Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection (original) (raw)
Abstract
Helicobacter pylori infects over half the world's population and causes a wide range of diseases, including gastritis, peptic ulcer, and two forms of gastric cancer. H. pylori infection elicits a variety of phenotypic responses in cultured gastric epithelial cells, including the expression of proinflammatory genes and changes in the actin cytoskeleton. Both of these responses are mediated by the type IV secretion system (TFSS) encoded by the cag pathogenicity island (cag PAI). We used human cDNA microarrays to examine the temporal transcriptional profiles of gastric AGS cells infected with H. pylori strain G27 and a panel of isogenic mutants to dissect the contributions of various genes in the cag PAI. Infection with G27 induced expression of genes involved in the innate immune response, cell shape regulation, and signal transduction. A mutant lacking the cagA gene, which encodes an effector molecule secreted by the TFSS and required for the host cell cytoskeletal response, induced the expression of fewer cytoskeletal genes. A mutant lacking cagE, which encodes a structural component of the TFSS, failed to up-regulate a superset of host genes, including the _cagA_-dependent genes, and many of the immune response genes. A mutant lacking the entire cag PAI failed to induce both the _cagE_-dependent genes and several transiently expressed cagE independent genes. Host cell transcriptional profiling of infection with isogenic strains offered a detailed molecular picture of H. pylori infection and provided insight into potential targets of individual virulence determinants such as tyrosine kinase and Rho GTPase signaling molecules.
Pathogenic bacteria have evolved elaborate mechanisms to manipulate host cells during infection. Helicobacter pylori, a human pathogen associated with a broad spectrum of gastric maladies, is no exception, possessing a number of virulence determinants that modulate its interaction with its host (1). These virulence factors include the secreted vacuolating cytotoxin VacA and the gene products of the pathogenicity island (cag PAI). The presence of these genetic loci is correlated with the more severe _H. pylori_-associated pathologies, peptic ulcer and gastric cancer.
The cag PAI encodes a type IV secretion system (TFSS) that facilitates the injection of bacterial proteins into eukaryotic cells. H. pylori attach to the surface of the cells and deliver the cag _PAI_-encoded protein, CagA, into the host cells (2–6). In a simplified tissue culture model of infection, using transformed gastric epithelial AGS cells, CagA is tyrosine phosphorylated by a host cell kinase of the Src family (7, 8). The phosphorylation of CagA and its subsequent interaction with SHP-2 phosphatase (9) are necessary for signaling events that lead to a dramatic cellular elongation (5). The molecular events that lead to this alteration of the actin cytoskeleton are not yet known, although the involvement of small GTPases has been suggested (10).
A second cellular phenotype associated with the cag PAI, but independent of the cagA gene product, is the activation of NF-κB and the expression of proinflammatory cytokines such as IL-8 (11). Although IL-8 expression does not require CagA delivery, it does require a functional TFSS. Thus disruption of the cagE gene, which encodes the homologue of the Agrobacterium VirB4 ATPase that powers the translocation process, or deletion of the entire cag PAI, obliterates _H. pylori_-induced IL-8 synthesis. Systematic mutagenesis studies of the cag PAI members have failed to identify a putative cytokine-inducing effector but have revealed other genes in the island that are not required for either CagA delivery or IL-8 induction, such as cagN (12–15). Other _cagA-_independent effector functions of the cag PAI remain to be discovered.
The host cell response to infection in a number of models of pathogenesis has been characterized at the transcriptional level by using DNA microarrays (reviewed in ref. 16). A common finding in many of these investigations is up-regulation of genes involved in the inflammatory response (17–19). This pattern is not restricted to cells of the immune system but can be elicited in epithelial cells as well (20, 21). Beyond a common innate immune response to infection, different cells may have characteristic signatures to different pathogens, often as the result of the highly specific activities of a particular pathogen's virulence determinants. Such phenotypic differences can be detected at the level of the host transcriptional responses to isogenic mutants (22). Several groups have examined the global transcriptional response of gastric epithelial cells to H. pylori (23–27). Here we have used microarray transcriptional profiling of gastric epithelial cells infected with a panel of isogenic strains to characterize in detail the temporal response of gastric epithelial cells to H. pylori infection and to dissect the contributions of individual virulence determinants.
Materials and Methods
Bacterial and Cell Culture.
