HIF-2α regulates Oct-4: effects of hypoxiaon stem cell function, embryonic development, and tumor growth (original) (raw)
Abstract
The division, differentiation, and function of stem cells and multipotent progenitors are influenced by complex signals in the microenvironment, including oxygen availability. Using a genetic “knock-in” strategy, we demonstrate that targeted replacement of the oxygen-regulated transcription factor HIF-1α with HIF-2α results in expanded expression of HIF-2α-specific target genes including Oct-4, a transcription factor essential for maintaining stem cell pluripotency. We show that HIF-2α, but not HIF-1α, binds to the Oct-4 promoter and induces Oct-4 expression and transcriptional activity, thereby contributing to impaired development in homozygous Hif-2α KI/KI embryos, defective hematopoietic stem cell differentiation in embryoid bodies, and large embryonic stem cell (ES)-derived tumors characterized by altered cellular differentiation. Furthermore, loss of HIF-2α severely reduces the number of embryonic primordial germ cells, which require Oct-4 expression for survival and/or maintenance. These results identify Oct-4 as a HIF-2α-specific target gene and indicate that HIF-2α can regulate stem cell function and/or differentiation through activation of Oct-4, which in turn contributes to HIF-2α’s tumor promoting activity.
Keywords: HIF, hypoxia, HIF-2α, Oct-4, VEGF, TGF-α, stem cells, cancer
Stem cells are characterized by the ability to self-renew and maintain pluripotency. The POU transcription factor Oct-4 (also known as Oct-3/4 and Pou5F1) is essential for maintaining an undifferentiated cell fate in embryonic stem (ES) cells, the embryonic epiblast, and primordial germ cells (PGCs) (Scholer et al. 1990b; Nichols et al. 1998). Oct-4 also plays a critical role in regulating the differentiation of ES cells and maintaining the pluripotent nature of the blastocyst inner cell mass (ICM). In vitro experiments have demonstrated the importance of maintaining strict Oct-4 expression levels. For example, twofold changes in Oct-4 expression cause ES cells to lose pluripotency and differentiate into trophectoderm, mesoderm, neuroectoderm, or endoderm lineages (Niwa et al. 2000; Shimozaki et al. 2003). Recently, Oct-4 has been shown to function in a complex with Nanog and Sox2 to activate and repress genes controlling stem cell identity and differentiation (Boyer et al. 2005).
Oct-4 expression is tightly controlled throughout embryogenesis and postnatal life. For example, down-regulation of Oct-4 is required for differentiation of trophectoderm and primitive endoderm lineages. At the egg cylinder stage, Oct-4 is expressed in the epiblast and subsequently down-regulated during gastrulation, although expression is maintained in PGCs. Furthermore, correlative evidence suggests that when Oct-4 expression is dysregulated, cloned embryos do not develop normally beyond post-implantation stages (Boiani et al. 2002). In the adult, Oct-4 is generally believed to be expressed exclusively in germ cells; however, recent evidence suggests Oct-4 is also present in adult stem cell populations, such as multipotent adult progenitor cells (MAPCs) and hematopoietic stem cells (Jiang et al. 2002; Tai et al. 2005). Finally, ectopic Oct-4 expression contributes to tumor growth and also drives reversible epithelial dysplasia in transgenic mice (Gidekel et al. 2003; Cheng et al. 2004; Hochedlinger et al. 2005). Currently, the mechanisms and factors regulating Oct-4 expression have not been fully elucidated.
Stem cells, as well as germ cells and other multipotent progenitors, reside in complex microenvironments, termed niches (Spradling et al. 2001). Stromal cell contacts, extracellular matrix proteins, temperature, and oxygen (O2) levels in the immediate microenvironment can influence stem cell function and differentiation. In particular, low O2 levels (hypoxia) promote the survival of neural crest stem cells, hematopoietic stem cells, and human ES cells (Morrison et al. 2000; Studer et al. 2000; Danet et al. 2003; Ezashi et al. 2005). Hypoxia also regulates the number of ICM cells in bovine blastocysts and enhances hemangioblast and hematopoietic progenitor development. (Adelman et al. 1999; Harvey et al. 2004; Ramirez-Bergeron et al. 2004). It is therefore possible that hypoxia mediates its effects on stem cell function by altering Oct-4 expression.
Hypoxia occurs in a number of physiological and pathophysiological settings, particularly when rapid tissue growth exceeds blood supply. For example, embryogenesis occurs in a physiologic “hypoxic” environment (3%–5% O2). The primary transcriptional regulators of both cellular and systemic hypoxic adaptation in mammals are hypoxia-inducible factors (HIFs). HIFs are heterodimers consisting of a regulated subunit (HIFα) and a constitutive subunit (HIFβ, also known as ARNT [aryl hydrocarbon nuclear translocator]) (Wenger and Gassmann 1997). HIFs regulate the expression of at least 180 genes involved in metabolism, cell survival, erythropoiesis, and vascular remodeling (Semenza 2000) by binding to _cis_-acting hypoxia response elements (HREs) located in the enhancers and/or promoters of these genes (Pugh et al. 1991).
The first HIFα subunit identified was HIF-1α, which is expressed ubiquitously in human and mouse tissues and considered to be the primary regulator of hypoxic responses. The more recent identification of HIF-2α (also known as endothelial PAS domain protein 1 [EPAS 1], HIF-1-like factor [HLF], and HIF-1-related factor [HRF]) has raised important questions about the relative activity of these factors in mediating hypoxic adaptation. HIF-1α and HIF-2α share a high degree of sequence identity, underscored by their shared ability to heterodimerize with ARNT and bind HREs to activate transcription in various in vitro reporter assays (Wiesener et al. 1998). Whereas HIF-1α is broadly expressed, HIF-2α transcripts are restricted to particular cell types, including vascular endothelial cells, neural crest cell derivatives, lung type II pneumocytes, liver parenchyma, and interstitial cells in the kidney (Ema et al. 1997; Flamme et al. 1997; Tian et al. 1997; Compernolle et al. 2002; Wiesener et al. 2003). Gene targeting experiments in mice have shown that loss of HIF-1α or HIF-2α results in strikingly different phenotypes, suggesting that each protein performs unique physiological functions. It is possible that HIF-1α and HIF-2α proteins are functionally equivalent but perform different physiological roles due to their distinct expression patterns; alternatively, HIF-1α and HIF-2α may regulate overlapping but nonidentical target genes. In studies designed to identify unique HIF-1α and HIF-2α target genes, we and others showed that HIF-1α and HIF-2α activate a number of common genes; however, HIF-1α exclusively induces the hypoxic transcription of glycolytic genes such as phosphoglycerate kinase 1 (Pgk1) and aldolase A (Alda) (Hu et al. 2003; Wang et al. 2005). Expression profiling of a renal clear cell (RCC) carcinoma line expressing HIF-2α, but not HIF-1α, identified a number of putative HIF-2α target genes, including Vascular endothelial growth factor (Vegf) and Transforming growth factor α (_Tgf_-α) (Gunaratnam et al. 2003; Hu et al. 2003; Wykoff et al. 2004).
To dissect the unique and overlapping roles of HIF-1α and HIF-2α more rigorously, we generated “knock-in” ES cells in which a cDNA encoding HIF-2α was targeted to the Hif-1α locus, generating a Hif-1αHif-2αKI allele, hereafter referred to as Hif-2α KI (Covello et al. 2005). This allele results in expanded HIF-2α expression under the regulatory control of the Hif-1α locus and replaces HIF-1α with HIF-2α, thereby allowing a direct comparison of HIF-1α and HIF-2α function. Previous experiments revealed that teratomas derived from homozygous Hif-2α KI/KI ES cells were significantly larger and more vascularized than wild-type tumors, and exhibited increased expression of HIF-2α target genes, including Tgf-α and Vegf (Covello et al. 2005). Interestingly, Hif-2α KI/KI tumors displayed a large proportion of undifferentiated cells in addition to a distinctive spectrum of differentiated mesodermal and neural cells.
In this report, we investigate the effects of expanded HIF-2α expression on embryonic development by generating Hif-2α KI mice. Surprisingly, homozygous Hif-2α KI/KI embryos were recovered at markedly reduced numbers at embryonic day 6.5–7.5 (E6.5–E7.5), exhibited developmental patterning defects, and displayed increased expression of Vegf, Tgf-α, and Oct-4. Increased Oct-4 mRNA levels in Hif-2α KI/KI embryoid bodies (EBs) correlated with enhanced Oct-4 protein levels and transcriptional activity, and was associated with defects in generating hematopoietic progenitors. Down-regulation of Oct-4 activity in Hif-2α KI/KI cells partly rescued the hematopoietic phenotypes; moreover, Oct-4 inhibition altered the cellular differentiation and diminished the growth of Hif-2α KI/KI teratomas. Finally, _Hif-2α_−/− embryos displayed strikingly reduced numbers of PGCs, which require Oct-4 expression for survival and/or maintenance. Together, our data identify HIF-2α as an upstream regulator of Oct-4 expression, and suggest a specific mechanism whereby HIF-2α can affect stem cell function and promote tumor growth.
