NOSTRIN: A protein modulating nitric oxide release and subcellular distribution of endothelial nitric oxide synthase (original) (raw)
Abstract
Activity and localization of endothelial nitric oxide synthase (eNOS) is regulated in a remarkably complex fashion, yet the complex molecular machinery mastering stimulus-induced eNOS translocation and trafficking is poorly understood. In a search by the yeast two-hybrid system using the eNOS oxygenase domain as bait, we have identified a previously uncharacterized eNOS-interacting protein, dubbed NOSTRIN (for eNOS traffic inducer). NOSTRIN contains a single polypeptide chain of 506-aa residues of 58 kDa with an N-terminal cdc15 domain and a C-terminal SH3 domain. NOSTRIN mRNA is abundant in highly vascularized tissues such as placenta, kidney, lung, and heart, and NOSTRIN protein is expressed in vascular endothelial cells. Coimmunoprecipitation experiments demonstrated the eNOS–NOSTRIN interaction in vitro and in vivo, and NOSTRIN's SH3 domain was essential and sufficient for eNOS binding. NOSTRIN colocalized extensively with eNOS at the plasma membrane of confluent human umbilical venous endothelial cells and in punctate cytosolic structures of CHO-eNOS cells. NOSTRIN overexpression induced a profound redistribution of eNOS from the plasma membrane to vesicle-like structures matching the NOSTRIN pattern and at the same time led to a significant inhibition of NO release. We conclude that NOSTRIN contributes to the intricate protein network controlling activity, trafficking, and targeting of eNOS.
Nitric oxide (NO) is a potent mediator in biological processes such as neurotransmission, inflammatory response, and vascular homeostasis (1). The prime source of NO in the cardiovascular system is endothelial NO synthase (eNOS), which is tightly regulated with respect to activity and localization. For example, coordinated phosphorylation contributes to activity control of eNOS because of activating and inhibiting phosphorylation at S1179 and T495, respectively (2–6). Myristoylation and dual palmitoylation at its extreme N terminus target eNOS to the cytoplasmic face of the Golgi complex and to the plasma membrane (7), where eNOS is thought to be fully capable of activation (8, 9). Misrouting of acylation-deficient eNOS impairs NO production (10, 11), indicating that correct subcellular targeting is critical for stimulus-dependent activation of the enzyme (8). Posttranslational modifications are efficiently complemented by multiple protein–protein interactions that help regulate eNOS activity with respect to time and space. For instance, chaperone hsp90 bound to eNOS may mediate vascular endothelial growth factor-induced eNOS phosphorylation by promoting the interaction between eNOS and Akt (12, 13). At the plasma membrane, eNOS is complexed to and inhibited by the master components of caveolae, i.e., caveolin-1 in endothelial cells (9, 14) and caveolin-3 in cardiac myocytes (15). After stimulus-induced [Ca2+]i increase, the Ca2+–calmodulin complex displaces eNOS from caveolin (16), stimulates eNOS to produce NO, and subsequently leads to the redistribution of eNOS from plasma membrane caveolae (17). The complexity of the protein network governing eNOS activity and trafficking has been highlighted by the recent identification of the eNOS-interacting protein (NOSIP), which binds to the oxygenase domain of eNOS (18). Overexpression of NOSIP triggers eNOS redistribution from the plasma membrane, thereby modulating eNOS activity, most likely by interfering with the membrane-bound caveolin-eNOS complex.
Dependent on cell type and/or mode of stimulation, eNOS has been found to reside in various locales of the cell, including plasma membrane, Golgi apparatus, endoplasmic reticulum, vesicular structures, and even nucleus (19–22), suggesting that an extensive meshwork of regulatory proteins may surround eNOS. In our quest for previously uncharacterized eNOS partners, we have identified a protein termed eNOS traffic inducer (NOSTRIN) which binds to eNOS and triggers the translocation of eNOS from the plasma membrane to vesicle-like subcellular structures, thereby strongly attenuating eNOS-dependent NO production.
Materials and Methods
Yeast Two-Hybrid (Y2H) System.
The Y2H screening was carried out as described by using a human placenta cDNA library and the oxygenase domain of human eNOS (positions 1–486 of the protein sequence) as the bait (18). To define the region(s) of the eNOS oxygenase-domain binding to NOSTRIN by Y2H analysis, the following pEG202 (pJG4–5) derivatives containing fragments of the indicated cDNAs were created (subscripts identify positions of the corresponding protein sequence): pEG-eNOS98–486, pEG-eNOS1–366, pEG-eNOS98–366, and pEG-eNOS241–486; pJG-NOSTRIN250–434, pJG-NOSTRIN323–470, pJGNOSTRIN433–506, and pJG-NOSTRIN242–506. For control, eNOS (NOSTRIN) cDNA fragments were subcloned into pJG4–5 (pEG202), and the constructs were used for mutual interaction analyses, which gave identical results. Empty vectors served as negative controls. For determination of relative strength of interaction, clones were plated on X-Gal- and Leu-deficient-medium plates; blue color intensity and growth, respectively, were assessed.
cDNA Cloning and Sequence Analysis.
Initially, we picked a partial cDNA sequence for NOSTRIN encoding the C-terminal portion of the protein. To obtain the full-size sequence, we PCR-screened cDNA pools of various human tissues (RZPD, Heidelberg) and hybridized corresponding high-density filters with a 32P-labeled NOSTRIN probe prepared by random priming amplification (Prime-it II, Stratagene). A single clone from small intestine cDNA was identified that covered a large portion (positions 304–1,605) of the full-size sequence. To obtain the 5′ end of the cDNA, we used the SMART RACE cDNA amplification kit and a human placenta mRNA (CLONTECH). Sequence comparisons, domain analyses, and EST searches were done by using CLUSTALW, PROSITE, SMART, PFAM, and BLAST software. Database analyses revealed that the sequence of our clone is identical to a cDNA sequence for an unknown protein deposited by the New Energy and Industrial Technology Development Organization (NEDO) human cDNA sequencing project (GenBank accession no. AK002203).
