Analysis of the role of Caenorhabditis elegans GC-AG introns in regulated splicing (original) (raw)
Abstract
GC-AG introns represent 0.7% of total human pre-mRNA introns. To study the function of GC-AG introns in splicing regulation, 196 cDNA-confirmed GC-AG introns were identified in Caenorhabditis elegans. These represent 0.6% of the cDNA- confirmed intron data set for this organism. Eleven of these GC-AG introns are involved in alternative splicing. In a comparison of the genomic sequences of homologous genes between C.elegans and Caenorhabditis briggsae for 26 GC-AG introns, the C at the +2 position is conserved in only five of these introns. A system to experimentally test the function of GC-AG introns in alternative splicing was developed. Results from these experiments indicate that the conserved C at the +2 position of the tenth intron of the let-2 gene is essential for developmentally regulated alternative splicing. This C allows the splice donor to function as a very weak splice site that works in balance with an alternative GT splice donor. A weak GT splice donor can functionally replace the GC splice donor and allow for splicing regulation. These results indicate that while the majority of GC-AG introns appear to be constitutively spliced and have no evolutionary constraints to prevent them from being GT-AG introns, a subset of GC-AG introns is involved in alternative splicing and the C at the +2 position of these introns can have an important role in splicing regulation.
INTRODUCTION
Accurate removal of introns from precursor messenger RNAs (pre-mRNAs) is dependent upon sequence signals at the 5′ and 3′ ends of the introns (1). These splicing consensus sequences help to guide spliceosome assembly at the proper junctions. In the early steps of spliceosome assembly, U1 snRNP is recruited and the region at the 5′ end of the intron base pairs with the 5′ end of U1 snRNA to establish the 5′-splice site (2). The 3′ end of the intron is recognized by the U2AF heterodimer binding to the polypyrimidine tract and the AG dinucleotide found at the 3′ end of introns (3–5). This leads to recruitment of U2 snRNP which base pairs with the branch point sequence upstream of the AG dinucleotide and thus to the identification of the branch point and 3′-splice site (2). It is through these types of RNA–RNA and RNA–protein interactions that the exact splice junctions are delineated.
Analysis of cDNA-confirmed introns in humans shows that the vast majority of introns begin with the dinucleotide GT and end with the dinucleotide AG. This GT-AG intron class constitutes 99.24% of human introns (6–8). In humans it has been reported that 0.7% of introns begin with GC instead of the canonical GT and these introns end with AG (7,9). The GC-AG intron splice donors generally contain a strong consensus match to the 5′-splice site sequence, with the exception of the C at the +2 position (6,7). Experiments have shown that of the possible mutations to the consensus T at the +2 position of an intron, only a change to a C allows for in vitro splicing to occur, although with less efficiency (10,11). Based on overall consensus sequences, it is thought that the major spliceosome which removes GT-AG introns also removes GC-AG introns (8,11). However, the reason why GC-AG introns are so rare is not clear. There is another class of pre-mRNA introns that is removed by a different spliceosome termed the minor spliceosome (1,12). This class of introns begins with AT and often ends with AC and assembles a different set of snRNPS including U11, U12 and U6atac/U4atac. This class represents 0.05% of cDNA-confirmed human introns (7).
Alternative pre-mRNA splicing generates multiple protein isoforms from a single gene (13,14). Analysis of human alternative splicing indicates that at least 33% of human genes are alternatively spliced (15–17). Alternative splicing is often highly regulated and this regulation involves protein factors that bind to the pre-mRNA at specific regulatory sequences, found in both introns and exons (18). These splicing factor proteins act to promote or inhibit spliceosome assembly. Regulated splicing often occurs where weak 5′ and 3′ consensus splice sequences are present (14). A recent study analyzed whether GC-AG introns are involved in alternative splicing regulation. Thanaraj and Clark (19) found that in humans, 5% of alternatively spliced introns are GC-AG introns. They also found evidence that 60% of these GC-AG introns are alternatively spliced. This is a striking result given that only 0.7% of introns are GC-AG introns. One interpretation of the finding that GC-AG introns are enriched in alternatively spliced regions is that the splice donors of GC-AG introns are weak consensus splice sites that can therefore be easily subjected to regulation by alternative splicing regulatory proteins (19).
We chose to study the nematode Caenorhabditis elegans as a model system for understanding the function and evolutionary conservation of GC-AG introns. The entire genome has been sequenced (20), and we have previously made a web-based browser called Intronerator for viewing our database of >30 000 cDNA-confirmed introns aligned with the genome (21). We have also assembled a database of 844 alternatively spliced C.elegans genes (21). The sequencing of the genome of a related nematode, Caenorhabditis briggsae, is nearing completion. We have previously reported a method for genomic alignments between these two species (22). We and others have found that comparative genomics in this system allows for the identification of conserved, non-coding regulatory elements (22–24). The first example of a GC-AG intron involved in alternative splicing was described for the C.elegans let-2 gene in 1993 (25).