Results
Expanded HIF-2α is deleterious to embryonic development
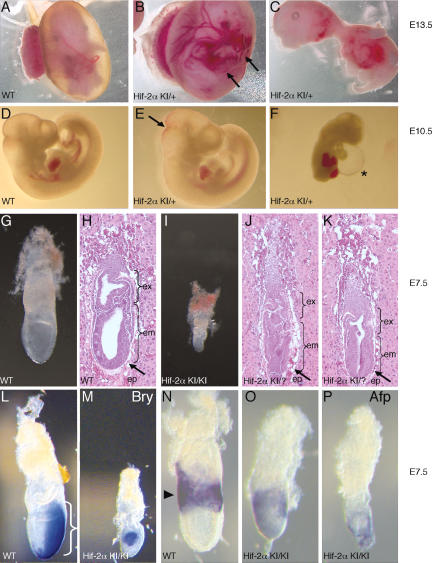
The Hif-2α KI genetic model was designed as a stringent genetic test of the degree to which HIF-2α could rescue a _Hif-1α_-null mutation. Briefly, the Hif-2α KI allele was generated by introducing a cDNA encoding a c-Myc epitope-tagged murine HIF-2α protein into the first coding exon of the murine Hif-1α gene via homologous recombination in ES cells. ES cells homozygous for Hif-2α KI express no HIF-1α protein, but instead express c-Myc-tagged HIF-2α under the control of the ubiquitously expressed Hif-1α locus (Covello et al. 2005). Chimeric mice were generated by blastocyst injection, and the Hif-2α KI allele was transmitted through the germline. Two separate Hif-2α KI ES cell lines were used to generate independent Hif-2α KI heterozygous mouse lines, and identical results were obtained from each line. When heterozygous Hif-2α KI/+ mice were intercrossed, no live-born homozygous Hif-2α KI/KI mice were recovered, and heterozygous Hif-2α KI/+ mice were recovered in nearly Mendelian ratios (63.1%, cf. 66.7% expected) (Supplementary Table 1). Embryos from timed matings between Hif-2α KI/+ mice were dissected beginning at E9.5. We predicted that if the Hif-2α KI allele failed to complement the loss of HIF-1α expression, then homozygous Hif-2α KI/KI embryos would die between E9.5 and E11.5, thereby phenocopying _Hif-1α_-null mutant embryos (Iyer et al. 1998; Ryan et al. 1998; Compernolle et al. 2003). Surprisingly, we were unable to recover any homozygous Hif-2α KI/KI embryos between E8.5 and E9.5 (Supplementary Table 1, n = 92). To control for possible effects of genetic background, we backcrossed the _Hif-1α−/−_-null mutant allele four times to the SvEvTac strain in which the Hif-2α KI mice were generated. Mutant Hif-1α−/− embryos with characteristic phenotypes were recovered in Mendelian ratios between E8.5 and E9.5 in crosses between heterozygous Hif-1α+/− mice (Supplementary Fig. 1A,B). Furthermore, whereas heterozygous Hif-1α+/− embryos were phenotypically indistinguishable from wild-type, heterozygous Hif-2α KI/+ embryos exhibited a range of phenotypes between E8.5 and E13.5 (Fig. 1A–F; data not shown). Approximately 50% of Hif-2α KI/+ embryos were indistinguishable from wild type, whereas the remainder exhibited various defects, including minor hemorrhaging in the embryo and yolk sac as well as more severe phenotypes such as cardiac effusion and death (Fig. 1C,F). The segregation data in Supplementary Table 1 indicate, however, that the majority of the phenotypes observed in heterozygous Hif-2α KI/+ embryos did not result in lethality.
Figure 1.
Disrupted development in Hif-2α KI embryos. (A_–_F) Between E10.5 and E13.5, ∼50% of Hif-2α KI/+ heterozygous embryos demonstrated a range of phenotypes. At E13.5, Hif-2α KI/+ heterozygous yolk sacs and embryos display hemorrhaging (B, arrows) and gross abnormalities (C), compared with wild-type (WT) controls (A). (C) Several embryos disintegrated upon dissection, suggesting they were dead or severely abnormal. At E10.5, Hif-2α KI/+ heterozygous embryos displayed phenotypes ranging from subtle hemorrhaging (E, arrow) to cardiac effusion (F, asterisk). A wild-type (WT) E10.5 embryo control is shown in D. (G_–_K) Comparison of wild-type (WT) (G,H) and homozygous Hif-2α KI/KI (I) and presumed homozygous Hif-2α KI/? (J,K) E7.5 embryos. Genotypes of embryos in J and K were inferred from morphological similarity to confirmed Hif-2α KI/KI embryos (shown in I) and frequency (10% of total embryos). Hif-2α KI/KI embryos displayed improper formation of the epiblast (ep, arrows in H,J,K), and embryonic (em) and extraembryonic (ex) cavities, compared with wild type (WT). (L–P) Whole-mount RNA in situ hybridization on E7.5 embryos using probes for the mesodermal marker Brachyury (Bry) (L,M) and visceral endodermal marker α-fetoprotein (AFP) (N–P). In contrast to wild-type (WT) littermate controls (L,N), which display normal mesoderm migration through the primitive streak (bracket in L), Hif-2α KI/KI embryos exhibit patterning defects (M) consistent with abnormal gastrulation. Similarly, AFP expression is restricted to the extraembryonic visceral endoderm in wild-type embryos (arrow in N), but maintained in embryonic visceral endoderm in Hif-2α KI/KI embryos (O,P).
By extending our analysis to earlier timepoints, we recovered morphologically aberrant homozygous Hif-2α KI/KI embryos at E6.5–E7.5 (Fig. 1I), although significantly fewer (10%) than the 25% expected based on Mendelian ratios (n = 120). In contrast, Hif-2α KI/KI blastocysts (E3.5) were recovered at the expected ratios (Supplementary Table 1). These data, coupled with the observation that 18% of the total E6.5–E7.5 decidua from Hif-2α KI/+ heterozygous crosses did not contain a detectable egg cylinder, suggested that the missing Hif-2α KI/KI embryos were resorbed. Histological analysis of intact E6.5–E7.5 decidua also revealed that 10% of the embryos exhibited an expansion of the epiblast and failed to form typical extraembryonic and embryonic cavities (Fig. 1J,K). Aberrant temporal and spatial expression of mesodermal and visceral endoderm markers (Brachyury and α_-Fetoprotein_, respectively) was observed in the few Hif-2α KI/KI embryos we recovered, suggesting that they failed to maintain normal patterning of embryonic and extraembryonic tissues (Fig. 1L–P). Taken together, these data indicate that ∼2/3 of homozygous Hif-2α KI/KI embryos cannot be recovered at E6.5–E7.5, whereas the remainder display defects associated with aberrant embryonic patterning. The abnormalities observed in Hif-2α KI/KI embryos are distinct from, and occur earlier than, those attributable to loss of HIF-1α expression (Supplementary Fig. 1A–E), and appear to result from a gain of HIF-2α function.
Expanded HIF-2α expression results in up-regulationof Vegf, Tgf-α, and Oct-4
We hypothesized that the developmental defects observed in homozygous Hif-2α KI/KI embryos were caused by expanded expression of specific HIF-2α target genes. Published studies performed on RCC carcinoma lines that express HIF-2α, but not HIF-1α, provided a list of target genes whose misregulation might contribute to the Hif-2α KI embryonic phenotype, including Vegf, _Tgf_-α, Adrenomedullin (Adm), and N-myc downstream-regulated-1 (NDRG-1) (Gunaratnam et al. 2003; Hu et al. 2003; Wykoff et al. 2004). Intriguingly, our analysis revealed that these RCC cells also up-regulated Oct-4 (Pou5f1) in response to hypoxia (Hu et al. 2003), raising the possibility that misregulation of Oct-4 might contribute to the developmental phenotypes observed in the Hif-2α KI/KI embryos. As dose-dependent functions of Oct-4 in ES cells have previously been described (Scholer et al. 1990b; Niwa et al. 2000), we hypothesized that aberrant Oct-4 expression accounted for the low number of post-implantation Hif-2α KI/KI embryos recovered in our experiments, as well as their morphological defects.
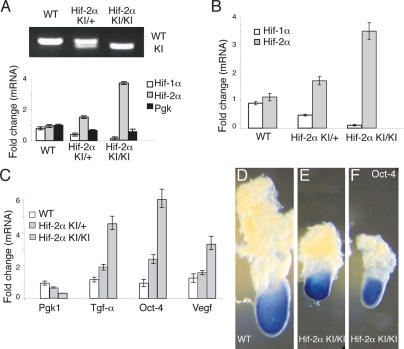
To determine whether Oct-4 and other HIF-2α target genes were overexpressed in Hif-2α KI/KI embryos, mRNAs from individual E6.5–E7.5 embryos were analyzed by quantitative real-time PCR for transcripts encoding Hif-1α, Hif-2α, Pgk1, Vegf, Tgf-α, and Oct-4 (Fig. 2). Genotyping was conducted on embryo DNA and correlated to mRNA transcript levels for Hif-1α, Hif-2α, and Pgk1, a HIF-1α-specific target gene (Fig. 2A; Hu et al. 2003). Homozygous Hif-2α KI/KI embryos exhibited <0.25-fold wild-type levels of Hif-1α and Pgk1, and a greater than twofold increase in the expression of Hif-2α (Fig. 2A,B). Figure 2A shows a single embryo per each genotype, and Figure 2B illustrates the average levels of Hif-1α and Hif-2α transcripts from pooled embryos (n = 9) of a given genotype. Compared with wild-type embryos, homozygous Hif-2α KI/KI embryos exhibited significantly increased Vegf, Tgf-α, and Oct-4 expression, which correlated with a 3.5-fold increase in Hif-2α transcripts (Fig. 2C). Transcript levels were normalized to 18S rRNA transcripts in each sample. Heterozygous Hif-2α KI/+ embryos showed intermediate expression levels of Tgf-α and Oct-4. Together, these data indicate that Vegf, Tgf-α, and Oct-4 mRNA are elevated by expanded HIF-2α expression in Hif-2α KI embryos.