Cell Culture.
Chinese hamster ovary (CHO) cells stably expressing eNOS (CHO-eNOS) were used (18). Transient expression was done with PolyFect (Qiagen, Chatsworth, CA). For expression with the Semliki Forest virus (SFV) system, cDNA was cloned into the pSFV2 vector, followed by in vitro transcription and packaging of recombinant viruses (23). Human microvascular endothelial cells (HMVEC) (from lung) were cultured until passage number 7 (Clonetics, San Diego). Human umbilical cord vein endothelial cells (HUVEC) were a generous gift of the Busse laboratory (University of Frankfurt, Frankfurt) and were maintained as detailed (24).
Antibody Production.
The following antisera were raised by using standard immunization protocols (subscripts identify positions in the corresponding protein sequence): antiserum AS619 (referred to as α-NOSTRIN619) directed to a GST-fusion protein with NOSTRIN242–506, generated in mouse; AS604, α-NOSTRIN604, directed to GST-NOSTRIN1–506 (rabbit); AS574, α-NOSTRIN574, against synthetic peptide LNK-17 (NOSTRIN170–186) coupled to keyhole limpet hemocyanin (KLH) (Pierce) via 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (rabbit); AS532, α-NOSTRIN532, against GST-NOSTRIN242–506 (rabbit); and AS468, α-eNOS468, to synthetic peptide PYN-16 (eNOS599–614) coupled to maleimide-activated KLH via an additional C-terminal Cys (rabbit). Sources for other antibodies are indicated.
Expression Analyses.
Equal amounts of HMVECs, HUVECs, and SFV-infected CHO-eNOS cells were lysed in 63 mM Tris⋅HCl, pH 6.8/2.5% SDS/5% glycerol/5% β-mercaptoethanol/0.005% bromophenol blue (sample buffer), subjected to SDS/10% PAGE and transferred to nitrocellulose. The membranes were probed with monoclonal antibody α-eNOSm (transduction) or α-NOSTRIN574 at 1:1,000, followed by chemiluminescence detection. The human 12-lane MTN blot (CLONTECH) was hybridized with a 32P-labeled DNA probe prepared from a NOSTRIN cDNA fragment (positions 724–1,522) by random priming (Stratagene).
Subcellular Fractionation.
Subcellular fractionation of SFV-NOSTRIN and SFV-GFP infected Chinese hamster ovary (CHO)-eNOS cells was done (25) with minor modifications. Briefly, cells were lysed in 10 mM Tris⋅HCl, pH 7.4/10 mM NaCl/100 μM Pefabloc, homogenized by 30 strokes in a Dounce homogenizer; lysates were then subjected to stepwise centrifugation at 1,000 × g for 10 min, 10,000 × g for 20 min, and 100,000 × g for 1 h at 4°C. The sediment after each centrifugation and the 100,000 × g supernatant were separated, dissolved in sample buffer, and analyzed by immunoblotting. To verify subcellular fractionation samples were probed for marker proteins with anti-α-actin, anti-β-COP, and with the Organelle Sampler Kit (Becton Dickinson).
Immunoprecipitation and Immunoblotting.
CHO-eNOS cells were transfected with full-length NOSTRIN cDNA cloned into vector pME18SFL3. After 48 h, the cells were washed with ice-cold PBS, lysed for 1 h on ice with RIPA buffer (1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/50 mM Tris⋅HCl, pH 7.4/150 mM NaCl/1 mM EDTA), and insoluble material was removed by centrifugation at 13,000 × g for 20 min at 4°C. The lysates were diluted 1:3 with washing buffer (50 mM Tris⋅HCl, pH 7.4/150 mM NaCl/1 mM EDTA) and incubated for 1 h at 4°C under rotation with 10 μl of α-eNOS468, α-NOSTRIN532, or the corresponding preimmune sera. Immune complexes were precipitated with Pansorbin (Calbiochem) at 8,000 × g for 2 min at 4°C. The pellets were washed three times, dissolved in sample buffer, and immunoblotted by using α-eNOSm at 1:1,000 or α-NOSTRIN619 at 1:5,000.
Immunochemistry and Immunofluorescence.
Paraffin-embedded human tissue samples (4 μm) were antigen-demasked (26) and incubated for 1 h at 37°C with α-NOSTRIN604 or preimmune serum at 1:25, followed by a biotinylated secondary antibody (BioGenex Laboratories, San Ramon, CA) at 1:20 for 30 min at 20°C and FITC- or AMCA-conjugated avidin (Vector Laboratories) and analyzed by a Zeiss Axioskop II. HUVEC and HMVEC were fixed in methanol, blocked for 15 min with BPT (1% BSA/0.1% Tween 20 in PBS), incubated for 30 min with α-NOSTRIN532 1:100 in BPT, and probed with Cy3-coupled secondary antibody at 1:500 (Sigma). CHO-eNOS cells were grown on chamber slides for 24 h and infected with SFV for 7 h. The cells were fixed in methanol, blocked with BPT, incubated with α-NOSTRIN574 or α-NOSTRIN532 and α-eNOSm at 1:100, followed by secondary antibodies coupled to Cy2 (anti-mouse Ig) or Cy3 (anti-rabbit Ig) at 1:500 (Sigma). Immunolabeling was documented by a Zeiss Axiovert 2000. Images were deconvoluted and further analyzed by OPENLAB3 software (Improvision, London).
