Decrease in hnRNP A/B expression during erythropoiesis mediates a pre-mRNA splicing switch (original) (raw)
Abstract
A physiologically important alternative pre-mRNA splicing switch, involving activation of protein 4.1R exon 16 (E16) splicing, is required for the establishment of proper mechanical integrity of the erythrocyte membrane during erythropoiesis. Here we identify a conserved exonic splicing silencer element (CE16) in E16 that interacts with hnRNP A/B proteins and plays a role in repression of E16 splicing during early erythropoiesis. Experiments with model pre-mRNAs showed that CE16 can repress splicing of upstream introns, and that mutagenesis or replacement of CE16 can relieve this inhibition. An affinity selection assay with biotinylated CE16 RNA demonstrated specific binding of hnRNP A/B proteins. Depletion of hnRNP A/B proteins from nuclear extract significantly increased E16 inclusion, while repletion with recombinant hnRNP A/B restored E16 silencing. Most importantly, differentiating mouse erythroblasts exhibited a stage-specific activation of the E16 splicing switch in concert with a dramatic and specific down-regulation of hnRNP A/B protein expression. These findings demonstrate that natural developmental changes in hnRNP A/B proteins can effect physiologically important switches in pre-mRNA splicing.
Keywords: alternative splicing/exonic splicing silencer/hnRNP A and B/protein 4.1R
Introduction
Alternative splicing of pre-mRNAs from a single gene facilitates expression of multiple protein isoforms that can have different functional characteristics (reviewed in Chabot, 1996; Smith and Valcarcel, 2000). Recent analysis of the Human Genome Project predicts that the majority of human genes undergo some form of alternative splicing (Lander et al., 2001), indicating that this is a major mechanism for regulating gene expression. These alternative splicing events are often developmentally regulated and/or exhibit tissue-specific variations. One of the major challenges in the field is to define the molecular mechanisms whereby splicing factors with splicing enhancer or silencer activity help regulate alternative pre-mRNA splicing in the appropriate developmental patterns.
In many cases, the primary sequence of an alternative exon plays a dual role: not only does the sequence perform an obvious protein-coding function, but it also often serves as binding sites for specific splicing factor proteins that regulate post-transcriptional processing. Alternatively spliced exons commonly are associated with both positively and negatively _cis_-acting elements that determine the usage of the (typically weak) flanking splice sites. A great deal of work over the past several years has been devoted to identification of exonic splicing enhancer (ESE) elements in both alternative exons (reviewed in Reed, 1996; Hertel et al., 1997; Wang and Manley, 1997) and constitutive exons (Mayeda et al., 1999; Schaal and Maniatis, 1999).
Many alternative exons also possess negatively acting sequences termed exonic splicing silencer (ESS) elements that can antagonize splicing enhancer function and prevent exon inclusion until the appropriate developmental stage. Recently, a number of alternative exons have been shown to contain ESS elements (Graham et al., 1992; Amendt et al., 1995; Del Gatto and Breathnach, 1995; Staffa et al., 1997; Konig et al., 1998; Si et al., 1998; Caputi et al., 1999; Kan and Green, 1999; Mayeda et al., 1999). Among the _trans_-acting factors that appear to be capable of acting through silencer elements to mediate a repressive effect are snRNP complexes (Nelson and Green, 1990; Siebel et al., 1992; Kan and Green, 1999) and members of the hnRNP family such as the A/B proteins or hnRNP I/polypyrimidine tract binding protein (PTB) (Ashiya and Grabowski, 1997; Chan and Black, 1997; Caputi et al., 1999; Del Gatto-Konczak et al., 1999; Matter et al., 2000; Bilodeau et al., 2001; Tange et al., 2001; Zhu et al., 2001).
The cytoskeletal protein 4.1R gene exhibits several developmentally regulated alternative splicing events in erythroid, epithelial and muscle cell types (Conboy, 1999). In particular, a regulated splicing switch involving exon 16 (E16) during erythroid differentiation plays a critical physiological role in establishing the appropriate red cell membrane material properties. E16 is skipped in early erythroid progenitors but included efficiently in mature erythroblasts (Chasis et al., 1993), leading to synthesis of 4.1R protein isoforms that bind spectrin and actin and assemble stably into the membrane skeleton (Horne et al., 1993; Discher et al., 1995; Schischmanoff et al., 1995). E16 also encodes part of a nuclear localization signal and is thus an important determinant of subcellular localization in certain nucleated cell types (Luque et al., 1998; Gascard et al., 1999).
We have developed a heterologous system in which the alternative splicing of E16 can be reconstituted in the context of a three-exon model pre-mRNA, and the sequence determinants for efficient inclusion can be explored (Gee et al., 2000). These studies yielded evidence for an ordered splicing model in which the intron downstream of E16 is removed preferentially prior to excision of the upstream intron. Coordinated interaction of regulatory elements in a specific spatial and temporal manner is postulated for proper regulation of E16 inclusion/exclusion during erythrocyte development. One of these key regulatory elements is the suboptimal 5′ splice site (5′ss) adjacent to E16 (Gee et al., 2000), which prevents constitutive splicing of the exon. Recent parallel studies have indicated the involvement of additional regulatory elements in exon 16 itself as well as the flanking intron sequences (Deguillien et al., 2001). In this report, we characterize an exonic RNA element that is involved in E16 splicing regulation, a phylogenetically conserved silencer element in E16 that functions through the binding of the hnRNP A/B proteins to repress E16 inclusion. Importantly, down-regulation of hnRNP A/B protein expression temporally correlates with the activation of E16 splicing during erythropoiesis, suggesting that this is the functional switch for activation of E16 expression in the 4.1R gene during erythropoiesis.
Results
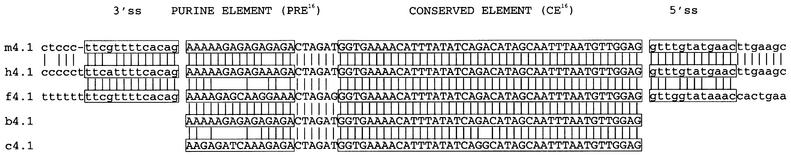
Alternative splicing of 4.1R E16 is conserved among three vertebrate orders (Winardi et al., 1995; Conboy, 1999). With the assumption that splicing regulatory sequences might be phylogenetically conserved, we compared the nucleotide sequences around E16 in the mammalian (human, mouse and bovine), amphibian (frog) and avian (chicken) genes (Figure 1). Several candidate regulatory elements were identified, including a conserved 3′ss, a weak 5′ss (Gee et al., 2000), a purine-rich element within the 5′ portion of E16 (PRE16) and a highly conserved element (CE16) of ∼40 nucleotides spanning most of the remainder of the exon. As shown below, some of these elements appear to play a cooperative role in regulating the ordered splicing of the introns flanking E16.
Fig. 1. Conservation of 4.1R E16 and flanking intron sequences. Conserved sequence elements are boxed. M, mouse; h, human; f, frog; c, chicken; b, bovine; introns are in lower case and exons in upper case.