H. pylori strains used in this study are described in Table 1. _vacA_− and cagA− derivatives of G27 were obtained by natural transformation with 10 μg of pVacKanSacB or pCagKan and selection of kanamycin-resistant clones as described (28). Disruptions of the vacA and cagA loci were confirmed by PCR and Western blotting (not shown). For time course (TC)1, AGS cells [American Type Culture Collection (ATCC) CRL-1739] were grown in 90% DMEM 10% FBS (Invitrogen Life Technologies) to ≈70% confluence in T175 flasks. H. pylori strains were grown microarobically at 37°C overnight in 90% Brucella broth (Fisher Scientific)/10% FBS and used to inoculate the cells at a multiplicity of infection of ≈10:1. At each time point, AGS cells were collected by scraping, isolated by centrifugation, and stored at −80°C for subsequent total RNA isolation by using Trizol Reagent according to the manufacturer's instructions (Invitrogen Life Technologies). For time course 2 (TC2), H. pylori strains were maintained in continuous culture with Madin–Darby canine kidney (MDCK) tissue culture cells (ATCC CCL-34) in DB media (81% DMEM/9% Brucella broth/10% FBS) in a 37°C incubator equilibrated with 5% CO2 and 95% air. Bacteria were subcultured by replacing the DB media daily. Under these culture conditions, the bacteria grew in the aqueous layer above but not attached to the MDCK cells, were consistently highly motile and helical, and exhibited maximum potential to translocate CagA (M. Amieva, personal communication). For the TC2 infection, AGS cells were grown in DB media in 150-mm dishes to ≈70% confluence. Bacteria in DB media were added at time 0 to a multiplicity of infection of ≈25:1 (range 18–50:1, determined by dilution plating of zero time point). At 0, 1, 3, 6, 12, and 24 h, media were removed, and bacteria and host cells were lysed directly in the dishes by addition of Trizol reagent. The cell suspension was stored at −80°C for subsequent total RNA isolation as above. Total RNA was then further purified with RNeasy kits (Qiagen, Chatsworth, CA). Reference RNA was generated from a single passage of uninfected AGS cells.
Table 1.
Strains
| Strain | Genotype | Ref. |
|---|---|---|
| G27 | Wild type, PAI+ | 43 |
| cagA− | G27, cagA:aphA3 | This study |
| vacA− | G27, vacA:aphA3 | This study |
| cagE− | G27, cagE:Tn3Kan | 12 |
| cagN− | G27, cagN:Tn3Kan | 12 |
| Δ_PAI_ | G27, Δ_PAI_ | 10 |
Microarray Hybridization.
Detailed protocols for probe synthesis and DNA microarray hybridization are given at http://cmgm.stanford.edu/pbrown/protocols. For each experimental sample, 40 μg of total RNA was used for single-stranded cDNA probe synthesis, incorporating dUTP conjugated with either Cy3 (for the reference) or Cy5 (for the experimental sample). Experimental and reference samples were combined and hybridized at 65°C in 3.4× SSC and 0.3% SDS to a 22,571-element human cDNA microarray (described at www.microarray.org).
Data Analysis.
Arrays were scanned by using a GenePix 4000A scanner (Axon Instruments, Foster City, CA) and images analyzed with genepix pro software. Microarray data were stored in the Stanford Microarray Database (29) and are publicly available (www.dnachip.org). Data were filtered to remove poor-quality measurements and log2 transformed. For hierarchical clustering in Fig. 1, the data for each TC were zero-transformed by averaging the zero time point measurements for each gene and subtracting these values from later time points followed by analysis using cluster and treeview (30). Statistical analysis was done on nonzero-transformed data with significance analysis of microarrays (SAM) (31). Enrichment of functional classes of genes in the SAM output was assessed by sorting for keywords: (i) “tyrosine kinase” or “tyrosine phosphatase”; (ii) “cdc42,” “rac,” or “rho”; (iii) “actin,” not “acting”; (iv) “junction,” “claudin,” or “cadherin” in the gene description for each clone, comparing the number of clones recovered from the entire filtered dataset versus the number recovered from the SAM output list by using a standard χ2 test.
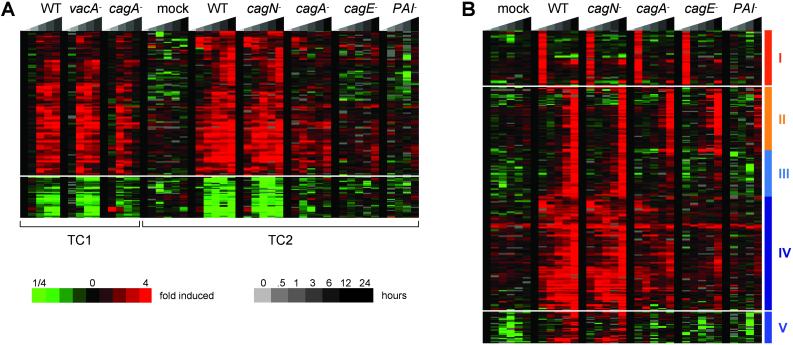
Fig 1.