In Vitro Binding Assay.
Full-length NOSTRIN cDNA and a cDNA fragment encoding amino acids 1–440 (NOSTRINΔSH3) were ligated into pGEX2T vector and expressed in E. coli BL21. The resultant GST fusion proteins were purified on glutathione-Sepharose (Amersham Pharmacia). [35S]Methionine-labeled eNOS was prepared by in vitro transcription/translation using T7-polymerase and pcDNA3.1-eNOS or empty vector for control (Promega). For assessment of in vitro interaction, 450 pmol (450 nM) of GST or GST fusion protein was incubated overnight at 4°C with 50 μl of reticulocyte lysate containing [35S]eNOS in 50 mM Tris⋅HCl, pH 7.4/150 mM NaCl/1 mM EDTA/100 μM Pefabloc (Roche Diagnostics). Matrix-bound proteins were eluted with sample buffer and analyzed by SDS/PAGE and autoradiography (27).
NO Release Assay.
CHO-eNOS cells grown on 6-well plates were infected for 6 h with SFV-NOSTRIN or SFV-GFP (control). After equilibration in Hepes buffer, pH 7.4, including 100 μM l-arginine for 30 min at 37°C, the cells were stimulated with 1 μM A23187 (Alexis, Gruenberg, Germany) for 90 min at 37°C in the presence or absence of 100 μM NG-nitro-l-arginine (L-NNA). After reduction with sodium iodide, the NO concentration in the supernatant was quantified by a chemiluminescence detector (Sievers, Boulder, CO) using ozone (18).
Results
Identification and Characterization of NOSTRIN.
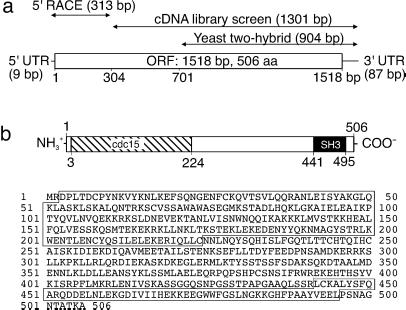
To search for previously uncharacterized eNOS-interacting proteins, we used the Y2H system and identified a cDNA clone encoding the C-terminal fragment of an interacting protein. We identified the full-length cDNA by screening human cDNA libraries followed by 5′-RACE. The resultant clone had an ORF of 1,518 bp, preceded by a 5′-untranslated region of 9 bp and followed by a 3′-untranslated region of 87 bp, totaling 1,614 bp (Fig. 1a). The ORF encodes a protein of 506 amino acids having a predicted molecular mass of 58 kDa and containing an N-terminally located cdc15 domain (positions 3–224) and a C-terminally located SH3 domain (441–495) that may mediate specific functions of the protein (Fig. 1b). In the course of our study, we designated this protein NOSTRIN (see below). Searches of the human genome databases revealed that the gene for NOSTRIN of 62 kb is located on locus 2q31.1 of chromosome 2. EST databases revealed several short ESTs of murine, porcine, or bovine origin that show 82–89% sequence identity on the cDNA level, suggesting the existence of well conserved homologues in other mammals.
Fig 1.
Human NOSTRIN is a protein of 506 amino acid residues. (a) The initial Y2H screen identified a fragment of 904 bp. Screening of human tissue cDNA libraries revealed an overlapping fragment of 1,301 bp that was further extended by 5′-RACE to yield the full-length clone of 1,614 bp. (b Upper) Predicted modular structure of NOSTRIN with an N-terminal cdc15 domain (amino acids 3–224, hatched) and a C-terminal SH3 domain (441–495, black). (b Lower) Predicted amino acid sequence of NOSTRIN, with the cdc15 and SH3 domains boxed.
NOSTRIN Is Expressed in Vascular Endothelial Cells.
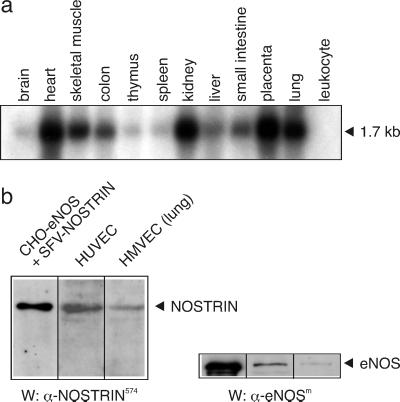
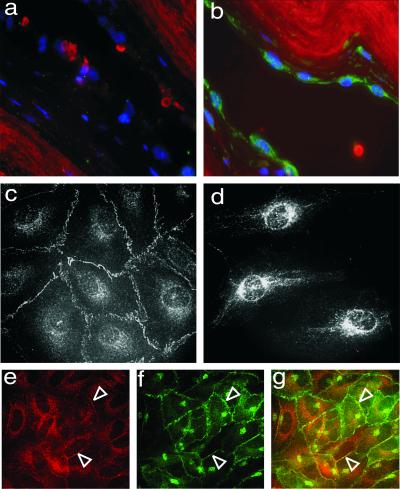
We analyzed the expression pattern of NOSTRIN both on mRNA and protein level. For Northern blot analyses, we used a [32P]NOSTRIN probe to screen mRNAs from various human tissues and found a single transcript of ≈1.7 kb. NOSTRIN mRNA levels were strongest in heart, kidney, placenta, and lung, modest in skeletal muscle, colon, liver, and small intestine, and lowest in brain, thymus, and spleen (Fig. 2a). The specific distribution pattern, i.e., abundance of NOSTRIN mRNA in highly vascularized tissues and low mRNA levels in skeletal muscle, central nervous system, and defense system, was confirmed with a human multiple tissue expression array and by Western blot analyses of murine tissue lysates (not shown). Because NOSTRIN expression was prominent in highly vascularized tissues, we analyzed by Western blotting the specific expression patterns in human microvascular endothelial cells (HMVEC) and human umbilical vein endothelial cells (HUVEC). The presence of a major band of ≈58 kDa indicated that NOSTRIN is endogenously expressed in primary cells together with eNOS (Fig. 2b). By contrast, cultured cell lines had only very low levels of NOSTRIN mRNA and no significant amounts of NOSTRIN protein (not shown). We also analyzed the left ventricle of the human heart by immunochemistry and found that NOSTRIN was prevalent in the flattened endothelial cells lining the inner face of the vessel wall (Fig. 3b), whereas it was absent from the adjacent cells of the intima. Controls with the corresponding preimmune serum were negative (Fig. 3a).