We previously reported that the 5′ss of 4.1R E16 in the mouse gene was suboptimal, and that substitution of a consensus 5′ss sequence resulted in greatly increased E16 inclusion (Gee et al., 2000). Here we show (Figure 1) that the 5′ss in the mouse, human and frog genes is diverged similarly from consensus by virtue of having pyrimidine nucleotides at the +3 (frog) or +3 and +4 positions (human and mouse) of the intron. Therefore, a weak 5′ss is a consistent feature of E16 in several species.
Identification of an exonic splicing silencer element
Two distinct domains are evident within E16: a 15 nucleotide purine-rich element (PRE16), whose primary sequence varies among species but whose purine-rich nature is conserved; and a 42 nucleotide element (CE16) in which the primary sequence is extraordinarily conserved (Figure 1). Indeed, the sequences are invariant in four of the five species, with a single transition present in the chicken exon. This strict sequence conservation suggests that constraints in addition to coding for protein must govern the evolution of this sequence. Functional experiments shown below support the hypothesis that the CE16 domain of E16 functions as an ESS.
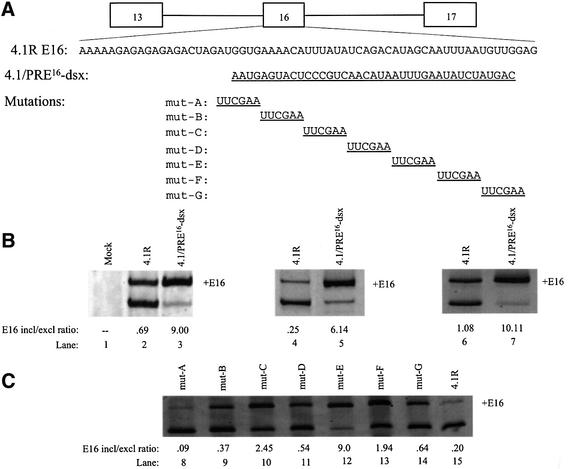
As an initial attempt to determine whether CE16 contains splicing regulatory elements, a substitution mutation was made in the 4.1R minigene (Gee et al., 2000), in which most of CE16 was replaced with an equal length of dsx exon 4 sequence to generate construct 4.1/PRE-dsx (Figure 2A). This dsx sequence was demonstrated in previous studies to lack active enhancer or silencer elements (Watakabe et al., 1993; Lynch and Maniatis, 1995). Splicing of the modified 4.1R pre-mRNA was tested in three different splicing assays. Transfection of HeLa cells showed that the level of E16 inclusion in the wild-type minigene (Figure 2B, lane 2) was greatly enhanced in the dsx substitution mutant (lane 3). Since the substituted region of dsx exon 4 lacks enhancer activity, the improved inclusion of E16 is most probably due to the loss of an ESS in CE16. Similar results were obtained in in vitro splicing assays and in microinjected Xenopus oocytes (compare lanes 4 and 5 and lanes 6 and 7, respectively), suggesting that the regulatory machinery has been evolutionarily conserved, and that the process is reproduced accurately in cell-free systems amenable to more detailed analysis.
Fig. 2. Mutation of CE16 activates E16 inclusion. (A) 4.1R minigene and E16 wild-type sequence with 4.1/PRE16-dsx, and linker-scan sequence substitutions (underlined) in E16 aligned below. All mutant constructs are the same length as the wild-type minigene. (B) RT–PCR assay of spliced products from wild-type 4.1R minigene and the PRE16-dsx substitution mutation. In vivo splicing in transfected HeLa cells (lanes 1–3), in HeLa nuclear extract (lanes 4 and 5) and in Xenopus oocytes (lanes 6 and 7) with the E16 inclusion/exclusion ratio. (C) RT–PCR results of in vitro splicing of CE16 linker-scanning mutations of the 4.1R minigene in HeLa nuclear extract (lanes 8–15).
To define boundaries of the ESS element further, linker-scanning mutagenesis of CE16 was performed in the context of the 4.1R minigene. Seven mutants were generated, each containing three to five nucleotide substitutions (Figure 2A). In vitro splicing reactions in HeLa cell nuclear extract revealed that one of these mutants (mutE) almost completely destroyed silencer activity, resulting in very efficient inclusion of exon 16 (lane 12). Notably, this mutation altered a sequence motif (UAG) that is characteristic of binding sites for the splicing silencer protein hnRNP A1 (Del Gatto et al., 1996), and its disruption quantitatively mimicked the effect of complete substitution of CE16 sequences observed in pre-mRNA substrate PRE16-dsx. Together, these observations demonstrate that the region altered in mutE comprises a critical part of the silencer element. However, the strong evolutionary conservation of CE16 and the finding that other mutations in this region partially disrupt silencer activity (Figure 2C, lanes 5–12) suggest that additional sequences affect the function of the putative A1-binding site.
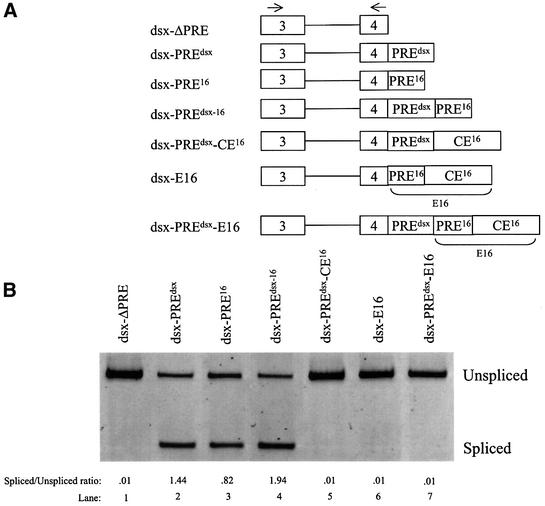
The next experiments tested whether CE16 silencer activity can function in a heterologous context, in _dsx_-based pre-mRNAs used previously to demonstrate splicing enhancer activity (Figure 3A; Watakabe et al., 1993; Lynch and Maniatis, 1995). dsx ΔPRE, an enhancerless construct from which the endogenous enhancer elements have been deleted (Lynch and Maniatis, 1995), has a weak 3′ss and splices poorly unless provided with a splicing enhancer such as the endogenous dsx E4 purine-rich element (_dsx_-PREdsx) (Figure 3B, lanes 1 and 2). When tested in parallel under identical conditions, the PRE16 element of E16 also significantly enhanced splicing of dsx pre-mRNA (lane 3). A double enhancer PREdsx-16 (lane 4) had even higher levels of splicing. Importantly, juxtaposition of CE16 to the enhancer(s) in all three of these test constructs led to complete repression of splicing activity (lanes 5–7). The repressive activity of CE16 can thus counteract at least two distinct enhancers, and can function up to 143 nucleotides from an upstream 3′ss (the distance in pre-mRNA _dsx_-PREdsx-E16). Similar results were obtained in two different assay systems employing microinjected oocytes (Figure 3) and HeLa cell nuclear extract (not shown), indicating that the CE16 silencer can function in different cell types and species. Moreover, recent studies have shown that a partial silencing effect is observed when this E16 region is inserted into a heterologous β-globin exon (Deguillien et al., 2001).