In this paper we investigate GC-AG introns in nematodes. We find that 0.6% of C.elegans introns (196 out of 32 513 in our database) are GC-AG introns, and we have evidence that 11 of these are involved in regulated splicing. By interspecies genome comparisons between C.elegans and C.briggsae, we find that there is a poor conservation of GC splice donors between species. One of the GC-AG intron splice donors that is conserved is from the let-2 gene and it is involved in developmentally regulated alternative splicing. We tested whether this GC splice donor is important for alternative splicing regulation. We found that the regulated splicing in this system is dependent on the GC splice donor because it is a weaker 5′ splice donor than that found at the alternative splice site. Mutations that turn this into a GT splice donor can strengthen the recognition of the 5′-splice site and change the pattern of alternative splicing. A weak GT splice donor can function equivalently to the GC splice donor, indicating that these weak GC splice donors can be an important factor in the regulation of alternative splicing.
MATERIALS AND METHODS
Mutation of let-2 gene and generation of GFP/let-2 splicing reporter construct
A modified version of pBluescript KS+ containing an _Age_1 site in the polylinker was used to clone exons 7 to 12 of the let-2 genomic DNA. Site-directed mutagenesis was done to produce each let-2 5′-splice site mutation (26). The exon 7 to 12 region with the 5′-splice site mutation of the let-2 genomic DNA was cloned into the _Age_1 site of the Fire vector pPD93.65. This vector contains the unc-54 minimal promoter driving expression of a green fluorescent protein (GFP) fusion with a nuclear localization signal derived from SV40 T antigen. This promoter is active only in body wall muscles through all stages of development (27). The let-2 region contains the last 148 nt of exon 7 to the first 62 nt of exon 12, maintaining the open reading frame of the GFP. Injection/transformation procedures were used to generate N2 worms containing these constructs as extrachromosomal arrays (28). Transformed animals were identified by GFP expression in body wall muscle cell nuclei.
Growth of C.elegans strains and isolation of RNA
Caenorhabditis elegans strains were grown on plates using standard methods (29). Gravid adult C.elegans were axenized in order to isolate embryos for synchronization. Embryos were then synchronously grown at 20°C for 24 or 48 h to isolate L2 and L4 animals, respectively. To prepare embryo RNA, embryos were isolated from gravid adult hermaphrodites 68–70 h post-synchronization. RNA was isolated from packed worm pellets using a protocol developed by Rebecca Burdine and Michael Stern and described by Roller et al. (30).
Generation of complementary DNAs
Complementary DNAs (cDNAs) for the let-2 gene were made in 25 µl reaction mixtures. Reaction mixtures contained 5 µg of total RNA, 25 pmol of oligodeoxynucleotide primer complementary to the GFP construct (5′-TGGGACAA CTCCAGTGAAAAG-3′) or to let-2 exon 12, 1 mM each of dATP, dCTP, dGTP and dTTP, 1 U RNase inhibitor (Promega, Madison, WI), 1× AMV reverse transcriptase buffer (Promega) and 10 U AMV reverse transcriptase (Promega). Reaction mixtures were incubated at 37°C for 1.5 h and stored at –20°C. Five microliters of the reaction mixtures were treated with 0.1 µg of RNase A. One microliter of the RNase A treated reaction mixture was then added directly to 25 µl polymerase chain reaction (PCR) mixtures.
Polymerase chain reactions
The same oligonucleotides used in reverse transcription were added to PCR reaction mixtures with a 32P-labeled let-2 exon 8 5′ primer (5′-CTCCGACCTAGGGGTCCTTTGGGT-3′), Taq DNA polymerase and let-2 cDNAs. The exon 8 primer was 5′ end-labeled with [γ-32P]ATP by T4 polynucleotide kinase. One microliter of PCR products was restriction digested with _Dde_1. Digested product DNA was resuspended in formamide and loaded onto 0.4 mm thick 6% polyacrylamide urea gels in TBE buffer. After electrophoresis, the gels were dried onto filter paper and visualized with a Molecular Dynamics (Sunnyvale, CA) PhosphorImager. Quantitation of relative splice site usage was done using ImageQuant software. An equal sized box was drawn around the band for exons 9 and 10 and the integrated volume determined. Another box of equal size in a non-band region of each lane had its integrated volume determined and this value was used as the local background for each lane. This background value was subtracted from both the value of the exon 9 and exon 10 band for each lane, and then the fraction of exon 9 and exon 10 usage was determined for each reaction.
RESULTS
Identification and characterization of GC-AG introns in C.elegans
We used the searchable intron database of Intronerator (21) (http://www.cse.ucsc.edu/∼zahler/scanIntrons/scanIntrons.html) to identify GC-AG introns in C.elegans. There are 32 513 cDNA-confirmed C.elegans introns in this database, and 196 of these are GC-AG introns. This represents 0.6% of cDNA-confirmed C.elegans introns, which matches to the data for humans showing that 0.7% of human introns start with GC (7).