Fig 2.
NOSTRIN is prevalent in highly vascularized tissues. (a) Northern blot of equal amounts of human mRNA hybridized with a [32P]NOSTRIN cDNA probe. (b) Western blot of lysates from CHO-eNOS cells overexpressing NOSTRIN and from primary cells expressing endogenous NOSTRIN. Representatives of several independent experiments are shown.
Fig 3.
NOSTRIN is expressed in vascular endothelial cells. (a and b) Immunocytochemistry of tissue sections of human heart (left ventricle) treated with preimmune serum (a) or with α-NOSTRIN604 (b) and an FITC-conjugated secondary antibody. Shown are DAPI staining for nuclei (blue) and autofluorescence of elastic fibers (red). (c and d) Immunostaining of confluent (c) and subconfluent (d) HMVECs with α-NOSTRIN532. (e_–_g) Confluent HUVECs double-immunolabeled with α-NOSTRIN532 (e) and α-eNOSm (f). (g) Overlay of e and f. Open arrowheads point to plasma membrane. Representatives of three independent experiments are shown.
Interaction with eNOS Depends on NOSTRIN's SH3 Domain.
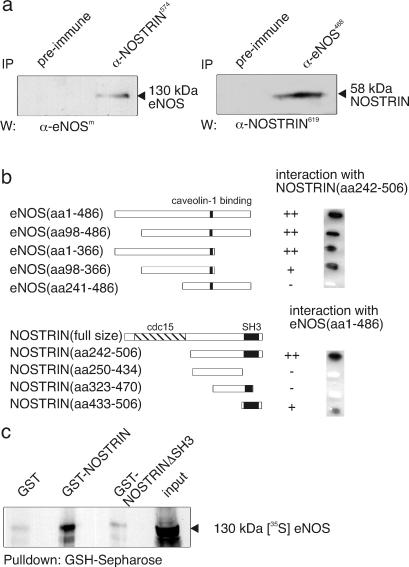
To explore the interaction between NOSTRIN and eNOS, we used CHO-eNOS cells showing the typical distribution and regulation pattern of human eNOS (18) and transiently transfected them with NOSTRIN cDNA. Fig. 4a demonstrates that antibodies to eNOS coprecipitate NOSTRIN and vice versa, indicating that the two proteins likely interact in vivo. Remarkably, the interaction was partially resistant to stringent lysis conditions that were necessary because NOSTRIN was hardly soluble in mild detergents such as Triton X-100. To map the mutual interaction sites of NOSTRIN and eNOS precisely, we used the Y2H system. Reporter strain EGY48/pSH18–34 was cotransformed with NOSTRIN242–506 and progressive deletion constructs of the eNOS oxygenase domain. Deletion of residues 1–97 or 367–486 from the oxygenase domain did not impair interaction with NOSTRIN (Fig. 4b), indicating that the eNOS-binding site for NOSTRIN is harbored by segment 98–366, which also comprises the caveolin-binding site at 350–358 (28). Further truncation (eNOS241–486) resulted in a loss of NOSTRIN binding. Unlike the other truncation mutants retaining their capacity to dimerize, eNOS241–486 showed impaired homodimerization, possibly reflecting incorrect folding. Thus, we cannot unambiguously conclude from the lack of interaction that the NOSTRIN-binding site lies on eNOS98–240. Applying the same strategy, we cotransformed EGY48/pSH18–34 with the full-size oxygenase domain and progressive deletion constructs of NOSTRIN. Deletion of residues 1–241 or 1–432 did not prevent binding to the oxygenase domain, whereas deletion of residues 434–506 or 471–506 nullified NOSTRIN binding to eNOS (Fig. 4b). Hence, NOSTRIN433–506, which in large part represents its SH3 domain, mediates complex formation with eNOS.
Fig 4.
NOSTRIN interacts via its SH3 domain with eNOS in vivo and in vitro. (a) Mutual coimmunoprecipitation of eNOS and NOSTRIN from lysates of NOSTRIN overexpressing CHO-eNOS cells. (b) Mapping of the interaction sites by the Y2H system. NOSTRIN and eNOS deletion constructs are identified by the relative positions of their amino acid (aa) sequence segments (Left). Colony color intensity on X-Gal plates (++, dark blue; +, blue; −, white) is presented (Right). Empty vectors served as negative controls. (c) Autoradiograph of an in vitro pull-down assay of [35S]eNOS with GST-NOSTRIN, GST-NOSTRINΔSH3, and GST. Representatives of three independent experiments are shown.