Fig. 3. 4.1R E16 sequence elements were tested for exonic splicing enhancer (ESE) and exonic splicing silencer (ESS) activities in a heterologous pre-mRNA context. (A) Schematic diagram of the dsx constructs with the position of primers used for RT–PCR indicated by arrows. (B) RT–PCR assay of splicing for each of the dsx constructs. Splicing was performed in microinjected oocytes.
Addition of competitor CE16 RNA or depletion of CE16 RNA-binding proteins leads to increased E16 inclusion in vitro
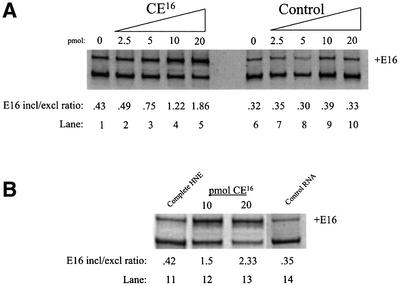
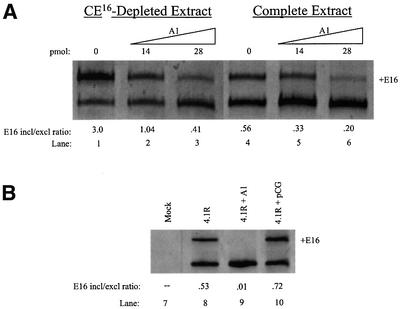
To determine whether the CE16 ESS functions through interaction with _trans_-acting protein factors, biotinylated CE16 RNA was utilized in splicing assays as a competitive inhibitor of silencer activity. In the absence of competitor, E16 skipping was the predominant pathway (Figure 4A, lane 1). Addition of competitor CE16 RNA to in vitro splicing assays increased E16 inclusion in a dose-dependent manner up to 4.5-fold (lanes 2–5). Addition of equal amounts of a control RNA known to bind hnRNP H protein (Bagga et al., 1995) had no effect on E16 inclusion (lanes 7–10). These results suggest that a titratable _trans_-acting silencing factor is necessary for CE16 ESS function. To confirm this finding, varying concentrations of biotinylated CE16 competitor RNA were pre-incubated in nuclear extract, then removed together with any bound proteins to generate depleted extract. 4.1R pre-mRNA spliced in these depleted extracts exhibited a competitor concentration-dependent increase in E16 inclusion (Figure 4B, lanes 12 and 13), similar to levels seen in the competition assay (Figure 4A, lanes 4 and 5). Depletion with control RNA had no effect on E16 inclusion (compare lanes 11 and 14).
Fig. 4. 4.1R pre-mRNA splicing in the presence of excess CE16 RNA competitor (A) or in HeLa cell nuclear extract depleted with CE16 RNA (B). (A) Splicing was performed in the absence of competitor (lanes 1 and 6) or in the presence of increasing amounts of CE16 RNA competitor (lanes 2–5) or control RNA (lanes 7–10). Amounts of competitor added are indicated above each lane (pmol), and the E16 inclusion/exclusion ratio is shown below each lane. (B) Splicing was performed in complete nuclear extract (lane 11), in nuclear extract depleted by pre-incubation with 10 or 20 pmol of CE16 RNA (lanes 12 and 13), or in extract mock depleted with control RNA (lane 14).
HnRNP A/B proteins bind specifically to CE16 RNA
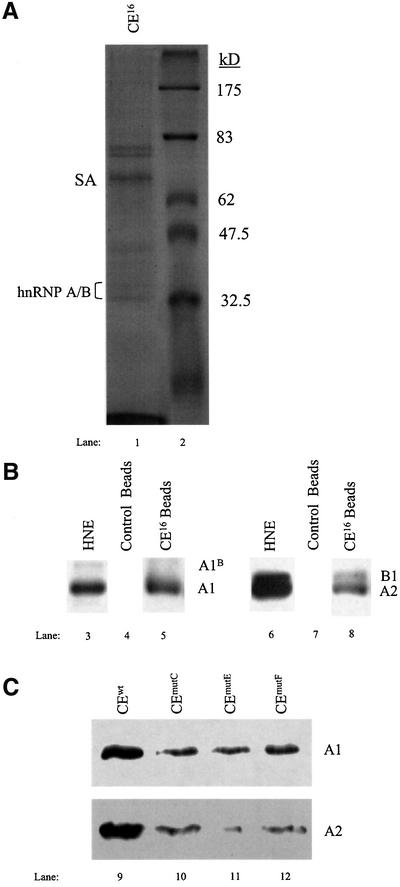
The CE16 RNA affinity-captured proteins were resolved by SDS–PAGE and characterized by immunological and biophysical techniques to identify candidate silencer-binding proteins. Coomassie Blue staining revealed the presence of several distinct bands (Figure 5A, lane 1). Intriguingly, nanospray mass spectrometry indicated that the major ∼34 kDa protein is hnRNP A1, an abundant nuclear protein that has been implicated previously as a regulator of alternative splicing in vitro and in vivo (Fu et al., 1992; Mayeda and Krainer, 1992; Mayeda et al., 1993; Caceres et al., 1994; Yang et al., 1994; Bai et al., 1999). Western blot analysis confirmed the identity of hnRNP A1 (Figure 5B, lane 5), and also showed that the related proteins hnRNP A2/B1 bound to CE16 RNA (lane 8). Neither hnRNP A1 nor hnRNP A2/B1 bound to control RNA beads used in the binding assays (lanes 4 and 7). As shown below, hnRNP A/B proteins have substantial silencer activity in 4.1R pre-mRNA splicing assays. In contrast, although hnRNP H and hnRNP I (PTB) were also detected in the CE16 eluates, no evidence for their functional relevance was obtained in later experiments (data not shown).
Fig. 5. CE16 RNA affinity isolation of candidate silencer protein(s). (A) Coomassie Blue stain of proteins eluted from CE16 RNA after incubation in HeLa nuclear extract. Bands of ∼33–35 kDa are hnRNP A/B proteins (lane 1) as identified by nanospray mass spectrometry. SA, streptavidin from magnetic beads. Lane 2 shows molecular weight standards. (B) Western blot analysis of HeLa nuclear extract (lanes 3 and 6) and of proteins eluted from CE16 and control RNAs performed with antibody against hnRNP A1/A1B (lanes 3–5) and hnRNP A2/B1 proteins (lanes 6–8). (C) Western blot of proteins eluted from biotinylated CE16 and silencer mutant RNAs performed with antibodies against hnRNP A1 and A2.
If binding of hnRNP A/B proteins to CE16 is essential for silencer activity, then mutations that interfere with silencer activity should exhibit reduced binding of A/B proteins. Using wild-type CE16 RNA and three mutant RNAs, a good correlation was observed between silencer activity and hnRNP A1 binding (Figure 5C). The strongest silencer mutation (mutE) bound ∼23% as much hnRNP A1 in nuclear extract as did wild-type CE16; the second strongest silencer mutant (mutC) bound ∼32% as much hnRNP A1; and a weak silencer mutant (mutF) bound ∼66% as much A1. Significant reduction in binding of hnRNP A2 to RNAs with silencer mutations was also observed (Figure 5C).