Gastric epithelial cell transcriptional response to isogenic H. pylori strains. The expression profiles of AGS cells cocultured with isogenic strains of H. pylori (TC1: G27, vacA −, cagA − TC2: mock, G27, cagN −, cagA −, cagE −, Δ_PAI_) for various lengths of time (TC1: 0, 0.5, 3, 6, 12 h; TC2: 0, 1, 3, 6, 12, 24) were determined for 22,571 human cDNAs arrayed on glass slides. The data were filtered to remove poor-quality spots. Genes were selected that had good data for 80% of the experiments and whose expression level changed by 3-fold or greater in at least two experiments. The gene profiles were organized by hierarchical clustering and displayed in matrices in which each row corresponds to an array element and each column corresponds to an experimental condition. The experiments are organized by increasing time of coculture with each strain, as shown in the key. Each measurement is the ratio of the treated sample vs. a common reference sample from uninfected AGS cells and is represented relative to the averaged time 0 measurements of each TC. The level of induction is indicated by color, as shown in the key (missing data are indicated in gray). (A) _H. pylori_-responsive genes. One hundred twenty-one induced genes (above) and 34 repressed genes (below) responded to coculture with H. pylori with similar kinetics in two independent experiments. Larger versions of these clusters, with gene names, can be found in Figs. 3 and 4. (B) _cag PAI_-dependent gene induction. Hierarchical clustering of the TC2 data revealed genes whose induction depended on an intact TFSS (clusters I and II, indicated by orange vertical bars) or cagA (clusters III, IV, and V, indicated by blue vertical bars). Larger versions of these clusters, with gene names, can be found in Figs. 5–10.
Results and Discussion
A Canonical Transcriptional Response to H. pylori Infection.
To characterize host cell responses to H. pylori infection and to identify responses specific to individual virulence determinants, we used human cDNA microarrays to monitor the relative abundance of gastric epithelial cell transcripts over time during coculture with a panel of isogenic H. pylori strains. Initially we characterized the transcriptional response of gastric epithelial AGS cells infected for 0, 0.5, 3, 6, and 12 h with a cag _PAI_-positive wild-type strain (G27) or isogenic mutants with disruptions in either the vacuolating cytotoxin VacA gene or the cag _PAI_-encoded gene for the effector CagA (TC1). Phenotypically the _vacA_− infected cells resembled those infected with wild-type H. pylori, exhibiting the characteristic cell elongation (5) apparent by 3 h postinfection. In addition, the wild-type infected AGS cells did not exhibit any excessive vacuolation relative to the _vacA_− mutant, a phenotype seen in other cell types or under different growth conditions. In contrast, the _cagA−_-infected cells failed to elongate. Cellular transcripts from each time point were compared with a common reference RNA from uninfected AGS cells by using a 22,571-element spotted human cDNA.
We used unsupervised hierarchical clustering (30) to identify genes whose expression changed during the parallel infections (Fig. 1A, TC1). This method allowed us to identify genes whose measured levels appeared biologically meaningful and reproducible across the parallel infections. Indeed, a distinct cluster of host genes was readily apparent whose expression increased during the TC of infection with all three strains, although some genes were less induced on coculture with the _cagA_− mutant. In addition, a distinct cluster of genes exhibited decreased levels over the course of the infections, with less repression observed in the cagA− infection.
To confirm the biological reproducibility of the results, the infection TC experiment was repeated. We modified the experiment to improve our temporal resolution by examined transcript levels at time points 0, 1, 3, 6, 12, and 24 h postinfection. In addition, we used a larger number of isogenic mutants to further dissect the contributions of the cag PAI to the host cell response. Six parallel experiments were performed by using a mock infection and infections with G27 and isogenic strains with mutations in cagN, cagA, cagE, or a deletion of the entire cag PAI (TC2). The transcriptional profiles were determined as for TC1.
When the entire dataset for TC1 and TC2 was subjected to hierarchical clustering, again clusters of induced and repressed genes were readily apparent (Fig. 1A and Figs. 3 and 4, which are published as supporting information on the PNAS web site, www.pnas.org). Using only genes whose level changed by 3-fold or more in at least two experiments across the dataset, we identified 127 induced clones corresponding to 106 unique genes. We designate these genes the canonical H. pylori responsive genes (Table 2, which is published as supporting information on the PNAS web site). Of the genes with assigned functions, the two most abundant functional classes of induced genes encoded proteins involved in the innate immune response and the regulation of cell shape and adhesion, consistent with the characterized AGS cell responses to H. pylori of cytokine expression and cell elongation.
H. pylori's proinflammatory properties have been well studied in the AGS cell model, including their ability to induce neutrophil-attracting chemokines (11). We measured induction of the chemokine GRO1, previously reported to be up-regulated by H. pylori infection, as well as three other cytokines (IL-8 was also induced in our experiments but to lower levels, excluding it from our induced set). Cytokine induction by H. pylori is known to be mediated by NF-κB signaling. Correspondingly we observed induction of several NF-κB signaling genes including NF-κB1 (p105) and RelB. Another proinflammatory cytokine associated with H. pylori infection is tumor necrosis factor (TNF)α (11). The induced gene set contained several targets of TNFα regulation, including four genes whose function is to protect cells against TNFα-induced apoptosis. In addition, among the transcription factors in the induced gene set were several jun transcription factor family members, components of the AP-1 transcription complex that mediates H. pylori's cytokine induction in response to NF-κB activation.
