Regulation of Hypoxia-Inducible Factor 1α Expression and Function by the Mammalian Target of Rapamycin (original) (raw)
Abstract
Hypoxia-inducible factor 1 (HIF-1) is a heterodimeric transcription factor containing an inducibly expressed HIF-1α subunit and a constititutively expressed HIF-1β subunit. Under hypoxic conditions, the HIF-1α subunit accumulates due to a decrease in the rate of proteolytic degradation, and the resulting HIF-1α-HIF-1β heterodimers undergo posttranslational modifications that promote transactivation. Recent studies suggest that amplified signaling through phosphoinositide 3-kinase, and its downstream target, mTOR, enhances HIF-1-dependent gene expression in certain cell types. In the present study, we have explored further the linkage between mTOR and HIF-1 in PC-3 prostate cancer cells treated with hypoxia or the hypoxia mimetic agent, CoCl2. Pretreatment of PC-3 cells with the mTOR inhibitor, rapamycin, inhibited both the accumulation of HIF-1α and HIF-1-dependent transcription induced by hypoxia or CoCl2. Transfection of these cells with wild-type mTOR enhanced HIF-1 activation by hypoxia or CoCl2, while expression of a rapamycin-resistant mTOR mutant rendered both HIF-1α stabilization and HIF-1 transactivating function refractory to inhibition by rapamycin. Studies with GAL4-HIF-1α fusion proteins pinpointed the oxygen-dependent degradation domain as a critical target for the rapamycin-sensitive, mTOR-dependent signaling pathway leading to HIF-1α stabilization by CoCl2. These studies position mTOR as an upstream activator of HIF-1 function in cancer cells and suggest that the antitumor activity of rapamycin is mediated, in part, through the inhibition of cellular responses to hypoxic stress.
Tumor development is characterized by an initial phase of rapid expansion, followed by a period of slowed growth as the proliferating malignant cells outstrip the local supply of oxygen and nutrients. In the absence of a dedicated blood supply, early-stage tumors attain steady-state volumes of only a few cubic millimeters, at which time the rate of cell death, due to oxygen and nutrient depletion, equals the rate of cell division (19). To resume growth, these microtumors must adapt to hypoxic stress through alterations in cellular metabolism and the stimulation of neovascularization, which provides the additional blood needed to sustain cellular proliferation. Accordingly, cellular adaptation to growth during hypoxic stress contributes to malignant progression and is correlated with a poor clinical outcome in several types of cancer (3, 4, 18). Two hallmark features of hypoxic adaptation are increased rates of anaerobic glycolysis and the secretion of proangiogenic factors, such as vascular endothelial growth factors (VEGFs) (28, 39). The molecular mechanisms that underlie cellular responses to hypoxic stress are therefore of considerable relevance to cancer biology and therapy.
A key regulator of the cellular response to oxygen deprivation is the transcription factor, hypoxia-inducible factor 1 (HIF-1). Originally identified as an oxygen-responsive activator of erythropoietin gene transcription, HIF-1 is now known to play a central role in the maintenance of oxygen homeostasis in virtually all bodily tissues (42, 43). The predominant form of HIF-1 is a heterodimer consisting of HIF-1α and HIF-1β subunits, both of which are members of the basic helix-loop-helix family of transcription factors. Although HIF-1β is a constitutively expressed nuclear protein, the expression of the HIF-1α subunit is tightly coupled to the ambient oxygen tension. Under normoxic conditions, the HIF-1α gene is continuously transcribed and translated; however, the HIF-1α protein is expressed at very low levels due to rapid destruction via the ubiquitin-proteasome pathway. In addition to its DNA-binding and transactivating motifs, HIF-1α contains a stretch of ca. 200 amino acids, termed the oxygen-dependent degradation (ODD) domain. As its name implies, the ODD domain mediates the interaction between HIF-1α and the E3 ubiquitin ligase complex that mediates continuous poly ubiquitination of HIF-1α in normoxic cells.
The oxygen-dependent turnover of HIF-1α is governed by a novel family of prolyl 4-hydroxylases (PHDs) that specifically modify HIF-1α at two conserved proline residues (Pro-402 and Pro-564), both located in the ODD domain (5, 15, 27, 41). Prolyl hydroxylation triggers the recognition of HIF-1α by the product of the VHL tumor suppressor gene, which serves as the targeting subunit of an E3 ubiquitin ligase complex (20). Although the exact mechanism remains unclear, a decrease in ambient oxygen tension leads to a correlative decrease in HIF-1α prolyl hydroxylation, which in turn leads to decreased rates of HIF-1α polyubiquitination and proteolysis. In addition to regulating the abundance of HIF-1α, hypoxia stimulates the transactivating function of this protein by suppressing a distinct amino acid hydroxylase that modifies a conserved Asn located in the carboxyl-terminal transactivation domain of HIF-1α (25).
The tumor suppressor function of VHL provided an early clue that deregulated expression of HIF-1α promoted tumor development (47). Subsequent immunohistochemical studies revealed high-level expression of HIF-1α in various tumors, with particular localization in the perinecrotic zones that mark areas of avascularity and hypoxia (28, 45, 49, 51). Although oxygen tension plays a determinant role in the process of HIF-1 activation, the amplitude of this response is modulated by growth factor-dependent signaling pathways, including the Ras-Erk and phosphoinositide 3-kinase (PI 3-kinase)/AKT cascades (23, 50, 52). The contributions of the PI 3-kinase pathway to tumorigenesis have attracted considerable interest, due in part to the discovery that a major tumor suppressor gene, PTEN, is frequently inactivated in human cancers (12). A recent study demonstrated that PTEN deficient cells display an exaggerated HIF-1 activation response to hypoxia, a finding that may explain in part the aggressive growth and metastatic characteristics of these tumors (52).
The observations reported above have fueled speculation that inhibitors of signaling through the PI 3-kinase/AKT pathway might have therapeutic efficacy against a broad spectrum of human cancers, particularly advanced-stage neoplasms that typically respond poorly to conventional chemotherapeutic regimens. One such agent, the rapamycin derivative CCI-779, has already entered clinical cancer trials (30, 34). Rapamycin is an exquisitely specific inhibitor of a high molecular mass protein kinase termed the “mammalian target of rapamycin” (mTOR) (1, 16). Although the relationship between PI 3-kinase and mTOR is not precisely defined, two groups have shown that mTOR is phosphorylated by AKT in mitogen-stimulated and PTEN-deficient cells (32, 38). The evidence supporting a functional linkage between PI 3-kinase and mTOR has been buttressed by observations that PTEN-deficient cells display increased sensitivities to the antiproliferative and anticancer activities of rapamycin (2, 24, 30, 33, 35). An intriguing possibility is that tumor sensitivity to rapamycin is partially attributable to the suppression of HIF-1 function and the subsequent adaptive responses to hypoxia.
In the present study, we examined the role of mTOR in the hypoxia-induced activation of HIF-1 in PTEN-deficient prostate cancer (PC-3) cells. Our results demonstrate that the inhibitory effect of rapamycin on HIF-1-dependent transcription in PC-3 cells is mediated through the suppression of mTOR function. Furthermore, we provide evidence that rapamycin interferes with HIF-1 activation in hypoxic cells by increasing the rate of HIF-1α degradation. These findings suggest that the anticancer activity of rapamycin in vivo may be attributed, in part, to the inhibition of the hypoxic response program in developing tumors.
MATERIALS AND METHODS
Plasmids, reagents, and antibodies.
The pHRE-Luc reporter plasmid and hemagglutinin-tagged HIF-1α expression vector have been described elsewhere (29). The Renilla luciferase construct, pRL-TK, was purchased from Promega (Madison, Wis.). The expression vectors encoding AU1-tagged wild-type mTOR (AmTOR-WT), rapamycin-resistant mTOR (AmTOR-SI), and a rapamycin-resistant, catalytically inactive mTOR (AmTOR-SIDA) were described previously (8). The rapamycin-resistant versions of mTOR contain a Ser2035→Ile substitution in the FKBP12-rapamycin binding domain of mTOR. Unless otherwise indicated, the expression vectors encoding the GAL4 (G4)-HIF-1α fusion proteins were provided by Andrew Kung (Dana-Farber Cancer Institute, Boston, Mass.).