Recombinant hnRNP A/B proteins exhibit E16 splicing silencer activity
Because reproducible and dramatic changes in E16 inclusion/exclusion ratios were observed under conditions in which significant residual hnRNP A/B proteins remained in the depleted extracts (not shown), it was important to test whether A/B proteins indeed represented the major active silencer factor(s). Figure 6 shows that addition of recombinant hnRNP A1 protein to an in vitro splicing reaction restored the silencer effect in CE16-depleted HeLa cell nuclear extract in a concentration-dependent manner, reducing the levels of E16 inclusion (Figure 6A, lanes 1–3). These add-back experiments were performed with amounts of recombinant A/B proteins nearly identical to those employed in studies of the HIV tat exon 2 silencer, and are within the range of endogenous hnRNP A1 protein estimated to be present in HeLa cell nuclear extracts (Caputi et al., 1999). Addition of hnRNP A1 protein to complete extract reduced E16 inclusion to even lower levels (lanes 4–6). These results suggest that E16 inclusion is very sensitive to the levels of the hnRNP A/B family of proteins. Control experiments showed that substrates with a mutated CE16 domain and reduced A1 binding were much less sensitive to added A/B proteins (not shown).
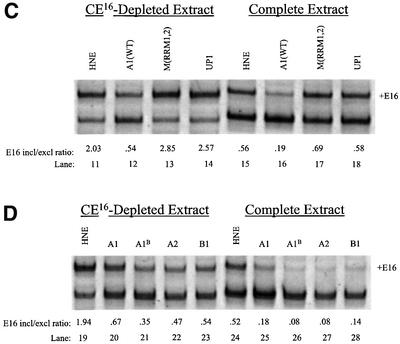
Fig. 6. Inhibition of E16 splicing by recombinant hnRNP A/B proteins. (A) In vitro splicing assay. Recombinant hnRNP A1 protein silences E16 splicing in CE16-depleted extract (lanes 1–3) and in complete extract (lanes 4–6). (B) In vivo splicing assay. HeLa cells were transfected with 4.1R minigene alone (lane 8) or co-transfected with 4.1R plus either pCG-hnRNP Al expression plasmid (lane 9) or the empty pCG expression vector (lane 10). Lane 7 is a mock transfection. (C) In vitro splicing assays with the indicated recombinant hnRNP A1 variants. Splicing was performed in HeLa nuclear extract in the absence of added A1 (lanes 11 and 15), or in the presence of wild-type A1 (lanes 12 and 16), A1 with mutant RRMs (lanes 13 and 17) or truncated UP1 variant (lanes 14 and 18). (D) In vitro splicing assays in the absence of added A1 (lanes 19 and 24) or with the indicated recombinant hnRNP A/B protein isoforms (lanes 20–23 and 25–28).
To validate the silencing effect of hnRNP A1 proteins observed in vitro, a comparable experiment was performed using transfection to alter hnRNP A1 levels in intact cells. Overexpression of hnRNP A1 in transfected HeLa cells significantly decreased inclusion of E16 in a co-transfected 4.1R minigene (compare lanes 8 and 9). Control experiments showed that co-transfection with the empty expression vector had little effect on E16 inclusion (lane 10).
To determine which domains of hnRNP A1 are required for silencer activity, variant hnRNP A1 proteins (Mayeda et al., 1994) were tested in splicing assays. In contrast to the strong silencer activity of wild-type hnRNP A1 (Figure 6C, lanes 12 and 16), hnRNP A1 protein with mutated RNA recognition motifs (RRMs) had no silencer activity (compare lanes 12 and 13 with lanes 16 and 17). The C-terminal truncation variant UP1, which lacks the glycine-rich protein–protein interaction domain of hnRNP A1 but has wild-type RRMs, was also inactive for silencer activity (lanes 14 and 18). These results indicate that both RNA–protein and protein–protein interactions are required for the splicing silencer activity of hnRNP A1.
Silencer activity among other members of the hnRNP A/B family of proteins was also investigated. In agreement with results reported for splicing of exon 2 in human immunodeficiency virus (HIV) tat pre-mRNA (Caputi et al., 1999; Bilodeau et al., 2001), the alternatively spliced isoform of hnRNP A1, hnRNP A1B, effectively silenced protein 4.1R E16 inclusion (lanes 21 and 26). Similarly, hnRNP A2 and its alternatively spliced isoform hnRNP B1 exhibited strong silencing activity in parallel assays (lanes 22 and 27, and lanes 23 and 28, respectively) compared with the control splicing reactions (lanes 19 and 24).
Down-regulated expression of hnRNP A/B proteins in developing erythroblasts correlates with E16 inclusion
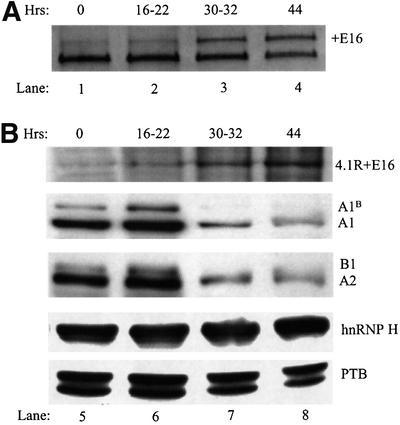
A mouse model system for erythropoiesis was used to examine whether a change in the expression of hnRNP A/B proteins could be responsible for the alternative splicing switch of E16 seen during erythroid differentiation. Immature erythroblasts isolated from the spleen of Friend virus-infected mice differentiate in culture in an erythropoietin-dependent fashion, with the majority of cells enucleating to reticulocytes between 40 and 48 h. These cells undergo the 4.1R E16 splicing switch and permit determination of temporal correlations in the level of candidate splicing factors. 4.1R RNA in immature erythroblasts at 0 h of culture showed very little inclusion of E16 (Figure 7A, lane 1). Inclusion remained low during a proliferative stage of growth through 16–22 h (lane 2). However, by 30–32 h, the erythroblasts have differentiated further and E16 inclusion can now be clearly seen (lane 3). E16 inclusion continued to increase at the 44 h time point (lane 4). Consistent with this analysis of 4.1R RNA, western blot analysis with an anti-peptide antibody showed that 4.1R protein isoforms containing the E16-encoded peptide increase over the same time period (Figure 7B, upper panel). In marked contrast, the levels of hnRNP A/B proteins exhibited a very different expression pattern (Figure 7B, middle panels). hnRNP A/B protein expression was strong at 0 h as assessed by blotting with anti-hnRNP A1 monoclonal antibody (9H10) and anti-hnRNP A2/B1 monoclonal antibody (DP3B3). A/B expression was maintained at a high level during the proliferative growth phase but declined sharply at the 30–32 and 44 h time points, coinciding with the observed increase in E16 inclusion. It should be noted that, while cells have begun to enucleate at 44 h, the released nuclei are still present in the cultures in intact (undegraded) form. Nuclear proteins are thus recovered together with the cellular proteins and analyzed in the western blots (Koury et al., 1989).
Fig. 7. Temporal relationship between 4.1R E16 splicing and the expression of hnRNP A/B proteins in differentiating mouse erythroblasts. (A) RT–PCR analysis of E16 splicing patterns in erythroblasts cultured for 0–44 h in the presence of erythropoietin (lanes 1–4). (B) Western blot analysis of expression of 4.1R + E16 and splicing factors in erythroblasts cultured for 0–44 h (lanes 5–8). Protein 4.1R isoforms including the peptide encoded by E16 were detected using anti-peptide antibody 10-1; splicing factors were assayed using antibodies against hnRNP A1, A2/B1, hnRNP H and hnRNP I/PTB.