Less is known about the molecular signaling events that lead to _H. pylori_-induced cell elongation. Among the induced genes were those that encoded the regulator of the actin cytoskeleton, Cdc42 effector protein 2 (CEP2 or Borg1) (32, 33), as well as molecules associated with the actin cytoskeleton (αV integrin) and intermediate filaments (keratin 17). Cell elongation, as well as inflammation, would be expected to cause changes in cells' connectivity with their substratum and their neighbors. The induced gene set encoded components of the extracellular matrix (laminin γ2, collagen VIα, matrix metalloproteinase 14), cell adhesion molecules (ICAM1), and a number of genes encoding components of cellular junctions (tight junction proteins claudins 1 and 4).
Coculture with H. pylori also caused specific repression in host gene expression. Clustering with the same filtering criteria used to identify up-regulated genes, we identified 34 clones, corresponding to 30 unique genes, repressed across both TCs (Fig. 1A and Table 2). One-fifth of these encoded signal transduction molecules, including the wingless signaling pathway members frizzled 7 and dickkopf, the antagonist of receptor tyrosine kinase signaling, disabled homologue 2, and the mitoattractant connective tissue growth factor. A different set of signaling molecules was induced by H. pylori infection, including the growth factors platelet-derived growth factor and prostate differentiation factor, a transforming growth factor family member. This suggests that H. pylori affects signaling in the host cell through both inductive and repressive mechanisms.
Differential Host Cell Responses to Infection with Wild Type and a Panel of cag PAI Mutants.
To better dissect the contribution of other cag PAI genes to the response of gastric cells to H. pylori infection, we analyzed additional cag PAI mutants in TC2. These included, (i) cagN− that does not disrupt CagA delivery or IL-8 expression and causes an infection that resembles that of wild-type H. pylori with elongated cells; (ii) _cagA_− that disrupts cell elongation but not IL-8 expression; (iii) _cagE_− that abolishes TFSS function, including cell elongation and IL-8 expression; and (iv) a deletion of the entire cag PAI.
Hierarchical clustering of the TC2 data with the larger panel of isogenic mutants and the increased temporal sampling revealed additional subtleties in the host cell transcriptional response to infection (Fig. 1B). Perhaps the most striking finding was that most of the H. pylori_-specific gene induction and repression was absent in the Δ_PAI infection, which resembled the mock infection. This suggests that the major response of AGS cells to H. pylori is mediated through the cag PAI.
The cagE− mutant also failed to up-regulate many of the _H. pylori_-responsive genes (Fig. 1B). However, at both the 1- and 24-h time points, which had not been sampled in the previous experiment, coculture with cagE_− induced transient expression of a number of genes also induced in the other infections, except with the Δ_PAI mutant. The genes transiently induced at 1 h (Fig. 1B, cluster I; Fig. 5, which is published as supporting information on the PNAS web site) included those involved in immediate early responses to stress or tissue damage such as the proinflammatory molecule IFN-γ, the transcription factors fos and transforming growth factor β-inducible early response, and a number of mediators of the inflammatory response, including plasminogen, endothelin 1, trefoil factor 1, tissue inhibitor of metalloprotease 3, and adipose most abundant 1. Additionally, a number of signal transduction genes were transiently induced, including the Src family tyrosine kinase, Lyn, the c-Jun-N-terminal kinase stimulated phosphatase mitogen-activated protein kinase phosphatase X, and the receptor tyrosine kinases HER2 and insulin-like growth factor 1 receptor. These may be involved in early host signaling events induced by H. pylori, although the fact that they are induced in the absence of CagA delivery (in the cagA− and cagE− mutants) would argue against a role in cagA_-dependent signaling events. Another intriguing group of the immediate transiently expressed genes may function in the cellular recognition event between bacteria and host cell. These included CD14, a component of the lipopolysaccharide-binding receptor, mucin 1, and α3, α6, and β4 integrins. The integrins have been proposed as possible receptors for the TFSS (34). That the cagE− mutant, unlike Δ_PAI, elicits this gene expression profile along with H. pylori strains with a functional TFSS (G27, cagN−, cagA−) demonstrates that host cells respond differently to H. pylori lacking the entire TFSS and those with a defective secretory system. The cagE− mutant may still harbor remnants of the secretion apparatus on its surface, which may allow it to engage in more intimate contact with host cells, eliciting a more proinflammatory response even in the absence of TFSS effector-mediated responses.