Figure 2.
Increased Oct-4, Vegf, and Tgf-α expression in Hif-2α KI/KI embryos. (A) Genotypes of individual E7.5 embryos were determined by PCR on genomic DNA and quantitative RT–PCR analysis of Hif-1α, Hif-2α, and Pgk transcripts. Transcript levels were normalized to 18S rRNA transcripts in each sample. Data are expressed as the ratio of target gene expression in each sample relative to a known wild-type (WT) litter (n = 8) that was stage matched. (B) Average levels of Hif-1α and Hif-2α mRNA in embryos pooled by genotype. (C) Average levels of Oct-4, Vegf, and Tgf-α mRNA in embryos pooled by genotype. (D–F) Whole-mount RNA in situ hybridization for Oct-4 mRNA demonstrated that Oct-4 is expressed in the embryonic epiblast in both wild-type (WT) and homozygous Hif-2α KI/KI E7.5 embryos.
At least two mechanisms could account for these results. First, the spatial expression pattern of HIF-2α target genes could be expanded, resulting in ectopic expression in atypical cell types. Alternatively, target gene transcripts could display normal spatial distribution but become more abundant. To explore these possibilities, we performed whole-mount in situ hybridization (WISH) to detect Oct-4 RNA in E7.5 embryos. Mutant embryos exhibited an expression pattern similar to wild type (Fig. 2D–F), with Oct-4 expression restricted to the embryonic epiblast. Therefore, the phenotypes observed in the Hif-2α KI embryos do not appear to correlate with dramatically expanded spatial regulation of Oct-4.
Induction of Vegf and Tgf-α by the Hif-2α KI allele in EBs
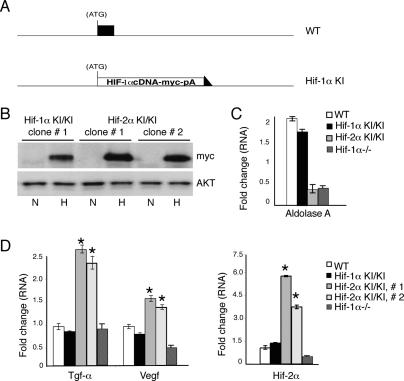
To further characterize the effects of the Hif-2α KI allele on cellular differentiation, we employed an in vitro ES cell-derived EB model. In this system, ES cells are cultured in methylcellulose to generate EBs that produce derivatives of all three germ layers (Keller et al. 1993). This system also provides sufficient material for biochemical analysis, in contrast to the limiting number of arrested E6.5–E7.5 embryos obtained from Hif-2α KI/+ crosses. Moreover, the EB model allowed us to assess the effects of the Hif-2α KI allele on hematopoietic stem cell function (see below). As an additional control for these experiments, we generated a corresponding Hif-1α KI allele in which a c-Myc-epitope-tagged HIF-1α cDNA was introduced into the first coding exon of the murine Hif-1α gene, using the same strategy employed to generate the Hif-2α KI allele (Fig. 3A; Supplementary Fig. 2). This Hif-1α KI allele controls for potential unforeseen effects of expressing a targeted cDNA from the genomic Hif-1α locus. Immunoprecipitation and immunoblot analysis revealed that the Hif-1α KI allele resulted in levels of c-Myc-tagged protein similar to the Hif-2α KI allele in hypoxic ES cells (Fig. 3B). Importantly, the Hif-1α KI allele showed hypoxic induction of the HIF-1α-specific target gene Aldolase A (Alda) to levels comparable to 3.5-d wild-type EBs (Fig. 3C). Embryos derived from the Hif-1α KI ES cells appeared normal at E9.5, in contrast to the Hif-2α KI embryos (Supplementary Fig. 1).
Figure 3.
Increased Vegf and Tgf-α expression in Hif-2α KI/KI 3.5 d EBs. (A) Schematic of HIF-1α control knock-in allele, in which a c-Myc-tagged Hif-1α cDNA was targeted to the first coding exon of the murine Hif-1α locus in ES cells. A detailed description is presented in Supplementary Figure 2. (B) Extracts from homozygous Hif-1α KI/KI and Hif-2α KI/KI ES cells were immunoprecipitated with c-Myc epitope antibodies and then analyzed on Western blots using c-Myc antibodies. Western blot analysis using AKT antibodies confirms that each sample in the immunoprecipitation experiment had equal amounts of input protein. (N) Normoxia (21% O2); (H) hypoxia (4 h at 1.5% O2). (C) Quantitative RT–PCR analysis in 3.5-d EBs of Alda A transcripts in wild-type (WT) and homozygous Hif-1α KI/KI, Hif-2α KI/KI, and _Hif-1α_−/− EBs. Fold change indicates increases in mRNA in EBs grown at 3.0% O2 relative to EBs grown at 21% O2. Transcript levels were normalized to 18S rRNA transcripts in each sample. (D) Real-time PCR analysis in 3.5-d EBs cultured at 21% O2 indicates increased Tgf-α, Vegf, and Hif-2α mRNA in two independently derived homozygous Hif-2α KI/KI clones. Changes in expression are specific to the Hif-2α KI allele, as Hif-1α KI/KI clones and Hif-1α−/− EBs behave similarly to wild type. (*) p < 0.005.
We focused initially on Vegf and Tgf-α expression in 3.5-d EBs generated from two independent Hif-2α KI/KI ES cell clones. In these experiments, EBs were cultured at 21% O2. However, given their three-dimensional structure in methylcellulose, they contain hypoxic subdomains (Adelman et al. 1999). Hif-2α KI/KI EBs displayed increased Vegf and Tgf-α expression compared with wild-type and Hif-1α KI/KI EBs (Fig. 3D), and the degree of target gene induction was proportional to the level of total Hif-2α transcript. Importantly, Vegf and Tgf-α induction was specific to Hif-2α KI/KI EBs, as the Hif-1α KI/KI control expressed Vegf and Tgf-α transcripts at levels similar to wild type. Immunological assays revealed a corresponding increase in TGF-α protein levels in Hif-2α KI/KI EBs derived from both ES cell clones: 5 ± 0.41-fold and 6 ± 0.35-fold, respectively. Expression of Tgf-α and Vegf in Hif-1α−/− EBs was similar to (or less than) that observed in wild type, indicating that elevated expression in Hif-2α KI/KI EBs was a consequence of expanded HIF-2α and not simply the lack of HIF-1α (Fig. 3D). In contrast to Vegf and Tgf-α, other HIF target genes including erythropoietin (Epo), Adm, adipose differentiation related protein (ADRP), and NDRG-1 were expressed at levels comparable to wild type (data not shown). Interestingly, Hif-2α transcript levels were decreased in Hif-1α−/− cells, suggesting that HIF-α subunits may regulate each other’s transcript levels, either directly or indirectly. These data indicate that the expanded expression of HIF-2α from the Hif-2α KI allele drives increased expression of specific HIF-2α target genes in EBs.
HIF-2α activates Oct-4 directly
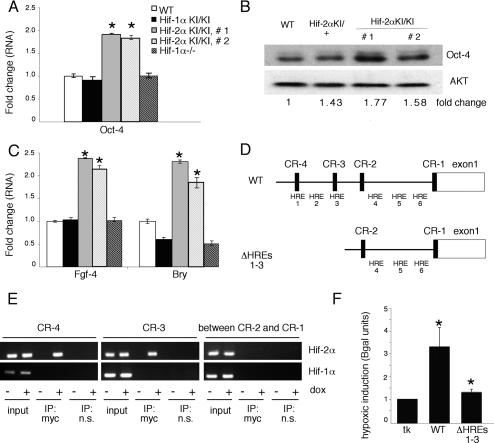
Hif-2α KI/KI 3.5 d EBs also displayed a twofold induction of Oct-4 transcripts compared with wild-type EBs (Fig. 4A), which correlated to enhanced Oct-4 protein levels (Fig. 4B). Furthermore, increased Oct-4 expression was not observed in Hif-1α KI/KI or _Hif-1α_−/− EBs, suggesting that up-regulation of Oct-4 was a specific consequence of expanded HIF-2α expression (Fig. 4A). Although subtle, the modest induction of Oct-4 protein levels is within the twofold range known to affect ES cell pluripotency and differentiation (Niwa et al. 2000). Oct-4 has been previously reported to activate Fgf-4, a preimplantation growth factor required for blastocyst viability (Feldman et al. 1995). Consistent with Oct-4 up-regulation, Hif-2α KI/KI EBs exhibited approximately twofold more Fgf-4 mRNA than wild type (Fig. 4C). It has also been reported that a twofold increase in Oct-4 levels induces ES cells to form mesoderm and endoderm lineages in vitro (Niwa et al. 2000). Indeed, Hif-2α KI/KI EBs demonstrated a twofold induction of the mesodermal marker Bry (Fig. 4C), as well as increases in endodermal markers Gata-binding protein 4 (Gata-4) and Forkhead box A2 (FoxA2) (data not shown). The increased expression of mesoderm markers in Hif-2α KI/KI EBs is distinct from the decreased mesoderm marker expression observed in Hif-2α KI/KI embryos. These apparently contradictory results likely reflect the fact that the Hif-2α KI/KI embryos arrest development around the time of mesoderm formation, whereas Hif-2α KI/KI EBs continue to grow in culture.