We next asked whether any of these GC introns correspond to alternatively spliced regions of genes. By examining the Intronerator displays for each of the 196 GC-AG introns, and by comparing with our database of 844 cDNA confirmed alternatively spliced C.elegans genes (21), we identified 11 examples of alternatively spliced genes in C.elegans in which the alternative splicing decision is based on the presence of a GC splice donor. The criteria for this category is the required presence of spliced cDNAs (evidence of intron removal when compared with genomic sequence) that confirm both forms of the alternative splicing event. In our findings, only 1.3% of alternative splicing decisions involve GC-AG introns. This differs from a recent report that suggests 5% of human alternative splice sites involve GC-AG introns (19).
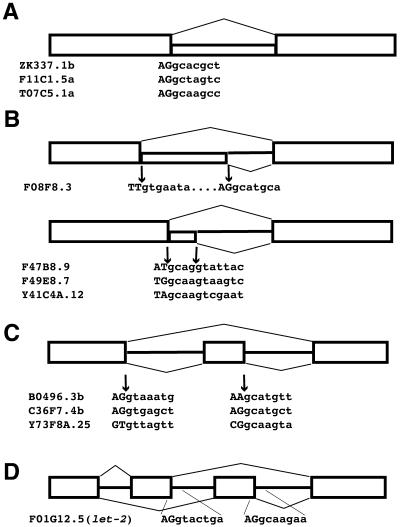
We found four general classes of alternative splice sites involving GC-AG introns: alternative introns, alternative 5′-splice sites, alternative exons and mutually exclusive exons. These results are summarized in Figure 1. We found three examples of retained introns (in the genes ZK337.1b, F11C1.5a and T07C5.1a) where the alternative splicing decision involves removal or retention of a GC-AG intron in the mature message. The expressed sequence tag (EST) data for retained introns came from messages that showed removal of flanking introns, indicating that the messages which retained the introns were spliced. The formal possibility exists that the ESTs with the retained introns may be from partially spliced messages. However, in each case multiple examples of retained introns are found in the C.elegans EST database indicating that these are reproducible and less likely to represent partial splicing. We found four examples of alternative 5′-splice sites with a single 3′-splice site, where one of the alternative 5′-splice site introns starts with GC and the other with GT. In the gene F08F8.3, the GC splice donor is downstream and it is located 51 nt from the GT splice donor. For the other three alternative 5′-splice sites, the GC splice donor is located 4 nt upstream of the GT splice donor. In these cases (F47B8.9, F49E8.7 and Y41C4A.12) it appears that the first two bases of the GT splice donor serve as the consensus GT at the +5 and +6 positions of the GC intron. These two nucleotides match the consensus for the C.elegans major spliceosome (AGgtaagtt) (31) at these sites. In Saccharomyces cerevisiae, the SRC1 message has been confirmed to be spliced at alternative 5′ sites 4 nt apart, where the intron of the upstream 5′-splice site begins gcaagt (32). Thus, in yeast, the ability to have this type of overlapping 5′-splice site organization where the GT at the +5 and +6 positions alternatively function as the GT at the +1 and +2 positions is also seen. Because this type of alternative splicing generates an out of frame message for one of the products, this will always lead to truncated messages. Perhaps these represent a type of splicing mistake that is tolerated by the cells at some level, or these may represent an important form of regulated splicing. Three of the alternatively spliced GC-AG introns are found flanking alternative exons. These exons are either spliced into the mature message or skipped over by the splicing machinery. In all three cases (B0496.3b, C36F7.4b and Y73F8A.25) the intron downstream of the alternative exon begins with GC. The final example of a GC-AG intron involved in alternative splicing had been previously identified. In the let-2 gene, there are two mutually exclusive exons, exons 9 and 10 (25). One or the other is included in the mature message. The intron downstream of exon 10 is a GC-AG intron.
Figure 1.
Examples of GC-AG introns involved in alternative splicing in C.elegans. (A) Examples of alternative GC-AG introns. Gene name and sequence at splice donor are indicated. Upper case letters are exons, lower case are intronic sequences. (B) Examples of GC splice donors as alternative 5′-splice sites. 5′-Splice sites in the upper part are separated by 51 nt. In the lower part, 5′-splice sites are separated by 4 nt. (C) GC splice donors involved in alternative exon splicing. (D) Mutually exclusive exons.
Evolutionary conservation of GC-AG introns
To test whether GC-AG introns serve an important regulatory role in splicing, we asked whether the rare C at the +2 position of GC-AG introns is conserved between the nematodes C.elegans and C.briggsae. These two species are separated by ∼50 million years of evolution (33). In a previous study, we described the creation of an algorithm, WABA, that we used to align 8 million bases of C.briggsae cosmid sequence to the C.elegans genome (22). Based on the results of this alignment, we concluded that coding regions were highly conserved between these two species, with on average a divergence at every other wobble position. Intergenic regions and introns were very poorly conserved with the exception of patches of sequence that represent regulatory regions such as promoters. In addition, we noted that some small regions in introns near alternatively spliced exons were conserved and suggested that these may function in splicing regulation. In the summer of 2001 we updated this alignment to include newly released C.briggsae sequences totaling 13 million bases; this alignment is available through the Intronerator web site (http://www.cse.ucsc.edu/∼zahler/intronerator/index.html).