Fugene 6 transfection reagent was purchased from Roche Molecular Biochemicals (Indianapolis, Ind.). GSH-Sepharose 4B was obtained from Amersham Pharmacia Biotech (Piscataway, N.J.). Rapamycin (Sigma, St. Louis, Mo.) was prepared as a 100 μM stock solution in ethanol and stored at −70°C. LY294002 (Sigma) was dissolved in dimethyl sulfoxide to yield a 50 mM stock solution. The LY294002 stock solution was aliquoted and stored as described above. The cobalt chloride (CoCl2; Sigma) stock solution (150 mM in water) was prepared before each experiment. LLnV (Sigma) was dissolved in dimethyl sulfoxide to yield a 10 mM stock solution, and aliquots were stored at −70°C.
The 12CA5 (α-HA) monoclonal antibody (MAb) was purchased from Babco (Richmond, Calif.). The α-AU1 MAb was purchased from Covance (Richmond, Calif.). The α-G4 MAb was purchased from Santa Cruz (Santa Cruz, Calif.). The α-HIF1α MAb was purchased from Transduction Labs (Lexington, Ky.). Anti-GST antibody conjugated to horseradish peroxidase was purchased from Sigma. The α-mTOR monoclonal antibody, 26E3, was a generous gift from Peter Houghton (St. Jude Children's Research Hospital, Memphis, Tenn.). Polyclonal antibodies directed against phospholipase C-γ1 (PLCγ1) were described previously (46).
Cell culture and stable transfections.
The human prostate cancer cell line, PC-3, was cultured in standard growth medium (RPMI 1640 supplemented with 10% [vol/vol] fetal bovine serum [FBS] and 10 mM HEPES [pH 7.4]). Stably transfected PC-3 cells were prepared by seeding 106 cells per 10-cm culture dish in 10 ml of standard growth medium. The cells were cultured overnight and transfected with 8 μg of _Sal_I-linearized AmTOR-encoding plasmid DNA and 13 μl of Fugene 6. At 36 h posttransfection, the cells were transferred into standard growth medium containing 1.5 mg of G418 (Gibco-BRL) per ml. Drug-resistant clones were isolated in cloning cylinders and were maintained in standard growth medium containing 0.8 mg of G418 per ml. The clones were screened for expression of the recombinant mTOR by immunoblotting of detergent-soluble proteins with α-AU1 MAb.
Luciferase assays.
PC-3 cells were seeded into 24-well plates (30,000 cells/well) in 0.5 ml of standard growth medium. After an overnight culture, the cells were transfected with 100 ng of pHRE-Luc, 50 ng of TK-Renilla luciferase, and 50 ng of empty plasmid DNA as filler. Transfections were performed with 1.2 μl of Fugene 6 per well. At 24 h posttransfection, the cells were washed and then transferred into low-serum (0.1 or 2%) medium. After 6 h, the cells were pretreated for 30 min with 50 μM LY294002, 100 nM rapamycin, or the solvent vehicle only. The cells were then incubated for 16 h in medium containing 150 μM CoCl2 or in an InVivO2 400 hypoxia workstation (Ruskinn Technology, Ltd., Leeds, United Kingdom) set to an ambient oxygen tension of 1% O2. The cells were then prepared for the dual-luciferase reporter assay (Promega, Madison, Wis.) according to the manufacturer's instructions. Briefly, the cells were lysed at ambient temperature for 15 min with 200 μl of 1× passive lysis buffer, and lysates were cleared of insoluble material by centrifugation. A total of 50 μl of the cleared extracts was assayed for both HRE-dependent (firefly) and control (Renilla) luciferase activities with a Berthold Lumat LB9507 luminometer. Samples were normalized for transfection efficiency based on the Renilla luciferase activity.
Transient transfections and immunoblot analyses.
For HIF-1α immunoblots, PC-3 cells and AmTOR-expressing PC-3 cells were plated in standard growth medium at a density of 5 × 105 cells per 60-mm dish. After an overnight culture, the cells were washed and incubated for 6 to 16 h in low-serum-containing medium as indicated in the figure legends. The cells were then pretreated for 45 min with 50 μM LY294002, 100 nM rapamycin, or 10 μM LLnV, followed by the addition of 150 μM CoCl2 or transfer into hypoxic conditions. After 6 to 16 h, the cells were harvested and lysed in HIF-1α lysis buffer (25 mM Tris-HCl, pH 7.4; 300 mM NaCl; 10% [wt/vol] glycerol; 3 mM EDTA; 1 mM MgCl2; 50 mM β-glycerophosphate; 25 mM NaF; 1% Triton X-100; 20 nM microcystin-LR; 100 μg of phenylmethylsulfonyl fluoride per ml, and protease inhibitor cocktail [10 μg of leupeptin per ml, 10 μg of aprotinin per ml, and 1 μM pepstatin]). The lysates were cleared of insoluble material, and the resulting extracts were assayed for total protein contents (Bio-Rad, Hercules, Calif.). Equivalent amounts of protein from each sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) through 6% gels. The proteins were transferred to an Immobilon-P membrane and probed with 1 μg per ml of α-HIF-1α MAb in Tris-buffered saline-0.2% Tween 20 (TBST) containing 5% (wt/vol) milk. Immunoreactive proteins were detected with sheep anti-mouse ximmunoglobulin G coupled to horseradish peroxidase (Amersham Pharmacia Biotech) and the Renaissance reagent (New England Nuclear, Boston, Mass.). Densitometric analysis was performed with a Molecular Dynamics PDSI densitometer (Sunnyvale, Calif.) and ImageQuant software. To control for variations in sample loading, the membrane was reprobed with rabbit polyclonal anti-PLCγ1 antibodies (1:10,000 dilution) in TBST supplemented with 2% bovine serum albumin. The membrane was then incubated with horseradish peroxidase-coupled protein A (Amersham Pharmacia Biotech), and stained proteins were detected as described above. For AmTOR immunoblots, the cells were lysed in HIF-1α lysis buffer, and proteins were separated by SDS-PAGE as described above. The membranes were probed with anti-AU1 mouse MAb (1:1,000 dilution) in TBST containing 5% milk.
To examine the effect of rapamycin on HIF-1α synthesis, PC-3 cells were precultured for 16 h in medium containing 2% FBS. The proteasome inhibitor LLnV (10 μM) was added to the medium, together with rapamycin (100 nM) or vehicle control, under normoxic conditions. The cells were harvested at the indicated times and processed for HIF-1α immunblot analyses as described above. To determine the effect of rapamycin on HIF-1α degradation, PC-3 cells were precultured for 8 h in medium containing 2% FBS, after which the cells were cultured for an additional 16 h under hypoxic conditions. The cells were then treated with 30 μg of cycloheximide (CHX) per ml, in the absence or presence of rapamycin, and were maintained under hypoxic conditions. Cells were harvested at the indicated times for HIF-1α immunoblot analyses.
To examine the effects of CoCl2 and rapamycin on the expression of G4-HIF-1α fusion proteins, PC-3 cells were plated into six-well plates (8.6 × 104 cells per well). After 2 days in culture, each sample well received a mixture of 4 μl of Fugene 6 and 2.5 μg of plasmid DNA encoding one of the indicated the G4-HIF-1α fusion proteins. After 24 h, the transfected cells were stimulated for 3 h with 150 μM CoCl2. The cells were treated for 14 h with 100 nM rapamycin, 10 μM LLnV, or solvent vehicle only and then lysed in HIF-1α lysis buffer as described above. The cleared extracts were subjected to SDS-PAGE in 8% gels, and the fusion proteins were detected by immunoblotting with 200 ng of α-G4 MAb per ml in TBST containing 5% milk. The membrane was then stripped and reprobed with anti-PLCγ1 antibody to ensure equivalent loading of each sample lane.
Reverse transcription-PCR (RT-PCR) analyses.
