of pAJ709 with the GAL1–10 region amplified by PCR, using oligonucleotides 5′-CGATCGGTACCGACCT TTTTTCTCCTTGACG and 5′-GCGCTCC CGGGATCGAATTCCATGGT CGTCGACGA GCTCAATTCCTTGAATTTTCAAAAATTC. pAJ1233 was made by cloning ski3 (amino acids 2–349) as an EcoRI–XmaI fragment from PCR amplification of yeast genomic DNA using oligonucleotides AJO370 and AJO376 (5′-CGCGGATCCCCCGGGCTTT TTATGGCCTTCAGG) into the same sites of pAJ1231. pAJ1234 and pAJ1235 were made similarly to pAJ1233 except the 3′-oligonucleotides for PCR amplification were AJO377 (5′-CGCGGATCCCCCGGGA ACCTCTTCCTCTAATAA) for pAJ1234, AJO378 (5′-CGCGGATCCCCCGGGATAAT AGTGGCAATAAAT) for pAJ1235. pAJ1238 was made by cloning ski3 (amino acids 1207–1432) as an EcoRI–XmaI fragment from PCR amplification of yeast genomic DNA using oligonucleotides AJO375 and AJO758 (5 ′-CGCGGATCCCCCGGGGAAACATTCG TTTAGCGC) into the same sites of pAJ1231. pAJ1241 was made by replacing NMD3 in pAJ544 with ski2(amino acids 1–279) as an NcoI–PacI fragment from PCR amplification of genomic Ski using oligonucleotides 5′-GCGCCCGGGGTCGACCTTTCTCTTCTTT GGGATCCGCTGCAGAAGTTGGTCTCTG TAGT (AJO794) and 5′-CGGTTAATTAACC CGGGTAATAGTTCAT CGATTTC (AJO793).
Directed yeast two hybrid
Pairwise interactions were tested by transforming different combinations of bait and prey plasmids into strain pJ69–4A (James et al. 1996). Leu+ Trp+ transformants were selected on appropriate plates and then plated onto leu− trp− his− triple-dropout medium supplemented with 6 mM 3-AT.
Immunoprecipitation experiments
All cell extracts were prepared by growing cells to mid-log phase in 100 mL of appropriate selective medium. Cells were collected and washed once with ice-cold extraction buffer (20 mM Tris-HCl at pH 7.5, 40 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM PMSF, 1 μM leupeptin, 1 μM pepstatin A) and resuspended in 0.5 mL of ice-cold extraction buffer, and protein extracts were prepared by disruption with glass beads. Immunoprecipitation and Western blot analysis were carried out as previously described (Brown et al. 2000).
RNA analysis
Cells were grown at 30°C to mid-log phase (OD600 = 0.5) in galactose-containing selective medium. RNA was prepared as described (Ho and Johnson 1999). Then 10 μg of each RNA sample was separated on a 7 M urea/polyacrylamide gel and transferred to a Zeta probe membrane (BioRad) by electroblotting. Blots were probed with radiolabeled oligonucleotide oRP140 (Jacobs Anderson and Parker 1998) and visualized by phosphorimaging.
Acknowledgments
This work was supported by NSF grant MCB-0321553 to A.W.J.
REFERENCES
- Aloy, P., Bottcher, B., Ceulemans, H., Leutwein, C., Mellwig, C., Fischer, S., Gavin, A.C., Bork, P., Superti-Furga, G., Serrano, L., et al. 2004. Structure-based assembly of protein complexes in yeast. Science 303: 2026–2029. [DOI] [PubMed] [Google Scholar]
- Araki, Y., Takahashi, S., Kobayashi, T., Kajiho, H., Hoshino, S., and Katada, T. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20: 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C.A. and Parker, R. 1995. Degradation of mRNA in eukaryotes. Cell 81: 179–183. [DOI] [PubMed] [Google Scholar]
- Benard, L., Carroll, K., Valle, R.C., and Wickner, R.B. 1998. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 18: 2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard, L., Carroll, K., Valle, R.C., Masison, D.C., and Wickner, R.B. 1999. The ski7 antiviral protein is an EF1-α homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae. J. Virol. 73: 2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.T. and Johnson, A.W. 2001. A _cis_-acting element known to block 3′ mRNA degradation enhances expression of polyA-minus mRNA in wild-type yeast cells and phenocopies a ski mutant. RNA 7: 1566–1577. [PMC free article] [PubMed] [Google Scholar]
- Brown, J.T., Bai, X., and Johnson, A.W. 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G. and Parker, R. 1996. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 60: 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Liu, Y., Wang, C., Parker, R., and Song, H. 2004. Crystal structure of Ski8p, a WD-repeat protein with dual roles in mRNA metabolism and meiotic recombination. Protein Sci. 13: 2673–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1994. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem. Sci. 19: 336–340. [DOI] [PubMed] [Google Scholar]
- de la Cruz, J., Kressler, D., and Linder, P. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families.Trends Biochem. Sci. 24: 192–198. [DOI] [PubMed] [Google Scholar]
- Frischmeyer, P.A., van Hoof, A., O’Donnell, K., Guerrerio, A.L., Parker, R., and Dietz, H.C. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261. [DOI] [PubMed] [Google Scholar]
- Goebl, M. and Yanagida, M. 1991. The TPR snap helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16: 173–177. [DOI] [PubMed] [Google Scholar]
- Ho, J.H. and Johnson, A.W. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 2389–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C.L. and Stevens, A. 1993. Yeast cells lacking 5′→ 3′ exoribo-nuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson, J.S. and Parker, R. 1996. RNA turnover: The heli-case story unwinds. Curr. Biol. 6: 780–782. [DOI] [PubMed] [Google Scholar]
- ———. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A.W. and Kolodner, R.D. 1995. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol. Cell. Biol. 15: 2719–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrona, A.Y. and Wilson, D.K. 2004. The structure of Ski8p, a protein regulating mRNA degradation: Implications for WD protein structure. Protein Sci. 13: 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, Y., Sarkar, G., Sommer, S.S., and Wickner, R.B. 1993. A yeast antiviral protein, SKI8, shares a repeated amino acid sequence pattern with β-subunits of G proteins and several other proteins. Yeast 9: 43–51. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2000a. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10**:** 193–198. [DOI] [PubMed] [Google Scholar]
- ———. 2000b. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7: 843–846. [DOI] [PubMed] [Google Scholar]
- ———. 2003. An NMD pathway in yeast involving accelerated dead-enylation and exosome-mediated 3′ → 5′ degradation. Mol. Cell 11: 1405–1413. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D. and Parker, R. 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581. [DOI] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O’Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., and Wilusz, J. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A.M., Davis, K., Molineux, C., Kolodner, R.D., and Johnson, A.W. 1998. Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucleic Acids Res. 26: 3707–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S.K., Icho, T., and Wickner, R.B. 1989. Structure and nuclear localization signal of the SKI3 antiviral protein of Saccharomyces cerevisiae. Yeast 5: 149–158. [DOI] [PubMed] [Google Scholar]
- Ridley, S.P., Sommer, S.S., and Wickner, R.B. 1984. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 4: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Araki, Y., Sakuno, T., and Katada, T. 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 22: 3951–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e, A., Guerry, P., and Wickner, R.B. 1978. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 136: 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Lennertz, P., and Parker, R. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Frischmeyer, P.A., Dietz, H.C., and Parker, R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264. [DOI] [PubMed] [Google Scholar]
- Wang, Z. and Kiledjian, M. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762. [DOI] [PubMed] [Google Scholar]
- Widner, W.R. and Wickner, R.B. 1993. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol. Cell. Biol. 13: 4331–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]