We searched the 196 GC-AG introns in C.elegans for ones that are present in regions of the genome covered by C.briggsae homology. We identified 26 such introns (Table 1). The last two bases of the preceding exon and the first seven bases of the intron are shown. It is apparent that the C at the +2 position of the intron is not conserved in C.briggsae for the majority of these introns. For five of the 26, the GC splice donor is conserved, and for the first four of these listed, the entire nine base 5′-splice site region is identical after ∼50 million years of evolution. For the next 20 of these C.elegans GC-AG introns, the intron in the C.briggsae homolog begins with GT. For the last C.elegans GC-AG intron listed in Table 1, there is no corresponding intron in the C.briggsae homolog. We have previously described over 250 examples of introns found in C.elegans but not in C.briggsae and vice versa (22). Therefore, this result is not surprising except for the implication that this particular GC-AG intron is not essential for regulation of gene expression.
Table 1. GC-AG intron-containing genes from C.elegans for which homologous genes have been identified in C.briggsae.
In this set of 26 GC-AG introns for which the homologous C.briggsae sequence is also available, we found only three examples of homologous genes that are alternatively spliced such that the GC-AG splice donor is part of the alternative splicing decision. Given that the EST coverage of the C.elegans genome is low for many genes, there exists the possibility that more examples of GC-AG alternative splicing may be present in this group of 26 homologous introns. Two examples of the GC intron being involved in an alternative splicing decision are in the group of GC introns whose 5′-splice site regions are completely conserved with C.briggsae. These two examples are the genes F11C1.5, an alternative intron, and let-2 (F01G12.5) whose alternative splicing involves a choice between mutually exclusive exons. One example of alternative splicing in which the C at the +2 position of the splice donor is not conserved is for Y41C4A.12. This is a member of the group of alternative 5′-splice sites separated by 4 nt described in Figure 1B. The ability to alternatively splice this substrate in the same way has presumably been conserved in C.briggsae, albeit with a choice between two GT splice donors spaced 4 nt apart.
Let-2 alternative splicing has been previously described (25). This alpha(2) type IV collagen contains two mutually exclusive exons, exons 9 and 10. What is interesting about this splicing is that this is a rare example of a C.elegans alternative splicing event for which developmental control is known (25). Exon 9 is included in 95% of embryonic let-2 messages while exon 10 is included in 90% of adult messages, with a gradual shifting between these two ratios during the four larval stages. In the case of this alternatively spliced gene, part of the regulation of alternative splicing may involve a competition between the two 5′-splice sites found at the ends of exons 9 and 10. The splice site that is recognized by the splicing machinery first may be the one whose exon gets spliced into the mature message. The intron between exons 10 and 11 begins with GC (AGgcaagaa) and it has three mismatches with the C.elegans consensus for base pairing the 5′ end of U1 snRNA (AGgtaagtt) over this region. The splice donor for exon 9 is AGgtactgt and has three mismatches with the 5′ end of U1 snRNA. Two of these mismatch positions are different from those for the exon 10 splice donor.
A functional test for the role of a GC-AG intron in alternative splicing
The known developmental splicing regulation of let-2 makes it an attractive candidate for analysis of regulated splicing and for testing models about the role of the GC splice donor in this regulation. In order to test the regulation of this splicing we have transformed worms with a plasmid construct derived from this gene (Fig. 2A) in which the alternatively spliced region is placed upstream of an open-reading frame for GFP (34). The fusion protein for let-2 contains the nuclear localization signal from SV40 T antigen at its N-terminus which concentrates GFP in the nucleus for easier visualization by fluorescence microscopy. This initial construct is derived from one of the Andrew Fire Vector Kits that his laboratory has made freely available to C.elegans researchers (28). Expression of the fusion protein is driven by the unc-54 promoter which is active in body wall muscle cells (27). This is also the primary site of let-2 expression (35). We have previously demonstrated the use of splicing reporter constructs in C.elegans to study splicing regulation by the sup-39 gene (30).
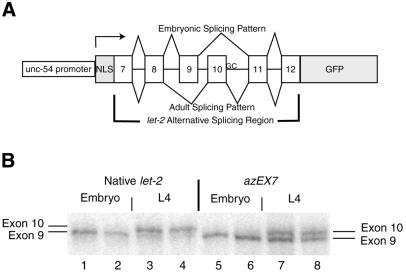
Figure 2.