For hypoxic stimulation, 5 × 105 PC-3 cells were seeded into 6-cm dishes in standard growth medium. After 24 h, the cells were placed in fresh medium containing 2% serum. Cells were pretreated with 100 nM rapamycin for 30 min and then exposed for 8 h to hypoxia. For CoCl2 stimulation, 7 × 105 PC-3 cells were plated into 6-cm dishes as described above. After 24 in culture, the growth medium was replaced with fresh medium containing 0.1% serum. The cells were cultured for an additional 8 h, pretreated with rapamycin, and then stimulated for 18 h with 150 μM CoCl2 prior to harvest.
For RT-PCR analysis, total cellular RNA was extracted with Trizol reagent (Life Technologies). First-strand cDNA was synthesized with an oligo(dT) primer and Superscript II reverse transcriptase (Life Technologies). The primers for PCR of human Glut 1 mRNA (GenBank accession number K031950) were as follows: forward primer, 5′-GCAAGTCCTTTGAGATGCTGATCC-3′; and reverse primer, 5′-GCCGACTCTCTTCCTTCATCTCC-3′. This primer combination amplifies a 402-bp sequence from the Glut1 mRNA. Primers for amplification of β-actin mRNA were described previously (31). The PCR conditions for both primer sets were as follows: hot start at 94°C for 2 min; 23 amplification cycles, each consisting of 94°C for 15 s, 60°C for 30 s, and 72°C for 1 min; and a final extension step at 72°C for 7 min. PCR products were separated on 2% agarose gels and visualized by staining with ethidium bromide.
RESULTS
Inhibition of HIF-1 activation by pharmacologic agents.
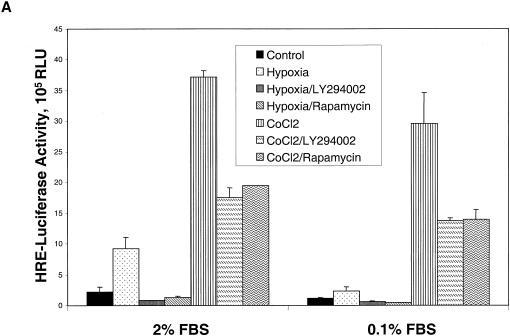
As a first step toward defining the role of the mTOR signaling pathway in HIF-1 function, we examined the effects of pharmacologic inhibitors of PI 3-kinase and mTOR on the activation of HIF-1 by hypoxia or the hypoxia-mimetic agent, CoCl2, in PC-3 prostate cancer cells. The cells were transiently transfected with a pHRE-Luc reporter plasmid, which contains four concatamerized HIF-1 binding sites (termed hypoxia response elements [HREs]). The transfected cells were exposed to hypoxia or CoCl2 in culture medium containing either 2 or 0.1% serum. The magnitude of the HIF-1-dependent transcriptional response induced by hypoxia was strongly influenced by the concentration of serum present during hypoxic stimulation (Fig. 1A). In contrast, CoCl2 treatment provoked similar increases in HRE-dependent luciferase expression in the presence of either 2 or 0.1% serum. The increase in HIF-1-dependent reporter gene transcription observed in hypoxic cells was completely blocked by cellular pretreatment with either LY294002, an inhibitor of both PI 3-kinase and mTOR kinase activities (9, 36), or rapamycin, a specific inhibitor of mTOR-dependent signaling events in mammalian cells. In contrast, the substantially stronger increase in reporter gene expression triggered by CoCl2 exposure was reduced by ca. 50% in LY294002- or rapamycin-treated cells. To rule out potential nonspecific inhibitory effects of rapamycin on the translation of the luciferase reporter mRNA, we repeated the CoCl2 induction experiments with a simian virus 40-regulated luciferase reporter plasmid. Rapamycin treatment caused no significant decrease in simian virus 40 luciferase activity in transfected PC-3 cells (results not shown), a finding which argues against the notion that the results in Fig. 1A resulted from a global suppression of mRNA translation by rapamycin.
FIG. 1.
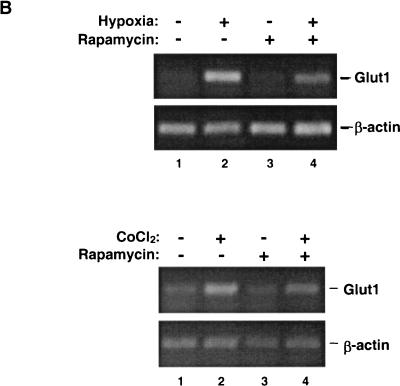
Stimulation of HIF-1 transcriptional activity and HIF-1α protein stabilization by hypoxia or CoCl2. (A) PC-3 cells were transfected with a pHRE-Luc reporter plasmid. After 24 h, the cells were cultured for 6 h in either 2 or 0.1% FBS and then pretreated for 30 min with solvent vehicle, 50 μM LY294002, or 100 nM rapamycin. The cells were then exposed for 19 h to normoxic conditions, hypoxia, or 150 μM CoCl2. Luciferase activities were measured with a luminometer, and are presented as relative light units (RLU). Error bars represent the variance of duplicate samples. (B) Effect of rapamycin on GLUT1 mRNA expression. PC-3 cells were cultured in medium containing 2% serum for hypoxic samples (upper panel) or in medium containing 0.1% serum for CoCl2 stimulation (lower panel). The cells were exposed for 8 h to hypoxia or 150 μM CoCl2 before harvest of total cellular RNA for semiquantitative RT-PCR analysis.
To confirm the results of the reporter gene assays described above, we examined the effect of rapamycin on the expression of GLUT1, a known HIF-1 target gene, (14). PC-3 cells were exposed for 16 h to either hypoxia or CoCl2, and total cellular mRNA was used as a template in semiquantitative RT-PCR assays. The results presented in Fig. 1B demonstrate that rapamycin treatment substantially inhibited the increase in Glut1 mRNA induced by hypoxia or CoCl2. In contrast, the expression of a control gene, β_-ACTIN_, was neither increased by these agents nor inhibited by rapamycin.
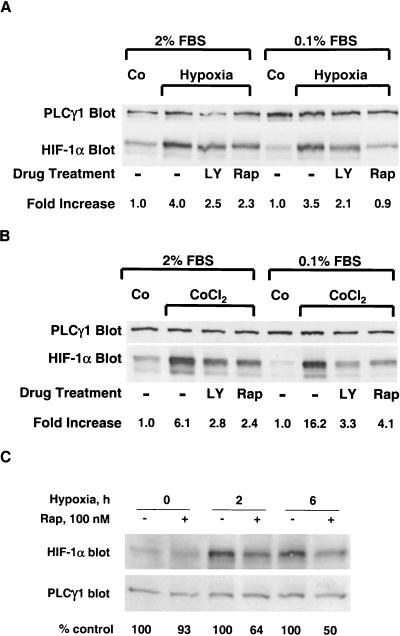
The level of HIF-1-dependent transcriptional activity is limited by the expression of the HIF-1α subunit, which is subject to rapid proteasomal degradation in normoxic cells (40). To examine the contribution of the mTOR signaling pathway to hypoxia-induced HIF-1α stabilization, we pretreated PC-3 cells with either LY294002 or rapamycin and then exposed the cells to hypoxia or CoCl2 in medium containing 2 or 0.1% serum. The steady-state levels of HIF-1α protein increased by 3.5- to 4-fold in hypoxic cells cultured at either concentration in serum, and the accumulation of this protein was reduced by ca. 50% in cells pretreated with LY294002 or rapamycin (Fig. 2A). Similar results were obtained with CoCl2 as the stimulus for HIF-1α accumulation (Fig. 2B). Although the fold induction of HIF-1α expression by CoCl2 was apparently increased when the serum concentration was dropped from 2 to 0.1%, this change is largely attributable to the lower basal level of HIF-1α expression in cells cultured in 0.1% serum. As a control, the immunoblots were reprobed with PLCγ1-specific antibodies. We observed no consistent effect of either drug on the level of PLCγ1 protein in the cellular extracts, indicating that these agents did not provoke global changes in protein turnover in PC-3 cells.
FIG. 2.