Because erythroid cells are undergoing many alterations in nuclear function at the late stages of erythropoiesis, it was important to examine whether the observed decrease in hnRNP A/B proteins was specific. Control immunoblots on subcellular fractions demonstrated that essentially all of the A/B protein in the differentiating erythroid cells was in the nucleus, supporting the hypothesis that a real decrease in nuclear A1 levels correlated with the splicing switch. Western blot analysis of other hnRNP proteins with known splicing regulatory activity, including hnRNP H and PTB, revealed no obvious changes in expression that correlated temporally with the E16 splicing switch (Figure 7B, lower panels). Together, these data implicate the modulated expression of the hnRNP A/B proteins as the developmental switch responsible for activation of exon 16 inclusion during erythropoiesis.
Discussion
The present study provides insight into the molecular mechanism responsible for a pre-mRNA alternative splicing switch that operates during erythroid differentiation to induce physiologically important changes in the structure and function of protein 4.1R. Activation of E16 inclusion is critical for establishment of normal membrane mechanical stability during erythropoiesis, as red cell membranes assembled with protein 4.1R isoforms possessing a complete spectrin–actin-binding domain (translated from mRNAs including E16) exhibit considerable resistance to shear forces. In contrast, membranes assembled with 4.1R bearing a truncated spectrin–actin-binding domain (translated from mRNAs excluding E16) are unstable in shear assays, and may result in hemolytic anemia due to increased fragmentation in the circulation (Takakuwa et al., 1986; Conboy et al., 1991b; Discher et al., 1993; Horne et al., 1993).
Analysis of 4.1R pre-mRNA splicing supports a model in which exon 16 splicing is regulated primarily by modulation of hnRNP A/B protein levels in differentiating erythroid cells. In early erythroid progenitors, high levels of hnRNP A/B proteins are present, leading to repression of exon 16 splicing. Then, as the cells differentiate into mature erythroblasts, a dramatic decrease in levels of hnRNP A/B proteins leads to derepression of splicing and activation of exon 16 inclusion in mature 4.1R mRNA. This model is supported by in vitro and in vivo data showing that hnRNP A/B proteins bind to a conserved splicing silencer element in exon 16, that the efficiency of exon 16 inclusion in functional splicing assays is correlated directly with the levels of hnRNP A/B proteins and that a specific decrease in hnRNP A/B proteins correlates with the activation of exon 16 inclusion in differentiating erythroid cells. These findings are consistent with previous studies demonstrating that hnRNP A/B proteins can function as ESS-responsive negative regulators of splicing (Caputi et al., 1999; Del Gatto-Konczak et al., 1999; Matter et al., 2000; Bilodeau et al., 2001; Zhu et al., 2001), and that changes in the relative amounts of hnRNP A/B proteins and SR proteins can alter either the alternative splice site choice or the inclusion/exclusion ratio of selected alternative exons (Fu et al., 1992; Mayeda and Krainer, 1992; Mayeda et al., 1993; Caceres et al., 1994; Yang et al., 1994; Bai et al., 1999; Blanchette and Chabot, 1999). The most novel finding in the current study is the demonstration that a natural developmental change in the level of endogenous hnRNP A/B proteins in differentiating erythroid cells can mediate an important pre-mRNA splicing switch.
Proper regulation of exon 16 ordered splicing, in which downstream splicing precedes excision of the upstream intron (Gee et al., 2000), requires mechanisms to prevent inappropriate splicing of the flanking introns. We propose that the CE16–hnRNP A/B interaction plays an important role in regulating both upstream and downstream intron splicing. This is demonstrated most easily in the case of upstream splicing: in vitro experiments with model 4.1R pre-mRNAs (Gee et al., 2000) and heterologous dsx pre-mRNAs (Figure 3) indicate that upstream splicing is inefficient in the presence of intact CE16. In vivo, this activity may be critical to prevent premature splicing of the upstream intron in nascent 4.1R pre-mRNA transcripts, because disruption of the normal ‘downstream first’ ordered splicing would lead to inappropriate activation of a cryptic splice site and to the generation of aberrant mRNAs (Gee et al., 2000). With regard to splicing of the downstream intron, we have shown previously that this step is inefficient in the absence of a strong exon 16 3′ss (Gee et al., 2000). Therefore, inhibition of 3′ss function by hnRNP A/B bound at the splicing silencer may indirectly block activation of the downstream 5′ss, perhaps by interference with critical exon-bridging interactions (Berget, 1995).
The finding that CE16 is essentially identical among mammalian, avian and amphibian species clearly indicates that this region of exon 16 is critical to proper 4.1R gene expression. At least two specific functions can now be ascribed to this domain. First, CE16 is important for protein function, because it encodes a protein domain that is essential for interaction of 4.1R protein with spectrin and actin in the erythroid membrane skeleton (Horne et al., 1993; Discher et al., 1995). Secondly, CE16 plays a key role in 4.1R pre-mRNA processing by negatively regulating E16 splicing in cells where it may be important to prevent expression of an intact spectrin–actin-binding domain. A similar dual constraint on exon sequence evolution has been proposed earlier in the case of exonic splicing enhancer elements (Liu et al., 1998; Schaal and Maniatis, 1999).
Future studies will focus on identification of the mechanism by which hnRNP A/B protein(s) binding to the CE16 silencer can block recognition and/or splicing of E16 by the nuclear spliceosomal machinery, and what role these interactions play in the ordered splicing of introns flanking E16 (Gee et al., 2000). Most probably, proper regulation requires additional interactions among hnRNP A/B proteins (Ding et al., 1999) or between hnRNP A/B proteins and other splicing factors, acting in a concerted fashion to orchestrate the developmental switch in E16 splicing during erythropoiesis. In particular, it will be important to clarify the possible function of SR proteins in E16 splicing, the role of enhancer activity in PRE16 and potential interaction(s) between PRE16 and CE16. Moreover, important _cis_-acting elements are likely to be located not only in E16, but also in the flanking intron sequences (Gee et al., 1998; Deguillien et al., 2001). Further characterization of these elements and their cognate interacting proteins will be necessary to gain a better understanding of the mechanism(s) by which E16 splicing is regulated during erythropoiesis.
Materials and methods
Construction of plasmids
4.1R minigene substitution mutation, 4.1/PRE16-dsx and 4.1/PRE16-dsx(pcDNA) for transfection. Splice overlap extension PCR was used to replace the E16 silencer element with an equal length of Drosophila dsx exon 4 sequence containing no known splicing enhancer or silencer activity. A 5′ fragment containing 4.1R exon 13, the upstream intron and part of exon 16 fused to dsx sequences was amplified using oligonucleotides T7, 5′-TAATACGACTCACTATAGG-3′ and CE-DSX 3′, 5′-GTCATAGATATTCAAATTATGTTGACGGGAGTACTCATT ATCTAGTCTCTCTCTCTTTTT-3′. The 3′ half of the construct containing dsx sequences linked to the last two nucleotides of exon 16, plus the downstream intron and exon 17, was amplified using primers CE-DSX 5′, 5′-AATGAGTACTCCCGTCAACATAATTTGAATATCT ATGACGAGGTTTGTATGAACTTGAAG-3′, and E17.1, 5′-GCG AATTCCCGGGATTCAGT-3′. A mixture of the two fragments, containing an overlap of 39 nucleotides (underlined), was amplified with primers T7 and E17.1 to create the full minigene (4.1/PRE16-dsx) with a substituted silencer region.