The pattern of PAI_-dependent expression was also apparent at the 24-h time point for a group of genes strongly up-regulated in all infections except for weak induction with the Δ_PAI mutant (Fig. 1B, cluster II; Fig. 6, which is published as supporting information on the PNAS web site). Again, a number of stress-related genes were induced, including the Hsp40 homologue DnaJ, the hypoxia sensor, hypoxia inducible factor (HIF) prolyl hydroxylase 3, and a HIF-responsive gene, as well as reexpression, from the 1-h time point, of trefoil factor 1, mucin 1, and the myc inhibitory MAX-interacting protein. Strikingly, the most abundant functional class of genes induced transiently at 24 h consisted of genes encoding the enzymes of the cholesterol biosynthesis pathway. This up-regulation cannot be attributed simply to an increase in the host cell membrane, because it occurred in infected cells regardless of whether they underwent cell elongation. Possibly cholesterol up-regulation functions as a host defense mechanism against bacterial insult by altering membrane fluidity or increasing concentrations of endotoxin neutralizing lipoproteins (35). Cholesterol, as the major component of membrane rafts, is emerging as an important component of the host cells' recognition of bacterial constituents, including lipopolysaccharide via CD14 (36). In addition, membrane cholesterol plays a specific role in the pathogenicity of the H. pylori cytotoxin VacA (37, 38), although the vacA dependence of cholesterol gene expression was not tested in these experiments, as the vacA− mutant was not analyzed at the 24-h time point. H. pylori induction of cholesterol production in host cells could provide a clue to the putative association between chronic H. pylori infection and atherosclerosis (39).
A second class of genes regulated in a _cagA_-dependent manner late in the TC suggests cellular processes that occur as a consequence of CagA injection. A cluster of induced genes (Fig. 1B, cluster III; Fig. 7, which is published as supporting information on the PNAS web site) was enriched for genes encoding cytoskeleton-associated proteins, such as the F-actin-binding proteins feminization 1A, cylindromatosis protein, and αV integrin, and the adhesion junction-associated proteins plakin family member bullous perphigoid antigen and AF-6. The kinetics of these genes' expression is consistent with a role in the downstream events of CagA signaling associated with the cytoskeletal changes in the elongated G27 and cagN− infected cells. Among the _cagA_-dependent repressed genes was a cluster of late repressed genes that function in DNA replication and cell cycle progression (Fig. 8, which is published as supporting information on the PNAS web site). This transcriptional profile is consistent with the observation that H. pylori induces cell cycle arrest in AGS cells (40). Although the mechanism of _H. pylori_-mediated cell cycle arrest is not well understood, our observation of _cagA_-dependent repression of cell proliferation genes suggests a role, direct or indirect, for CagA in this process.
The final group of genes was induced early in the infection at a much higher level in the G27 and _cagN_− infections than in the others (Fig. 1B, clusters IV and V; Figs. 9 and 10, which are published as supporting information on the PNAS web site). These contained many of the genes shared in common with TC1 and included many innate immune response, cell shape, and signal transduction genes. The level of induction of many of these genes in cluster IV is intermediate for the cagA− infection relative to the strong induction with G27 and _cagN_− and the weak induction with cagE_− and Δ_PAI. Ranking the average induction across the TC for all of the genes in this cluster revealed that the genes disrupted least in their induction in the cagA− profile were those involved in the immune response including the cytokine MIP1β, natural killer cell transcript 4, and the regulators of NF-κB signaling RelB, BIRC3 and TNFAIP3. This is consistent with the known _cagA_-independent proinflammatory activity of the cag PAI. Cluster V consists of genes that were not induced at all in the cagA− infection. Interesting, this cluster is enriched for genes encoding molecules involved in tyrosine kinase signal pathways, including growth factor receptor-bound protein 10, MAPKK5, and the Src family member Lck. CagA has been shown to be a substrate for Src tyrosine kinases (7, 8); the _cagA_-specific induction of Lck makes it an intriguing candidate for the CagA kinase in these cells.
_cagA_- and _cagE_-Dependent Host Cell Responses.
The cluster analysis identified similarly regulated genes that exhibited subtle differences in their level of expression between different isogenic strains. To identify statistically significant difference in gene expression between infections with different strains, we used SAM (31). SAM uses a t test algorithm to find genes whose levels differ significantly between conditions relative to the variation within replicates, thus allowing the identification of genes whose absolute level of induction could be small (and thus excluded from the cluster analysis). Because of the similarity in the expression profiles at 1, 3, 6, 12, and 24 h for the majority of the H. pylori_-responsive genes, we treated these points as replicates and used an unpaired t test to compare them between different parallel infections. Comparing the mock and Δ_PAI infections, which appeared very similar by cluster analysis, we identified only seven significantly different genes with a minimum false-positive rate of 28.6% (two false positives). Similarly, comparing the G27 and _cagN_− infections, no significantly different genes could be identified.
We next compared gene expression between the G27 and _cagN_−-infected cells (which we treated as replicates) and those infected with either the _cagA_− mutant or the _cagE_− mutant (Table 3, which is published as supporting information on the PNAS web site). The _cagA_− infection differed from wild-type and _cagN_− in the expression of 21 clones (corresponding to 18 genes), with a minimal false discovery rate of 4.6% (one false positive). The most abundant class of genes in this group were those involved in cell shape changes, including the actin-interacting protein pleckstrin homologue domain1, the small GTPase RhoB, and the Cdc42 effector protein 2, as well as the junctional proteins claudin 4 and enigma. The comparison of G27 and _cagN_−- with _cagE−_-infected cells revealed a larger group of 68 significantly different clones (60 genes), with a minimum false discovery rate of 1.2% (0.8 false positives). With the exception of three clones, the _cagE_-dependent induced genes constituted a superset of the _cagA_-dependent genes, consistent with the fact that CagA requires CagE to be translocated into host cells. In the set of genes that differed significantly between the wild-type and _cagE_− infections, but not between the wild-type and _cagA_− infections, were many genes involved in inflammation, including NF-κB and TNFα signaling.