(A) PC-3 cells were precultured in medium containing 2 or 0.1% FBS, treated with the indicated drugs, and then exposed to normoxia (Co) or hypoxia as described in the Fig. 1A legend. The cells were lysed, and detergent-soluble proteins were resolved by SDS-PAGE. The membrane was immunoblotted sequentially with α-HIF-1α MAb and α-PLCγ1 antibodies. The immunoblot was subjected to densitometric analysis, and values were normalized to that obtained in the normoxic control sample. (B) PC-3 cells were treated as described above, except that cells were stimulated with 150 μM CoCl2. (C) Cells were exposed to hypoxia for 2 or 6 h in the absence of presence of rapamycin, and samples were processed as described in panel A. The band intensities were quantitated by densitometry, and numbers below the bottom panel indicate the percentage of the non-drug-treated control for each time point.
Rapamycin treatment slows the proliferation of many types, including PC-3 cells (1, 13; also results not shown). The antiproliferative effects of rapamycin raise the possibility that the suppression of HIF-1α expression is an indirect consequence of the perturbation in cell cycle progression at the relatively long drug exposure time (16 h) used in the studies described above. To address this issue, we repeated the experiment shown in Fig. 2A and examined the effect of rapamycin on HIF-1α protein levels after only 4 or 6 h of exposure to hypoxia (Fig. 2C). The results indicated that rapamycin treatment also suppressed the accumulation of HIF-1α triggered by short-term hypoxic stress, which argues against the idea that the delay in G1 progression imposed by rapamycin interferes nonspecifically with the events, leading to HIF-1α accumulation in hypoxic cells.
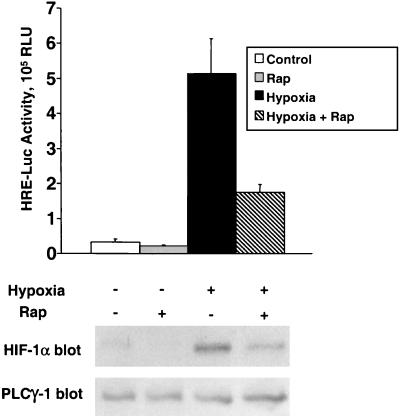
In a subsequent study, we examined the correlation between the suppressive effect of rapamycin on HIF-1α expression and the reduction in HIF-1-dependent transcription induced by this drug. The test population of PC-3 cells was transfected with pHRE-Luc reporter and then divided into parallel samples. The samples were exposed to hypoxia in 2% serum-containing medium, in the absence or presence of 100 nM rapamycin. After 16 h in culture, one sample set was harvested and processed for HIF-1α immunoblotting, and the other was prepared for luciferase assays (Fig. 3). The results demonstrate that the suppressive effects of rapamycin on HIF-1α protein expression and HIF-1 transactivating function are well correlated. Moreover, we noted in this and many other experiments that rapamycin treatment inhibited the basal expression of HIF-1α in normoxic cells, as well as the hypoxia-induced accumulation of this protein. The latter results suggest that mTOR activity increases the steady-state level of HIF-1α in both normoxic and hypoxic cells.
FIG. 3.
Correlative suppressive effects of rapamycin on HIF-1 activity and HIF-1α expression. PC-3 cells were transfected with the pHRE-Luc reporter plasmid and, after 24 h, the cells were placed in 2% serum-containing medium. After 8 h, the indicated samples were treated with 100 nM rapamycin and were cultured for an additional 16 h under either normoxic or hypoxic conditions. (Upper panel) Samples were harvested for determinations of luciferase activities. Bars and error flags represent the means ± the standard deviation from triplicate samples. (Lower panel) Parallel samples were harvested, and detergent-soluble proteins were immunoblotted with α-HIF-1α MAb. The blot was stripped and reprobed with PLCγ1-specific antibodies to control for sample loading.
Effect of rapamycin on HIF-1α turnover.
The steady-state level of HIF-1α is a function of the relative rates of HIF-1α synthesis and degradation. We reasoned that treatment of normoxic PC-3 cells with the proteasome inhibitor, LLnV, should block the major HIF-1α degradative pathway, thereby rendering the rate of HIF-1α accumulation largely a function of the rate of HIF-1α synthesis. To further probe the relative roles of altered HIF-1α synthesis versus proteolysis in the inhibitory mechanism of rapamycin, we exposed the cells to hypoxia for 16 h (left panel) or to CoCl2 for 6 h (right panel) in the absence or presence of LLnV. Hypoxia induced a 5.4-fold increase in HIF-1α expression, whereas CoCl2 treatment increased the level of this protein by 3.7-fold (Fig. 4A). Under these conditions, the accumulation of HIF-1α provoked by each stimulus was reduced by 30 to 50% in rapamycin-treated cells. Pretreatment of the cells with LLnV alone (i.e., with no rapamycin) enhanced the stimulatory effects of hypoxia and CoCl2 on HIF-1α expression by approximately twofold. Notably, the addition of LLnV to the cultures clearly blunted the inhibitory effects of rapamycin on hypoxia- or CoCl2-induced HIF-1α accumulation.
FIG. 4.
Effect of rapamycin on HIF-1α turnover. (A) Proteasome inhibition antagonizes the suppressive effect of rapamycin on hypoxia-induced HIF-1α expression. (Left panel) PC-3 cells were incubated for 6 h in 2% FBS before pretreatment for 45 min with solvent vehicle, rapamycin (Rap), or LLnV. Cells were incubated under normoxic conditions (−) or hypoxia (H). The cells were harvested after 16 h, and soluble proteins were resolved by SDS-PAGE. (Right panel) PC-3 cells were precultured for 16 h in 0.1% FBS prior to the indicated drug treatments and were then stimulated for 6 h with 150 μM CoCl2 (C). The blotted proteins were probed sequentially with α-HIF-1α MAb and α-PLCγ1 antibodies. Expression of HIF-1α protein was quantitated by densitometry and was normalized to the value obtained in the corresponding normoxic control samples. (B) LLnV-induced HIF-1α accumulation. PC-3 cells were treated for the indicated times with 10 μM LLnV under normoxic conditions, in the absence or presence of 100 nM rapamycin. HIF-1α expression was determined by immunoblotting, and the blot was stripped and reprobed with α-PLCγ1 antibody to control for sample loading. The results shown are representative of those obtained in three independent trials. (C) Effect of rapamycin on HIF-1α degradation. Cells were cultured in medium containing 2% serum and exposed to hypoxia overnight. The cells were then treated with CHX to block new protein synthesis, and the indicated samples were concomitantly exposed to 100 nM rapamycin. The cells were maintained under hypoxic conditions for the indicated times, and HIF-1α levels were determined by immunoblotting as described in panel A. PLCγ1 served as a sample loading control as described above.
In order to focus more specifically on the impact of rapamycin on HIF-1α synthesis, we treated cells with LLnV and monitored the time-dependent accumulation of HIF-1α under normoxic conditions. Treatment with the proteasome inhibitor alone resulted in a progressive increase in the level of HIF-1α, presumably due to the backup of ubiquitinated intermediates that results from obstruction of the downstream mechanism that mediates their elimination (Fig. 4B). Once again, rapamycin exerted little or no inhibitory effect on the accumulation of HIF-1α in LLnV-treated PC-3 cells, suggesting that suppression of HIF-1α gene transcription and/or translation does not underlie the reduced accumulation of HIF-1α observed in hypoxic cells after treatment with rapamycin.
To determine more directly the impact of rapamycin on the degradation rate of HIF-1α, we first induced expression of this protein by exposing PC-3 cells to hypoxia. The cells were then treated with the protein synthesis inhibitor, CHX, to block further translation of HIF-1α mRNA and were maintained for up to 4 h under hypoxic conditions. In the presence of CHX alone, HIF-1α expression declined by ca. 50% after 2 h (Fig. 4C). Concomitant exposure of these cells to rapamycin decreased the levels of HIF-1α by 40 to 50% at all four time points examined in the present study. Collectively, these results indicate that the suppressive effect of rapamycin on HIF-1α expression in hypoxic cells primarily reflects impaired functioning of the mechanism responsible for the stabilization of this protein at decreased oxygen tensions.
HIF-1 activation in mTOR-transfected PC-3 cell lines.