Figure 4.
Expanded HIF-2α expression promotes Oct-4 expression and activity in 3.5-d EBs, and HIF-2α protein directly binds the Oct-4 promoter. (A) Real-time PCR analysis of Oct-4 demonstrates increased Oct-4 mRNA in normoxic 3.5-d EBs generated from two separately derived homozygous Hif-2α KI/KI clones. Increased Oct-4 expression is specific to the Hif-2α KI/KI EBs, as Hif-1α KI/KI and _Hif-1α_−/− EBs behave similarly to wild type (WT). (*) p < 0.005. (B) Western blot analysis shows increased Oct-4 protein in independent homozygous Hif-2α KI/KI EB clones. Levels were normalized to the AKT loading control, and fold change was determined by densitometry. (C) Real-time PCR analysis for mesodermal markers Fgf-4 and Bry mRNA demonstrates elevation of Oct-4 target genes specifically in independent homozygous Hif-2α KI/KI EBs. (*) p < 0.005. (D) Schematic diagram of the Oct-4 promoter displaying the relative position of putative HREs, as well as regions of notable sequence conservation between mouse and human (CR1–CR4). (E) ChIP on 293 cells shows that HIF-2α binds the putative HREs in CR4 and CR3 of the Oct-4 promoter. PCR products are obtained only when immunoprecipitated with HIF-2α and not when immunoprecipitated with an isotype control nonspecific antibody. No PCR product was observed in the HIF-2α ChIP using prim ers between CR2 and CR1 of the Oct-4 gene. Furthermore, HIF-1α does not appear to bind these regions in the Oct-4 promoter. (F) Transient transfection of Oct-4 promoter reporters into 786-O WT-8 renal carcinoma cells demonstrates that Oct-4 is induced by hypoxia (1.5% O2), but this induction is abolished when the HREs in CR4 and CR3 are removed. Normoxic basal expression was unchanged when comparing full-length Oct-4 promoter constructs (GOF-9) and Oct-4 promoter constructs with the HREs deleted (GOF-6).
HIF-2α activation of Oct-4 could be direct or indirect. Sequence analysis of the mouse and human Oct-4 upstream promoter and enhancer region (Nordhoff et al. 2001) revealed several putative HREs, some of which reside in regions previously reported as conserved between mice and humans (CR1–CR4). We identified one putative HRE each in CR4 and CR3, one HRE between CR4 and CR3, and three HREs between CR2 and CR1 in the human and mouse promoter (Fig. 4D). Chromatin immunoprecipitation (ChIP) experiments revealed that HIF-2α occupied the HREs in CR4 and CR3, but not the HREs between CR2 and CR1, in hypoxic human RCC cells (786-O WT-8) that express stabilized HIF-2α, but not HIF-1α (data not shown; Maxwell et al. 1999). ChIP experiments in 293 cells that express doxycycline-inducible forms of HIF-1α and HIF-2α demonstrated that HIF-2α, but not HIF-1α, occupies the HREs in the Oct-4 promoter and enhancer region (Fig. 4E). Importantly, deletion of these HREs abrogates the hypoxic induction of Oct-4 (Fig. 4F). Taken together, these data indicate that endogenous HIF-2α binds to HREs in Oct-4 regulatory regions in cells and is consistent with the hypothesis that HIF-2α directly activates Oct-4 expression in our KI model.
TGF-α inhibition and Oct-4 knock-down rescues Hif-2α KI hematopoietic defects.
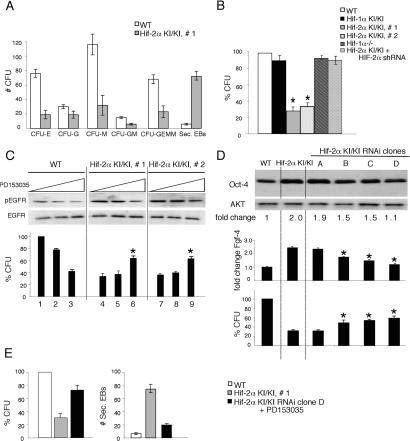
Next, we employed an in vitro system of hematopoietic differentiation to investigate the effects of expanded HIF-2α target gene expression on stem cell function. As stated above, ES cells form three-dimensional EBs in methylcellulose, which generate hypoxic subdomains even when maintained in normoxic conditions (Adelman et al. 1999). After 9 d, the EBs can be disaggregated and plated in the presence of specific growth factors that allow the expansion and differentiation of various hematopoietic colony-forming units (CFUs) representing erythroid (CFU-E), macrophage (CFU-M), granulocyte (CFU-G), and mixed lineage (CFU-GM, CFU-GEMM) progenitors (Keller et al. 1993). Compared with wild-type ES cells, independent homozygous Hif-2α KI/KI ES clones formed significantly fewer CFUs of all hematopoietic lineages (Fig. 5A), giving rise to ∼30% of the total CFUs generated by wild-type cells (Fig. 5B). Interestingly, the Hif-2α KI/KI ES cells formed 7.8-fold more secondary EBs compared with wild type, consistent with maintenance of an undifferentiated state. The inability to generate proper numbers of CFUs was specific to EBs derived from Hif-2α KI/KI ES cells, as both _Hif-1α_−/− and Hif-1α KI/KI EBs formed a similar number of CFUs compared with wild type (Fig. 5B). Furthermore, knock-down of HIF-2α in the Hif-2α KI/KI clones rescued the ability to form colonies (Fig. 5B). We observed no difference in proliferation or apoptosis in the wild-type and Hif-2α KI/KI EBs as measured by staining for Ki67 and cleaved caspase-3, respectively (data not shown), suggesting that the Hif-2α KI allele was affecting the formation and/or differentiation of hematopoietic progenitors.
Figure 5.
Reversal of Hif-2α KI hematopoietic phenotypes by TGF-α and Oct-4 inhibition. (A) Normoxic day 9 Hif-2α KI/KI EBs form fewer hematopoietic CFUs than wild-type (WT) EBs. Numbers of CFU-E, CFU-M, CFU-G, and CFU-GM, CFU-GEMM progenitors are shown. Homozygous Hif-2α KI/KI EBs form more secondary EBs, compared with wild type. (B) Comparison of total CFU formation in EBs derived from ES cells of different genotype, expressed as a percentage of wild type (WT). Note the statistically significant decrease in the ability of homozygous Hif-2α KI/KI ES cells to form CFUs. This phenotype is specific to the two independent homozygous Hif-2α KI/KI clones, as Hif-1α KI/KI clones, _Hif-1α_−/− EBs, and Hif-2α KI/KI + HIF-2α shRNA clones behave similarly to wild type. (*) p < 0.005. (C) Western blot analysis for phosphorylated EGFR revealed inhibition of TGF-α activity by PD153035 in wild-type (WT) and in homozygous Hif-2α KI EBs. Hif-2α KI/KI EBs exhibited hyperphosphorylation of EGFR, consistent with increased TGF-α activity. Reduction in EGFR phosphorylation in wild-type EBs correlated to a decrease in total CFU formation. In contrast, reduction of EGFR phosphorylation in Hif-2α KI/KI EBs correlated to increased CFU formation. Restoration of CFU formation in Hif-2α KI/KI EBs was most notable in samples displaying nearly wild-type levels of EGFR phosphorylation. Total CFU formation is expressed as a percentage of that obtained with EBs derived from wild-type ES cells. (*) p < 0.005. (D) Western blot analysis indicating variable knock-down of Oct-4 protein and activity (Fgf-4 RNA expression) in EBs derived from homozygous Hif-2α KI/KI ES cell clones expressing Oct-4 shRNAs. EBs derived from Hif-2α KI/KI ES clones with Oct-4 protein levels similar to wild type (WT) displayed reduced expression of Fgf-4 RNA, and partial restoration of CFU formation compared with Hif-2α KI/KI. (*) p < 0.005. (E) EBs derived from Hif-2α KI/KI ES clones doubly inhibited for Oct-4 and TGF-α show even greater restoration of CFU formation and reduced secondary EB formation compared with Hif-2α KI/KI.
To determine the contribution of Vegf, Tgf-α, and Oct-4 to this hematopoietic defect, we used multiple approaches to selectively inhibit Vegf, Tgf-α, or Oct-4 function in Hif-2α KI/KI EBs. Treatment of Hif-2α KI/KI EBs with a previously characterized neutralizing antibody against VEGF (Gunaratnam et al. 2003) did not rescue the CFU defect of Hif-2α KI/KI EBs (data not shown). Next, the compound PD153035 was used to inhibit TGF-α signaling by reducing phosphorylation of the epidermal growth factor receptor (EGFR) (Gunaratnam et al. 2003) in a dose-dependent manner. Consistent with increased TGF-α secretion, Hif-2α KI/KI EBs exhibit hyperphosphorylation of the EGFR (Fig. 5C). When TGF-α activity was inhibited in Hif-2α KI/KI EBs to levels equivalent to wild-type EBs, there was an increased ability to form CFUs relative to the untreated controls (∼60% of wild type, cf. 30%) (Fig. 5C, lanes 6,9). In contrast, treatment of wild-type EBs with PD153035 inhibited their ability to form CFUs. Importantly, TGF-α inhibition did not affect Oct-4 or Hif-2α levels (data not shown). This partial restoration of CFU-forming ability suggests that up-regulation of TGF-α is partly responsible for the Hif-2α KI hematopoietic phenotype in EBs.