By using a pull-down strategy, we examined the binding of purified GST-NOSTRIN, deletion mutant GST-NOSTRINΔSH3 lacking the SH3 domain, and GST to _in vitro_-transcribed [35S]eNOS (Fig. 4c). Our results demonstrate the in vitro interaction between [35S]eNOS and GST-NOSTRIN but not with GST-NOSTRINΔSH3 or GST. Collectively, these findings indicate that NOSTRIN and eNOS specifically interact in vitro and most likely in vivo, i.e., in intact cells, and that NOSTRIN's SH3 domain is necessary and sufficient for eNOS binding. The interaction is most likely direct, although we cannot entirely exclude that bridging protein(s) are engaged.
NOSTRIN Colocalizes with eNOS in Intact Cells.
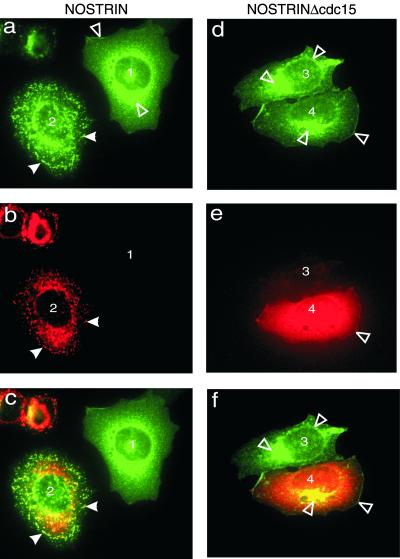
Immunofluorescence studies of primary macrovascular endothelial cells showed that eNOS and NOSTRIN colocalize extensively at the plasma membrane of confluent HUVEC monolayers (Fig. 3 e–g). NOSTRIN immunoreactivity is also associated with vesicle-like structures in the cytosol of these cells. Of note, the distribution patterns of NOSTRIN are distinctly different in confluent monolayers (Fig. 3c) vs. subconfluent cell populations (Fig. 3d) of microvascular HMVECs. In the latter case, we found a prominent staining for NOSTRIN of perinuclear compartments and fibrous structures that seem to be associated with the cytoskeleton. We also treated CHO-eNOS cells with SFV-encoding NOSTRIN at a low titer such that only a small fraction of cells was infected. After 7 h, the cells were fixed and immunostained for NOSTRIN and eNOS. NOSTRIN localized in punctate cytosolic structures of infected cells (Fig. 5b, cell 2), whereas noninfected cells were virtually devoid of NOSTRIN immunostaining (Fig. 5b, cell 1). In noninfected cells, eNOS showed the typical localization at the plasma membrane and more prominently at the Golgi region (Fig. 5a, cell 1). In NOSTRIN-expressing cells, eNOS localization was dramatically distinct, showing the same punctate pattern as NOSTRIN (Fig. 5 a_–_c, cell 2), whereas transient expression of SFV-encoded GFP did not alter eNOS distribution (not shown). Thus, overexpressed NOSTRIN seems to change the subcellular distribution of eNOS by inducing its translocation from the plasma membrane to intracellular compartments.
Fig 5.
NOSTRIN colocalizes with eNOS. CHO-eNOS cells were infected by SFV-NOSTRIN (a_–_c) or SFV-NOSTRINΔcdc15 (d_–_f) and double-immunolabeled with α-eNOSm (a and d) and α-NOSTRIN574 (b) or α-NOSTRIN532 (e). (c) Overlay of a and b. (f) Overlay of d and e. Noninfected cells (1 and 3) and infected cells (2 and 4) are marked; filled arrowheads point to vesicular structures (a_–_c), and open arrowheads identify plasma membrane and Golgi. Representatives of three independent experiments are shown.
To explore the role of NOSTRIN's cdc15 domain, we studied the subcellular distribution of mutant NOSTRINΔcdc15 lacking residues 1–240. Deletion of the cdc15 domain resulted in a complete loss of the punctate cytosolic-staining characteristic for full-length NOSTRIN and produced a rather uniform distribution with a marked staining of plasma membrane and Golgi apparatus in CHO-eNOS cells (Fig. 5e, cell 4). As would be expected for a construct retaining the SH3 domain, NOSTRINΔcdc15 colocalizes with eNOS (Fig. 5 d and f, cell 4). Thus, the cdc15 deletion mutant has selectively lost the ability to induce eNOS trafficking.
NOSTRIN Overexpression Alters eNOS Subcellular Distribution.
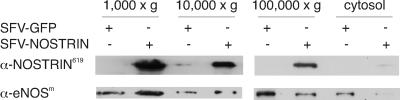
We studied the influence of NOSTRIN overexpression on the distribution of eNOS after subcellular fractionation of SFV-NOSTRIN and SFV-GFP infected CHO-eNOS cells. The majority of NOSTRIN protein was present in the 1,000 × g pellet (Fig. 6), which also contained cytoskeletal marker proteins (not shown). A considerable amount of NOSTRIN was detected in the 10,000 × g pellet, mainly representing the Golgi apparatus, and only a minor fraction was in the 100,000 × g pellet, mainly representing the plasma membrane; we found hardly any NOSTRIN in the cytosolic fraction (Fig. 6). Compared with GFP, NOSTRIN overexpression enriched eNOS in the cytoskeletal fraction and diminished the amount of eNOS both in the plasmalemmal and cytosolic fractions. These results are consonant with our immunofluorescence studies demonstrating that NOSTRIN induces trafficking of eNOS from the plasma membrane to intracellular compartments, most likely associated with the cytoskeleton.
Fig 6.
NOSTRIN induces subcellular redistribution of eNOS. Subcellular fractionation of SFV-NOSTRIN- and SFV-GFP-infected CHO-eNOS cells. The 1,000, 10,000, and 100,000 × g pellets represent mainly cytoskeleton, Golgi apparatus, and plasma membrane, respectively, and the supernatant represents cytosol. (Bottom) The exposure time for the panel at left was shorter than for the panel at right to allow for discrimination of subtle intensity differences. A representative of three independent experiments is shown.