The interplay between mTOR signaling and HIF-1 function was further explored via a genetic approach, which was based on the introduction of either wild-type mTOR (AmTOR) or a rapamycin-resistant mTOR mutant (AmTOR-SI) into PC-3 cells. The AmTOR-SI polypeptide contains a Ser2035→Ile substitution that decreases the affinity of the mutated protein for the inhibitory FKBP12-rapamycin complex (8, 11). In AmTOR-SI-expressing cells, rapamycin treatment suppresses endogenous mTOR functions; however, these functions should be rescued by the drug-resistant AmTOR-SI mutant. Both the AmTOR-WT and the AmTOR-SI constructs contained AU1 epitope tags at their amino termini to facilitate detection of the recombinant proteins. In preliminary studies, we confirmed that the activation of a known mTOR target protein, p70S6 kinase, was resistant to rapamycin in the AmTOR-SI-expressing PC-3 subclones but not in the PC-3 subclones that stably expressed the rapamycin-sensitive AmTOR-WT protein (results not shown).
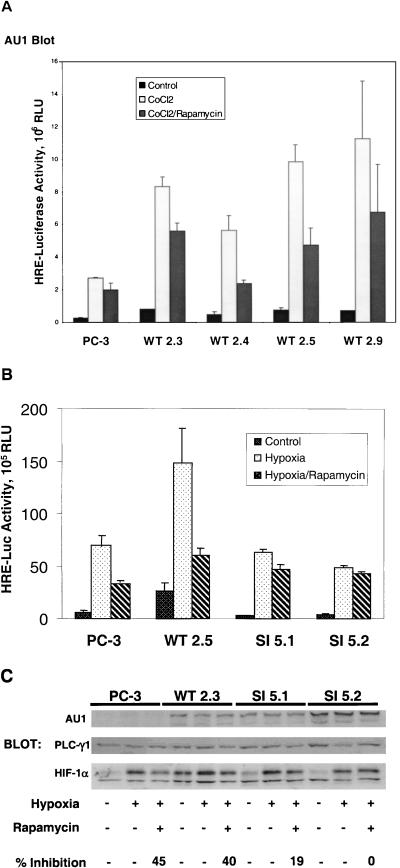
In the initial studies, we stably expressed AmTOR-WT in PC-3 cells and examined the effect of increased mTOR expression on HIF-1-dependent transcriptional activity. Four independently isolated, AmTOR-WT-expressing sublines were transiently transfected with the pHRE-luc reporter plasmid, together with a Renilla luciferase reporter plasmid, which permits correction for variations in transfection efficiency among the various sublines. The transfected cells were pretreated with rapamycin and then stimulated with CoCl2 to induce HIF-1-dependent transcription. Interestingly, the introduction of AmTOR-WT increased the levels of HIF-1-regulated luciferase expression in PC-3 cells under both basal conditions and after CoCl2 stimulation (Fig. 5A). As observed in previous experiments with CoCl2 as the stimulus, expression of the HRE-regulated luciferase reporter gene was reduced by 30 to 50% in rapamycin-treated cells. These findings indicate that an increase in the level of mTOR activity in PC-3 cells results in a corresponding increase in the HIF-1-mediated transcriptional response to CoCl2.
FIG. 5.
Activation of HIF-1 in stably transfected PC-3 sublines. PC-3 cells were stably transfected with an expression plasmid encoding either AU1-tagged, wild-type AmTOR (WT), or a rapamycin-resistant (Ser2035→Ile) AmTOR mutant (SI). Clonal sublines were cotransfected with the pHRE-luc reporter, together with a Renilla luciferase reporter (as a control for transfection efficiency). After 24 h, the culture medium was changed to RPMI 1640 supplemented with 0.1% FBS, and the cells were cultured for an additional 6 h. The cells were pretreated for 30 min with solvent vehicle or 100 nM rapamycin and then were stimulated for 16 h with 150 μM CoCl2 or hypoxia as indicated. Cell lysates were subjected to the dual luciferase assay, and sample values were normalized for variations in transfection efficiency based on the Renilla luciferase activity. Error bars represent the variance of duplicate samples. Aliquots of cell extracts were assayed for expression of the recombinant AmTOR protein by α-AU1 immunoblotting (upper panels). (A) AmTOR-WT-transfected subclones. (B) Comparison of AmTOR-WT- and AmTOR-SI-transfected subclones. (C) PC-3 cells and stably transfected WT or SI sublines wereprecultured for 8 h in medium containing 0.1% serum and then treated with either vehicle or rapamycin. After 30 min, the cells were stimulated for 16 h with hypoxia. Detergent-soluble proteins were immunoblotted sequentially with α-HIF-1α MAb and α-PLCγ1 antibodies. HIF-1α levels were determined by densitometric analysis, and the percent inhibition by rapamycin of hypoxia-induced HIF-1α expression was calculated for each sample set. In the upper panel, separate aliquots of the cell extracts were immunoblotted with α-AU1 MAb to detect the expression of the AmTOR-WT or AmTOR-SI proteins.
To obtain genetic evidence that mTOR functions as a positive modulator of HIF-1 activation in hypoxic PC-3 cells, we next tested the effects of rapamycin on HIF-1-dependent reporter gene expression in two AmTOR-SI-transfected PC-3 subclones. As shown in Fig. 5B, rapamycin exposure only marginally suppressed hypoxia-induced luciferase expression in cells transfected with the drug-resistant mTOR mutant. These results substantiate the hypothesis that mTOR is an upstream regulator of HIF-1 function in this prostate cancer cell line. An unexpected but consistent observation was that the AmTOR-SI-expressing subclones, in contrast to the corresponding AmTOR-WT transfectants, showed no elevation of HIF-1-dependent luciferase activity relative to the control PC-3 cells. The latter observation hints that the Ser2035→Ile substitution that renders the AmTOR-SI mutant resistant to FKBP12-rapamycin also alters the ability of this protein to hyperstimulate HIF-1 function in the host cells.
In subsequent studies, we examined the effects of AmTOR-WT or AmTOR-SI expression on CoCl2-induced HIF-1α accumulation in PC3 cells. As expected, this response remained sensitive to rapamycin in the AmTOR-WT-expressing PC-3 cells (Fig. 5C). Interestingly, however, the AmTOR-WT-expressing PC-3 subclone consistently displayed an elevated basal level of HIF-1α under normoxic conditions. This finding correlated with the observation that the basal level of HIF-1-dependent reporter gene expression was also elevated in these cells (Fig. 5A and B). The HIF-1α immunoblot results obtained with the two AmTOR-SI-expressing subclones were also consistent with those found in the HIF-1-dependent reporter gene assays. While the hypoxia-induced expression of HIF-1α protein was slightly lower than that observed in the AmTOR-WT-expressing cell line, this response was clearly resistant to rapamycin in the SI5.1 and SI5.2 clones. Thus, these results demonstrate that the inhibition of hypoxia-induced HIF-1α accumulation by rapamycin was directly related to the disruption of mTOR-dependent signaling functions.
Role of the ODD domain in mTOR-dependent HIF-α stabilization.
The oxygen-regulated signals that govern the stability of HIF-1α converge on the ODD domain (41). To examine the role of the ODD domain in the regulation of HIF-1α stability by mTOR, we transiently transfected PC-3 cells with expression vectors encoding the DNA-binding domain of G4 fused to HIF-1α residues 498 to 603. This fragment of HIF-1α comprises the carboxyl-terminal half of the ODD domain (Fig. 6A) and contains the critical Pro-564 residue, which is targeted for hydroxylation and subsequent VHL binding in normoxic cells (27, 48). As a control, we used the G4-HIF-1α (residues 700 to 826) fusion protein, which spans the carboxyl-terminal transactivation domain of HIF-1α. After transfection, the PC-3 cells were stimulated with hypoxia in the presence or absence of 100 nM rapamycin, and expression levels of the G4 fusion proteins were determined by immunoblotting with a G4-specific MAb (Fig. 6B). As expected, the G4-HIF-1α (residues 700 to 826) fusion protein was constitutively expressed in PC-3 cells, and the expression level was neither increased by hypoxia nor decreased by exposure to rapamycin. Strikingly different results were obtained with cells transfected with the G4-HIF-1α (residues 498 to 603) expression plasmid. Exposure of these cells to hypoxic conditions strongly increased the steady-state level of this fusion protein, and the expression the ODD domain-containing protein was sensitive to inhibition by rapamycin under both normoxic and hypoxic conditions. These results indicate that the isolated ODD domain of HIF-1α is sufficient to confer both targeting to the proteasome under normoxic conditions, and mTOR-dependent stabilization during cellular exposure to hypoxia-mimetic agents.