Analysis of alternative splicing of the let-2 gene of C.elegans. (A) Diagram of let-2 alternative splicing region in the context of the GFP expression reporter construct designated azEX7. The intron between exons 10 and 11 starts with GC instead of the canonical GT. NLS, nuclear localization signal; GFP, green fluorescent protein. (B) Developmental regulation of splicing of the let-2 gene and the let-2/GFP fusion protein splicing reporter constructs. Total RNA was isolated from staged animals carrying the azEX7 let-2/GFP fusion construct described in (A) (lanes 1–8). 32P RT–PCR was performed on these RNAs with primers specific for the flanking region of the native let-2 gene (lanes 1–4) or the flanking sequences in the extrachromosomal array (lanes 5–8). These PCR products were separated on a 6% acrylamide sequencing gel and visualized with a Molecular Dynamics PhosphorImager. Exon 9 is 3 nt shorter than exon 10 and thus its inclusion by splicing results in a PCR product with higher mobility.
We transformed worms with the construct shown in Figure 2A, selected animals that expressed GFP, and maintained this as an extrachromosomal array which we have designated as azEX7. We then monitored splicing of the construct through 32P RT–PCR analysis of developmentally staged animals. We also monitored splicing of the native gene with a similar reaction using the same 32P-labeled 5′ primer, but with a different 3′ RT and PCR primer specific for the native gene and located the same distance from the 5′ PCR primer. Exons 9 and 10 differ by 3 nt so the RT–PCR products amplified from the alternatively spliced forms of the genes can be separated on a sequencing gel. Using staged animals, we compared the signal from our transformed extrachromosomal array with the signal from the endogenous let-2 gene to determine whether alternative splicing is regulated in the same way. We found that for both the let-2 gene and the reporter construct, the exon 9-containing form of the message represents >95% of the let-2 messages in embryos (Fig. 2B, lanes 1, 2, 5, 6). In L4 animals, exon 10 was found to be used in 90% of the native let-2 genes and in only 35% of the messages arising from the array (Fig. 2B, lanes 3, 4, 7, 8). This difference between the alternative splicing ratios of the native gene and the alternative splicing reporter construct in L4 animals can be explained in several ways; use of the unc-54 gene promoter may lead to expression in inappropriate cells of L4 animals which lack the proper regulatory factors to promote usage of exon 10 or overexpression of this message has occurred to the point where the regulatory factors are not at a high enough concentration to promote the expected splicing. However, a clear developmental switch in the alternative splicing of this reporter construct can be detected, which makes it useful for answering questions about the role of the GC splice donor in alternative splicing.
We tested the hypothesis that this GC-AG intron was involved in regulation of splicing by changing the base at the +2 position of the intron after exon 10 from C to T in the reporter construct and transforming this into worms. The resulting extrachromasomal array is designated as azEX8 and analysis of the alternative splicing of this reporter is presented in Figure 3B. In Figure 3B we restriction digested the 32P RT–PCR products with the enzyme _Dde_1 to generate easily separable bands for the two alternative splicing products. This C to T change in azEX8 resulted in the inclusion of only exon 10 in mRNAs derived from our constructs, both in L4s and in embryos (Fig. 3B, lanes 4–6). The effect of the ‘C to T mutation’ on alternative splicing is striking in that a single point mutation eliminated the developmental regulation of this splicing. Mutation of the second base of intron 10 from a C to a T creates a GT splice donor with a fairly strong 5′-splice site with (AGgtaagaa), with seven consecutive bases that can pair with the 5′ end of U1 snRNA.
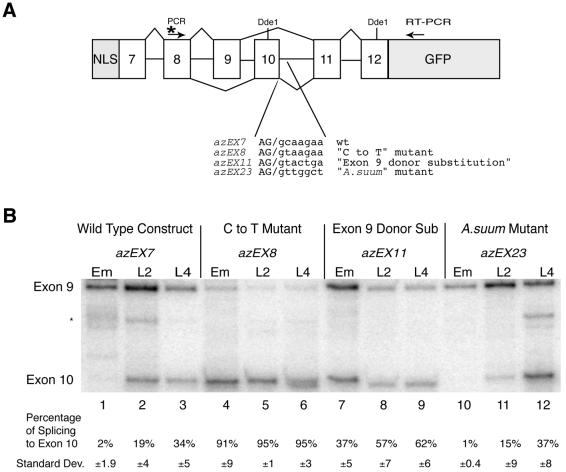
Figure 3.
Effects of GC splice donor mutations on let-2 alternative splicing. (A) Diagram of let-2 reporter construct showing location of RT/3′ PCR primer and 32P-labeled 5′ PCR primer. Locations of _Dde_1 sites are indicated. After RT–PCR, products are digested with _Dde_1 to yield different sized bands for the two alternative splice forms that are easily separable. The sequence of each of the mutant splice sites tested is indicated. (B) Analysis of developmental regulation of alternative splicing of let-2 5′-splice site mutants. RNA from the indicated mutants at the indicated developmental stage (embryo or L2 or L4 larvae) were isolated and the products of the reporter gene were amplified by RT–PCR. Products of RT–PCR reactions were digested with _Dde_1, separated on a 6% polyacrylamide gel and visualized with a Molecular Dynamics PhosphorImager. Quantitation of the percentage of exon 10 in the amplified message is shown. The quantitation is based on assays from a minimum of two independent embryonic RNA isolations and three independent L2 and L4 RNA isolations for each strain. The asterisk marks a PCR artifact band that appears occasionally. It cannot be digested with _Dde_1 and is therefore not part of the let-2 reporter construct.