NOSTRIN Overexpression Attenuates NO Release.
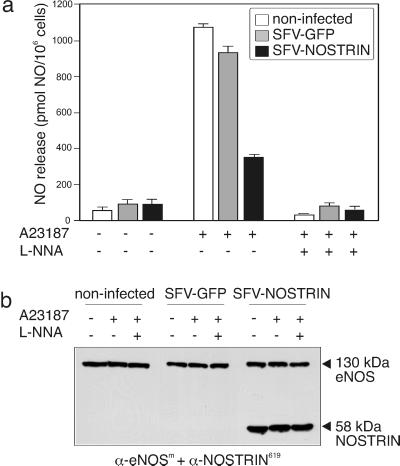
Because NOSTRIN has the potential to alter eNOS distribution, we asked for the effect of NOSTRIN on NO release. CHO-eNOS cells were infected with SFV-NOSTRIN or SFV-GFP for 8 h, and noninfected cells served as control. The immanent viral cytotoxicity mildly reduced eNOS expression levels in infected cells (Fig. 7b), whereas application of the calcium ionophore A23187 in the absence or presence of L-NNA did not change eNOS levels. Hence, differences in NO release in response to stimulation do not reflect alterations in the eNOS protein level. Stimulation of noninfected cells with A23187 robustly increased NO production (measured as NO release into the medium) by a factor of 17, and this effect was completely reversed in the presence of the eNOS inhibitor L-NNA (Fig. 7a, white bars). A23187-mediated stimulation of SFV-GFP-infected cells increased NO production almost to the level of wild-type cells (Fig. 7a, gray bars). In sharp contrast, overexpression of NOSTRIN prevented A23187-induced NO production to a great extent and caused a strong reduction in NO release by 62% or 67%, compared with SFV-GFP-infected cells or noninfected cells (set 100% each), respectively (Fig. 7a, black bars). Hence, overexpression of NOSTRIN induces redistribution of eNOS from the plasma membrane to intracellular vesicular structures, and at the same time attenuates Ca2+-induced NO release.
Fig 7.
NOSTRIN inhibits NO release. (a) NO release from noninfected (white bars), SFV-GFP-infected (gray bars), and SFV-NOSTRIN-infected CHO-eNOS (black bars) cells. Means ± SEM for three consecutive measurements are given. (b) Immunoblot of lysates of NO-producing cells (from a). Cells were treated with 1 μM A23187 in the absence (−) or presence (+) of 100 μM L-NNA, as indicated. A representative of three independent experiments is shown.
Discussion
eNOS is a key enzyme of the cardiovascular system that contributes to vascular homeostasis through tightly regulated NO production. Multilateral control of eNOS activity affects transcriptional activity, mRNA stability, posttranslational modifications, reversible protein–protein interactions, as well as routing of eNOS to subcellular targets such as plasma membrane and Golgi apparatus (1, 29–31). Dynamic changes of the intracellular eNOS localization seem to be well coordinated, and because external stimuli induce cycling of eNOS back and forth from the plasma membrane to intracellular compartments, translocation has been implied in the regulation of the enzyme's activity in vitro and in vivo (29). For example, targeting of eNOS to plasma membrane and cell–cell junctions upon cells reaching confluency is accompanied by a strong increase in NO production (32). Further, oxidized LDL inhibits agonist-induced NO formation in HUVEC caused by subcellular redistribution of eNOS from the plasma membrane, while substrate and cofactor supply as well as eNOS expression are unchanged (33). To date, the molecular machinery targeting eNOS to various intracellular locales, the factors and mechanisms governing this translocation, and the physiological consequences ensuing from eNOS trafficking have remained largely unknown.
Here, we report the identification and characterization of a previously uncharacterized protein, NOSTRIN, affecting the subcellular localization of eNOS. Display of a single cdc15 and SH3 domain each characterize the protein as a member of the Pombe cdc15 homology (PCH) family of proteins (34), including CD2BP1 (35), PACSIN1 (36), PSTPIP (37), as well as the nonmammalian homologue CG4040. The functions of PCH proteins are not fully understood, but they have been implied with dynamic rearrangements of the cytoskeleton (34). The dependence on the integrity of the cdc15 domain of the typical punctate subcellular distribution pattern as well as the poor solubility of NOSTRIN might indicate a common property among NOSTRIN and other PCH family members with respect to their cytoskeletal association.
Analysis of the expression profile of NOSTRIN revealed that the corresponding mRNA is present in a variety of human tissues, including lung and kidney, that is, two highly vascularized organs known to be associated with endothelium-dependent NO production. Because NOSTRIN is also present in primary endothelial cells and in the endothelium of cardiac vessels expressing eNOS (20, 21, 38), it seems reasonable to assume that NOSTRIN and eNOS may interact in these cells under physiological and/or pathological conditions. By contrast, NOSTRIN mRNA is almost absent from tissue samples derived from the human central nervous system where the neuronal isoform nNOS is the major NO-producing enzyme (38). In keeping with this notion, our Y2H analyses indicate that NOSTRIN does not interact with the oxygenase domain of nNOS, nor does it bind to iNOS (N.O., unpublished observations). NOSTRIN associates via its C-terminal domain (positions 433–506), mainly consisting of the SH3 domain, with the oxygenase domain of eNOS both in vitro and in vivo. Consistent with this notion, the minimum sequence segment of eNOS sufficient for NOSTRIN binding (positions 98–366) exposes a Pro-rich segment which may serve as an SH3-binding site.