FIG. 6.
Role of the ODD domain in the regulation of HIF-1α expression by mTOR. (A) Schematic diagram of HIF-1α functional domains. The domain structure of HIF-1α includes the basic helix-loop-helix (bHLH) and PER-ARNT-SIM (PAS) motifs, the carboxyl-terminal transactivation domains (TAD N and C), and the ODD domain. The regulatory Pro-402 and Pro-564 residues (see the text) are also highlighted. (B) Effect of rapamycin on CoCl2-induced stabilization of G4-HIF-1α proteins. PC-3 cells were transiently transfected with empty vector (mock) or with plasmids encoding the indicated G4-HIF-1α fusion proteins. After 24 h, the transfected cells were cultured for 2 h in 2% serum-containing medium, and the indicated samples were exposed to hypoxia for 16 h, in the absence or presence of 100 nM rapamycin. Detergent-soluble proteins were separated by SDS-PAGE and immunoblotted with α-G4 MAb, followed by the addition of PLCγ1-specific antibody as a control.
DISCUSSION
The signal transduction events that modulate the expression of HIF-1α, as well as the subsequent expression of VEGF and other HIF-1-regulated genes, are currently under intensive scrutiny. The results of the present study confirm and extend the earlier report that rapamycin inhibits both the stabilization of HIF-1α and the transcriptional activity of HIF-1 in hypoxic cancer cells (50). Furthermore, we provide genetic evidence to support the conclusion that the rapamycin target protein, mTOR, functions as a positive regulator of HIF-1 activation by hypoxia or the hypoxia-mimetic agent, CoCl2.
A synthesis of the available data indicates that at least two integrated signaling pathways promote the accumulation of HIF-1α in mammalian cells. The first pathway is triggered by hypoxia or CoCl2 and involves the inhibition of a family of PHDs that modify Pro-564 and Pro-402 of HIF-1α (5, 21, 22, 27, 41). The second pathway is triggered by polypeptide growth factors or oncogenic mutations (e.g., PTEN gene loss) and leads to the activation of PI 3-kinase and its downstream targets, AKT and the Rac GTPases (17, 23, 26, 29, 50, 52). Although it seems clear that hypoxia regulates HIF-1α turnover at the level of degradation rather than synthesis (26, 44), PI 3-kinase-dependent signals appear to regulate both the synthesis and stabilization of HIF-1α in hypoxic tumor cells (26, 52).
The findings presented here substantially strengthen previous observations that mTOR-dependent signals stimulate HIF-1α accumulation and HIF-1-mediated transcription in cells exposed to hypoxia or hypoxia-mimetic agents (50). As appears to be the case for PI 3-kinase, our results indicate that the rapamycin-sensitive functions of mTOR are not essential for the accumulation of HIF-1α but are needed for maximal expression of this protein, as well as for optimal HIF-1-dependent gene expression under hypoxic conditions. The notion that PI 3-kinase and mTOR both serve as amplifiers rather than essential triggers of HIF-1 activation is consistent with the model that these two signaling kinases reside in the same signaling pathway (33, 35, 38). However, the evidence that supports a functional linkage between PI 3-kinase and mTOR is not definitive and, at this stage, we cannot exclude the alternative possibility that PI 3-kinase and mTOR converge on the HIF-1 regulatory machinery through parallel pathways. The proposed function of mTOR as a nutrient sensor (16, 37) may be particularly relevant to HIF-1 function, since decreased oxygen tensions are almost inevitably accompanied by limited supplies of glucose and amino acids in mammalian tissues.
Our initial studies demonstrated that pretreatment of PC-3 cells with rapamycin strongly inhibited the increase in HIF-1-dependent reporter gene expression provoked by hypoxia and CoCl2. These results are in general agreement with those reported by Zhong et al., who observed that rapamycin exposure inhibited epidermal growth factor and phorbol myristate acetate-induced secretion of VEGF, a HIF-1-regulated gene product, from TSU prostate cancer cells (50). On the other hand, our finding that rapamycin suppresses HIF-1-mediated gene transcription in PC-3 cells seems at odds with an earlier study, which demonstrated that rapamycin exposure had no effect on hypoxia-induced VEGF promoter activity in Ha-_Ras_-transformed NIH 3T3 cells (29). The latter findings suggest that mTOR is not an obligate intermediate in the transmission of activating signals to HIF-1 and that the host cell type, together with the particular oncogenic background, play determinant roles in the cellular response to mTOR inhibition by rapamycin. Further understanding of the signaling inputs that govern HIF-1α turnover will be critical in the event that rapamycin or other PI 3-kinase/mTOR inhibitors are approved for clinical use as cytostatic and/or cytotoxic agents in patients with solid tumors. This information may facilitate the selection of patients who are most likely to benefit from therapy with rapamycin or related drugs.
As previously reported, we observed that hypoxia- or CoCl2-induced HIF-1 activation was strongly suppressed by cellular treatment with the PI 3-kinase inhibitor, LY294002 (50). The inhibitory effects of LY294002 on both HIF-1α accumulation (50) and HIF-1-dependent transcription (Fig. 1 in the present study) are consistent with the idea that PI 3-kinase is involved in hypoxia-induced signaling to HIF-1 (10, 23, 29, 52). However, LY294002 is not a specific inhibitor of PI 3-kinase; indeed, this drug suppresses mTOR kinase activity at concentrations similar to those required for PI 3-kinase inhibition (9). Therefore, although PI 3-kinase activity is strongly implicated in the stimulation of HIF-1α by growth factors (26), its role in hypoxia-induced HIF-1α stabilization requires further investigation.
The inhibitory effect of rapamycin on HIF-1α accumulation indicated that this drug either decreased the rate of HIF-1α synthesis or increased the rate of HIF-1α degradation in hypoxic cells. By using a proteasome inhibitor to block the major pathway of HIF-1α degradation, we found that rapamycin treatment had little effect on the accumulation of HIF-1α in PC-3 cells cultured under reduced serum (either 2 or 0.1% FBS) conditions. On the other hand, rapamycin exposure significantly decreased the stability of HIF-1α in hypoxic PC-3 cells, when ongoing synthesis of HIF-1α protein was blocked with CHX. Collectively, these results suggest that rapamycin decreases the steady-state level of HIF-1α in PC-3 cells primarily through interference with the mechanism that promotes the stabilization of this protein under hypoxic conditions.
The negative effect of rapamycin on HIF-1α stability was somewhat unexpected, based on the recent report by Semenza and coworkers (26). These investigators demonstrated that stimulation of HER2 receptors in normoxic cells increased the expression of HIF-1α via a pathway that involved PI 3-kinase, AKT, and mTOR. However, in contrast to the present findings in hypoxic PC-3 cells, HER2 receptor-mediated HIF-1α accumulation occurred primarily through the upregulation of HIF-1α synthesis, due largely to the stimulation of HIF-1α mRNA translation. These authors further demonstrated that the 5′-untranslated region of the HIF-1α mRNA contains a translational control element that is upregulated by HER2 receptor occupancy and is sensitive to inhibition by rapamycin. The latter result strongly implicates mTOR as a positive regulator of HIF-1α translation, a scenario that is highly reminiscent of the stimulatory roles of mTOR in the synthesis of ribosomal and other polypyrimidine tract-containing proteins in mitogen-stimulated cells (16). Because hypoxia affects the turnover of HIF-1α at the level of degradation rather than synthesis (26, 44), the translational effect of mTOR might support, but should not drive, the increase in HIF-1α expression in hypoxic cells, particularly under conditions of limiting growth factor and nutrient availability. Our finding that rapamycin interferes with the stabilization of HIF-1α under hypoxic conditions is therefore of particular importance and strongly suggests that mTOR-dependent signals promote HIF-1α expression at the levels of both synthesis and proteolytic degradation. The nature of the stimulus (i.e., polypeptide hormone stimulation versus hypoxia), together with the cellular background, likely determine which of these signaling inputs from mTOR exerts dominant control over the increase in HIF-1α protein in different tumors.