The shift to complete exon 10 splicing in the azEX8 construct, even in embryos, indicates that alternative 5′-splice site choice for this splice donor is dependent on a competition between the 5′-splice sites at the end of exon 9 and exon 10. To address this hypothesis we replaced the 5′ splice donor at the end of exon 10 with a sequence identical to the 5′ splice donor of exon 9. Therefore, the two 5′-splice sites have equal ability to base pair with U1 snRNA. The splice donor sequence AGgtactgt contains three mismatches in the region that base pairs with U1 snRNA, and is therefore predicted to be a weaker 5′-splice site than the ‘C to T mutant’ which contains seven consecutive complementary bases. We transformed this new construct into C.elegans and isolated a line bearing the extrachromosomal array designated azEX11. We tested the splicing of this array and compared it with the wild-type and C to T mutant reporters at different developmental stages (Fig. 3B, lanes 7–9). For this new construct, exon 10 splicing is detected in embryos, L2, and L4 stages at a level that is considerably higher than that found for the wild-type substrate (lanes 1–3), but not as high as that found for the ‘C to T mutant’ substrate (lanes 4–6). There is also a change in splicing ratio during development, although it is not as dramatic as for the wild-type substrate. This indicates that the 5′-splice site of exon 9 is a stronger splice donor than the wild-type exon 10 splice donor, but not nearly as strong as the ‘C to T mutant’ exon 10 splice donor.
We next sought to determine whether a GT splice donor equivalent in strength to the GC splice donor exists by looking at a roundworm much further removed evolutionarily from C.elegans and C.briggsae. Ascaris suum is a 0.5 m long parasite commonly found in porcine intestines. It has been shown to contain an alpha(2) type IV collagen gene similar to the one in C.elegans. Like the C.elegans gene, the A.suum gene undergoes developmentally regulated alternative splicing involving the same use of mutually exclusive exons 9 and 10 (36). What is striking is that in A.suum, the splice donor for exon 10 is AGgttggct, not a GC splice donor. This sequence differs from the U1 snRNA base-pairing consensus sequence at three positions. We created a construct in which we changed the wild-type C.elegans exon 10 splice donor to the exon 10 splice donor sequence from the Ascaris gene. We transformed this construct into C.elegans, isolated a stable line containing this extrachromosomal array designated azEX23, and tested for the alternative splicing of this reporter construct. This particular GT splice donor has the same developmental regulation of splicing and the same level of alternative splice site usage as the wild-type C.elegans GC splice donor (Fig. 3B, lanes 10–12 versus lanes 1–3). Taken together, these data suggest that the GC splice donor of exon 10 serves as a weak 5′-splice site that allows for regulation of alternative splicing, and that there are equivalent GT splice donors which can functionally replace a GC splice donor and still provide the same regulation of alternative splicing.
Discussion
In this paper we have taken both a bioinformatics and an experimental approach to analyze the rare class of GC-AG introns in C.elegans. Our analysis indicates that GC-AG introns are found in C.elegans at roughly the same frequency as in humans (0.6 versus 0.7%) (7). Previous work in humans has indicated that GC-AG introns are represented at the sites of alternative splicing decisions at a much higher level than their total representation in the intron population. Thanaraj and Clark (19) concluded that 5% of alternative human splice donors come from GC-AG introns, a 7-fold enrichment over the level of GC-AG introns found in the genome. In our analysis of 844 C.elegans alternatively spliced genes, we could only identify 11 examples out of 844 alternatively spliced genes (1.3%) where a splice donor with C at the +2 position of the intron was involved in the alternative splicing decision. Given that the majority of these alternative splicing events involved two splice donors competing for spliceosome assembly, the number of 1.3% needs to be divided by two to account for multiple splice donors at each alternative splicing event. Therefore, our results indicate that the GC-AG splice donors in C.elegans are not overly enriched at alternative splice donor sites, which is different from the case in humans.
Our analysis of comparative genomics between C.elegans and C.briggsae indicates that only 20% of the +2 position Cs of the GC-AG introns are conserved. Since our original hypothesis was that C.elegans GC-AG donors might represent important regulatory elements for splicing, this result was surprising because for other regulatory elements, promoters for example, we saw a strong evolutionary conservation in non-coding regions (22). Although our data set of homologous introns is relatively small, it is interesting that two of the three intron splice donors in this set known to be involved in alternative splicing are completely conserved at the splice donor sequence. One interpretation is that the majority of GC-AG splice donors are involved in constitutive splicing. This group might be easily converted between GC and GT splice donors with little effect, and perhaps are not subject to evolutionary constraints. In contrast, a small subset of the GC-AG splice donors are involved in alternative splicing decisions, and for at least one of these, the GC splice donor plays an important role in alternative splicing regulation.