Our dataset sampled a large number of cells that had been exposed to H. pylori in the presence or absence of the CagA effector. We extended the SAM analysis to compare the 3-h or later responses of all cells that received CagA (wild type, _vacA_−, and cagN_− infections, n = 17) to all cells that did not (cagA−, cagE−, and Δ_PAI infections, n = 17). This dataset included a great deal of biological variation, but the replication allowed us to identify a much larger set of genes that differed significantly, although not necessarily dramatically, between the two groups: 603 elevated and 59 repressed (with a minimum false discovery rate of 0.09%) (Table 4, which is published as supporting information on the PNAS web site). The expression data for all of the genes identified by this analysis were hierarchically clustered and are shown in Fig. 2 (expanded versions are shown in Figs. 11 and 12, which are published as supporting information on the PNAS web site). Although some of the selected genes merely reflect differences between TC1 and TC2, due to the fact that more samples not exposed to CagA came from TC2, the majority exhibit significant differences in the level of gene expression between cells exposed to CagA or not. The list offers an intriguing picture of molecules and cellular functions associated with CagA activity, including possible downstream effectors of CagA signaling whose transcriptional levels may be expected to fluctuate only subtly in these experiments. For example, among all genes annotated to have tyrosine kinase or phosphatase activity, the only Src family tyrosine kinase up-regulated in the presence of CagA was Lck, and the only nonreceptor tyrosine phosphatase up-regulated was SHP2, known to associate with phospho-CagA (9). CagA's activity on the actin cytoskeleton has been suggested to be mediated through the Rho family of small GTPases (10). Among this class, both Cdc42 and the Cdc42 effector protein 2 were up-regulated, making them good candidates for this activity. The CagA-induced changes on the actin cytoskeleton were suggested by the CagA-specific up-regulation of actin itself (γ2 and α2) and a large number of actin-associated genes that are known to play a role in cytoskeleton remodeling, including Arp2/3 subunit 5, α-actinin, capping protein, filamin, and coronin. Finally, among the induced genes were a striking number of junctional proteins, including the adhesion junction protein E-cadherin and its associated proteins α and β catenin; as well as 10 of the 19 tight junction-associated clones on the array, including claudins 1, 3, and 4; occludin; and junctional adhesion molecule. In each of these cases, enrichment in the induced class over the total set of clones on the array was significant by a χ2 test (P < 0.05). The _cagA_-dependent expression of tight junction genes has led to the investigation of the role of cell junctions as an important site for _H. pylori_–epithelium interactions. Indeed, this striking interaction between H. pylori and cellular junctions appears to be an important feature of H. pylori pathogenesis, as revealed by data from biopsy specimens (41) and cell biological experiments on the function of CagA in polarized epithelia (M. R. Amieva, R. Vogelmann, A. Covacci, L.S.T., W. J. Nelson, and S.F., unpublished data).
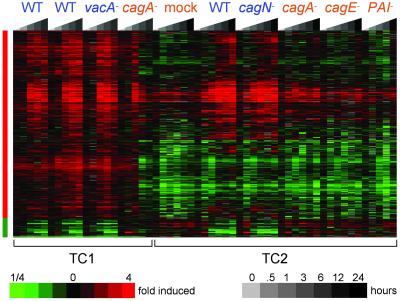
Fig 2.
CagA-dependent gene expression profiles of gastric epithelial cells. The gene expression levels of all samples infected for 3 h or longer were compared between cells that received CagA (wild type, _vacA_−, and cagN_− infections, indicated with blue lettering) vs. cells that did not (cagA −, cagE −, and Δ_PAI infections, indicated with orange lettering), using SAM analysis. The nonzero transformed expression profiles of the genes from the resulting gene list were hierarchically clustered and are displayed as in Fig. 1. Replicate measurements of the expression levels of the TC1 wild-type infection are shown. The 603 genes identified as elevated in CagA-exposed cells and the 59 genes identified as decreased in CagA-exposed cells are indicated by vertical red and green bars, respectively. Larger versions of the elevated and decreased clusters can be found in Figs. 11 and 12.