4.1/PRE16-dsx(pcDNA) was made by amplifying the above minigene with primers E13, 5′-AGCCATTGCTCAGAGTCAGG-3′, and E17.1 using Pfu polymerase (Stratagene) to make a blunt-ended insert. After T7 polynucleotide kinase treatment, the insert was ligated into the plasmid pcDNA 3.1 (Invitrogen) that had been digested with _Bam_HI and _Eco_RV and the ends made blunt with mung bean nuclease.
dsx plasmid constructs dsx-PRE16, dsx-E16, dsx-PREdsx-CE16, dsx-PREdsx-16 and dsx-PREdsx-E16. A series of modified dsx constructs containing elements PRE16 or CE16 was generated by annealing complementary oligonucleotides into the _Cla_I site of plasmid p_dsx_T7 (Lynch and Maniatis, 1995). For _dsx_-PREdsx-16, _dsx_-PREdsx-CE16 and _dsx_-PREdsx-E16, the same annealed oligonucleotides were made blunt with mung bean nuclease and then subcloned into pCSC-PU (Lynch and Maniatis, 1995) linearized with _Sma_I. The following oligonucleotides were used: PRE16 sense, 5′-CGAAAAAGAGAGAGAGA-3′; PRE16 antisense, 5′-CGT CTCTCTCTCTTTTT-3′; CE16 sense, 5′- CGGTGAAAACATTTATA TCAGACATAGCAATTTAATGTTGGAG-3′; CE16 antisense, 5′-CGC TCCAACATTAAATTGCTATGTCTGATATAAATGTTTTCAC-3′; E16 sense, 5′-CGAAAAAGAGAGAGAGACTAGATGGTGAAAACATTT ATATCAGACATAGCAATTTAATGTTGGAG-3′; and E16 antisense, 5′-CGCTCCAACATTAAATTGCTATGTCTGATATAAATGTTTTC ACCATCTAGTCTCTCTCTCTTTTT-3′.
4.1R CE16 linker-scanning mutants. A _Bst_BI restriction site (TTCGAA) was engineered into the 4.1R minigene at 6 bp increments across CE16 using the QuikChange kit described above. A representative oligonucleotide primer pair used to generate the 4.1R mut-A mutation is given. Mutations mutB through mutG used the same methodology and primer design outlined. A1, 5′-TTCACAGAAAAAGAGAGAGAGACTAG TTCGAAAAAACATTTATATCAGACATAGCAAT-3′; and A2, 5′- ATTGCTATGTCTGATATAAATGTTTTTTCGAACTAGTCTCTCT CTCTTTTTCTGTGAA-3′.
Genomic sequencing of Xenopus 4.1R E16
A Xenopus genomic library (λFIX II, Stratagene) was screened by hybridization to radiolabeled Xenopus 4.1R cDNA. A 5 kb DNA fragment hybridizing to E16 was subcloned and flanking introns were sequenced using _Xenopus_-specific primers located within E16. Mouse and human genomic sequences at the intron/exon flanking E16 (Huang et al., 1993; Baklouti et al., 1997) and avian E16 sequence (Yew et al., 1987) have been published previously.
Synthesis of pre-mRNAs and microinjection into oocytes
Synthesis of capped RNA transcripts was done using the mMESSAGE mMACHINE (Ambion, Inc., Austin, TX) in vitro transcription kit according to the manufacturer’s protocols. 4.1R and β-globin plasmids were linearized and transcribed as described previously (Gee et al., 2000). dsx constructs were linearized with _Bam_HI and transcribed using the T7 promoter kit. Transcripts were purified using RNeasy (Qiagen, Valencia, CA) columns and microinjected into oocytes as described previously (Gee et al., 2000).
In vitro splicing assays
HeLa cell nuclear extract was prepared as described (Mayeda and Krainer, 1999b). The 25 µl splicing reactions, containing 6.25 fmol of RNA substrate in 40% HeLa cell nuclear extract, 3.2 mM MgCl2, 1 mM ATP, 20 mM creatine phosphate, 3.1% polyvinyl alcohol and 40 U of RNasin (Promega Corp., Madison, WI) were incubated 2 h at 30°C (Mayeda and Krainer, 1999a). All in vitro splicing experiments were performed at least three times.
Competitor splicing assays were performed by adding a 46 nucleotide 5′-biotinylated RNA (Dharmacon Research, Inc., Lafayette, CO) containing the conserved element of E16 (CE16) plus the two flanking nucleotides on each end with 2′_O_-methyl group modifications. A biotinylated RNA containing a high-affinity hnRNP H-binding site, 5′-AAGGGGGAGGUGUGGGUC-3′ (Bagga et al., 1995), was used as a control. Depleted nuclear extract was generated by pre-incubating the splicing mixture with up to 28 pmol of biotinylated RNA for 30 min at 4°C, then removing RNA and bound proteins by a 15 min capture step at 4°C with 150 µl of streptavidin MagneSphere paramagnetic particles (Promega) pre-washed in buffer D [20 mM HEPES–KOH pH 8, 100 mM KCl, 0.2 mM EDTA, 20% (v/v) glycerol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT)]. Recombinant hnRNP A1, A1B, A2, B1 and mutants A1-M(RRM1,2) and UP1 proteins were expressed in Escherichia coli and purified as described previously (Mayeda and Krainer, 1992; Mayeda et al., 1994).
Transient transfection of HeLa cells
HeLa cells were grown to ∼80% confluency in six-well plates and transfected using lipofectamine (Gibco-BRL, Gaithersburg, MD) according to the manufacturer’s protocol. In brief, 1 µg of each plasmid DNA was used in 10 µl of lipofectamine and transfected for 18 h before removal and addition of fresh media. Total RNA was then isolated after 48 h of incubation. The plasmid pCG-hnRNP A1 was used for overexpression of hnRNP A1 as described previously (Caceres et al., 1994). Control co-transfection experiments were performed using the empty expression vector pCG.
RT–PCR analysis of spliced pre-mRNA
Analysis of splicing reactions was carried out as described previously (Gee et al., 2000) using DNA primers in protein 4.1R exons 13 (forward) and 17 (reverse). Twenty-five cycles of PCR were used routinely to analyze the products of the splicing reactions; this condition was in the linear range of amplification since similar exon inclusion/exclusion ratios were obtained even after 35 cycles. As described previously (Gee et al., 2000), duplicate splicing reactions exhibited very little intra-experimental variability when processed in parallel under identical conditions. Although inter-experimental variability in absolute splicing efficiencies was observed, relative splicing efficiencies among various pre-mRNAs remained extremely consistent. Moreover, the relative splicing efficiencies of various pre-mRNA substrates was very similar in splicing assays with microinjected oocytes or transfected HeLa cells. Together, these results strongly support the validity of the RT–PCR results for in vitro splicing assays.