Experimental results in the human system indicate that substituting a C for a T at the +2 position of the intron still allows for splicing, but the splicing is weaker (10,11). This is consistent with the U1 snRNA of the major spliceosome having weaker base pairing with the 5′-splice site. The mismatch at the +2 position of the intron is almost in the center of the region of base pairing and would have a disruptive effect on U1 snRNA duplex formation. This has been demonstrated experimentally for HIV regions known to bind U1 snRNP but not necessarily promote splicing (37). Results in humans indicate that GC-AG intron splice donors have enhanced consensus nucleotides at other donor positions besides the +2 position (7). We asked whether other bases in the 5′-splice site region of C.elegans also compensate for this mismatch at +2 by having stronger ability for base pairing with the 5′ end of U1 snRNA. Consistent with the human result, we found that the 26 GC splice donors in Table 1 had on average 2.4 mismatches from the optimal U1 snRNA base pairing sequence. In contrast, the 19 homologous GT splice donors in C.briggsae had on average 2.8 mismatches.
Our experimental system for analysis of the alternative splicing of the let-2 reporter construct allowed us to test the importance of GC introns in alternative splicing and the relative 5′-splice site strength for GC and GT splice donors. Our results indicate that the naturally occurring GC splice donor serves as a weak splice donor that is both evolutionarily conserved and required for regulated splicing. Mutation of just the C at the +2 position to a T (azEX8) creates a strong 5′-splice site with only two mismatches to the U1 consensus and seven consecutive bases for base pairing with U1 snRNA. This leads to constitutive exon 10 usage and no developmentally regulated splicing. Duplication of the exon 9 splice donor (azEX11) creates a transcript in which two equivalent splice donor sites give almost equivalent usage of the two alternative exons and only a modest developmental switch in the splice site usage. This is again consistent with alternative splicing being controlled in part by the relative strengths of the two splice donors. Substitution of the exon 10 donor from A.suum allows for the wild-type alternative splicing, even though this splice donor defines an intron beginning with GT. This result suggests that given sufficient evolutionary distance, a GT splice donor equivalent in strength to a GC donor can be identified.
Our results suggest that the majority of GC-AG introns are constitutively spliced, and for these introns the GC splice donor has no important regulatory function. Interconversion between GT-AG and GC-AG introns in these cases may require only a single base mutation. For a small subset of GC-AG introns, the GC splice donor is functionally important and allows for regulation of alternative splicing through its weak U1 snRNA base pairing. This suggests that there may be an evolutionary constraint to maintain the splice donor sequence. Interconversion of a regulatory GC-AG donor with a GT-AG donor of equivalent strength is possible, but it may take longer in evolutionary terms because interconversion will require more than one mutation to evolve an equivalently weak GT splice donor from a GC donor. This may be due to the fact that GC donors have higher consensus with the optimal U1 snRNA binding sequence outside of the +2 position. This is also consistent with our finding that when GC donors are conserved between species, there is a strong trend to conserve the entire splice donor sequence (Table 1). This implies that where the relative level of strength of a splice donor is important for alternative splicing regulation, multiple mutations may be required to evolve a GT splice donor equivalent in strength to a GC splice donor. For regulated splice donors such as the let-2 gene, the time since the divergence of C.elegans and C.briggsae may not yet have been long enough for these changes to occur.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Ron Patterson, Jennifer Kabat and Mary Ann Dassah for helpful discussions and critical reading of the manuscript, and Dr Manny Ares for very helpful discussions. This work is supported by grant 1R01GM61646 from the National Institutes of Health.