The importance of mTOR in the transduction of hypoxia/CoCl2-initiated signals to HIF-1α was underscored by the results of genetic experiments involving the expression of wild-type or mutated mTOR constructs in PC-3 cells. Overexpression of wild-type mTOR significantly increased the level of HIF-1-dependent reporter gene expression provoked by exposure of the engineered cell lines to the hypoxia-mimetic agent, CoCl2 (Fig. 4A), as well as hypoxia itself (unpublished results). These results suggest that the endogenous level of mTOR activity in PC-3 cells limits the magnitude of the HIF-1-dependent transcriptional response to these stimuli. Furthermore, expression of the rapamycin-resistant AmTOR-SI mutant in the same cells abrogates the inhibitory effects of rapamycin on CoCl2-stimulated HIF-1α accumulation and HIF-1-dependent transcription. Thus, the results obtained with AmTOR-SI-transfected PC-3 sublines provide genetic evidence to support the conclusion that rapamycin suppresses hypoxia/CoCl2-induced HIF-1 function through inhibition of a single target protein, mTOR.
Transfection experiments with a panel of G4-HIF-1α fusion proteins pinpointed the ODD domain as an important target for the mTOR signaling pathway that amplifies HIF-1α stabilization at low oxygen tensions. In normoxic PC-3 cells, expression of the G4-HIF-1α (residues 498 to 603) construct was relatively low due to proteasome-mediated degradation of the fusion protein. This construct contains the carboxyl-terminal half of the ODD domain and includes the critical Pro-564 residue, which undergoes PHD-dependent hydroxylation in hypoxic cells (41). Exposure of the cells to hypoxia or to the PHD inhibitor CoCl2 (unpublished results) led to significant stabilization of HIF-1α, which is consistent with the model that the ODD domain, and specifically the region surrounding Pro-564, confers instability on the HIF-1α protein at normal oxygen tensions (5, 6, 21, 27, 48). The hypoxia-induced increase in G4-HIF-1α (residues 498 to 603) expression was strongly suppressed by rapamycin, whereas the expression of G4 fusion proteins containing amino- or carboxyl-terminal fragments of HIF-1α was neither hypoxia-inducible nor sensitive to rapamycin (see Fig. 6B and unpublished results). We conclude from these findings that the mTOR-dependent signals that promote HIF-1α stabilization under hypoxic conditions impinge largely, if not entirely, on the ODD domain. Moreover, the suppressive effect of rapamycin on the expression of the chimeric G4-HIF-1α (residues 498 to 603) protein substantiates the conclusion that, in the setting of hypoxia/CoCl2 stimulation, the drug is not simply acting as a 5′-untranslated-region-dependent inhibitor of HIF-1α mRNA translation.
The exact mechanism whereby mTOR contributes to the stabilization of HIF-1α in hypoxic cells remains unclear. Given the complexity of the oxygen-responsive machinery that governs the stability of HIF-1α, the list of potential mTOR substrates in this pathway is extensive. Based on the results described above, we considered the ODD domain a potential site of intersection for both mTOR- and oxygen-dependent signals that govern HIF-1α stability. The ODD domain is serine- and threonine-rich and contains numerous Ser/Thr-Pro sites that represent potential targets for the mTOR kinase domain (7, 8). Consequently, we examined the ability of a series of glutathione _S_-transferase-HIF-1α fusion proteins to serve as substrates for mTOR in immune complex kinase assays. The results revealed that the ODD domain was the only region of HIF-1α that was detectably phosphorylated by mTOR in vitro (unpublished results). However, the stoichiometry of ODD phosphorylation by mTOR was quite low, and additional studies will be required to determine the significance of this finding. We also cannot rule out the possibility that mTOR modulates the activities of the PHDs that modify HIF-1α or interferes with the recognition of proline-hydroxylated HIF-1α by the VHL-ubiquitin E3 ligase complex.
In summary, this study identifies mTOR as a positive regulator of HIF-1-dependent gene transcription in cells exposed to hypoxia or hypoxia-mimetic agents. Our results suggest that mTOR plays a key role in the transmission of signals that lead to the adaptation of mammalian cells to oxygen- and nutrient-poor environmental conditions. It is noteworthy that studies in budding yeast have established the mTOR orthologs, TOR1p and TOR2p, as central regulators of the pathway that coordinates nutrient availability with yeast growth (37). The particular demands placed on nutrient-deprived cells sequestered in bodily tissues may have provided the evolutionary driving force for the insertion of mTOR into the hypoxia response pathway in metazoan organisms. Finally, our findings add further support to the idea that mTOR is an intriguing target for anticancer therapy (30). If rapamycin or other mTOR inhibitors prove to be effective inhibitors of hypoxic adaptation in developing tumors, these drugs could have dramatic effects on tumor growth, invasiveness, and metastatic potential in human cancer patients.
Acknowledgments
We thank Mark Dewhirst, Chuan-Yuan Li, and Yiting Cao for their assistance with the hypoxia experiments; Andrew Kung for the G4-HIF-1α expression vectors; and Peter Houghton for supplying the 26E3 α-mTOR MAb.
This work was supported by grants CA76193 (to R.T.A.) and CA67166 (to A.J.G.) from the National Cancer Institute and by a Department of Defense Prostate Cancer Research Program Idea Award DAMD17-01-1-0052 (to R.T.A.). C.C.H. is the recipient of postdoctoral fellowship PF-99-100-01-TBE from the American Cancer Society.
REFERENCES
- 1.Abraham, R. T., and G. J. Wiederrecht. 1996. Immunopharmacology of rapamycin. Annu. Rev. Immunol. 14**:**483-510. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, M., E. Blazek, and P. K. Vogt. 2001. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc. Natl. Acad. Sci. USA 98**:**136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brizel, D. M., S. P. Scully, J. M. Harrelson, L. J. Layfield, J. M. Bean, L. R. Prosnitz, and M. W. Dewhirst. 1996. Tumor oxygenation predicts for the likelihood of distant metastases in human soft tissue sarcoma. Cancer Res. 56**:**941-943. [PubMed] [Google Scholar]
- 4.Brizel, D. M., G. S. Sibley, L. R. Prosnitz, R. L. Scher, and M. W. Dewhirst. 1997. Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 38**:**285-289. [DOI] [PubMed] [Google Scholar]
- 5.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294**:**1337-1340. [DOI] [PubMed] [Google Scholar]
- 6.Bruick, R. K., and S. L. McKnight. 2002. Transcription: oxygen sensing gets a second wind. Science 295**:**807-808. [DOI] [PubMed] [Google Scholar]
- 7.Brunn, G. J., P. Fadden, T. A. J. Haystead, and J. C. Lawrence, Jr. 1997. The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J. Biol. Chem. 272**:**32547-32550. [DOI] [PubMed] [Google Scholar]
- 8.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, J. Hosoi, P. J. Houghton, J. C. Lawrence, and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277**:**99-101. [DOI] [PubMed] [Google Scholar]
- 9.Brunn, G. J., J. Williams, C. Sabers, G. Wiederrecht, J. C. Lawrence, Jr., and R. T. Abraham. 1996. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 15**:**5256-5267. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, E. Y., N. M. Mazure, J. A. Cooper, and A. J. Giaccia. 1915. Hypoxia activates a platelet-derived growth factor receptor/phosphatidylinositol 3-kinase/Akt pathway that results in glycogen synthase kinase-3 inactivation. Cancer Res. 61**:**2429-2433. [PubMed] [Google Scholar]
- 11.Chen, J., X. F. Zheng, E. J. Brown, and S. L. Schreiber. 1995. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 92**:**4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Cristofano, A., and P. P. Pandolfi. 2000. The multiple roles of PTEN in tumor suppression. Cell 100:387-390. [DOI] [PubMed] [Google Scholar]