Overexpression of NOSTRIN in CHO cells mobilized eNOS such that the membrane-bound fraction of eNOS was considerably reduced, and, at the same time, eNOS was found to be associated with cytosolic vesicle-like structures, where it colocalized with NOSTRIN. Redistribution of eNOS in NOSTRIN-expressing CHO-eNOS cells was paralleled by a drastic decrease in NO release, down to 33–38% of control cells (100%). This marked inhibitory effect could be brought about by (i) direct inhibition of eNOS activity, (ii) modulation of regulatory mechanisms such as phosphorylation or protein association indirectly affecting eNOS activity, or (iii) intracellular redistribution of eNOS. At present, we cannot rule out that NOSTRIN interferes with eNOS activity through similar allosteric mechanisms such as Ca2+-calmodulin, although the eNOS-binding sites for the two proteins are clearly distinct.
In vascular endothelial growth factor (VEGF)-triggered endothelial cells, hsp90 mediates the interaction between eNOS and Akt by inducing the transition from the “early” Ca2+-dependent to the “late” phosphorylation-dependent activation of eNOS (13). It is conceivable that NOSTRIN may modulate eNOS (de)phosphorylation, thereby affecting subcellular distribution and, thus, activity of eNOS. Indeed, “correct” compartmentalization of eNOS is an absolute requirement for VEGF-driven phosphorylation (39) and full-blown activation of eNOS (8, 10). Distinct localization patterns in subconfluent microvascular endothelial cells vs. confluent monolayers have been reported for eNOS, and establishment of cell–cell contacts has been implied in its recruitment to intercellular junctions, followed by a significant increase in NO release (32). As demonstrated in this article, NOSTRIN is also targeted to the plasma membrane and possibly to intercellular junctions upon cells reaching confluency, so it is tempting to speculate that NOSTRIN induces translocation of eNOS to these sites, allowing agonist-induced eNOS activation to occur in “proper” locations.
Recently, we identified an eNOS-interacting protein, NOSIP, which likely forms part of the hypothetical eNOS translocation machinery (18). Given that both NOSIP and NOSTRIN partake in the intracellular reshuffling of eNOS, one may ask how these two proteins differ. First, the proteins are structurally unrelated. Second, NOSIP is in the cytosol and also in the nucleus (40), whereas NOSTRIN is found exclusively in extranuclear locations. Third, NOSIP overexpression induces eNOS translocation into the proximity of the Golgi apparatus and cytoskeletal structures, whereas NOSTRIN overexpression targets the enzyme to vesicle-like structures spread all over the cytosol. Hence, NOSIP and NOSTRIN may share some of their functional features but clearly differ in many other respects. Also, it is very possible that NOSIP and NOSTRIN act in concert; however, we have not addressed this intriguing possibility. Elucidation of the precise function and timing of eNOS-interacting proteins will provide fresh insights into the intricate molecular machinery delivering NO synthase capacity to the various locales of the cell.
Acknowledgments
We thank Dr. Eberhard Hildt, Robert-Koch-Institut, Berlin, for critical discussion, and the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie for support.
Abbreviations
- Y2H, yeast two-hybrid
- CHO, Chinese hamster ovary
- eNOS, endothelial NO synthase
- NOSIP, eNOS-interacting protein
- SFV, Semliki Forest virus
- HMVEC, human microvascular endothelial cells
- HUVEC, human umbilical cord vein endothelial cells
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 16510.
References
- 1.Bredt D. S. & Snyder, S. H. (1994) Annu. Rev. Biochem. 63**,** 175-195. [DOI] [PubMed] [Google Scholar]
- 2.Dimmeler S., Fleming, I., Fisslthaler, B., Hermann, C., Busse, R. & Zeiher, A. M. (1999) Nature 399**,** 601-605. [DOI] [PubMed] [Google Scholar]
- 3.Fulton D., Gratton, J. P., McCabe, T. J., Fontana, J., Fujio, Y., Walsh, K., Franke, T. F., Papapetropoulos, A. & Sessa, W. C. (1999) Nature 399**,** 597-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michell B. J., Chen, Z., Tiganis, T., Stapleton, D., Katsis, F., Power, D. A., Sim, A. T. & Kemp, B. E. (2001) J. Biol. Chem. 276**,** 17625-17628. [DOI] [PubMed] [Google Scholar]
- 5.Harris M. B., Ju, H., Venema, V. J., Liang, H., Zou, R., Michell, B. J., Chen, Z. P., Kemp, B. E. & Venema, R. C. (2001) J. Biol. Chem. 276**,** 16587-16591. [DOI] [PubMed] [Google Scholar]
- 6.Fleming I., Fisslthaler, B., Dimmeler, S., Kemp, B. E. & Busse, R. (2001) Circ. Res. 88**,** 68-75. [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Hughes, T. E. & Sessa, W. C. (1997) J. Cell Biol. 137**,** 1525-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sessa W. C., Garcia-Cardena, G., Liu, J., Keh, A., Pollock, J. S., Bradley, J., Thiru, S., Braverman, I. M. & Desai, K. M. (1995) J. Biol. Chem. 270**,** 17641-17644. [DOI] [PubMed] [Google Scholar]
- 9.Shaul P. W., Smart, E. J., Robinson, L. J., German, Z., Yuhanna, I. S., Ying, Y., Anderson, R. G. & Michel, T. (1996) J. Biol. Chem. 271**,** 6518-6522. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Garcia-Cardena, G. & Sessa, W. C. (1996) Biochemistry 35**,** 13277-13281. [DOI] [PubMed] [Google Scholar]
- 11.Sakoda T., Hirata, K., Kuroda, R., Miki, N., Suematsu, M., Kawashima, S. & Yokoyama, M. (1995) Mol. Cell. Biochem. 152**,** 143-148. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Cardena G., Fan, R., Shah, V., Sorrentino, R., Cirino, G., Papapetropoulos, A. & Sessa, W. C. (1998) Nature 392**,** 821-824. [DOI] [PubMed] [Google Scholar]
- 13.Brouet A., Sonveaux, P., Dessy, C., Balligand, J. L. & Feron, O. (2001) J. Biol. Chem. 276**,** 32663-32669. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Cardena G., Oh, P., Liu, J., Schnitzer, J. E. & Sessa, W. C. (1996) Proc. Natl. Acad. Sci. USA 93**,** 6448-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feron O., Belhassen, L., Kobzik, L., Smith, T. W., Kelly, R. A. & Michel, T. (1996) J. Biol. Chem. 271**,** 22810-22814. [DOI] [PubMed] [Google Scholar]
- 16.Michel J. B., Feron, O., Sase, K., Prabhakar, P. & Michel, T. (1997) J. Biol. Chem. 272**,** 25907-25912. [DOI] [PubMed] [Google Scholar]
- 17.Feron O., Saldana, F., Michel, J. B. & Michel, T. (1998) J. Biol. Chem. 273**,** 3125-3128. [DOI] [PubMed] [Google Scholar]
- 18.Dedio J., König, P., Wohlfart, P., Schroeder, C., Kummer, W. & Müller-Esterl, W. (2001) FASEB J. 15**,** 79-89. [DOI] [PubMed] [Google Scholar]
- 19.Feron O. & Michel, T. (2000) in Nitric Oxide and the Cardiovascular System, eds. Loscalzo, J. & Vita, J. A. (Humana, Totowa, NJ), Vol. 4.