Our analysis of the temporal transcriptional response of gastric epithelial cells to infections with isogenic strains of H. pylori demonstrates that host cells sense and respond to even subtle genetic differences in the bacteria they encounter. Interestingly, infection with the Δ_PAI_ mutant failed to elicit a response much different from the mock infection. Possibly H. pylori lacking a cag PAI actively suppress host cell innate immune responses, as has been shown for live vs. dead Bordetella (17) and for commensal Salmonella (42). The cagE− mutant induced transient expression of certain stress and possible adhesion genes, suggesting that the cagE− mutant can achieve a more intimate and proinflammatory interaction with host cells, although lacking a functional TFSS. Much of the canonical host transcriptional response to H. pylori, including expression of innate immune response, signal transduction, and cytoskeletal genes, required the TFSS structural component CagE. Our analysis revealed a subset of the host cell response that depended on the TFSS effector CagA that was enriched for genes encoding regulators of cell shape. Temporal profiling uncovered subtleties in the kinetics of the host response to H. pylori virulence determinants, including the late _PAI_-dependent induction of cholesterol biosynthesis genes and the late _cagA_-dependent repression of cell proliferation genes. Statistical analysis revealed consistent low-level differences in the host response to different H. pylori mutants, including the _cagA_-associated up-regulation of genes encoding cell junction proteins. Our analysis also provides specific candidate molecules, such as Lck and Cdc42 effector protein 2, that can be tested to be involved in general classes of CagA-mediated host responses, such as CagA signal transduction and small GTPase regulation of the actin cytoskeleton. Host cell transcriptional profiling as a method to dissect the differences between mutant and wild-type bacteria thus provides information about both the nature of bacterial virulence factors and host cell processes in the normal and diseased state.
Supplementary Material
Supporting Information
Acknowledgments
We thank Antonello Covacci (IRIS Chiron, Siena, Italy) for H. pylori strains; the staff of the Stanford Functional Genomics Facility and the Stanford Microarray Database for help with DNA microarray experiments; Manuel Amieva for help with experimental design; and Pat Brown, David Botstein, David Relman, and members of the Falkow laboratory for useful discussions. We acknowledge support from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation [Fellowship DRG-1509 (to K.G.)] and National Institutes of Health Grant AI38459 (to L.S.T.). K.G. is a recipient of a Bourroughs Wellcome Fund Career Award in the Biomedical Sciences.
Abbreviations
- PAI, pathogenicity island
- SAM, significance analysis of microarrays
- TC, time course
- TFSS, type IV secretion system
- VacA, vacuolating cytotoxin A
- TNF, tumor necrosis factor
References
- 1.Montecucco C. & Rappuoli, R. (2001) Nat. Rev. Mol. Cell. Biol. 2**,** 457-466. [DOI] [PubMed] [Google Scholar]
- 2.Asahi M., Azuma, T., Ito, S., Ito, Y., Suto, H., Nagai, Y., Tsubokawa, M., Tohyama, Y., Maeda, S., Omata, M., Suzuki, T. & Sasakawa, C. (2000) J. Exp. Med. 191**,** 593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backert S., Ziska, E., Brinkmann, V., Zimny-Arndt, U., Fauconnier, A., Jungblut, P. R., Naumann, M. & Meyer, T. F. (2000) Cell Microbiol. 2**,** 155-164. [DOI] [PubMed] [Google Scholar]
- 4.Odenbreit S., Puls, J., Sedlmaier, B., Gerland, E., Fischer, W. & Haas, R. (2000) Science 287**,** 1497-1500. [DOI] [PubMed] [Google Scholar]
- 5.Segal E. D., Cha, J., Lo, J., Falkow, S. & Tompkins, L. S. (1999) Proc. Natl. Acad. Sci. USA 96**,** 14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein M., Rappuoli, R. & Covacci, A. (2000) Proc. Natl. Acad. Sci. USA 97**,** 1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selbach M., Moese, S., Hauck, C. R., Meyer, T. F. & Backert, S. (2002) J. Biol. Chem. 277**,** 6775-6778. [DOI] [PubMed] [Google Scholar]
- 8.Stein M., Bagnoli, F., Halenbeck, R., Rappuoli, R., Fantl, W. J. & Covacci, A. (2002) Mol. Microbiol. 43**,** 971-980. [DOI] [PubMed] [Google Scholar]
- 9.Higashi H., Tsutsumi, R., Muto, S., Sugiyama, T., Azuma, T., Asaka, M. & Hatakeyama, M. (2002) Science 295**,** 683-686. [DOI] [PubMed] [Google Scholar]
- 10.Covacci A. & Rappuoli, R. (2000) J. Exp. Med. 191**,** 587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crabtree J. E. (1998) Dig. Dis. Sci. 43**,** 46S-55S. [PubMed] [Google Scholar]
- 12.Censini S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93**,** 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer W., Puls, J., Buhrdorf, R., Gebert, B., Odenbreit, S. & Haas, R. (2001) Mol. Microbiol. 42**,** 1337-1348. [DOI] [PubMed] [Google Scholar]