An RNA interference (RNAi) approach was used to reduce Oct-4 expression in Hif-2α KI/KI EBs. Stable cell lines were generated in Hif-2α KI/KI ES cells and used to form EBs. As expected, the clones exhibited variable levels of Oct-4 inhibition: Some displayed modest reduction of Oct-4 levels compared with parental Hif-2α KI/KI EBs (Fig. 5D, clone A), some demonstrated Oct-4 levels comparable to wild type (Fig. 5D, clone D), and others displayed an intermediate reduction in Oct-4 expression (Fig. 5D, clones B and C). Interestingly, a few clones displayed Oct-4 levels significantly lower than wild type, but appeared morphologically similar to trophectoderm cells and expressed the trophectoderm markers _Hand1_and Cdx2 (Supplementary Fig. 4), consistent with published data (Niwa et al. 2000).
Oct-4 knock-down in Hif-2α KI/KI EBs correlated with a reduction of Fgf-4 levels (Fig. 5D), as well as reduced Bry expression (data not shown). Next, we measured CFUs in EBs derived from Hif-2α KI/KI clones expressing different Oct-4 levels. As Oct-4 levels were reduced, Hif-2α KI/KI clones A–D exhibited an increased ability to form CFUs in this assay (∼55% of wild type) (Fig. 5D). Of note, inhibition of Oct-4 did not affect Tgf-α or Hif-2α levels (data not shown). Similar to TGF-α inhibition, Oct-4 knock-down only partly rescued the CFU phenotype, suggesting that TGF-α and Oct-4 function independently to alter hematopoietic cell fate in the Hif-2α KI model. Inhibition of both TGF-α and Oct-4 resulted in a greater number of CFUs (70% of wild type) derived from Hif-2α KI/KI EBs than inhibition of either TGF-α or Oct-4 alone (60% and 55%, respectively) (Fig. 5E). Furthermore, inhibition of both TGF-α and Oct-4 reduced the ability of Hif-2α KI/KI cells to form secondary EBs (Fig. 5E). Taken together, these data suggest that up-regulation of TGF-α and Oct-4 is primarily responsible for the Hif-2α KI hematopoietic phenotype observed in EBs.
HIF-2α promotes tumor growth through Oct-4 induction
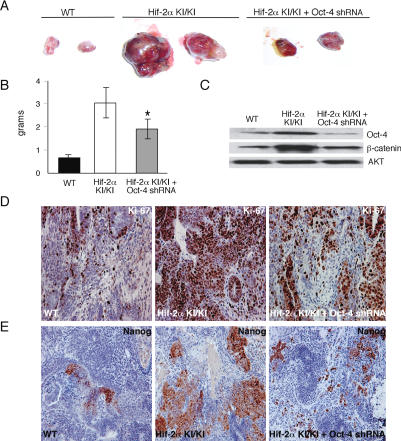
We reported previously that teratomas derived from homozygous Hif-2α KI/KI ES cells were more proliferative than their wild-type controls, and were characterized by abundant regions of undifferentiated cells as well as regions marked by differentiated mesoderm and neural cell types (Covello et al. 2005). Further analysis revealed that these Hif-2α KI/KI teratomas exhibited a fourfold increase in Oct-4 expression (data not shown). To investigate whether Oct-4 up-regulation by HIF-2α contributes to tumorigenesis, we inhibited Oct-4 in Hif-2α KI/KI teratomas using short hairpin RNAs (shRNA) (Fig. 6A). Wild-type, homozygous Hif-2α KI/KI, and Hif-2α KI/KI ES cells stably expressing Oct-4 shRNA hairpins (“_Hif-2α KI/KI_-_Oct-4 shRNA_”) were injected subcutaneously into NIH-III-immunodeficient mice. As expected, teratomas derived from Hif-2α KI/KI ES cells grew more rapidly, exhibited increased mass, and appeared more hemorrhagic than wild-type controls (Fig. 6A,B). In contrast, _Hif-2α KI/KI_-Oct-4 shRNA teratomas exhibited reduced mass (p = 0.012) and growth rates compared with Hif-2α KI/KI teratomas, but still displayed similar hemorrhaging (Fig. 6A,B).
Figure 6.
HIF-2α mediates tumor growth through up-regulation of Oct-4. (A) Gross appearance of teratomas resected from nude mice 21 d after subcutaneous injection with willd-type (WT), Hif-2α KI/KI ES cells, and Hif-2α KI/KI ES cells with Oct-4 shRNA knock-down (_Hif-2α KI/KI_-Oct-4 shRNA). (B) Final teratoma masses of wild-type (WT), Hif-2α KI/KI, and _Hif-2α KI/KI_-Oct-4 shRNA teratomas formed after 22 d of growth. Error bars measure standard error of the mean. (*) p = 0.012. (C) Western blot analysis of Oct-4 and β-catenin protein in wild-type (WT), Hif-2α KI/KI, and _Hif-2α KI/KI_-Oct-4 shRNA teratomas. Data are representative of three independent teratomas per genotype. (D) Ki67 staining of wild-type (WT), Hif-2α KI/KI, and _Hif-2α KI/KI_-Oct-4 shRNA teratoma sections. Final magnifications are 200×. (E) Nanog staining of wild type (WT), Hif-2α KI/KI, and Hif-2α KI/KI with Oct-4 shRNA knock-down teratoma sections.
To assess differences in proliferation or apoptosis, teratomas were analyzed by immunostaining for Ki67 and cleaved caspase-3, respectively (Fig. 6D; data not shown). Both Hif-2α KI/KI and _Hif-2α KI/KI_-Oct-4 shRNA teratomas demonstrated an increase in Ki67 staining compared with wild type (Fig. 6D). However, the percentage of Ki-67-positive cells in _Hif-2α KI/KI_-Oct-4 shRNA teratomas was only 1.9-fold greater than wild type, whereas Hif-2α KI/KI teratomas contained 2.5-fold more Ki-67-positive cells than wild type. No quantitative differences in cleaved caspase-3 staining were observed between the three genotypes (data not shown).
Teratoma mRNA and protein analysis confirmed that the elevated levels of Oct-4 in Hif-2α KI/KI tumors were reduced by expression of Oct-4 shRNAs (Fig. 6C). Furthermore, introduction of Oct-4 shRNA was specific, as _Hif-2α KI/KI_-Oct-4 shRNA teratomas exhibited similar levels of Vegf, and Tgf-α transcripts compared with the Hif-2α KI/KI tumors (data not shown). Interestingly, Oct-4 inhibition also reduced elevated β-catenin levels in the Hif-2α KI/KI tumors, supporting a recent report that places β-catenin downstream of Oct-4 in tumorigenesis (Fig. 6C; Hochedlinger et al. 2005).
We have demonstrated previously that Hif-2α KI/KI tumors contained undifferentiated cell types, as well as an abundance of mesodermal and neural cells, compared with wild type (Covello et al. 2005). To investigate the role of Oct-4 in altering the spectrum of cell types present, _Hif-2α KI/KI_-Oct-4 shRNA tumor sections were evaluated by standard histological methods. Consistent with the hypothesis that Oct-4 is influencing the Hif-2α KI/KI tumors cell fate, we observed fewer undifferentiated cell populations as revealed by Nanog immunostaining (Fig. 6E; Supplementary Fig. 4), as well as less bone, cartilage, Schwann cells, and ganglia when Oct-4 levels were inhibited by shRNA (Fig. 6D,E; Supplementary Fig. 4).
HIF-2α activation of Oct-4 is necessary for PGC development
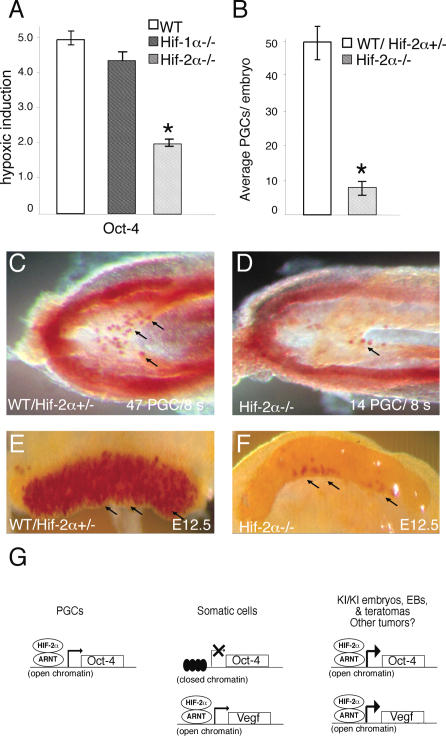
Taken together, our data suggest that expanded HIF-2α expression is sufficient for up-regulation of Oct-4. To determine whether HIF-2α maintains Oct-4 expression in normal physiological settings, we examined Oct-4 expression in cells devoid of HIF-2α function. Oct-4 transcripts were hypoxically induced in wild-type and Hif-1α−/− 3.5-d EBs, but this response was dramatically attenuated in Hif-2α−/− EBs (Fig. 7A). Next, we asked whether HIF-2α plays a regulatory role in PGCs, which retain Oct-4 expression during embryonic development. Hif-2α−/− embryos display a significant decrease in PGC numbers, as visualized by alkaline phosphatase (AP) staining of E8.5 embryos (Fig. 7B–D). Wild-type and heterozygous E8.5 Hif-2α+/− embryos (s7–s11) displayed 49.8 ± 4.4 PGCs per embryo, whereas littermate somite-matched _Hif-2α_−/− embryos exhibited only 8.1 ± 2.2 PGCs (Fig. 7B). A dramatic defect was also observed in genital ridges of E12.5 mutant embryos (Fig. 7E,F). This defect may be caused by decreased PGC survival, as previously reported for PGCs in which Oct-4 was deleted (Kehler et al. 2004). In summary, HIF-2α is required for normal Oct-4 expression in EBs, and loss of HIF-2α negatively affects PGC formation and/or maintenance in embryos.