Image and densitometry measurements were done using the IS 1000 digital imaging system and software (Alpha Innotech Corp., San Leandro, CA). All labeled bands in the figures have been confirmed by sequencing. DNA primers used for analysis of dsx RNAs were located in exon 3 (sense) and exon 4 (antisense), as follows: E3, 5′-GGAGCTGATG CCACTCATGTATG-3′; and E4, 5′-GCTCACCCCCGTCATAGATA TTC-3′. The E4 primer was used for both the RT–PCRs.
Erythroblast cell procurement and culture
Erythroid cells obtained from the spleens of mice infected with the anemia-inducing strain of Friend erythroleukemia virus were isolated and cultured as described previously (Koury et al., 1984; Sawyer et al., 1987). Cells at t = 0 h are mainly proerythroblasts, which then differentiate over ∼48 h into late-stage erythroblasts and enucleated reticulocytes. Total RNA was isolated from cell pellets using RNeasy columns, and proteins prepared by lysing cells in 5× protein sample buffer (Conboy et al., 1991a) at a concentration of 20 000 cells/µl. Western blot analysis was done as described previously (Conboy et al., 1991a). Protein from 2 × 106 cells per sample was loaded for 4.1R + E16 (anti-SAB antibody) detection, and 5 × 105 cells per sample for hnRNP A/B (9H10 and DP3B3 monoclonal antibodies, respectively). Control blots were performed using anti-PTB and anti-hnRNP H antibodies (kindly provided by Doug Black).
Nanospray mass spectrometry
Bands of interest were excised and subjected to in-gel tryptic digestion (Hellman et al., 1995). Tryptic peptides were extracted from the gel pieces and cleaned using a gel-loader pipet tip filled with 100 nl of POROS C18 resin (PE Biosytems, Foster City, CA). The peptide mixture was eluted into a nanospray glass capillary (PROTANA, Odense, Denmark) using 500 nl of 60% methanol/5% formic acid, then peptide solutions were infused into an LCQ Iontrap mass spectrometer (FinniganMat, San Jose, CA) at a flow rate of 10 nl/min. Individual peptide ions were isolated and subjected to MS/MS analysis. The acquired MS/MS spectra were then subjected to protein and DNA database searches using the SEQUEST program (Eng et al., 1994).
Acknowledgments
Acknowledgements
We thank G.Dreyfuss for kindly providing the monoclonal antibodies 9H10 and DP3B3, D.Black for antibodies to hnRNP H and PTB, and T.Maniatis for providing the p_dsx_T7 and pCSC-PU plasmids. This work was supported by NIH grant HL45182 to J.G.C., by the Director, Office of Biological and Environmental Research, US Department of Energy under contract DE-AC03-76SF00098, and by a Merit Review Award from the Department of Veterans Affairs to M.J.K. A.M. is a research member of the Sylvester Comprehensive Cancer Center and supported by the Florida Biomedical Research Program Grant (BM031) from the FDH. A.R.K. acknowledges support from National Cancer Institute grant CA13106.
References
- Amendt B.A., Si,Z.H. and Stoltzfus,C.M. (1995) Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat–rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol., 15, 4606–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiya M. and Grabowski,P.J. (1997) A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA, 3, 996–1015. [PMC free article] [PubMed] [Google Scholar]
- Bagga P.S., Ford,L.P., Chen,F. and Wilusz,J. (1995) The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a _trans_-acting factor. Nucleic Acids Res., 23, 1625–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Lee,D., Yu,T. and Chasin,L.A. (1999) Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res., 27, 1126–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baklouti F., Huang,S.C., Vulliamy,T.J., Delaunay,J. and Benz,E.J.,Jr (1997) Organization of the human protein 4.1 genomic locus: new insights into the tissue-specific alternative splicing of the pre-mRNA. Genomics, 39, 289–302. [DOI] [PubMed] [Google Scholar]
- Berget S.M. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem., 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- Bilodeau P.S., Domsic,J.K., Mayeda,A., Krainer,A.R. and Stoltzfus,C.M. (2001) RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol., 75, 8487–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M. and Chabot,B. (1999) Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J., 18, 1939–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- Caputi M., Mayeda,A., Krainer,A.R. and Zahler,A.M. (1999) hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J., 18, 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B. (1996) Directing alternative splicing: cast and scenarios. Trends Genet., 12, 472–478. [DOI] [PubMed] [Google Scholar]
- Chan R.C. and Black,D.L. (1997) The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol., 17, 4667–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasis J.A., Coulombel,L., Conboy,J., McGee,S., Andrews,K., Kan,Y.W. and Mohandas,N. (1993) Differentiation-associated switches in protein 4.1 expression. Synthesis of multiple structural isoforms during normal human hematopoiesis. J. Clin. Invest., 91, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy J. (1999) The role of alternative pre-mRNA splicing in regulating the structure and function of skeletal protein 4.1. Proc. Soc. Exp. Biol. Med., 220, 73–78. [DOI] [PubMed] [Google Scholar]
- Conboy J.G., Chan,J., Chasis,J.A., Kan,Y.W. and Mohandas,N. (1991a) Tissue- and development-specific alternative RNA splicing regulates expression of multiple isoforms of erythroid membrane protein 4.1. J. Biol. Chem., 266, 8273–8280. [PubMed] [Google Scholar]
- Conboy J.G., Shitamoto,R., Parra,M., Winardi,R., Kabra,A., Smith,J. and Mohandas,N. (1991b) Hereditary elliptocytosis due to both qualitative and quantitative defects in membrane skeletal protein 4.1. Blood, 78, 2438–2443. [PubMed] [Google Scholar]
- Deguillien M., Huang,S.C., Moriniere,M., Dreumont,N., Benz,E.J.,Jr and Baklouti,F. (2001) Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood, 98, 3809–3816. [DOI] [PubMed] [Google Scholar]
- Del Gatto F. and Breathnach,R. (1995) Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol., 15, 4825–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto F., Gesnel,M.C. and Breathnach,R. (1996) The exon sequence TAGG can inhibit splicing. Nucleic Acids Res., 24, 2017–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Olive,M., Gesnel,M.C. and Breathnach,R. (1999) hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol., 19, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Hayashi,M.K., Zhang,Y., Manche,L., Krainer,A.R. and Xu,R.M. (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev., 13, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D., Parra,M., Conboy,J.G. and Mohandas,N. (1993) Mechanochemistry of the alternatively spliced spectrin–actin binding domain in membrane skeletal protein 4.1. J. Biol. Chem., 268, 7186–7195. [PubMed] [Google Scholar]