- 13.Dilling, M. B., P. Dias, D. N. Shapiro, G. S. Germain, R. K. Johnson, and P. J. Houghton. 1994. Rapamycin selectively inhibits the growth of childhood rhabdomyosarcoma cells through inhibition of signaling via the type I insulin-like growth factor receptor. Cancer Res. 54**:**903-907. [PubMed] [Google Scholar]
- 14.Ebert, B. L., J. D. Firth, and P. J. Ratcliffe. 1995. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct _cis_-acting sequences. J. Biol. Chem. 270**:**29083-29089. [DOI] [PubMed] [Google Scholar]
- 15.Epstein, A. C. R., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107**:**43-54. [DOI] [PubMed] [Google Scholar]
- 16.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 17.Hirota, K., and G. L. Semenza. 2001. Rac1 activity is required for the activation of hypoxia-inducible factor 1. J. Biol. Chem. 276**:**21166-21172. [DOI] [PubMed] [Google Scholar]
- 18.Hockel, M., and P. Vaupel. 2001. Biological consequences of tumor hypoxia. Semin. Oncol. 28**:**36-41. [PubMed] [Google Scholar]
- 19.Holmgren, L. 1996. Antiangiogenis restricted tumor dormancy. Cancer Met. Rev. 15**:**241-245. [DOI] [PubMed] [Google Scholar]
- 20.Ivan, M., and W. G. J. Kaelin. 2001. The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Gen. Dev. 11**:**27-34. [DOI] [PubMed] [Google Scholar]
- 21.Ivan, M., K. Kondo, H. F. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin. 2001. HIF alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O-2 sensing. Science 292**:**464-468. [DOI] [PubMed] [Google Scholar]
- 22.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292**:**468-472. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, B. H., G. Q. Jiang, J. Z. Zheng, Z. M. Lu, T. Hunter, and P. K. Vogt. 2001. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor. Cell Growth Differ. 12**:**363-369. [PubMed] [Google Scholar]
- 24.Klippel, A., M. A. Escobedo, M. S. Wachowicz, G. Apell, T. W. Brown, M. A. Giedlin, W. M. Kavanaugh, and L. T. Williams. 1998. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol. Cell. Biol. 18**:**5699-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain: A hypoxic switch. Science 295**:**858-861. [DOI] [PubMed] [Google Scholar]
- 26.Laughner, E., P. Taghavi, K. Chiles, P. C. Mahon, and G. L. Semenza. 2001. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1 alpha (HIF-1α) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell. Biol. 21**:**3995-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20**:**5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxwell, P. H., C. W. Pugh, and P. J. Ratcliffe. 2001. Activation of the HIF pathway in cancer. Curr. Opin. Gen. Dev. 11**:**293-299. [DOI] [PubMed] [Google Scholar]
- 29.Mazure, N. M., E. Y. Chen, K. R. Laderoute, and A. J. Giaccia. 1997. Induction of vascular endothelial growth factor by hypoxia is modulated by a phosphatidylinositol 3-kinase/Akt signaling pathway in Ha-ras-transformed cells through a hypoxia inducible factor-1 transcriptional element. Blood 90**:**3322-3331. [PubMed] [Google Scholar]
- 30.Mills, G. B., Y. Lu, and E. C. Kohn. 2001. Linking molecular therapeutics to molecular diagnostics: inhibition of the FRAP/RAFT/TOR component of the PI3K pathway preferentially blocks PTEN mutant cells in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98**:**10031-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatani, K., D. A. Thompson, A. Barthel, H. Sakaue, W. Liu, R. J. Weigel, and R. A. Roth. 1999. Upregulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J. Biol. Chem. 274**:**21528-21532. [DOI] [PubMed] [Google Scholar]
- 32.Nave, B. T., D. M. Ouwens, D. J. Withers, D. R. Alessi, and P. R. Shepherd. 1999. Mammalian target of rapamycin is a direct target for PKB: identification of a convergence point for opposing effects of insulin and amino acid deficiency on protein translation. Biochem. J. 344**:**427-431. [PMC free article] [PubMed] [Google Scholar]
- 33.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98**:**10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owa, T., H. Yoshino, K. Yoshimatsu, and T. Nagasu. 2001. Cell cycle regulation in the G1 phase: a promising target for the development of new chemotherapeutic anticancer agents. Curr. Med. Chem. 8**:**1487-1503. [DOI] [PubMed] [Google Scholar]
- 35.Podsypanina, K., R. T. Lee, C. Politis, I. Hennessy, A. Crane, J. Puc, M. Neshat, H. Wang, L. Yang, J. Gibbons, P. Frost, V. Dreisbach, J. Blenis, Z. Gaciong, P. Fisher, C. Sawyers, L. Hedrick-Ellenson, and R. Parsons. 2001. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten(+/−) mice. Proc. Natl. Acad. Sci. USA 98**:**10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powis, G., R. Bonjouklian, M. M. Berggren, A. Gallegos, R. Abraham, C. Ashendel, L. Zalkow, W. F. Matter, J. Dodge, and G. Grindey. 1994. Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res. 54**:**2419-2423. [PubMed] [Google Scholar]
- 37.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276**:**9583-9586. [DOI] [PubMed] [Google Scholar]
- 38.Sekulic, A., C. C. Hudson, J. L. Homme, P. Yin, D. M. Otterness, L. M. Karnitz, and R. T. Abraham. 2000. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 60**:**3504-3513. [PubMed] [Google Scholar]
- 39.Semenza, G. L. 2000. HIF-1 and human disease: one highly involved factor. Genes Dev. 14**:**1983-1991. [PubMed] [Google Scholar]
- 40.Semenza, G. L. 2001. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 13**:**167-171. [DOI] [PubMed] [Google Scholar]
- 41.Semenza, G. L. 2001. HIF-1, O-2, and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107**:**1-3. [DOI] [PubMed] [Google Scholar]
- 42.Semenza, G. L. 2000. HIF-1: using two hands to flip the angiogenic switch. Cancer Met. Rev. 19**:**59-65. [DOI] [PubMed] [Google Scholar]
- 43.Semenza, G. L. 1999. Regulation of mammalian O-2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15**:**551-578. [DOI] [PubMed] [Google Scholar]
- 44.Sutter, C. H., E. Laughner, and G. L. Semenza. 2000. Hypoxia-inducible factor 1α protein expression is controlled by oxygen-regulated ubiquitination that is disrupted by deletions and missense mutations. Proc. Natl. Acad. Sci. USA 97**:**4748-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talks, K. L., H. Turley, K. C. Gatter, P. H. Maxwell, C. W. Pugh, P. J. Ratcliffe, and A. L. Harris. 2000. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 157**:**411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ting, A. T., C. J. Dick, R. A. Schoon, L. M. Karnitz, R. T. Abraham, and P. J. Leibson. 1995. Interaction between Lck and Syk family tyrosine kinases in Fc gamma receptor-initiated activation of natural killer cells. J. Biol. Chem. 270**:**16415-16421. [DOI] [PubMed] [Google Scholar]
- 47.Yang, H. F., and W. G. Kaelin. 2001. Molecular pathogenesis of the von Hippel-Lindau hereditary cancer syndrome: implications for oxygen sensing. Cell Growth Differ. 12**:**447-455. [PubMed] [Google Scholar]
- 48.Yu, F., S. B. White, Q. Zhao, and F. S. Lee. 2001. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98**:**9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zagzag, D., H. Zhong, J. M. Scalzitti, E. Laughner, J. W. Simons, and G. L. Semenza. 2000. Expression of hypoxia-inducible factor 1α in brain tumors: association with angiogenesis, invasion, and progression. Cancer 88**:**2606-2618. [PubMed] [Google Scholar]
- 50.Zhong, H., K. Chiles, D. Feldser, E. Laughner, C. Hanrahan, M. M. Georgescu, J. W. Simons, and G. L. Semenza. 2000. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60**:**1541-1545. [PubMed] [Google Scholar]
- 51.Zhong, H., M. De, E. Laughner, M. Lim, D. A. Hilton, D. Zagzag, P. Buechler, W. B. Isaacs, G. L. Semenza, and J. W. Simons. 1999. Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59**:**5830-5835. [PubMed] [Google Scholar]
- 52.Zundel, W., C. Schindler, D. Haas-Kogan, A. Koong, F. Kaper, E. Chen, A. R. Gottschalk, H. E. Ryan, R. S. Johnson, A. B. Jefferson, D. Stokoe, and A. J. Giaccia. 2000. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 14**:**391-396. [PMC free article] [PubMed] [Google Scholar]