Figure 7.
HIF-2α is required for Oct-4 expression and normal PGC numbers. (A) Real-time PCR analysis demonstrates increased hypoxic induction (1.5% O2 for 16 h) of Oct-4 mRNA in wild-type (WT) and _Hif-1α_−/− 3.5-d EBs, which is diminished in _Hif-2α_−/− EBs. (B_–_D) PGCs in E8.5 embryos revealed by AP staining. Arrows point to representative PGCs in C and D. In contrast to wild-type (WT) and heterozygous Hif-2α+/− littermate controls (C), somite-matched _Hif-2α_−/− embryos exhibit reduced numbers of PGCs (D). (B) Quantification of PGC numbers in somite-matched E8.5 (s7–s11) embryos. (E,F) PGCs in E12.5 genital ridges dissected from embryos revealed by AP staining. (G) Proposed model for regulation of Oct-4 by HIF-2α. In PGCs, the Oct-4 locus is expressed and regulated by HIF-2α. The Oct-4 locus is also expressed in ES cells; however, HIF-2α activity is normally restricted or repressed in these cells (C.-J. Hu, S. Iyer, A. Sataur, K.L. Covello and M.C. Simon, in prep.). In differentiating somatic cells, the Oct-4 locus adopts a closed conformation and is not expressed. At this point, HIF-2α regulates other target genes (e.g., Vegf) without modulating Oct-4 levels. In homozygous Hif-2α KI/KI embryos, EBs, and teratomas, expanded HIF-2α expression results in up-regulation of Oct-4 and other targets. HIF-2α-mediated expression of an inappropriately derepressed Oct-4 locus in transformed cells could similarly modulate cancer stem cell identity and tumor progression.
Discussion
We demonstrate here that HIF-2α is a direct upstream regulator of Oct-4, a transcription factor essential for maintaining ES and ICM cells in an undifferentiated state. Our data also provide a molecular mechanism to explain how hypoxia can regulate stem cell function in both normal and pathophysiological settings. Normal Oct-4 expression is tightly regulated and generally restricted to cells of the embryonic epiblast, ES cells, PGCs, and possibly adult stem cells. Precise Oct-4 protein levels are critical for peri-implantation development (Nichols et al. 1998), and even subtle changes in Oct-4 expression can profoundly alter cellular differentiation in the early embryo (Boiani et al. 2002; Ramos-Mejia et al. 2005). Embryos homozygous for the Hif-2α KI allele were recovered in strikingly sub-Mendelian ratios, were morphologically aberrant, and exhibited increased expression of HIF-2α target genes, most notably Oct-4. A similar loss of post-implantation embryos was associated with aberrant Oct-4 expression in cloned mouse embryos (Boiani et al. 2002) or transgenic overexpression of HIF-2α (Supplementary Fig. 5). Increased expression of Oct-4 and Tgf-α also compromised the ability of Hif-2α KI/KI EBs to generate functional progenitors from hematopoietic stem cells. Our data demonstrate that HIF-2α regulates stem cell function by controlling Oct-4 expression in multiple settings. Interestingly, this effect appears to be specific for HIF-2α, as HIF-1α does not regulate Oct-4 in our experiments.
We also demonstrate that HIF-2α is required for production, survival, and/or maintenance of PGCs, the principal cell type known to express Oct-4 in post-implantation embryos (Scholer et al. 1990a). In fact, HIF-2α is expressed in multiple tissues (including blastocyst stage embryos, E7.5 embryos, and PGCs) where Oct-4 is known to play a pivotal role (data not shown). It is not yet clear whether HIF-2α-mediated Oct-4 expression also plays a role in adult stem cells, as the embryonic or perinatal lethality of most _Hif-2α_−/− mice precludes a direct test of this hypothesis. Interestingly, a small number of _Hif-2α_−/− mice bearing a specific genetic background survive for several weeks but ultimately succumb to complex pathologies (Scortegagna et al. 2003, 2005). These mice display decreased levels of hematopoietic progenitors, correlated to a decrease in Epo levels (Scortegagna et al. 2003, 2005). As Epo expression was similar in the Hif-2α KI/KI, wild-type, and Hif-1α KI/KI EBs, however, it appears unlikely that Epo plays a major role in the defects we observe.
Our data suggest the model shown in Figure 7G, which proposes that the Oct-4 promoter is maintained in an open conformation in particular cells of preimplantation embryos and PGCs, and is thereby sensitive to regulation by HIF-2α. Our model also predicts that the Oct-4 locus adopts a closed or “silent” conformation in differentiating embryonic somatic cells, making it refractory to regulation by HIF-2α, which can nevertheless regulate Vegf and Tgf-α and other important target genes. We propose that expanded expression of HIF-2α in homozygous Hif-2α KI/KI embryos results in inappropriate maintenance of Oct-4 transcription, which underlies the developmental defects we observe. As Oct-4 is expressed in ES cells, however, our results raise the question of how hypoxic ES cells avoid Oct-4 induction by stabilized HIF-2α and the consequent loss of pluripotency. We have recently discovered that although HIF-2α is clearly stabilized in hypoxic ES cells, it is rendered transcriptionally inactive by one or more specific repressors (C.-J. Hu, S. Iyer, A. Sataur, K.L. Covello, and M.C. Simon, in prep.). Repression of HIF-2α activity would protect against Oct-4 induction and consequent differentiation of blastocyst ICM cells in the hypoxic uterine microenvironment.
The relatively subtle phenotypes of heterozygous Hif-2α KI/+ embryos, which still express HIF-1α, suggest that induction of Oct-4 and Tgf-α by a single Hif-2α KI allele is insufficient to overcome a threshold necessary to arrest development. Alternatively, HIF-1α may directly inhibit the activation of Oct-4 through competition for cofactors or other mechanisms. However, this appears unlikely as embryos with transgenic HIF-2α overexpression show phenotypes similar to the Hif-2α KI embryos (Supplementary Fig. 5).
The degree to which increased expression of TGF-α VEGF and other specific HIF-2α targets also mediate the embryonic phenotypes we observe is currently unknown, although the fact that embryos overexpressing VEGF develop to E13.5 (Miquerol et al. 2000) suggests that it is not the primary factor effecting development in our model. Future experiments combining the Hif-2α KI allele with either Oct-4 or Tgf-α loss-of-function alleles in murine embryos (analogous to those in Fig. 5) will be required to determine the role of each gene in the Hif-2α KI model.
The specific induction of Oct-4 by HIF-2α provides a novel mechanism to explain recent observations that HIF-2α promotes tumor growth more effectively than HIF-1α. HIF-1α and HIF-2α are overexpressed in a large number of human tumor types, including brain, breast, RCC carcinomas, pheochromoctyomas, astrocytomas, and non-small-cell lung cancers (Favier et al. 2001; Giatromanolaki et al. 2001; Jaakkola et al. 2001; Khatua et al. 2003). Several experiments have shown that expression of HIF-2α promotes more rapid growth of RCC xenografts (Kondo et al. 2002, 2003; Maranchie et al. 2002; Raval et al. 2005) and ES cell-derived teratomas (Covello et al. 2005) than does expression of HIF-1α. The degree to which this generalization extends is not yet clear, but the differential activity of HIF-2α in these assays has usually been ascribed to expression of target genes, including Vegf and Tgf-α (Gunaratnam et al. 2003). Whereas expanded expression of HIF-2α in RCC and Hif-2α KI teratoma models is clearly correlated with more rapid tumor growth, Acker et al. (2005) provide evidence that HIF-2α can act instead as a tumor suppressor in rat glioma xenografts. The explanations for the apparent discrepancies with our data are not yet clear, but we suggest that subtle differences in the spectrum of HIF-2α targets expressed in specific cell types may produce significantly different growth phenotypes; future work will be required to clarify these points. Finally, we observed a subtle decrease in HIF-2α transcript levels in _Hif-1α_−/− ES cells, suggesting that the HIF-α subunits may regulate each other’s expression. Interestingly, Raval et al. (2005) report cross-regulation of HIF-α subunits at the protein level in RCC cell lines, although this is not reflected by changes in mRNA expression. Elucidation of the different mechanisms by which HIF-α subunits modulate the function of other family members in specific cell types will be an important question for future studies.