- 14.Li S. D., Kersulyte, D., Lindley, I. J., Neelam, B., Berg, D. E. & Crabtree, J. E. (1999) Infect. Immun. 67**,** 3893-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selbach M., Moese, S., Meyer, T. F. & Backert, S. (2002) Infect. Immun. 70**,** 665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diehn M. & Relman, D. A. (2001) Curr. Opin. Microbiol. 4**,** 95-101. [DOI] [PubMed] [Google Scholar]
- 17.Boldrick J. C., Alizadeh, A. A., Diehn, M., Dudoit, S., Liu, C. L., Belcher, C. E., Botstein, D., Staudt, L. M., Brown, P. O. & Relman, D. A. (2002) Proc. Natl. Acad. Sci. USA 99**,** 972-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Q., Liu, D., Majewski, P., Schulte, L. C., Korn, J. M., Young, R. A., Lander, E. S. & Hacohen, N. (2001) Science 294**,** 870-875. [DOI] [PubMed] [Google Scholar]
- 19.Nau G. J., Richmond, J. F., Schlesinger, A., Jennings, E. G., Lander, E. S. & Young, R. A. (2002) Proc. Natl. Acad. Sci. USA 99**,** 1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belcher C. E., Drenkow, J., Kehoe, B., Gingeras, T. R., McNamara, N., Lemjabbar, H., Basbaum, C. & Relman, D. A. (2000) Proc. Natl. Acad. Sci. USA 97**,** 13847-13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichikawa J. K., Norris, A., Bangera, M. G., Geiss, G. K., van't Wout, A. B., Bumgarner, R. E. & Lory, S. (2000) Proc. Natl. Acad. Sci. USA 97**,** 9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Detweiler C. S., Cunanan, D. B. & Falkow, S. (2001) Proc. Natl. Acad. Sci. USA 98**,** 5850-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach S., Makristathis, A., Rotter, M. & Hirschl, A. M. (2002) Infect. Immun. 70**,** 988-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiou C. C., Chan, C. C., Sheu, D. L., Chen, K. T., Li, Y. S. & Chan, E. C. (2001) Gut 48**,** 598-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox J. M., Clayton, C. L., Tomita, T., Wallace, D. M., Robinson, P. A. & Crabtree, J. E. (2001) Infect. Immun. 69**,** 6970-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda S., Otsuka, M., Hirata, Y., Mitsuno, Y., Yoshida, H., Shiratori, Y., Masuho, Y., Muramatsu, M., Seki, N. & Omata, M. (2001) Biochem. Biophys. Res. Commun. 284**,** 443-449. [DOI] [PubMed] [Google Scholar]
- 27.Sepulveda A. R., Tao, H., Carloni, E., Sepulveda, J., Graham, D. Y. & Peterson, L. E. (2002) Aliment. Pharmacol. Ther. 16 Suppl. 2**,** 145-157. [DOI] [PubMed] [Google Scholar]
- 28.Salama N. R., Otto, G., Tompkins, L. & Falkow, S. (2001) Infect. Immun. 69**,** 730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherlock G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29**,** 152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisen M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95**,** 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusher V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98**,** 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch D. S., Pirone, D. M. & Burbelo, P. D. (2001) J. Biol. Chem. 276**,** 875-883. [DOI] [PubMed] [Google Scholar]
- 33.Joberty G., Perlungher, R. R. & Macara, I. G. (1999) Mol. Cell. Biol. 19**,** 6585-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odenbreit S. & Haas, R. (2002) Curr. Top. Microbiol. Immunol. 264**,** 1-22. [PubMed] [Google Scholar]
- 35.Rauchhaus M., Coats, A. J. & Anker, S. D. (2000) Lancet 356**,** 930-933. [DOI] [PubMed] [Google Scholar]
- 36.Triantafilou M., Miyake, K., Golenbock, D. T. & Triantafilou, K. (2002) J. Cell Sci. 115**,** 2603-2611. [DOI] [PubMed] [Google Scholar]
- 37.Schraw W., Li, Y., McClain, M. S., Van Der Goot, F. G. & Cover, T. L. (2002) J. Biol. Chem. 277**,** 34642-34650. [DOI] [PubMed] [Google Scholar]
- 38.Patel H. K., Willhite, D. C., Patel, R. M., Ye, D., Williams, C. L., Torres, E. M., Marty, K. B., MacDonald, R. A. & Blanke, S. R. (2002) Infect. Immun. 70**,** 4112-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connor S., Taylor, C., Campbell, L. A., Epstein, S. & Libby, P. (2001) Emerg. Infect. Dis. 7**,** 780-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirin H., Weinstein, I. B. & Moss, S. F. (2001) Front. Biosci. 6**,** E104-E118. [DOI] [PubMed] [Google Scholar]
- 41.Hazell S. L., Lee, A., Brady, L. & Hennessy, W. (1986) J. Infect. Dis. 153**,** 658-663. [DOI] [PubMed] [Google Scholar]
- 42.Neish A. S., Gewirtz, A. T., Zeng, H., Young, A. N., Hobert, M. E., Karmali, V., Rao, A. S. & Madara, J. L. (2000) Science 289**,** 1560-1563. [DOI] [PubMed] [Google Scholar]
- 43.Covacci A., Censini, S., Bugnoli, M., Petracca, R., Burroni, D., Macchia, G., Massone, A., Papini, E., Xiang, Z., Figura, N., et al. (1993) Proc. Natl. Acad. Sci. USA 90**,** 5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information