REFERENCES
- 1.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosome. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds.), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- 2.Ares M. Jr and Weiser,B. (1994) Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog. Nucleic Acid Res. Mol. Biol., 50, 131–159. [DOI] [PubMed] [Google Scholar]
- 3.Zorio D.A. and Blumenthal,T. (1999) Both subunits of U2AF recognize the 3′ splice site in C. elegans. Nature, 402, 835–838. [DOI] [PubMed] [Google Scholar]
- 4.Merendino L., Guth,S., Bilbao,D., Martinez,C. and Valcarcel,J. (1999) Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature, 402, 838–841. [DOI] [PubMed] [Google Scholar]
- 5.Wu S.,Romfo,C.M., Nilsen,T.W. and Green,M.R. (1999) Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature, 402, 832–835. [DOI] [PubMed] [Google Scholar]
- 6.Senapathy P., Shapiro,M.B. and Harris,N.L. (1990) Splice junctions, branch point sites and exons: sequence statistics, identification and applications to genome project. Methods Enzymol., 183, 252–278. [DOI] [PubMed] [Google Scholar]
- 7.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res., 28, 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mount S.M. (2000) Genomic sequence, splicing and gene annotation. Am. J. Hum. Genet., 67, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res., 29, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aebi M., Hornig,H., Padgett,R.A., Reiser,J. and Weissmann,C. (1986) Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell, 47, 555–565. [DOI] [PubMed] [Google Scholar]
- 11.Aebi M., Hornig,H. and Weissmann,C. (1987) 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell, 50, 237–246. [DOI] [PubMed] [Google Scholar]
- 12.Mount S.M. (1996) AT-AC introns: an ATtACk on dogma. Science, 271, 1690–1692. [DOI] [PubMed] [Google Scholar]
- 13.Smith C.W.J., Patton,J.G. and Nadal-Ginard,B. (1989) Alternative splicing in the control of gene expression. Annu. Rev. Genet., 23, 527–577. [DOI] [PubMed] [Google Scholar]
- 14.Stamm S., Zhang,M.Q., Marr,T.G. and Helfman,D.M. (1994) A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res., 22, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- 17.Thanaraj T.A. (1999) A clean data set of EST-confirmed splice sites from Homo sapiens and standards for clean-up procedures. Nucleic Acids Res., 27, 2627–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black D.L. (1995) Finding splice sites within a wilderness of RNA. RNA, 1, 763–771. [PMC free article] [PubMed] [Google Scholar]
- 19.Thanaraj T.A. and Clark,F. (2001) Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Res., 29, 2581–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 21.Kent W.J. and Zahler,A.M. (2000) The intronerator: exploring introns and alternative splicing in Caenorhabditis elegans. Nucleic Acids Res., 28, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent W.J. and Zahler,A.M. (2000) Conservation, regulation, synteny and introns in a large-scale C. briggsae_–_C. elegans genomic alignment. Genome Res., 10, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 23.Thacker C., Marra,M.A., Jones,A., Baillie,D.L. and Rose,A.M. (1999) Functional genomics in Caenorhabditis elegans: an approach involving comparisons of sequences from related nematodes. Genome Res., 9, 348–359. [PMC free article] [PubMed] [Google Scholar]
- 24.Webb C.T., Shabalina,S.A., Ogurtsov,A.Y. and Kondrashov,A.S. (2002) Analysis of similarity within 142 pairs of orthologous intergenic regions of Caenorhabditis elegans and Caenorhabditis briggsae. Nucleic Acids Res., 30, 1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sibley M.H., Johnson,J.J., Mello,C.C. and Kramer,J.M. (1993) Genetic identification, sequence and alternative splicing of the Caenorhabditis elegans alpha 2(IV) collagen gene. J. Cell Biol., 123, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 27.Okkema P.G., Harrison,S.W., Plunger,V., Aryana,A. and Fire,A. (1993) Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics, 135, 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mello C. and Fire,A. (1995) DNA transformation. Methods Cell Biol., 48, 451–482. [PubMed] [Google Scholar]
- 29.Lewis J.A. and Fleming,J.T. (1995) Basic culture methods. Methods Cell Biol., 48, 3–29. [PubMed] [Google Scholar]
- 30.Roller A.B., Hoffman,D.C. and Zahler,A.M. (2000) The allele-specific suppressor sup-39 alters use of cryptic splice sites in C. elegans. Genetics, 154, 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blumenthal T. and Steward,K. (1997) RNA processing and gene structure. In Riddle,D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (eds), C. elegans II. Cold Spring Harbor Laboratory Press, Plainview, NY, pp. 117–145. [PubMed]
- 32.Davis C.A., Grate,L., Spingola,M. and Ares,M.,Jr (2000) Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res., 28, 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaxter M.L., De Ley,P., Garey,J.R., Liu,L.X., Scheldeman,P., Vierstraete,A., Vanfleteren,J.R., Mackey,L.Y., Dorris,M., Frisse,L.M., Vida,J.T. and Thomas,W.K. (1998) A molecular evolutionary framework for the phylum Nematoda. Nature, 392, 71–75. [DOI] [PubMed] [Google Scholar]
- 34.Chalfie M., Tu,Y., Euskirchen,G., Ward,W.W. and Prasher,D.C. (1994) Green fluorescent protein as a marker for gene expression. Science, 263, 802–805. [DOI] [PubMed] [Google Scholar]
- 35.Graham P.L., Johnson,J.J., Wang,S., Sibley,M.H., Gupta,M.C. and Kramer,J.M. (1997) Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J. Cell Biol., 137, 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettitt J. and Kingston,I.B. (1994) Developmentally regulated alternative splicing of a nematode type IV collagen gene. Dev. Biol., 161, 22–29. [DOI] [PubMed] [Google Scholar]
- 37.Kammler S., Leurs,C., Freund,M., Krummheuer,J., Seidel,K., Tange,T.O., Lund,M.K., Kjems,J., Scheid,A. and Schaal,H. (2001) The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA, 7, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]