Our data suggest that induction of Oct-4 by HIF-2α could maintain hypoxic tumor cells in a more undifferentiated and malignant state. For example, the Oct-4 locus may regain an open conformation in dedifferentiated transformed cells or cancer stem cells, thereby rendering it susceptible to regulation by HIF-2α. Hypoxic induction of Oct-4 may, in turn, promote expansion of an undifferentiated cell population, promote mesodermal identity, and/or stimulate dedifferentiation of terminally differentiated cells (Niwa et al. 2000; Jogi et al. 2002). Data from the Hif-2α KI/KI teratomas support this idea; specifically, Hif-2α KI/KI tumors exhibited increased Oct-4 and Nanog expression and contained undifferentiated cell types, as well as an abundance of mesodermal and neural cells (Covello et al. 2005). The range of cell types we observed in Hif-2α KI/KI teratomas is similar to that described in tumors derived from ES cells that express Oct-4 in a doxycycline-inducible manner (Gidekel et al. 2003). Oct-4 protein is also elevated in various types of testicular germ cell tumors, where it regulates growth in a dose-dependent manner (Gidekel et al. 2003). Our results predict that hypoxic Oct-4 expression may contribute to the progression of multiple tumor types including breast, pancreas, colon, and liver (Monk and Holding 2001; Tai et al. 2005). Notably, a recent report demonstrated that ectopic expression of Oct-4 in mice was sufficient to cause epithelial dysplasia, which represents an early stage in the development of carcinomas (Hochedlinger et al. 2005). Our findings suggest that hypoxia can regulate the activity and differentiation of cancer stem cells and their progeny through HIF-2α-mediated control of Oct-4 expression. If true, then inhibiting HIF-2α activity might reduce Oct-4 levels below a threshold required to maintain stem cell identity, thereby promoting cellular differentiation and/or tumor dormancy.
In conclusion, we identify Oct-4 as a novel HIF-2α target and demonstrate that expanded expression of HIF-2α is sufficient for up-regulation of Oct-4, Tgf-α, and Vegf in different biological contexts, including embryonic development and tumor growth (Covello et al. 2005). These data provide the initial connection between hypoxia (HIF-2α) and a factor critical for stem cell maintenance (Oct-4) and novel insight into the regulation of stem cells in normal development and tumor progression. Finally, these data suggest that targeting HIF-2α activity may be a useful strategy for modulating stem cell function, and possibly cancer stem cell differentiation, through alteration of Oct-4 expression.
Materials and methods
Embryo collection and WISH
Timed matings were performed on heterozygous Hif-2α KI/+ intercrosses. Embryos were harvested at E6.5–E13.5 and analyzed by gross morphology and H&E or WISH. For WISH, embryos were fixed in 4% PFA and dehydrated through a PBST/methanol series. WISH was performed using Costar 12 well inserts as previously described (Wilkinson 1992). Digoxigenin labeled RNA probes were synthesized using -Digoxigenin RNA kit according to manufacturer’s protocol (Roche). Embryos were photographed and then genotyped by PCR using primers specific for the Hif-2α KI allele. Briefly, to purify DNA, embryos were incubated for 6 h at 55°C in 50 mM Tris (pH 8.3), 7.5 mM MgCl2, 0.5 mg/mL gelatin, and 100 μg/mL Proteinase K.
Quantitative RT–PCR
Total RNA was purified from wild-type and KI ES cells, EBs, or embryos using Trizol and micro-RNAeasy kit (Qiagen). Mixed Oligo-d(T)15 and ribosomal 18S RNA-specific primers were used to generate single-stranded cDNAs, which were assayed for levels of Vegf-A, Pgk, Alda, Tgf-α, Oct-4, Fgf-4, and 18S transcripts as previously described (Covello et al. 2005).
Protein analysis
c-Myc epitope-tagged HIF-1α and HIF-2α protein was immunoprecipitated using a c-Myc polyclonal antibody (Cell Signaling, Inc.) according to the manufacturer’s protocol. Protein levels for Oct-4 (Santa Cruz), EGFR (Cell Signaling), phospho-EGFR (Cell Signaling), and β-catenin (Transduction Labs) were determined by Western blot as previously described (Covello et al. 2005).
ChIP and reporter assays
ChIP experiments were performed as previously described (Letting et al. 2003). Briefly, 1 × 107 786-O cells and 293 cells were grown for 16 h in hypoxic conditions. Cells were cross-linked in 0.4% formaldehyde for 10 min at room temperature. Cells were washed five times in PBS, sonicated, precleared, and immunoprecipated with HIF-2α or c-myc antibody. Beads were washed and DNA/protein complexes eluted. Cross-links were reversed, and protein was digested with 20 mg/mL proteinase K. DNA was purified by phenol/chloroform extraction and ethanol precipitation. Oct-4 reporter constructs and assays were performed as previously described (Yeom et al. 1996).
In vitro differentiation and replatings
EB and hematopoietic differentiation assays were performed as originally described (Adelman et al. 1999). Briefly, gelatin-adapted ES cells were plated in base methylcellulose containing 10% serum, 500 U/mL rhIL-1, 5 ng/mL rmIL-3, 10 μg/mL insulin, 200 μg/mL transferrin, and 104 M monothioglycerol and allowed to differentiate for 3.5–9 d. The cells were plated on day 0 and allowed to form EBs for 3.5 d (mRNA experiments) or 9 d (for replating experiments, see below). Exogenous TGF-α (1–500 pM) or inhibitors for TGF-α (1–10 μM) (Calbiochem) and VEGF (0–80 μM) (Calbiochem) were added as described (Gunaratnam et al. 2003). After 9 d, EBs were washed free of methylcellulose with PBS and disaggregated with trypsin and shearing, using a 21-gauge needle and syringe. Cells were replated into a secondary complete methylcellulose (Stem Cell Technologies, catalog no. 3434). The number of cells plated was equal to the number of cells in 50 EBs derived from wild-type ES cells. Hematopoietic colonies were counted after 6 d.
Oct-4 RNAi
As previously described for expression of shRNAs, annealed DNA oligonucleotides corresponding to nucleotides 670–688 of Oct-4 were cloned behind the human H1 RNA promoter in the pRetro-H1G vector (Velkey and O’Shea 2003). Upon electroporation of Hif-2α KI/KI ES cells, stable cell lines were selected by treatment at 2 μg/mL puromycin. GFP-positive Hif-2α KI/KI ES clones were picked under a microscope, expanded, and screened by Oct-4 RNA and Oct-4 protein.
Teratoma assay and immunohistochemistry
ES cells (5 × 106) in 100 μL of PBS were injected subcutaneously into the dorsal area of 4- to 6-wk-old NIH-III-immunodeficient mice (Charles River). Tumors were measured every 2–3 d with calipers in the two greatest dimensions to calculate tumor growth over a 21-d period. At 21 d, tumors were harvested, photographed, weighed, fixed in 4% paraformaldehyde, and processed for histological analysis. Tumor samples were sectioned and stained as previously described (Covello et al. 2005). Primary antibodies used were Nanog (Chemicon), cleaved caspase-3 (Cell Signaling, Inc.), Ki-67 (Novocastra), CD34 (Pharmingen), Oct-4 (Santa Cruz), and β-catenin (NeoMarkers).
PGC analysis
Embryos were isolated from litters of Hif-2α+/− intercrosses at E8.5–E12.5. All embryos used for analysis were somite matched. For AP staining, the posterior portions of E8.5 embryos and E12.5 genital ridges were fixed with 4% PFA, washed three times with PBS, and then stained with AP solution (naphthyl phosphate/fast red TR) at room temperature. They were then rinsed in water, post-fixed in 4% PFA, and photographed. PGC isolation and RT–PCR analysis was performed as previously described (Saitou et al. 2002). Briefly, E12.5 genital ridges were dissected from embryos. Genital ridges were placed in a saline/EDTA plus glucose solution to dissociate germ cells from the stroma.
Statistical analysis
Error bars represent standard error of the mean. Statistical analyses were done by χ2 analysis and Student’s _t_-test. Statistical significance was defined as P < 0.005.