- 20.Andries L. J., Brutsaert, D. L. & Sys, S. U. (1998) Circ. Res. 82**,** 195-203. [DOI] [PubMed] [Google Scholar]
- 21.Thuringer D., Maulon, L. & Frelin, C. (2002) J. Biol. Chem. 277**,** 2028-2032. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y., Venema, V. J., Venema, R. C., Tsai, N. & Caldwell, R. B. (1999) Biochem. Biophys. Res. Commun. 256**,** 192-197. [DOI] [PubMed] [Google Scholar]
- 23.Lundstrom K., Mills, A., Allet, E., Ceszkowski, K., Agudo, G., Chollet, A. & Liljestrom, P. (1995) J. Recept. Signal Transduct. Res. 15**,** 23-32. [DOI] [PubMed] [Google Scholar]
- 24.Busse R. & Lamontagne, D. (1991) Naunyn-Schmiedebergs Arch. Pharmacol. 344**,** 126-129. [DOI] [PubMed] [Google Scholar]
- 25.Ozols J. (1990) Methods Enzymol. 182**,** 225-235. [DOI] [PubMed] [Google Scholar]
- 26.Norton A. J., Jordan, S. & Yeomans, P. (1994) J. Pathol. 173**,** 371-379. [DOI] [PubMed] [Google Scholar]
- 27.Cao S., Yao, J., McCabe, T. J., Yao, Q., Katusic, Z. S., Sessa, W. C. & Shah, V. (2001) J. Biol. Chem. 276**,** 14249-14256. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Cardena G., Martasek, P., Masters, B. S., Skidd, P. M., Couet, J., Li, S., Lisanti, M. P. & Sessa, W. C. (1997) J. Biol. Chem. 272**,** 25437-25440. [DOI] [PubMed] [Google Scholar]
- 29.Michel T. & Feron, O. (1997) J. Clin. Invest. 100**,** 2146-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nathan C. & Xie, Q. W. (1994) Cell 78**,** 915-918. [DOI] [PubMed] [Google Scholar]
- 31.Marletta M. A. (2001) Trends Biochem. Sci. 26**,** 519-521. [DOI] [PubMed] [Google Scholar]
- 32.Govers R., Bevers, L., de Bree, P. & Rabelink, T. J. (2002) Biochem. J. 361**,** 193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuszkowski A., Grabner, R., Marsche, G., Unbehaun, A., Malle, E. & Heller, R. (2001) J. Biol. Chem. 276**,** 14212-14221. [DOI] [PubMed] [Google Scholar]
- 34.Lippincott J. & Li, R. (2000) Microsc. Res. Tech. 49**,** 168-172. [DOI] [PubMed] [Google Scholar]
- 35.Li J., Nishizawa, K., An, W., Hussey, R. E., Lialios, F. E., Salgia, R., Sunder-Plassmann, R. & Reinherz, E. L. (1998) EMBO J. 17**,** 7320-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plomann M., Lange, R., Vopper, G., Cremer, H., Heinlein, U. A., Scheff, S., Baldwin, S. A., Leitges, M., Cramer, M., Paulsson, M. & Barthels, D. (1998) Eur. J. Biochem. 256**,** 201-211. [DOI] [PubMed] [Google Scholar]
- 37.Spencer S., Dowbenko, D., Cheng, J., Li, W., Brush, J., Utzig, S., Simanis, V. & Lasky, L. A. (1997) J. Cell Biol. 138**,** 845-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Főrstermann U., Boissel, J. P. & Kleinert, H. (1998) FASEB J. 12**,** 773-790. [PubMed] [Google Scholar]
- 39.Fulton D., Fontana, J., Sowa, G., Gratton, J. P., Lin, M., Li, K. X., Michell, B., Kemp, B. E., Rodman, D. & Sessa, W. C. (2002) J. Biol. Chem. 277**,** 4277-4284. [DOI] [PubMed] [Google Scholar]
- 40.König P., Dedio, J., Müller-Esterl, W. & Kummer, W. (2002) Gastroenterology 123**,** 314-332. [DOI] [PubMed] [Google Scholar]