- Discher D.E., Winardi,R., Schischmanoff,P.O., Parra,M., Conboy,J.G. and Mohandas,N. (1995) Mechanochemistry of protein 4.1’s spectrin–actin-binding domain: ternary complex interactions, membrane binding, network integration, structural strengthening. J. Cell Biol., 130, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng Y., McCormack,A.L. and Yates,J.R.,III (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom., 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Fu X.D., Mayeda,A., Maniatis,T. and Krainer,A.R. (1992) General splicing factors SF2 and SC35 have equivalent activities in vitro and both affect alternative 5′ and 3′ splice site selection. Proc. Natl Acad. Sci. USA, 89, 11224–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascard P., Nunomura,W., Lee,G., Walensky,L., Krauss,S.W., Takakuwa,Y., Chasis,J.A., Mohandas,M. and Conboy,J.G. (1999) Deciphering the nuclear import pathway for the cytoskeletal protein 4.1R. Mol. Biol. Cell, 10, 1783–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee S.L., Parra,M., Willig,T., Hou,V.C., Chan,N., Wu,M. and Conboy,J.G. (1998) Alternative splicing of protein 4.1 exon 16 is regulated in part by downstream intron elements. Blood, 92, Suppl. 1, 5a. [Google Scholar]
- Gee S.L., Aoyagi,K., Lersch,R., Hou,V., Wu,M. and Conboy,J.G. (2000) Alternative splicing of protein 4.1R exon 16: ordered excision of flanking introns ensures proper splice site choice. Blood, 95, 692–699. [PubMed] [Google Scholar]
- Graham I.R., Hamshere,M. and Eperon,I.C. (1992) Alternative splicing of a human α-tropomyosin muscle-specific exon: identification of determining sequences. Mol. Cell. Biol., 12, 3872–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman U., Wernstedt,C., Gonez,J. and Heldin,C.H. (1995) Improvement of an in-gel digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem., 224, 451–455. [DOI] [PubMed] [Google Scholar]
- Hertel K.J., Lynch,K.W. and Maniatis,T. (1997) Common themes in the function of transcription and splicing enhancers. Curr. Opin. Cell Biol., 9, 350–357. [DOI] [PubMed] [Google Scholar]
- Horne W.C., Huang,S.C., Becker,P.S., Tang,T.K. and Benz,E.J.J. (1993) Tissue-specific alternative splicing of protein 4.1 inserts an exon necessary for formation of the ternary complex with erythrocyte spectrin and F-actin. Blood, 82, 2558–2563. [PubMed] [Google Scholar]
- Huang J.-P., Tang,C.-J., Kou,G.-H., Marchesi,T., Benz,E.J.,Jr and Tang,T.K. (1993) Genomic structure of the locus encoding protein 4.1. Structural basis for complex combinational patterns of tissue-specific alternative RNA splicing. J. Biol. Chem., 268, 3758–3766. [PubMed] [Google Scholar]
- Kan J.L. and Green,M.R. (1999) Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev., 13, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig H., Ponta,H. and Herrlich,P. (1998) Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J., 17, 2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury M.J., Sawyer,S.T. and Bondurant,M.C. (1984) Splenic erythroblasts in anemia-inducing Friend disease: a source of cells for studies of erythropoietin-mediated differentiation. J. Cell. Physiol., 121, 526–532. [DOI] [PubMed] [Google Scholar]
- Koury S.T., Koury,M.J. and Bondurant,M.C. (1989) Cytoskeletal distribution and function during the maturation and enucleation of mammalian erythroblasts. J. Cell Biol., 109, 3005–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Liu H.X., Zhang,M. and Krainer,A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque C.M., Lallena,M.J., Alonso,M.A. and Correas,I. (1998) An alternative domain determines nuclear localization in multifunctional protein 4.1. J. Biol. Chem., 273, 11643–11649. [DOI] [PubMed] [Google Scholar]
- Lynch K.W. and Maniatis,T. (1995) Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev., 9, 284–293. [DOI] [PubMed] [Google Scholar]
- Matter N., Marx,M., Weg-Remers,S., Ponta,H., Herrlich,P. and Konig,H. (2000) Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem., 275, 35353–35360. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Krainer,A.R. (1999a) Mammalian in vitro splicing assays. Methods Mol. Biol., 118, 315–321. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Krainer,A.R. (1999b) Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol., 118, 309–314. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Helfman,D.M. and Krainer,A.R. (1993) Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF [published erratum appears in Mol. Cell. Biol. (1993) 13, 4458]. Mol. Cell. Biol., 13, 2993–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Munroe,S.H., Caceres,J.F. and Krainer,A.R. (1994) Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J., 13, 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Screaton,G.R., Chandler,S.D., Fu,X.D. and Krainer,A.R. (1999) Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol., 19, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K.K. and Green,M.R. (1990) Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 87, 6253–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. (1996) Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev., 6, 215–220. [DOI] [PubMed] [Google Scholar]
- Sawyer S.T., Koury,M.J. and Bondurant,M.C. (1987) Large-scale procurement of erythropoietin-responsive erythroid cells: assay for biological activity of erythropoietin. Methods Enzymol., 147, 340–352. [DOI] [PubMed] [Google Scholar]
- Schaal T.D. and Maniatis,T. (1999) Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol., 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schischmanoff P.O., Winardi,R., Discher,D.E., Parra,M.K., Bicknese,S.E., Witkowska,H.E., Conboy,J.G. and Mohandas,N. (1995) Defining the minimal domain of protein 4.1 involved in spectrin–actin binding. J. Biol. Chem., 270, 21243–21250. [DOI] [PubMed] [Google Scholar]
- Si Z.H., Rauch,D. and Stoltzfus,C.M. (1998) The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol., 18, 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C.W., Fresco,L.D. and Rio,D.C. (1992) The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev., 6, 1386–1401. [DOI] [PubMed] [Google Scholar]
- Smith C.W. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- Staffa A., Acheson,N.H. and Cochrane,A. (1997) Novel exonic elements that modulate splicing of the human fibronectin EDA exon. J. Biol. Chem., 272, 33394–33401. [DOI] [PubMed] [Google Scholar]
- Takakuwa Y., Tchernia,G., Rossi,M., Benabadji,M. and Mohandas,N. (1986) Restoration of normal membrane stability to unstable protein 4.1-deficient membranes by incorporation of purified protein 4.1. J. Clin. Invest., 78, 80–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange T.O., Damgaard,C.K., Guth,S., Valcarcel,J. and Kjems,J. (2001) The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J., 20, 5748–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. and Manley,J.L. (1997) Regulation of pre-mRNA splicing in metazoa. Curr. Opin. Genet. Dev., 7, 205–211. [DOI] [PubMed] [Google Scholar]
- Watakabe A., Tanaka,K. and Shimura,Y. (1993) The role of exon sequences in splice site selection. Genes Dev., 7, 407–418. [DOI] [PubMed] [Google Scholar]
- Winardi R., Discher,D., Kelley,C., Zon,L., Mays,K., Mohandas,N. and Conboy,J.G. (1995) Evolutionarily conserved alternative pre-mRNA splicing regulates structure and function of the spectrin–actin binding domain of erythroid protein 4.1. Blood, 86, 4315–4322. [PubMed] [Google Scholar]
- Yang X., Bani,M.R., Lu,S.J., Rowan,S., Ben-David,Y. and Chabot,B. (1994) The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl Acad. Sci. USA, 91, 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew N.S., Choi,H.R., Gallarda,J.L. and Engel,J.D. (1987) Expression of cytoskeletal protein 4.1 during avian erythroid cellular maturation. Proc. Natl Acad. Sci. USA, 84, 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Mayeda,A. and Krainer,A.R. (2001) Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell, 8, 1351–1361. [DOI] [PubMed] [Google Scholar]