Acknowledgments
We thank Anja Runge, Mercy Gohil, and Kim Tremblay for technical assistance. Thanks to Hans Scholer for providing the Oct-4 reporter plasmids. Special thanks to Marisa Bartolomei, Dan Kessler, and members of the Simon laboratory for thoughtful discussions and reading of the manuscript. This research was supported by National Institutes of Health Grant HL66130 (M.C.S. and B.K.), the American Heart Association (K.L.C.) and the Abramson Family Cancer Research Institute. M.C.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
References
- Acker T, Diez-Juan A, Aragones J, Tjwa M, Brusselmans K, Moons L, Fukumura D, Moreno-Murciano M.P, Herbert J.M, Burger A, et al. Genetic evidence for a tumor suppressor role of HIF-2α. Cancer Cell. 2005;8:131–141. doi: 10.1016/j.ccr.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Adelman D.M, Maltepe E, Simon M.C. Multilineage embryonic hematopoiesis requires hypoxic ARNT activity. Genes & Dev. 1999;13:2478–2483. doi: 10.1101/gad.13.19.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Eckardt S, Scholer H.R, McLaughlin K.J. Oct4 distribution and level in mouse clones: Consequences for pluripotency. Genes & Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A, Lee T.I, Cole M.F, Johnstone S.E, Levine S.S, Zucker J.P, Guenther M.G, Kumar R.M, Murray H.L, Jenner R.G, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Thomas A, Roth L.M, Zheng W, Michael H, Karim F.W. OCT4: A novel biomarker for dysgerminoma of the ovary. Am. J. Surg. Pathol. 2004;28:1341–1346. doi: 10.1097/01.pas.0000135528.03942.1f. [DOI] [PubMed] [Google Scholar]
- Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F, et al. Loss of HIF-2α and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat. Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- Compernolle V, Brusselmans K, Franco D, Moorman A, Dewerchin M, Collen D, Carmeliet P. Cardia bifida, defective heart development and abnormal neural crest migration in embryos lacking hypoxia-inducible factor-1α. Cardiovasc. Res. 2003;60:569–579. doi: 10.1016/j.cardiores.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Covello K.L, Simon M.C, Keith B. Targeted replacement of hypoxia-inducible factor-1α by a hypoxia-inducible factor-2α knock-in allele promotes tumor growth. Cancer Res. 2005;65:2277–2286. doi: 10.1158/0008-5472.CAN-04-3246. [DOI] [PubMed] [Google Scholar]
- Danet G.H, Pan Y, Luongo J.L, Bonnet D.A, Simon M.C. Expansion of human SCID-repopulating cells under hypoxic conditions. J. Clin. Invest. 2003;112:126–135. doi: 10.1172/JCI17669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Nat. Acad. Sci. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T, Das P, Roberts R.M. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Nat. Acad. Sci. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier J, Kempf H, Corvol P, Gasc J.M. Coexpression of endothelial PAS protein 1 with essential angiogenic factors suggests its involvement in human vascular development. Dev. Dyn. 2001;222:377–388. doi: 10.1002/dvdy.1207. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou V.E, DeChiara T.M, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Flamme I, Frohlich T, von Reutern M, Kappel A, Damert A, Risau W. HRF, a putative basic helix–loop–helix–PAS-domain transcription factor is closely related to hypoxia-inducible factor-1α and developmentally expressed in blood vessels. Mech. Dev. 1997;63:51–60. doi: 10.1016/s0925-4773(97)00674-6. [DOI] [PubMed] [Google Scholar]
- Giatromanolaki A, Koukourakis M.I, Sivridis E, Turley H, Talks K, Pezzella F, Gatter K.C, Harris A.L. Relation of hypoxia inducible factor 1α and 2α in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br. J. Cancer. 2001;85:881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Gunaratnam L, Morley M, Franovic A, de Paulsen N, Mekhail K, Parolin D.A, Nakamura E, Lorimer I.A, Lee S. Hypoxia inducible factor activates the transforming growth factor-α/epidermal growth factor receptor growth stimulatory pathway in VHL−/− renal cell carcinoma cells. J. Biol. Chem. 2003;278:44966–44974. doi: 10.1074/jbc.M305502200. [DOI] [PubMed] [Google Scholar]
- Harvey A.J, Kind K.L, Pantaleon M, Armstrong D.T, Thompson J.G. Oxygen-regulated gene expression in bovine blastocysts. Biol. Reprod. 2004;71:1108–1119. doi: 10.1095/biolreprod.104.028639. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hu C.J, Wang L.Y, Chodosh L.A, Keith B, Simon M.C. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer N.V, Kotch L.E, Agani F, Leung S.W, Laughner E, Wenger R.H, Gassmann M, Gearhart J.D, Lawler A.M, Yu A.Y, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes & Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole D.R, Tian Y.M, Wilson M.I, Gielbert J, Gaskell S.J, Kriegsheim A, Hebestreit H.F, Mukherji M, Schofield C.J, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar B.N, Reinhardt R.L, Schwartz R.E, Keene C.D, Ortiz-Gonzalez X.R, Reyes M, Lenvik T, Lund T, Blackstad M, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- Jogi A, Ora I, Nilsson H, Lindeheim A, Makino Y, Poellinger L, Axelson H, Pahlman S. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc. Nat. Acad. Sci. 2002;99:7021–7026. doi: 10.1073/pnas.102660199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomeli H, Nagy A, McLaughlin K.J, Scholer H.R, et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G, Kennedy M, Papayannopoulou T, Wiles M.V. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatua S, Peterson K.M, Brown K.M, Lawlor C, Santi M.R, LaFleur B, Dressman D, Stephan D.A, MacDonald T.J. Overexpression of the EGFR/FKBP12/HIF-2α pathway identified in childhood astrocytomas by angiogenesis gene profiling. Cancer Res. 2003;63:1865–1870. [PubMed] [Google Scholar]
- Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin W.G., Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Kondo K, Kim W.Y, Lechpammer M, Kaelin W.G., Jr. Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letting D.L, Rakowski C, Weiss M.J, Blobel G.A. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranchie J.K, Vasselli J.R, Riss J, Bonifacino J.S, Linehan W.M, Klausner R.D. The contribution of VHL substrate binding and HIF1-α to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- Maxwell P.H, Wiesener M.S, Chang G.W, Clifford S.C, Vaux E.C, Cockman M.E, Wykoff C.C, Pugh C.W, Maher E.R, Ratcliffe P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Langille B.L, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;18:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20:8085–8091. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- Morrison S.J, Csete M, Groves A.K, Melega W, Wold B, Anderson D.J. Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J. Neurosci. 2000;20:7370–7376. doi: 10.1523/JNEUROSCI.20-19-07370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Nordhoff V, Hubner K, Bauer A, Orlova I, Malapetsa A, Scholer H.R. Comparative analysis of human, bovine, and murine Oct-4 upstream promoter sequences. Mamm. Genome. 2001;12:309–317. doi: 10.1007/s003350010279. [DOI] [PubMed] [Google Scholar]
- Pugh C.W, Tan C.C, Jones R.W, Ratcliffe P.J. Functional analysis of an oxygen-regulated transcriptional enhancer lying 3′ to the mouse erythropoietin gene. Proc. Nat. Acad. Sci. 1991;88:10553–10557. doi: 10.1073/pnas.88.23.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Bergeron D.L, Runge A, Dahl K.D, Fehling H.J, Keller G, Simon M.C. Hypoxia affects mesoderm and enhances hemangioblast specification during early development. Development. 2004;131:4623–4634. doi: 10.1242/dev.01310. [DOI] [PubMed] [Google Scholar]
- Ramos-Mejia V, Escalante-Alcalde D, Kunath T, Ramirez L, Gertsenstein M, Nagy A, Lomeli H. Phenotypic analyses of mouse embryos with ubiquitous expression of Oct4: Effects on mid-hindbrain patterning and gene expression. Dev. Dyn. 2005;232:180–190. doi: 10.1002/dvdy.20211. [DOI] [PubMed] [Google Scholar]
- Raval R.R, Lau K.W, Tran M.G, Sowter H.M, Mandriota S.J, Li J.L, Pugh C.W, Maxwell P.H, Harris A.L, Ratcliffe P.J. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan H.E, Lo J, Johnson R.S. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Barton S.C, Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Scholer H.R, Dressler G.R, Balling R, Rohdewohld H, Gruss P. Oct-4: A germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990a;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer H.R, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990b;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- Scortegagna M, Morris M.A, Oktay Y, Bennett M, Garcia J.A. The HIF family member EPAS1/HIF-2α is required for normal hematopoiesis in mice. Blood. 2003;102:1634–1640. doi: 10.1182/blood-2003-02-0448. [DOI] [PubMed] [Google Scholar]
- Scortegagna M, Ding K, Zhang Q, Oktay Y, Bennett M.J, Bennett M, Shelton J.M, Richardson J.A, Moe O, Garcia J.A. HIF-2α regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood. 2005;105:3133–3140. doi: 10.1182/blood-2004-05-1695. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. HIF-1 and human disease: One highly involved factor. Genes & Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- Shimozaki K, Nakashima K, Niwa H, Taga T. Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development. 2003;130:2505–2512. doi: 10.1242/dev.00476. [DOI] [PubMed] [Google Scholar]
- Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- Studer L, Csete M, Lee S.H, Kabbani N, Walikonis J, Wold B, McKay R. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J. Neurosci. 2000;20:7377–7383. doi: 10.1523/JNEUROSCI.20-19-07377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai M.H, Chang C.C, Olson L.K, Trosko J.E. Oct4 expression in adult human stem cells: Evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- Tian H, McKnight S.L, Russell D.W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes & Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- Velkey J.M, O’Shea K.S. Oct4 RNA interference induces trophectoderm differentiation in mouse embryonic stem cells. Genesis. 2003;37:18–24. doi: 10.1002/gene.10218. [DOI] [PubMed] [Google Scholar]
- Wang V, Davis D.A, Haque M, Huang L.E, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1α and hypoxia-inducible factor-2α in HEK293T cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- Wenger R.H, Gassmann M. Oxygen(es) and the hypoxia-inducible factor-1. Biol. Chem. 1997;378:609–616. [PubMed] [Google Scholar]
- Wiesener M, Turley H, Allen W, William C, Eckardt K, Talks K, Wood S, Gatter K, Harris A, Pugh C, et al. Induction of endothelial PAS domain protein-1 by hypoxia: Characterization and comparison with hypoxia-inducible factor-1α. Blood. 1998;92:2260–2268. [PubMed] [Google Scholar]
- Wiesener M.S, Jurgensen J.S, Rosenberger C, Scholze C.K, Horstrup J.H, Warnecke C, Mandriota S, Bechmann I, Frei U.A, Pugh C.W, et al. Widespread hypoxia-inducible expression of HIF-2α in distinct cell populations of different organs. FASEB J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. In situ hybridization: A practical approach. Oxford University Press; London: 1992. [Google Scholar]
- Wykoff C.C, Sotiriou C, Cockman M.E, Ratcliffe P.J, Maxwell P, Liu E, Harris A.L. Gene array of VHL mutation and hypoxia shows novel hypoxia-induced genes and that cyclin D1 is a VHL target gene. Br. J. Cancer. 2004;90:1235–1243. doi: 10.1038/sj.bjc.6601657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom Y.I, Fuhrmann G, Ovitt C.E, Brehm A, Ohbo K, Gross M, Hubner K, Scholer H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]